Abstract
Crepidiastrum sonchifolium, a flowering plant in the daisy family (Asteraceae), is native to East Asia. In Korea, this plant is a locally cultivated vegetable, and its market size is gradually growing. Since the plants with downy mildew infection were initially found at a private farm of Chuncheon city, the occurrences have continued in commercial farms of other regions, highlighting that this disease is spreading throughout Korea. The pathogen was attributed to a member of the genus Bremia that contains many specialized species, each of which displays a narrow host spectrum on Asteraceae. Based on morphological and molecular phylogenetic analyses, along with the high host specificity recently proven for Bremia species, the identity of the causal agent was confirmed as a so far undescribed species of Bremia. Here, we introduce Bremia itoana sp. nov., specific to C. sonchifolium.
Keywords: Barcoding, Cichorioideae, cox2 mtDNA, downy mildew, newly emerging disease
1. Introduction
Crepidiastrum sonchifolium (Maxim.) Pak & Kawano, called “sonchus-leaf crepidiastrum”, is native to East Asia. In Korea, it is an economically important vegetable, which is consumed to make kimchi, and its cultivated area is gradually expanding throughout the country [1–4]. In addition to its culinary value, this plant has been traditionally used as a folk medicine because of its digestive, diuretic, and anti-inflammatory activities [5].
Since downy mildew disease caused by the genus Bremia (Oomycota, Peronosporaceae) has been initially found on C. sonchifolium at a private farm in Chuncheon city, Korea in 1998, this disease has been continuously spreading to other regions, although it does not seem to have reached outside of Korea. On a large number of genera or species of the family Asteraceae, including this crop, downy mildew is a notorious disease, among which Bremia lactucae is one of the most well-known species, which causes devastating damage in the cultivation of lettuce (Lactuca sativa) [6]. According to the broad species concept that a downy mildew species is responsible for all infections occurring on a host family [7], the causal pathogen affecting C. sonchifolium has been presumed to be B. lactucae. However, recent phylogenetic studies with multigene sequences have found several well-supported groups in Bremia but also a better resolution power for discriminating them at the species level [8–13]. As a result, it revealed that Bremia is not monotypic, which consists of a dozen of highly host-specific species, and in addition, Choi et al. [10] found several previously overlooked lineages of Bremia, including Bremia sp. affecting C. sonchifolium, which was distant from other East Asian species of Bremia, parasitic to Crepidiastrum and two allied genera, Ixeris and Youngia. In the present study, we aimed to clarify the identity of the downy mildew pathogen, parasitic to C. sonchifolium, using both morphological and molecular phylogenetic approaches.
2. Materials and methods
2.1. Morphological analysis
C. sonchifolium plants with downy mildew symptoms were collected from various regions of Korea. For morphological investigation, conidiophores and conidia formed from the lower surface of the infected leaves were transferred onto a drop of lactic acid on a slide glass, covered with a coverslip, and gently warmed up using an alcohol lamp. A detailed microscopic examination was performed using an Olympus BX53F microscope (Olympus, Tokyo, Japan) equipped with a DigiRetina 16 M digital camera (Tucsen, Fuzhou, China). The following morphological characteristics were observed at 100–200× for conidiophores and at 400× for conidia and ultimate branchlets. Measurements were reported as maxima and minima in parentheses and the mean plus and minus the standard deviation of the number of measurements given in parentheses. Among the specimens examined morphologically, three specimens were selected for phylogenetic analyses (Table 1), and deposited at the Korea University Herbarium (KUS-F, Seoul, Korea) and the National Institute of Biological Resources (NIBR, Incheon, Korea).
Table 1.
Herbarium specimens of Bremia sp. parasitic to Crepidiastrum sonchifolium.
| Herb. No. (KUS-F) | Seq. ID | Collection year | Geographic origin | GenBank Acc. No. |
|
|---|---|---|---|---|---|
| 18S + ITS1/LSU D1-3/BrRxLR11 nDNA | cox2/cox1/cox2-1 spacer mtDNA | ||||
| 19257 | D216 | 2002 | Korea; Hongcheon | KT249120/ KT249315/ KT249705 | KT249510/ KP684694/ KP684894 |
| 19490 | D217 | 2003 | Korea; Chuncheon | KT249121/ KT249316/ KT249706 | KT249511/ KP684695/ KP684895 |
| 23946 | D543 | 2008 | Korea; Yangpyeong | MH665654/MH665655/MH665656 | MH665652/MH665651/MH665653 |
2.2. Molecular phylogenetic analysis
About 10–20 mg of infected leaves from herbarium specimens were disrupted with homogenizing pestles in 1.5-mL tube. Genomic DNA extraction was performed using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Three nuclear (ITS1, LSU D1-3 rDNA, BrRxLR11) and three mitochondrial (cox1, cox2, cox2-1 spacer mtDNA) markers were amplified by PCR as outlined previously [10–12]. The PCR products were purified and sequenced by a DNA sequencing service (Macrogen Inc., Seoul, Korea). For phylogenetic analysis, multigene sequences of Bremia species used in the previous studies [10–12] were retrieved from GenBank. Alignments for each of multigene datasets were done using the MAFFT 7 [14], by choosing the Q-INS-i algorithm [15]. After ensuring no strongly supported conflicting topologies among the trees inferred from individual loci, the alignments were concatenated using SequenceMatrix [16]. Two different phylogenetic inference methods were performed on the concatenated alignment. Minimum Evolution (ME) was done in MEGA6.0 [17] using the Tamura-Nei substitution model, and the robustness of internal branches was evaluated by performing 10,000 bootstrap replicates. Maximum Likelihood (ML) inference was computed using RAxML 7.0.3 [18], with default settings on the RAxML BlackBox web server [19].
3. Results and discussion
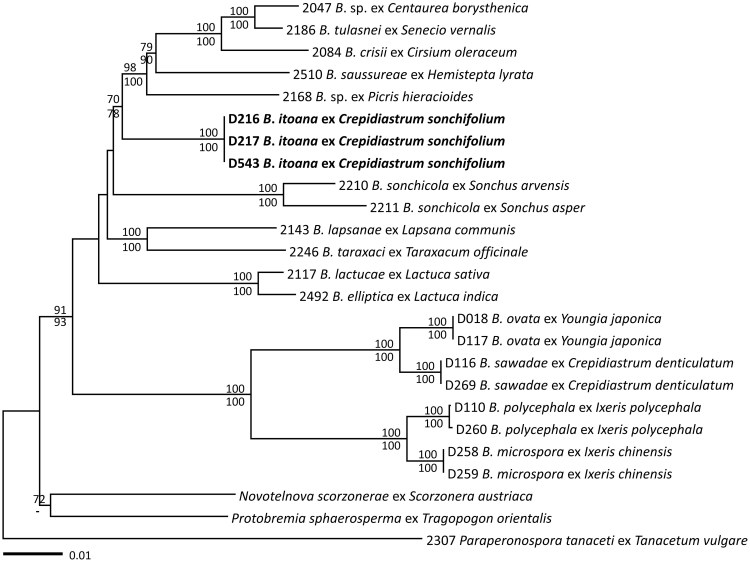
Trees based on each alignment of three nuclear (ITS, LSU D1-3 rDNA, BrRxLR11) and three mitochondrial (cox1, cox2, cox2-1 spacer mtDNA) loci showed no strong conflicting support with a phylogeny based on the concatenated alignment of all six loci. Thus, only the phylogeny based on concatenation of all loci was used for phylogenetic reconstruction. The final concatenated alignment displayed 4148 total characters, including 903 variable characters, 800 of which were parsimony-informative for Bremia species. Because the dataset revealed no significant conflicts in the topologies derived from ME and ML analyses, only the tree from the ME inference was shown in Figure 1. The overall topology and major groupings were in line with those of the previous studies [8–12], with unsupported replacements of a few branches. Three specimens affecting C. sonchifolium formed an independent lineage with maximum support and was separated from all previously accepted species of Bremia. Interestingly, this lineage is quite distant from another well-supported group consisting of four East Asian species, B. microspora, B. ovata, B. polycephala, and B. sawadae, despite their host plant affinity within the tribe Cichorieae (subfamily Cichorioideae). Instead, Bremia sp. affecting C. sonchifolium grouped with other Bremia species, parasitic to two subfamilies Asteroideae and Carduoideae, with moderate support of 70% in ME and 78% in ML analyses.
Figure 1.
Minimum evolution tree based on a concatenated alignment of ITS1, LSU nrDNA, cox2, cox1, the spacer region between cox2 and cox1 mtDNA, and BrRxLR11 sequences. Bootstrap support values higher than 70%, are displayed above (minimum evolution) or below (maximum likelihood) the corresponding branches. Branch lengths are proportional to the estimated number of nucleotide substitutions.
In agreement with the previous studies [8,11,20,21] that all Bremia species, parasitic to Cichorieae, have smaller conidia than other accepted species of Bremia, the present pathogen affecting C. sonchifolium could be characterized by small conidia of av. 15.9 × 14.1 µm, which is similar to B. microspora (av. 15.9 × 13.7 µm) and B. polycephala (av. 15.6 × 14.0 µm), but somewhat larger than B. ovata (av. 13.2 × 11.2 µm) and B. microspora (av. 13.4 × 12.7 µm). However, in line with the present phylogenetic results, this species exhibits several morphological differences. First, the length of conidiophores was markedly shorter as 200–410 (av. 305) µm than other four species (at least more than av. 400 µm). About 20–40% of conidiophores in Bremia sp. are shorter than 250 µm, but in other species, such short ones were <5%. As a related feature, the length of trunks was also shorter; av. 150 µm in Bremia sp. versus at least av. 237 µm in other species. In addition, the position of the first branching in Bremia sp. was somewhat lower as 4/10–6/10, thus often rendering the branching part occupying over half of the conidiophore’s length, while in other species they branched mostly at 3/10. As Bremia sp. and B. sawadae have the close host affinity [22], both parasitic to the genus Crepidiastrum, their morphological characteristics were compared in detail (Table 2). In addition to the differences listed above, the shapes of conidia and ultimate branchlets can discriminate between the two species; oblong to ovoid conidia in Bremia sp. versus ovoid conidia in B. sawadae, and often confluent vesicles at the end of ultimate branchlets in Bremia sp. versus mostly spherical vesicles in B. sawadae.
Table 2.
Comparison of morphological characteristics of Bremia species parasitic to Crepidiastrum species.
| Characteristics | B. sawadae | B. itoana |
|---|---|---|
| Host plant | Crepidiastrum denticulatum | Crepidiastrum sonchifolium |
| Conidiophores (n = 50) | ||
| Length | (355–)424–536–648(–718) µm | (150–)200–305–410(–500) µm |
| length of trunk | (224–)288–374–460(–514) µm | (60–)100–150–200(–300) µm |
| Width of trunk | (5.4–)6.2–7.1–8.0(–9.4) µm | (4.9–)6.5–7.7–9.3(–10.9) µm |
| Position of the first branch | 2/10–4/10 | 4/10–6/10 |
| Branching type | Dichotomous | Dichotomous |
| No. of branch orders | 4–6 | 3–5(–6) |
| Callose plugs | Often present in trunk, rarely in branches | Often present in trunk and branches |
| Ultimate branchlets (n = 50) | ||
| Shape | Curved | Slightly curved to substraight |
| Length | 8–20(–30) µm | 6–17 µm |
| Vesicles (n = 50) | ||
| Shape | Spherical | Spherical or confluent |
| Size (diam.) | 8–10 µm | 8–12 µm |
| No. of extensions | 4–6(–7) | 4–6 |
| Length of extensions | 5–7 µm | 6–8 µm |
| Shape of tips | Obtuse to somewhat swollen | Obtuse to somewhat swollen |
| Width of tips | 1.3–1.8 µm | 1.4–1.9 µm |
| Conidia (n = 100) | ||
| Shape | Ovoid | Oblong to ovoid |
| Colour | Hyaline | Hyaline |
| Length (µm) | (11.5–)12.5–13.4–14.3(–14.7) µm | (14.0–)15.1–15.9–16.7(–17.7) µm |
| Width (µm) | (10.6–)11.1–11.7–12.4(–14.1) µm | (13.0–)13.4–14.1–14.8(–15.7) µm |
| l/w ratio | (1.06–)1.1–1.14–1.19(–1.26) µm | (1.05–)1.09–1.13–1.17(–1.22) µm |
| Pedicel | Mostly present | Mostly present |
| Reference | Park et al. [12] | The present study |
The host range of Bremia species is restricted to the plant family Asteraceae, favouring three subfamilies, Asteroideae, Carduoideae, and Cichorioideae [10]. Especially, small conidia-possessing species of Bremia, including the present species, all colonize the three East Asian genera of the subtribe Crepidinae of Cichorioideae, Crepidiastrum, Ixeris, and Youngia [8,10,11]. However, although they infect the close host plants in the same area, Bremia sp. affecting C. sonchifolium seems to have evolved independently from other species. It is most likely that long after branching of the main group infecting Crepidinae, Bremia sp. has host-jumped from another lineage infecting Cichorioideae onto C. sonchifolium. Despite the similar morphology to Cichorioideae-infecting species but the close phylogenetic relationship to Asteroideae- and Carduoideae-infecting species, it is unlikely that this species served as an evolutionary bridge connecting the two groups because the host plant was always limited in East Asia.
The present study provided evidence that the causal agent of downy mildew occurring on the cultivated crop, Crepidiastrum sonchifolium, is independent from all previously known species of Bremia, for which we introduce a new species here.
Taxonomy
Bremia itoana Y.J. Choi & H.D. Shin, sp. nov. [MB827100] Figure 2.
Figure 2.
Morphological characteristics of Bremia itoana sp. nov. parasitic on Crepidiastrum sonchifolium. (A–C) Conidiophores; (D–E) Ultimate branchlets; (F–G) Conidia. Scale bars: 100 µm for conidiophores, 20 µm for ultimate branchlets, and 10 µm for conidia. Source: NIBRFG0000501456.
Etymology: named in honour of Seiya Ito for his outstanding studies on East Asian species of Bremia.
Lesions commonly causing discolouration of the tissues, pale green or yellow, later becoming dark brown, vein-limited, poly-angular, frequently covering larger areas by coalescing; infected tissues become necrotic. Down present on the under the surface of host leaf, whitish, consisting of scattered conidiophores, only one or two in a fascicle, sparse. Conidiophores emerging through stomata, colourless, straight, (150–)200–305–410(–500) µm; trunk straight, (60–)100–150–200(–300) µm long, (4.9–)6.5–7.7–9.3(–10.9)µm wide below the first branch; basal end not differentiated to slightly bulbous, (6.4–)8.6–10–11.4(–12.2) µm wide; callose plugs often present in trunk and branches; branches dichotomous, 3–5(–6) orders. Ultimate branchlets in pairs in most branchlets, rarely single, slightly curved to substraight, 6–17 µm long, obtuse to somewhat swollen; vesicles spherical or confluent, 8–12 µm diam., bearing 4–6 extensions with lengths of 6–8 µm. Conidia colourless, oblong to ovoid, almost symmetrical at the equatorial plane, (14.0–)15.1–15.9–16.7(–17.7) µm long, (13.0–)13.4–14.1–14.8(–15.7) µm wide, a l/w ratio of (1.05–)1.09–1.13–1.17(–1.22), greatest width median to submedian, tip round, base broadly round; pedicel mostly present, having thin-walled papilla. Germination directly with a germ tube only at the tip, up to 215 µm long, rarely branched. Resting organs not seen.
Typus: KOREA; Chuncheon-si, Dongnae-myeon, Goeun-ri (37°50'19"N 127°46'58"E), 21 May 2003, Y.J. Choi & H.D. Shin, NIBRFG0000501456 (holotypus).
Habitat: On living leaves of Crepidiastrum sonchifolium (Asteraceae).
Additional specimens examined for morphological investigation: KUS-F15622 (Nov. 1 1998, Chang-ri, Nam-myeon, Yanggu, Korea), 20946 (Nov. 4 2004, Goeun-ri, Dongnae-myeon, Chuncheon, Korea), 21516 (Oct. 17 2005, Jangjeonpyeong-ri, Hongcheon-eup, Hongcheon, Korea), 21684 (Nov. 11 2005, Goeun-ri, Dongnae-myeon, Chuncheon, Korea), 21688 (Nov. 12 2005, Seosang-ri, Seo-myeon, Chuncheon, Korea).
Funding Statement
YJC was supported by a grant from the National Institute of Biological Resources (NIBR), funded by of the Ministry of Environment (MOE), and the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT & Future Planning [Project No. 2016R1C1B2008013], Republic of Korea. YJC and JL were supported by a grant from the Agenda programme [Project No. PJ013476], Rural Development Administration, Republic of Korea.
References
- 1.Park MS, Chun Y-M. The usage of regional folk plants in Jeollanam-do. Korean J Plant Resourc. 2015;28:79–92. [Google Scholar]
- 2.Shin Y-H, Kim H-J, Jeong H-S, et al. . The folk plants in southern region of Chungcheongbuk-do, Korea. Korean J Plant Resourc. 2013;26:90–102. [Google Scholar]
- 3.Jeong H-R, Choi K, Park K-W. The regional folk plants in southern inland area of Gyeonggi-do. Korean J Plant Resourc. 2012;25:523–542. [Google Scholar]
- 4.Song M-J, Kim H, Brian H, et al. . Traditional knowledge of wild edible plants on Jeju Island, Korea. Indian J Tradit Knowldege. 2013;12:177–194. [Google Scholar]
- 5.Suh J, Jo Y, Kim ND, et al. . Cytotoxic constituents of the leaves of Ixeris sonchifolia. Arch Pharm Res. 2002;25:289–292. [DOI] [PubMed] [Google Scholar]
- 6.Lebeda A, Pink DAC, Astley D. Aspects of the interactions between wild Lactuca spp. and related genera and lettuce downy mildew (Bremia lactucae) In: Spencer-Phillips PTN, Gisi U, Lebeda A, editors. Advances in Downy Mildew Research. Vol. 1 Dordrecht (Boston): Kluwer Academic; 2002. p. 85–117. [Google Scholar]
- 7.Yerkes WD, Shaw CG. Taxonomy of the Peronospora species on Cruciferae and Chenopodiaceae. Phytopathology. 1959;49:499–507. [Google Scholar]
- 8.Choi YJ, Thines M, Runge F, et al. . Evidence for high degrees of specialisation, evolutionary diversity, and morphological distinctiveness in the genus Bremia. Fungal Biol . 2011;115:102–111. [DOI] [PubMed] [Google Scholar]
- 9.Thines M, Runge F, Telle S, et al. . Phylogenetic investigations in the downy mildew genus Bremia reveal several distinct lineages and a species with a presumably exceptional wide host range. Eur J Plant Pathol. 2010;128:81–89. [Google Scholar]
- 10.Choi YJ, Thines M. Host jumps and radiation, not co-divergence drives diversification of obligate pathogens. A case study in downy mildews and Asteraceae. PLoS One. 2015;7:e0133655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JH, Thines M, Lee HB, et al. . Bremia polycephala and Bremia sawadae spp. nov. (Peronosporaceae; Oomycota), parasitic to Northeast Asian Asteraceae. Nova Hedw. 2018;106:303–314. [Google Scholar]
- 12.Choi YJ, Wong J, Runge F, et al. . BrRxLR11 – a new phylogenetic marker with high resolution in the downy mildew genus Bremia and related genera. Mycol Prog. 2017;16:185–190. [Google Scholar]
- 13.Voglmayr H, Riethmüller A, Göker M, et al. . Phylogenetic relationships of Plasmopara, Bremia and other genera of downy mildews with pyriform haustoria based on Bayesian analysis of partial LSU rDNA sequence data. Mycol Res. 2004;108:1011–1024. [DOI] [PubMed] [Google Scholar]
- 14.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katoh K, Toh H. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinform. 2008;9:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27:171–180. [DOI] [PubMed] [Google Scholar]
- 17.Tamura K, Stecher G, Peterson D, et al. . MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. [DOI] [PubMed] [Google Scholar]
- 19.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–771. [DOI] [PubMed] [Google Scholar]
- 20.Skidmore DI, Ingram DS. Conidial morphology and the specialization of Bremia lactucae Regel (Peronosporaceae) on hosts in the family Compositae. Botanical J Linnean Soc (Lond). 1985;91:503–522. [Google Scholar]
- 21.Ling L, Tai MC. On the specialization of Bremia lactucae on compositae. Trans Brit Mycol Soc. 1945;28:16–25. [Google Scholar]
- 22.Peng Y-l, Zhang Y, Gao X-f, et al. . A phylogenetic analysis and new delimitation of Crepidiastrum (Asteraceae, tribe Cichorieae). Phytotaxa. 2014;159:241–255. [Google Scholar]