Abstract
Transient receptor potential (TRP) cation channel genes code for an extensive family of conserved proteins responsible for a variety of physiological processes, including sensory perception, ion homeostasis, and chemical signal transduction. The TRP superfamily consists of seven subgroups, one of which is the transient receptor potential vanilloid (trpv) channel family. While trpv channels are relatively well studied in adult vertebrate organisms given their role in functions such as nociception, thermoregulation, and osmotic regulation in mature tissues and organ systems, relatively little is known regarding their function during embryonic development. Although there are some reports of the expression of specific trpv channels at particular stages in various organisms, there is currently no comprehensive analysis of trpv channels during embryogenesis. Here, performing in situ hybridization, we examined the spatiotemporal expression of trpv channel mRNA during early Xenopus laevis embryogenesis. Trpv channels exhibited unique patterns of embryonic expression at distinct locations including the trigeminal ganglia, spinal cord, cement gland, otic vesicle, optic vesicle, nasal placode, notochord, tailbud, proctodeum, branchial arches, epithelium, somite and the animal pole during early development. We have also observed the colocalization of trpv channels at the animal pole (trpv 2/4), trigeminal ganglia (trpv 1/2), and epithelium (trpv 5/6). These localization patterns suggest that trpv channels may play diverse roles during early embryonic development.
1. Introduction
Transient receptor potential vanilloid (trpv) receptors are one of the seven subgroups of the transient receptor potential (TRP) channel superfamily, a family that is implicated in a wide range of physiological processes in sensory and chemical signal transduction (Nilius and Szallasi, 2014). Conserved in yeast, invertebrates, and vertebrates (Nilius and Owsianik, 2011), the trpv subfamily consists of six genes, trpv1–6. While trpv1–4 are Ca2+ permeable cationnonselective channels, trpv5/6 are highly Ca2+ selective and Mg2+ permeable (Courjaret et al., 2013). A variety of stimuli are known to activate the trpv1–4 channels, including moderate to noxious heat, chemical ligands, and even mechanical forces (Christensen and Corey, 2007; Vay et al., 2012; Kaneko and Szallasi, 2014), while trpv5/6 are constitutively active (Courjaret et al., 2013).
Given their known involvement in physiological processes in mature tissues, expression patterns of trpv channels in the adult are relatively well characterized (Vrenken et al., 2016). Trpv1 is expressed in a wide range of neuronal and non-neuronal adult tissues in mammals, and mediates nociception, synaptic plasticity, skin proliferation and homeostasis, (Szallasi et al, 2007; Baylie and Brayden, 2011, Cavanaugh et al., 2011, Ferrandiz-Huertas et al., 2014). Trpv2 is expressed in sensory neurons and plays a role in nociception, axon outgrowth, and immune responses (Shibasaki, 2016). In adult mice, trpv3 is predominantly expressed in the epithelium and is involved in skin barrier formation, hair growth, and cutaneous pain (Nilius and Bíró, 2013; Luo and Hu, 2014). Trpv4 is expressed in most adult organs (Zhan and Li, 2017) and its diverse functions include thermoregulation, maintaining osmotic homeostasis, and mechanotransduction during skeletal growth (Ferrandiz-Huertas et al., 2014; Moore and Liedtke, 2017). Trpv5 and trpv6 are vastly different from trpv1–4 in terms of both expression and function. While trpv5 expression is largely restricted to kidneys (Na and Peng, 2014), trpv6 is expressed in a wider array of tissues including the small intestine, kidney, and prostate (FecherTrost et al., 2014).
While most research on trpv channels has focused on adult physiological processes, this family of channels also displays embryonic expression. Despite their potentially critical role during development, there is still no comprehensive study on their expression patterns during embryogenesis. Rather, there are only limited analyses restricted to specific stages or tissues. or example, the spatiotemporal expression patterns of trpv1 is available for a few stages after E11.5 or 1dpf in mouse and zebrafish (Magdaleno et al., 2006; Caron et al., 2008; Shibasaki et al., 2010; Diez-Roux et al., 2011; Pan et al., 2012; Graham et al., 2013; Gau et al., 2017). The only available expression analysis on trpv2 is at E14.5 in mouse (Diez-Roux et al., 2011), while that on trpv3 is at E11.5 and E15.5 in mouse (Magdaleno et al., 2006). The embryonic expression of trpv4 is reported in zebrafish (Mangos et al., 2007) and partially available in chick and mouse (Magdaleno et al., 2006; Antin et al., 2007; Diez-Roux et al., 2011). Trpv5, on the contrary, only has reported expression data at E11.5 and E15.5 in mouse (Magdaleno et al., 2006). Data on trpv6 embryonic expression pattern is available only after E9.5 and at 3dpf in zebrafish (Magdaleno et al., 2006; Diez-Roux et al., 2011; Naguchi et al., 2012). Given the importance and versatility of trpv channels, a more comprehensive characterization during development will provide important insight into some of their earlier functions. Given the easy access to the earliest stages of development and its role as a classic developmental model system (Sive et al., 2000), we have characterized the spatiotemporal expression patterns of the six trpv channels at the transcriptional level during Xenopus laevis embryogenesis. The results indicated that transcripts for trpv channels have dynamic and diverse expression patterns throughout embryogenesis with some degree of overlap, and likely play critical roles during embryonic development.
2. Results
2.1. Trpv1 mRNA Expression
Trpv1 expression was first detected by ISH in the trigeminal ganglia and dorsal neural tube at the late neurula stage (St. 20) (Fig. 1 A, A’, A”), persisting through swimming tadpole stages (St. 35) (Fig. 1 B-D, B’-D”‘). Trpv1 showed bilateral expression localized to a small group of cells throughout the dorsal neural tube at late neurula (St. 20) (Fig. 1 A”) and early tailbud (St. 25) (Fig. 1 B”) stages. Trpv1 continued to localize to the dorsal spinal cord, namely Rohon Beard neurons at late tailbud (St. 30) (Fig. 1C”) and swimming tadpole stages (St. 35) (Fig. 1D”). At the swimming tadpole stage, in addition to trigeminal ganglia and spinal cord, trpv1 was also expressed in the somite (Fig. 1D ”’), with more posterior somites showing stronger signals. At swimming tadpole stages (St. 35), the signal also became more abundant (Fig. 1 D).This expression is consistent with the RNA-Seq data from Session et al. (2016).
Fig. 1.
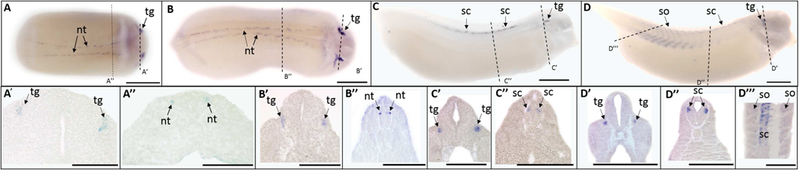
Whole mount (A-D) and histological (A’-D”’) expression of trpv1 transcripts in developing X. laevis embryos. (A-B) dorsal view, (C-D) lateral view, anterior to the right for all whole mount embryos; dorsal to the top for all histology images. (A, A’, A”) stage 20 (late neurula stage); (B, B’, B”) stage 25 (early tailbud stage); (C, C’, C”) stage 30 (late tailbud stage); (D, D’, D”, D”’) stage 35 (swimming tadpole stage). Arrows indicate regions of gene expression (nt, nural tube; sc, spinal cord; tg, trigeminal ganglia). Dashed lines represent positions of corresponding sections. Scale bars = 250µm.
2.2. Trpv2 mRNA Expression
Trpv2 mRNA was first detected as a maternal transcript localized to the animal pole in the unfertilized egg (Fig. 2 A, A’), St. 5 (16-cell stage) (Fig. 2 B, B’), and St. 8 (mid-blastula stage) (Fig. 2 C, C’). The level of maternal expression decreases as the embryo develops, and eventually showed no signal at the early gastrula stage (St. 10) (data not shown). Trpv2 signal was detected again at early tailbud stage (St. 25) in the cement gland, trigeminal ganglia, otic vesicle, and neural tube (Fig. 2 D’-D”). These expression patterns were maintained at late tailbud (St. 30) (Fig. 2 E’-E””) and swimming tadpole stages (St. 35) (Fig. 2 F’-F””) as well. Additionally, trpv2 expression was also detected in the optic vesicle at the swimming tadpole stage (St. 35) (Fig. 2 F’).
Fig. 2.
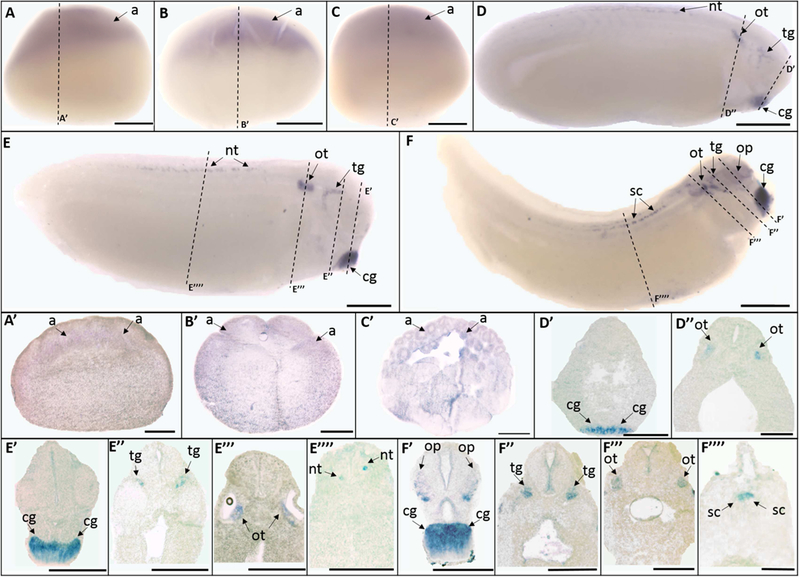
Whole mount (A-F) and histological (A’-F””) expression of trpv2 transcripts in X. laevis embryos. Lateral view for all whole mount embryos, animal pole to the top (A-C), anterior to the right (D-F); dorsal to the top for all histology images. (A, A’) unfertilized egg; (B, B’) stage 5 (16cell stage); (C, C’) stage 8 (mid-blastula stage); (D, D’, D”) stage 25 (early tailbud stage); (E, E’, E”, E”’, E””) stage 30 (late tailbud stage); (F, F’, F”, F”’, F””) stage 35 (swimming tadpole stage). Arrows indicate regions of gene expression (a, animal pole; cg, cement gland; nt, neural tube; op, optic vesicle; ot, otic vesicle; tg, trigeminal ganglia; sc, spinal cord). Dashed lines represent positions of corresponding sections. Scale bars = 250µm.
2.3. Trpv3 mRNA Expression
Current Xenopus laevis RNA-Seq data indicate that trpv3 expression is not detected throughout all stages of embryonic development (Session et al., 2016). Xenopus tropicalis RNASeq data also show that trpv3 has an extremely low number of transcripts per embryo (Collart et al., 2014; Owens et al., 2016). Consistent with these data, multiple RT-PCR attempts failed to detect trpv3 during embryonic stages. These data suggest that trpv3 is not expressed in Xenopus laevis during embryogenesis.
2.4. Trpv4 mRNA Expression
Similar to trpv2, ISH showed that trpv4 has highly abundant maternal expression, which was localized to the animal pole in the unfertilized egg (Fig. 3 A, A’), and in stage 5 (16-cell stage) (Fig. 3 B, B’), and stage 8 (mid-blastula stage) (Fig. 3 C, C’) embryos. The animal pole expression remained prominent at early gastrula stages (St. 10) (Fig. 3 D, D’). While no signal was detected at the mid-neurula stage (St. 15) (data not shown), at late the neurula stage (St. 20) trpv4 was found to be expressed diffusely throughout the tailbud region (Fig. 3 E, E’). The tailbud expression became more prominent at late tailbud stages (St. 30) (Fig. 3 F, F”). Trpv4 also exhibited strong signal at the notochord at late tailbud (St. 30) and swimming tadpole stages (St. 35) (Fig. 3 F, G, F’, G”). Moreover, at the swimming tadpole stage (St. 35), trpv4 was also expressed in the nasal placode (Fig. 3 G, G’).
Fig. 3.
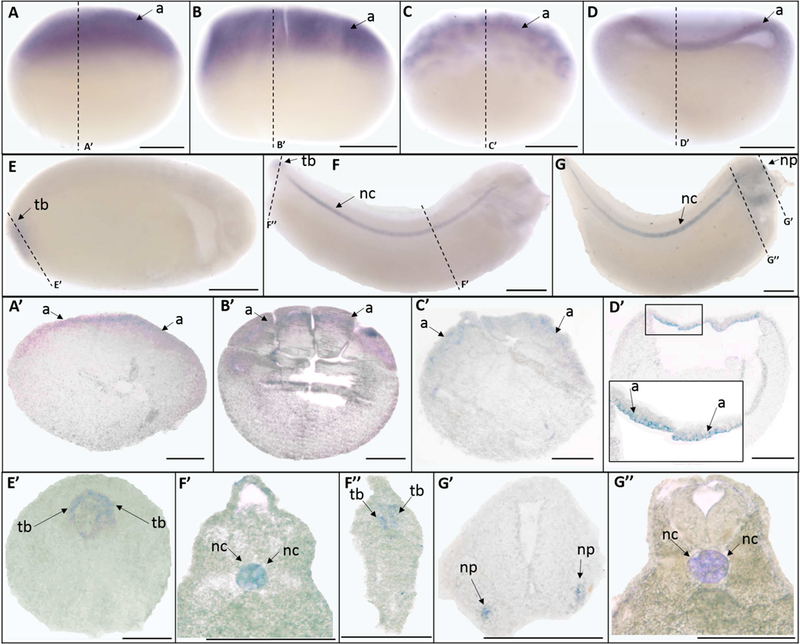
Whole mount (A-G) and histological (A’-G”) expression of trpv4 mRNA expression in X. laevis embryos. Lateral view for all whole mount embryos, animal pole to the top (A-D), anterior to the right (E-G); dorsal to the top for all histology images. (A, A’) unfertilized egg; (B, B’) stage 5 (16-cell stage); (C, C’) stage 8 (mid-blastula stage); (D, C’) stage 10 (early gastrula stage); (E, E’) stage 20 (late neurula stage); (F, F’, F”) stage 30 (late tailbud stage); (G, G’, G”) stage 35 (swimming tadpole stage). Arrows indicate regions of gene expression (a, animal pole; nc, notochord; np, nasal placode; tb, tailbud). Dashed lines represent positions of corresponding sections. Scale bars = 250µm.
2.5. Trpv5 mRNA Expression
The trpv5 ISH signals were first detectable via ISH at mid-neurula stages (St. 15). Trpv5 showed epidermal expression at mid-neurula (St. 15) (Fig. 4 A, A’), late neurula (St. 20) (Fig. 4 B, B’), early tailbud (St. 25) (Fig. 4 C, C’), late tailbud (St. 30) (Fig. 4 D, D’), and swimming tadpole (St. 35) stages (Fig. 4 E, E’). Trpv5 was not uniformly expressed in the epidermis, but rather, was present in some, but not all, epidermal cells. Trpv5 signal was also detected at the proctodeum at late neurula (St. 20) (Fig. 4 B, B”), early tailbud (St. 25) (Fig. 4 C, C”), and late tailbud (St. 30) stages (Fig. 4 D, D”). At the swimming tadpole stage (St. 35), trpv5 was also prominently expressed at the branchial arches (Fig. 4 E, E’).
Fig. 4.
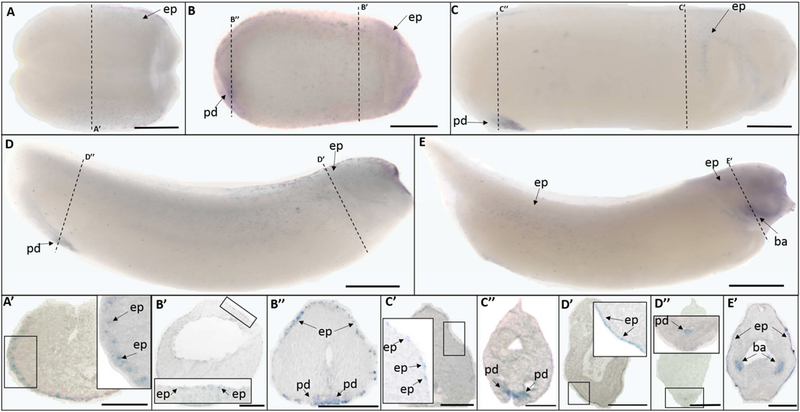
Whole mount (A-E) and histological (A’-E’) expression of trpv5 transcripts in developing X. laevis embryos. (A) dorsal view, (B-E) lateral view, anterior to the right for all whole mount embryos; dorsal to the top for all histology images. (A, A’) stage 15 (mid-neurula stage); (B, B’, B”) stage 20 (late neurula stage); (C, C’, C”) stage 25 (early tailbud stage); (D, D’, D”) stage 30 (late tailbud stage); (E, E’) stage 35 (swimming tadpole stage). Arrows indicate regions of gene expression (ba, branchial arches; ep, epidermis; pd, proctodeum). Dashed lines represent positions of corresponding sections. Scale bars = 250µm.
2.6. Trpv6 mRNA Expression
Trpv6 transcripts were present in a pattern very similar to that of trpv5. Trpv6 was expressed in the epidermis at mid-neurula (St. 15) (Fig. 5 A, A’), late neurula (St. 20) (Fig. 5 B, B’), early tailbud (St. 25) (Fig. 5 C, C’), late tailbud (St. 30) (Fig. 5 D, D’), and swimming tadpole (St. 35) stages (Fig. 5 E, E’). However, unlike trpv5, trpv6 signal was not detected at any other locations.
Fig. 5.
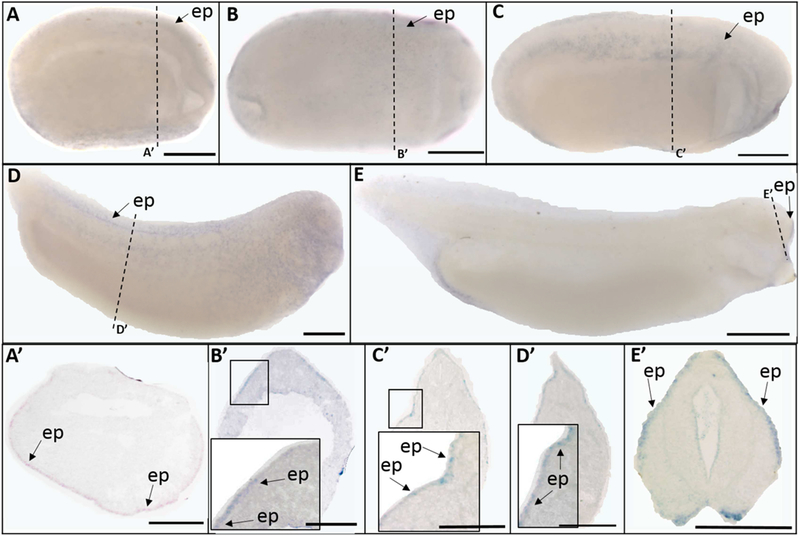
Whole mount (A-E) and histological (A’-E’) expression of trpv6 transcripts in X. laevis embryos. Lateral view for all whole mount embryos, anterior to the right; dorsal to the top for all histology images. (A, A’) stage 15 (mid-neurula stage); (B, B’) stage 20 (late neurula stage); (C, C’) stage 25 (early tailbud stage); (D, D’) stage 30 (late tailbud stage); (E, E’) stage 35 (swimming tadpole stage). Arrows indicate regions of gene expression (ep, epidermis). Dashed lines represent positions of corresponding sections. Scale bars = 250µm.
3. Discussion
Although there has been no comprehensive analysis of trpv channels expression during vertebrate embryogenesis, several studies do report on the embryonic expression patterns of trpv channels at specific stages or in individual tissues. While our data are generally consistent with the existing reports, there are some significant differences as well. In zebrafish, trpv1 was found to be expressed in trigeminal ganglia, Rohon-Beard neurons, and epidermis (Caron et al., 2008; Graham et al., 2013; Gau et al., 2017). In mouse, it was expressed in the trigeminal, dorsal root, and glossopharyngeal ganglia at E14.5 (Diez-Roux et al., 2011). Our results showed that trpv1 was expressed in the trigeminal ganglia and dorsal neural tube starting at late neurula stages and continued through swimming tadpole stages. At swimming tadpole stages, trpv1 was also detected in somitic tissue. While the trigeminal ganglia and spinal cord expression is consistent with previous data in other species at corresponding stages, and consistent with its role in nociception and thermal regulation, trpv1 expression in the somites has not been reported during embryonic development. Similar to mouse, no epidermal expression was detected.
Expressed in the central nervous system and placodal regions in mouse at E14.5 (DiezRoux et al., 2011), trpv2 has not been previously studied at early embryonic stages. Strikingly, we observed strong trpv2 signal localized to the animal pole of very early blastula-stage embryos. Additionally, trpv2 expression was detected in cement gland, optic vesicles, and otic vesicles, expression that has not been previously reported in other organisms. Its function at these early stages of development remains unknown. While trpv3 is widely expressed in the CNS of mouse after E11.5, it is not detectable in Xenopus during early development, according to RNA-Seq results in both X. laevis and X. tropicalis (Collart et al., 2014; Owens et al., 2016; Session et al., 2016) and using RT-PCR. The embryonic expression pattern of trpv4 is largely consistent among Xenopus laevis, zebrafish, and chick. Trpv4 is maternally expressed in both Xenopus laevis and zebrafish, though such data are not available in chick. It is also highly expressed in the notochord and presumptive head region in all three organisms following gastrulation (Antin et al., 2007; Mangos et al., 2007). However, we observed tailbud expression of trpv4, which initially becomes detectable at late neurula stages and has not been reported in other organisms. Whether trpv4 expression at these early stages is related to its known role in osmoregulation or skeletal development is unknown. Our data on trpv5 embryonic expression differs markedly from that in the mice. At E15.5 in mouse, trpv5 transcripts are detected throughout the CNS (Magdaleno et al., 2006), but In Xenopus laevis signal was observed only in the epidermis, proctodeum, and branchial arches. For trpv6, Xenopus laevis and zebrafish exhibited similar patterns of scattered expression in some but not all epidermal cells (Lin et al., 2011). This expression is consistent with the established role of trpv6 in calcium uptake in epithelial cells. Overall, while some differences may be attributable to the fact that our analysis included a more comprehensive series of stages, many species differences also exist.
Our results also indicate that the trpv channels have dynamic and diverse patterns of expression with some degree of overlapping expression during Xenopus laevis embryogenesis. The two pairs of trpv channels that show most dramatic overlap in expression are trpv1/2 and trpv5/6. Trpv1 and trpv2 are both expressed in the trigeminal ganglia and neural tube during tailbud and swimming tadpole stages. Both trpv5 and trpv6 are expressed in the epidermis starting at the mid-neurula stage (Nijenhuis et al., 2003). Such similarities in trpv1/2 and trpv5/6 embryonic expression patterns are not surprising given the trpv channels’ evolutionary history. Sequence analysis indicated that trpv1 and trpv2 are most closely related to one another, as are trpv5 and trpv6 (Saito and Shingai, 2006). Trpv2 and trpv4 are both prominently expressed maternally at the animal pole, suggesting their possible roles in the formation of the presumptive neuroectoderm. The expression of trpv1, trpv2, and trpv4 in neural structures like the trigeminal ganglion, neural tube, or the placodes is consistent with their roles in neuronal differentiation and axon outgrowth during early development (Andaloussi-Lilja et al., 2009; Shibasaki et al., 2010; Jang et al., 2012). Trpv4 expression in the tailbud is consistent with its role as a regulator of cell proliferation given the highly proliferative and stem-cell like character of the vertebrate tailbud (Hatano et al., 2013; Song et al., 2014). Moreover, trpv channels have long been known as regulators of ion homeostasis, especially that of calcium, during embryonic development (O’Neil and Brown, 2003; Vrenken et al., 2016). Given the importance of calcium activity during embryonic development, particularly for neuronal development (Rosenberg and Spitzer, 2011), somitogenesis (Webb and Miller, 2006), and notochord convergent extension (Wallingford et al., 2001), the expression of trpv channels in the nervous system, somites, and notochord suggests potential roles for this gene family during development. In conclusion, our study represents the first comprehensive analysis of the spatiotemporal expression patterns of trpv channels throughout embryogenesis in a vertebrate model organism. Our analysis reveals highly specific expression of the trpv channels in a diverse array of tissues from the earliest stages of embryogenesis through organogenesis.
4. Experimental Procedures
4.1. Embryo collection
Albino Xenopus laevis embryos were obtained with HCG injection as described by Sive et al. (2000). Embryos were staged according to Nieuwkoop and Faber (1994) and fixed in 1X MEMFA (Harland, 1991). All animal care and procedures were performed in accordance with the regulations set forth by the Institutional Animal Care and Use Committee (IACUC) at the College of William and Mary.
4.2. PCR cloning, sequence analysis, and ISH probe synthesis
Sequences for primer design were obtained from NCBI Reference Sequences (NCBI accession numbers: trpv1, XM_018246044.1; trpv2: XM_018248812.1; trpv4: XM_018261018.1; trpv5: XM_018224919.1; trpv6: XM_018224924.1). Primers for PCR cloning were as follows: trpv1, (5’-CGCACCTCTGGCGAGTTAAT-3’) and (5’-TTCACTGCTGGGATGTCTCT-3’); trpv2, (5’-GTTCACTCCCATCTCTTCGC-3’) and (5’-CCACCAACTGACTCCATCCT-3’); trpv4 (5’-GCTGTAGCCACAGACACTTCG-3’) and (5’-AGCCACCTTCATCCTTTGGTT-3’); trpv5, (5’-GCCCATTCTATATGCCGCAC-3’) and (5’-GGAAGCCTCAGACAAGTCCC-3’); trpv6, (5’-GCACAGAAGGCTCCATGAGT-3’) and (5’-TCACTAGCCAACCAGTGCTC-3’). The resulting PCR products were designed to ensure that the probe would not cross-hybridize with transcripts from other trpv channels. Every desired PCR product was also designed to hybridize against both of the homeologs of the allotetraploid Xenopus laevis, and all splicing isoforms of the gene (Session et al., 2016).
Total RNA was isolated from Xenopus laevis embryos (ThermoFisher MagMax kit), and cDNA was synthesized (Bio-Rad iScript cDNA synthesis kit). PCR was performed using total Xenopus laevis cDNA and the resulting PCR products were then ligated into pSC-A-amp/kan vectors and transformed into competent cells (StrataClone PCR cloning kit). Plasmids were purified using the NucleoBond Xtra midiprep kit from Macherey-Nagel, and the identity of each clone was confirmed by Sanger sequencing. Plasmid DNA was linearized with restriction enzymes, and both sense and antisense digoxigenin-labeled ISH probes were synthesized using in vitro transcription as follows: trpv1: sense XholI/T7, antisense NotI/T3; trpv2: sense EcoRV/T7, antisense BamHI/T3; trpv4 sense NotI/T3, antisense XhoI/T7; trpv5 sense XhoI/T7, antisense NotI/T3; trpv6 sense XhoI/T7, antisense NotI/T3.
4.3. Expression analysis
Whole mount in situ hybridization (ISH) was performed as described by Harland (1991) with slight modifications (Pownall and Saha, 2018). Following ISH, embryos were cleared as described by Sive et al. (2000) and imaged. ISH was performed with sense embryos in each experiment as a negative control; no signal was detected in sense embryos. Whole mount images were taken by a Nikon SMZ800N microscope with a Nikon DS-Ri2 camera. Embryos that underwent ISH were also embedded in paraffin, transversely sectioned at 18 or 30 µm, and transferred to slides. Images were taken using an Olympus MU100 camera with AmScope Imaging software. ISH was repeated at least three times with a minimum of ten embryos for each gene and stage. All images were excised using the Quick Selection tool, globally adjusted for color, brightness, and contrast, and placed on a uniform grey background in Photoshop.
Supplementary Material
Acknowledgements
This work was supported by NIH (1R15HD077624-01 to MSS), NSF (1257895 to MSS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors have no conflicts of interest to be declared in relation to this work.
References
- Andaloussi-Lilja JE, Lundqvist J, & Forsby A (2009). TRPV1 expression and activity during retinoic acid-induced neuronal differentiation. Neurochemistry international, 55(8), 768–774. [DOI] [PubMed] [Google Scholar]
- Antin PB, Kaur S, Stanislaw S, Davey S, Konieczka JH, Yatskievych TA, & Darnell DK (2007). Gallus expression in situ hybridization analysis: a chicken embryo gene expression database. Poultry science, 86(7), 1472–1477. [DOI] [PubMed] [Google Scholar]
- Baylie RL, & Brayden JE (2011). TRPV channels and vascular function. Acta Physiologica, 203(1), 99–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron SJ, Prober D, Choy M, & Schier AF (2008). In vivo birthdating by BAPTISM reveals that trigeminal sensory neuron diversity depends on early neurogenesis. Development, 135(19), 3259–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh DJ, Chesler AT, Jackson AC, Sigal YM, Yamanaka H, Grant R, O’Donnnell D, Nicoll RA, Shah NM, Julius D, Basbaum AI (2011). Trpv1 reporter mice reveal highly restricted brain distribution and functional expression in arteriolar smooth muscle cells. Journal of Neuroscience, 31(13), 5067–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AP, & Corey DP (2007). TRP channels in mechanosensation: direct or indirect activation?. Nature Reviews Neuroscience, 8(7), 510–521. [DOI] [PubMed] [Google Scholar]
- Collart C, Owens ND, Bhaw-Rosun L, Cooper B, De Domenico E, Patrushev I, Sesay AK, Smith JN, Smith JC, Gilchrist MJ (2014). High-resolution analysis of gene activity during the Xenopus mid-blastula transition. Development, 141(9), 1927–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courjaret R, Hubrack S, Daalis A, Dib M, & Machaca K (2013). The Xenopus TRPV6 homolog encodes a Mg2+-permeant channel that is inhibited by interaction with TRPC1. Journal of cellular physiology, 228(12), 2386–2398. [DOI] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, … and Lin-Marq N (2011). A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS biology, 9(1), e1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecher-Trost C, Weissgerber P, & Wissenbach U (2014). TRPV6 channels. In Mammalian Transient Receptor Potential (TRP) Cation Channels, pp. 359–384. [DOI] [PubMed] [Google Scholar]
- Ferrandiz-Huertas C, Mathivanan S, Wolf CJ, Devesa I, & Ferrer-Montiel A (2014). Trafficking of thermotrp channels. Membranes, 4(3), 525–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gau P, Curtright A, Condon L, Raible DW, & Dhaka A (2017). An ancient neurotrophin receptor code; a single Runx/Cbfβ complex determines somatosensory neuron fate specification in zebrafish. PLoS genetics, 13(7), e1006884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DM, Huang L, Robinson KR, & Messerli MA (2013). Epidermal keratinocyte polarity and motility require Ca2+ influx through TRPV1. J Cell Sci, 126(20), 4602–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM (1991). In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods in cell biology, 36, 685. [DOI] [PubMed] [Google Scholar]
- Hatano N, Suzuki H, Itoh Y, & Muraki K (2013). TRPV4 partially participates in proliferation of human brain capillary endothelial cells. Life sciences, 92(4), 317–324. [DOI] [PubMed] [Google Scholar]
- Jang Y, Jung J, Kim H, Oh J, Jeon JH, Jung S, … & Kim IB (2012). Axonal neuropathy-associated TRPV4 regulates neurotrophic factor-derived axonal growth. Journal of Biological Chemistry, 287(8), 6014–6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, & Szallasi A (2014). Transient receptor potential (TRP) channels: a clinical perspective. British journal of pharmacology, 171(10), 2474–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Tsai IL, Su CH, Tseng DY, & Hwang PP (2011). Reverse effect of mammalian hypocalcemic cortisol in fish: cortisol stimulates Ca2+ uptake via glucocorticoid receptor-mediated vitamin D3 metabolism. PloS one, 6(8), e23689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, & Hu H (2014). Thermally activated TRPV3 channels. In Current topics in membranes (Vol. 74, pp. 325–364). Academic Press. [DOI] [PubMed] [Google Scholar]
- Magdaleno S, Jensen P, Brumwell CL, Seal A, Lehman K, Asbury A, … & Norland SM (2006). BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS biology, 4(4), e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangos S, Liu Y, & Drummond IA (2007). Dynamic expression of the osmosensory channel trpv4 in multiple developing organs in zebrafish. Gene Expression Patterns, 7(4), 480484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C, & Liedtke WB (2017). Osmomechanical-Sensitive TRPV Channels in Mammals [PubMed]
- Na T, & Peng JB (2014). TRPV5: a Ca 2+ channel for the fine-tuning of Ca 2+ reabsorption. In Mammalian Transient Receptor Potential (TRP) Cation Channels (pp. 321–357). Springer, Berlin, Heidelberg. [Google Scholar]
- Naguchi TK, Ishimine H, Nakajima Y, Watanabe-Susaki K, Shigeta N, Yamakawa N, Asashima M, Wang PC, Kurisaki A (2012). Novel cell surface genes expressed in the stomach primordium during gastrointestinal morphogenesis of mouse embryos. Gene Expression Patterns, 12(3), 154–163. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, & Faber J (1994). Normal table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg till the end of Metamorphosis
- Nijenhuis T, Hoenderop JG, van der Kemp AW, & Bindels RJ (2003). Localization and regulation of the epithelial Ca2+ channel TRPV6 in the kidney. Journal of the American Society of Nephrology, 14(11), 2731–2740. [DOI] [PubMed] [Google Scholar]
- Nilius B, Owsianik G (2011). The transient receptor potential family of ion channels. Genome biology, 12(3), 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius B, & Bíró T (2013). TRPV3: a ‘more than skinny’channel. Experimental dermatology, 22(7), 447–452. [DOI] [PubMed] [Google Scholar]
- Nilius B, & Szallasi A (2014). Transient receptor potential channels as drug targets: from the science of basic research to the art of medicine. Pharmacological reviews, 66(3), 676–814. [DOI] [PubMed] [Google Scholar]
- O’Neil RG, & Brown RC (2003). The vanilloid receptor family of calcium-permeable channels: molecular integrators of microenvironmental stimuli. Physiology, 18(6), 226–231. [DOI] [PubMed] [Google Scholar]
- Owens ND, Blitz IL, Lane MA, Patrushev I, Overton JD, Gilchrist MJ, Cho KWY, Khokha MK (2016). Measuring absolute RNA copy numbers at high temporal resolution reveals transcriptome kinetics in development. Cell reports, 14(3), 632–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YA, Choy M, Prober DA, & Schier AF (2012). Robo2 determines subtype-specific axonal projections of trigeminal sensory neurons. Development, 139(3), 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pownall M and Saha MS (2018). Histological observation of teratogenic phenotypes induced in frog embryo assays. Methods Mol Biol 1797, 309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SS, & Spitzer NC (2011). Calcium signaling in neuronal development. Cold Spring Harbor perspectives in biology, 3(10), a004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, & Shingai R (2006). Evolution of thermoTRP ion channel homologs in vertebrates. Physiological genomics, 27(3), 219–230. [DOI] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, … and van Heeringen SJ (2016). Genome evolution in the allotetraploid frog Xenopus laevis. Nature, 538(7625), 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibasaki K (2016). Physiological significance of TRPV2 as a mechanosensor, thermosensor and lipid sensor. The Journal of Physiological Sciences, 66(5), 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibasaki K, Murayama N, Ono K, Ishizaki Y, Tominaga M (2010). TRPV2 enhances axon outgrowth through its activation by membrane stretch in developing sensory and motor neurons. Journal of Neuroscience, 30(13), 4601–4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, & Harland RM (2000). Early development of Xenopus laevis: a laboratory manual CSHL Press. [Google Scholar]
- Song Y, Zhan L, Yu M, Huang C, Meng X, Ma T, Zhang L, Li J (2014). TRPV4 channel inhibits TGF-β1-induced proliferation of hepatic stellate cells. PloS one, 9(7), e101179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szallasi A, Cortright DN, Blum CA, & Eid SR (2007). The vanilloid receptor TRPV1: 10 years from channel cloning to antagonist proof-of-concept. Nature reviews Drug discovery, 6(5), 357. [DOI] [PubMed] [Google Scholar]
- Vay L, Gu C, & McNaughton PA (2012). The thermo-TRP ion channel family: properties and therapeutic implications. British journal of pharmacology, 165(4), 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrenken KS, Jalink K, van Leeuwen FN, Middelbeek J (2016). Beyond ion-conduction: Channel-dependent and-independent roles of TRP channels during development and tissue homeostasis. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1863(6), 14361446. [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Ewald AJ, Harland RM, & Fraser SE (2001). Calcium signaling during convergent extension in Xenopus. Current Biology, 11(9), 652–661. [DOI] [PubMed] [Google Scholar]
- Webb SE, & Miller AL (2006). Ca2+ signaling during vertebrate somitogenesis. Acta Pharmacologica Sinica, 27(7), 781–790. [DOI] [PubMed] [Google Scholar]
- Zhan L, & Li J (2017). The role of TRPV4 in fibrosis. Gene 2018. February 5;642:1–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


