Abstract
Whole-genome sequencing of Flammulina ononidis, a wood-rotting basidiomycete, was performed to identify genes associated with carbohydrate-active enzymes (CAZymes). A total of 12,586 gene structures with an average length of 2009 bp were predicted by the AUGUSTUS tool from a total 35,524,258 bp length of de novo genome assembly (49.76% GC). Orthologous analysis with other fungal species revealed that 7051 groups contained at least one F. ononidis gene. In addition, 11,252 (89.5%) of 12,586 genes for F. ononidis proteins had orthologs among the Dikarya, and F. ononidis contained 8 species-specific genes, of which 5 genes were paralogous. CAZyme prediction revealed 524 CAZyme genes, including 228 for glycoside hydrolases, 21 for polysaccharide lyases, 87 for glycosyltransferases, 61 for carbohydrate esterases, 87 with auxiliary activities, and 40 for carbohydrate-binding modules in the F. ononidis genome. This genome information including CAZyme repertoire will be useful to understand lignocellulolytic machinery of this white rot fungus F. ononidis.
Keywords: Flammulina ononidis, genome, carbohydrate-active enzyme
1. Introduction
Flammulina ononidis was first described as a new species based on material from Germany and growth on Ononis spinosa [1]. In previous studies, F. ononidis differs from the popular edible mushroom F. velutipes in having wider spores. The minimum average spore width (SW) of F. ononidis is reported to be 4.2 μm, with a maximum SW for F. velutipes of 4 μm [2–4]. In addition, Flammulina species can be discriminated by species-specific motifs in the internal transcribed spacer (ITS) DNA region and there were variable regions of ITS1 ranging from 28-bp to 63-bp, and from 148-bp to 149-bp that were species-specific [5]. The authors demonstrated that both F. velutipes and F. ononidis lacked bases in the latter area [5].
Biological assay with mycelial cultures of the genus Flammulina has been investigated by several authors to provide fundamental data and for widespread application [6,7].
Interestingly, inconsistent results from earlier studies on Flammulina species have been reported, one of which concerns the laccase activity of Flammulina species. In cultural studies of Flammulina species, the similarity of F. ononidis and F. velutipes were observed in growth rate [7]. In addition, the authors reported that Flammulina species varied in laccase (EC 1.10.3.2) activity and F. velutipes and F. ononidis produced no laccase [7]. Other studies [8,9] also described the inability of F. velutipes to produce laccase. However, the presence of laccase activity has been reported from all Flammulina species, with significant variation between different isolates [10]. Moreover, laccase production can vary based on growth time and season [11]. It has been suggested that the morphological characteristics and habitat of Flammulina species are important for classification of the European species of the genus Flammulina and that F. ononidis generally grows on roots, gregarious or scattered, and sometimes occurs in clusters [12].
Several fungi possess lignocellulolytic activity and efficiently degrade lignocellulosic polymers derived from plants [13]. Thus, fungi are frequently found in nature, such as on softwoods, hardwoods, grasses, crops, and forest waste. Among them, fungi capable of decomposing lignin and polysaccharides are termed wood-rotting fungi. They are generally divided into white-rot fungi and brown-rot fungi [14]. These abilities of fungi are mainly due to their various enzymes, which include carbohydrate-active enzymes (CAZymes). CAZymes include glycoside hydrolases (GHs), carbohydrate esterases (CEs), polysaccharide lyases (PLs), glycosyltransferases (GTs), and enzymes with auxiliary activities (AA), as well as carbohydrate-binding modules (CBMs) having carbohydrate-binding activity. These CAZymes are classified into several families based on their functional amino acid sequences and structural similarity (CAZy database; http://www.cazy.org/) [15]. Understanding the mechanism of degradation of wood-rotting fungi that can degrade lignocellulosic polymers allows for a wide variety of use. Therefore, CAZymes have a great potential due to their diverse biotechnological and industrial applications [14].
To date, several studies have been carried out to uncover wood degrading machinery of the fungal genome. In addition, the excavation of enzymes capable of degrading biomass in the fungal genome has also become an important research area in the post-genome era. Here, we first report the genome sequence of F. ononidis for identification of CAZyme genes to increase utilization of these biotechnologically and industrially useful enzymes. The genomic information of F. ononidis will help to understand this fungus, and the gene information predicted to encode CAZymes will be useful for future biotechnology and industrial applications.
2. Materials and methods
2.1. Genome sequencing, gene modeling, and gene annotation of F. ononidis
F. ononidis KACC46186 was obtained from the National Agrobiodiversity Center (http://genebank.rda.go.kr/) and was grown at 26 °C on potato dextrose agar (PDA, 20 g dextrose, 4 g potato starch, 15 g agar per liter) for 14 days. F. ononidis genomic DNA was extracted by a previously described method [16]. Genome sequencing of F. ononidis was performed using the HiSeq 2000 platform according to the manufacturer’s protocol (Illumina Inc., USA). Sequence reads were deposited in the Sequence Read Archive (SRA) at the National Center for Biotechnology Information (NCBI; SRP152895). Raw sequencing reads were processed by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Trimmomatic (version 0.32) [17] for quality control and trimming of bad quality reads including sequencing adapters, respectively. De novo assembly of the resulting short-reads was performed by Velvet Optimiser [18] with kmer-size search range of 17–31. Gene structure modeling of F. ononidis de novo assembly was performed using the AUGUSTUS tool [19] trained in Laccaria bicolor. Functional annotation of the predicted genes was performed using DIAMOND [20] and BLASTP (v 2.2.31) softwares against a non-redundant database and fungal genome database from NCBI. Protein family search was further conducted using Pfam-scan software [21] against the protein family database (Pfam 31.0, http://pfam.xfam.org).
2.2. Ortholog analysis
Orthologous groups were clustered using OrthoFinder (version 2.2.1) software [22] based on all-versus-all protein comparison with fungal species including Aspergillus nidulans FGSC A4 [23], Botrytis cinerea B05.10 [24], Agaricus bisporus var. bisporus H97 [25], Coprinopsis cinerea okayama7#130 [26], Cordyceps militaris CM01 [27], Cryptococcus neoformans var. grubii H99 [28], F. velutipes KACC42780 [16], L. bicolor S238N-H82 [29], Lentinula edodes [30], Neurospora crassa OR74A [31], Phanerochaete chrysosporium RP78 [32], Saccharomyces cerevisiae S288C [33], Schizophyllum commune H4-8 [34], Trichoderma reesei QM6a [35], and Ustilago maydis 521 [36].
2.3. CAZyme identification
CAZyme genes associated with GHs, GTs, PLs, CEs, and AAs, and CBMs were predicted using HMMER 3.0 package software (http://hmmer.org/) with the dbCAN CAZyme database [37] and obtained from DOE Joint Genome Institute (JGI Fungi Portal database; https://genome.jgi.doe.gov/programs/fungi/index.jsf) and CAZy database (http://www.cazy.org/).
3. Results and discussion
3.1. General features of F. ononidis genome
The total number of quality-checked and trimmed reads from the total 25,964,380 reads (100-bp paired-end) was 24,132,250 (>Q30). The resulting short reads were assembled using Velvet assembly tool with a kmer-size search range of 17–31. The resulting optimized assembly (31 kmer) consisted of 12,825 sequence contigs with a total length of 34,524,258 bp (49.76% GC) and N50 length of 73,451 bp (Table 1). Gene structure modeling was performed using the AUGUSTUS tool. A total of 12,269 gene models were predicted with an average gene length of 2,009 bp (Table 1). The predicted genes contained exons and introns averaging 234.09 bp and 68.14 bp in length, respectively. Of the 12,269 predicted genes, 85.8% (10,529) and 82% (10,068) had significant sequence similarity (0.001 > e-value) to documented proteins in NCBI-NR and fungal genome database, respectively (Supplementary data S1). In genome comparison with other basidiomycetes, the general features of F. ononidis genome were similar with those of its nearest sequenced species, F. velutipes [16], except average intron size (Table 2).
Table 1.
Statistics of genome sequencing of F. ononidis.
| Hiseq 2000 NGS analysis | |
| Total reads (100 bp) | 25,964,380 |
| Reads after trimming (%), >Q30 | 24,132,250 (92.94) |
| Velvet de novo assembly | |
| Optimised Velvet hash value (kmer) | 31 |
| Total number of contigs | 12,825 |
| Number of contigs (>1 kb) | 1586 |
| Contig N50 (bp) | 73,451 |
| Length of longest contig (bp) | 808,623 |
| Total bases in contigs (bp) | 34,524,258 |
| Total bases in contigs (>1 kb) | 32,738,129 |
| GC content (%) | 49.76 |
| AUGUSTUS gene prediction | |
| Predicted gene | 12,269 |
| Average gene length (bp) | 2009 |
| Average protein length (aa) | 535.53 |
| Average exon per gene | 6.86 |
| Average exon size (bp) | 234.09 |
| Average intron size (bp) | 68.14 |
Table 2.
Comparison of genome characteristics between F. ononidis and other basidiomycetes.
| Genome characteristics | F. ononidis | F. velutipes | L. bicolar | C. cinerea | P. chrysosporium | U. maydis | S. commune |
|---|---|---|---|---|---|---|---|
| Strain | KACC46186 | KACC42780 | S238N-H82 | Okayama7#130 | RP78 | 521 | H4-8 |
| Genome assembly (Mb) | 34.5 | 35.6 | 64.9 | 37.5 | 35.1 | 19.7 | 38.5 |
| Number of protein-coding genes | 12,269 | 12,218 | 20,614 | 13,544 | 10,048 | 6522 | 13,181 |
| GC contents (%) | 49.76 | 48.99 | 46.6 | 51.6 | 53.2 | 54.0 | 56.6 |
| Average gene length (bp) | 2009 | 2,294 | 1533.0 | 1679.0 | 1667.0 | 1935.0 | 1794.9 |
| Average exon size (bp) | 234.09 | 231.4 | 210.1 | 251.0 | 232.0 | 1051.0 | 249.3 |
| Average intron size (bp) | 68.14 | 190.3 | 92.7 | 75.0 | 117.0 | 127.0 | 79.0 |
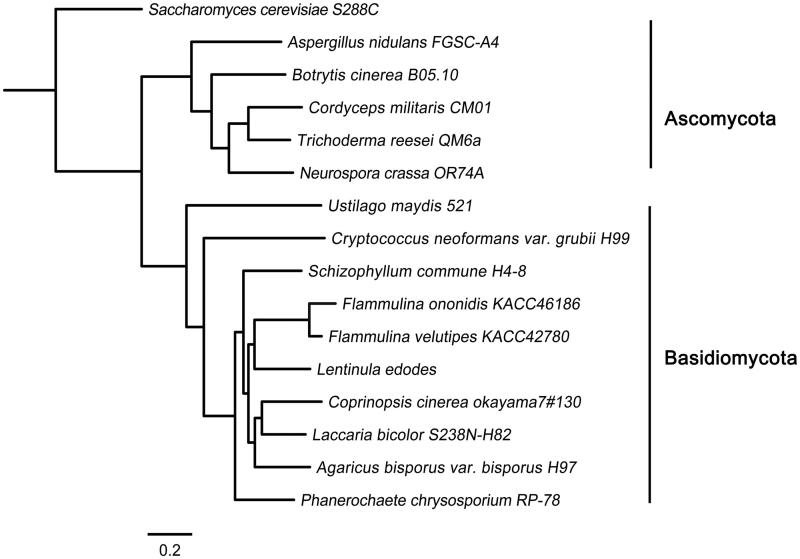
In a protein family search against the Pfam 31.0 database, 6,964 and 1,206 genes were annotated as functional proteins and as multi-domain protein families, respectively (Supplementary data S2). Cluster analysis with other sequenced fungal species identified 7,051 groups containing at least one F. ononidis protein (Table 3). Analysis of these clusters suggested that 58.7% of F. ononidis proteins had orthologs among the Dikarya and were thus conserved in basidiomycetes and ascomycetes (Table 3). Among the set of homologous genes, 592 single-copy orthologs were detected and F. ononidis contained 8 species-specific genes, of which 5 were paralogous. Additionally, F. ononidis was classified into one group with F. velutipes by ortholog-clustering analysis (Figure 1).
Table 3.
Ortholog analysis of F. ononidis with other sequenced fungal species.
| Taxon | Fungal species | No. of genes | No. of genes in orthogroups (%) | No. of unassigned genes (%) | No. of orthogroups containing species (%) | No. of species-specific orthogroups | No. of genes in species-specific orthogroups (%) |
|---|---|---|---|---|---|---|---|
| Basidiomycota | Flammulina ononidis KACC46186 | 12,269 | 11,249 (92) | 1020 (8) | 7051 (58.7) | 3 | 8 (0.1) |
| Flammulina velutipes KACC42780 | 12,218 | 10,460 (86) | 1758 (14) | 6674 (55.6) | 5 | 12 (0.1) | |
| Agaricus bisporus var. bisporus H97 | 10,448 | 9077 (87) | 1371 (13) | 5,893 (49.1) | 24 | 186 (1.8) | |
| Coprinopsis cinerea okayama 7#130 | 13,356 | 10,780 (81) | 2576 (19) | 6,585 (54.8) | 44 | 247 (1.8) | |
| Cryptococcus neoformans var. grubii H99 | 7,826 | 6,515 (83) | 1,311 (17) | 4,898 (40.8) | 38 | 131 (1.7) | |
| Laccaria bicolor S238N-H82 | 18,215 | 12,502 (69) | 5713 (31) | 6376 (53.1) | 66 | 448 (2.5) | |
| Lentinula edodes | 14,079 | 11,605 (82) | 2474 (18) | 6879 (57.3) | 49 | 297 (2.1) | |
| Phanerochaete chrysosporium RP78 | 13,602 | 10,411 (77) | 3,191 (24) | 6437 (53.6) | 34 | 186 (1.4) | |
| Schizophyllum commune H4-8 | 13,194 | 10,895 (83) | 2,299 (17) | 6397 (53.3) | 50 | 337 (2.6) | |
| Ustilago maydis 521 | 6783 | 5656 (83) | 1127 (17) | 4845 (40.3) | 9 | 22 (0.3) | |
| Ascomycota | Aspergillus nidulans FGSC-A4 | 9561 | 8442 (88) | 1119 (12) | 5817 (48.4) | 5 | 24 (0.3) |
| Botrytis cinerea B05.10 | 16,389 | 9329 (57) | 7060 (43) | 6389 (53.2) | 14 | 43 (0.3) | |
| Cordyceps militaris CM01 | 9651 | 8382 (87) | 1269 (13) | 6254 (52.1) | 5 | 15 (0.2) | |
| Neurospora crassa OR74A | 10,812 | 8649 (80) | 2163 (20) | 6341 (52.8) | 8 | 22 (0.2) | |
| Saccharomyces cerevisiae S288C | 6002 | 4631 (77) | 1371 (23) | 3572 (29.7) | 13 | 34 (0.6) | |
| Trichoderma reesei QM6a | 9115 | 8331 (91) | 784 (9) | 6417 (53.4) | 4 | 9 (0.1) |
Figure 1.
Phylogenetic tree of fungal species based on ortholog clustering. The amino acid sequences in each orthologous group were aligned by MAFFT (https://mafft.cbrc.jp/alignment/software/) and phylogenetic tree was constructed with FastTree (http://www.microbesonline.org/fasttree/) with 1000 bootstraps.
3.2. GTs in F. ononidis genome
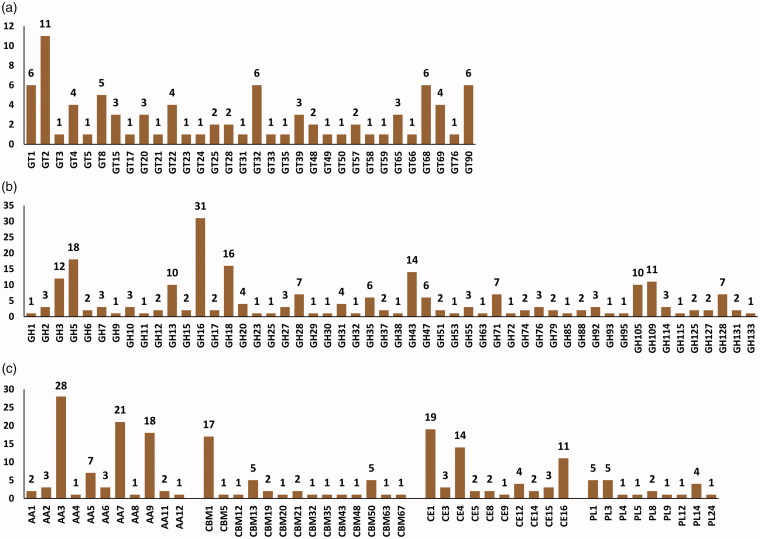
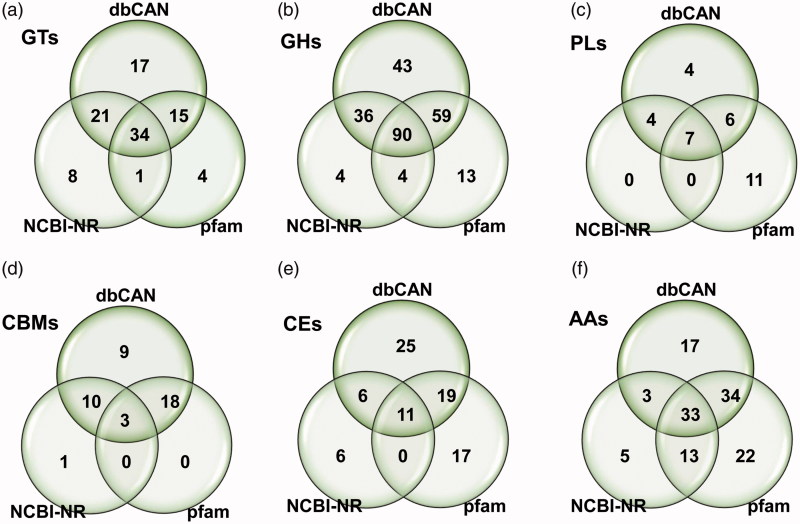
GTs (EC 2.4.x.y) catalyze the formation of a glycoside involved in the biosynthesis of oligosaccharides, polysaccharides, and glycoconjugates [38,39]. GTs also catalyze glycosyl group transfer to specific acceptor molecules, forming glycosidic bonds [39–41]. In CAZyme identification based on a dbCAN database search, a total of 32 GT families including 87 GTs were identified from F. ononidis genome, of which about 50% (15 families) were identified as a family of one GT gene (Figure 2(a) and Supplementary data S3). It has been reported that a large number of GTs (approximately 1 − 2% of the gene products) are found in completely sequenced organisms [15]. Among the GT families, GT2 and GT4 account for approximately half of the total number of GTs in the CAZy database (http://www.cazy.org/). Presently, the GT2 family was prominently present, with 11 genes in the F. ononidis genome (Figure 2(a) and Supplementary data S3). Additionally, 64 and 54 GTs were identified by BLASTP (NCBI-NR) and protein family database (Pfam 31.0) search, respectively (Supplementary data S4). Thus, a total of 100 GTs was identified from three different databases, of which 34 GTs were commonly identified. Among the total 100 GTs, 17, 8, and 4 GTs were uniquely identified by dbCAN, NCBI-NR, and pfam databases searches, respectively (Figure 3(a) and Supplementary data S4).
Figure 2.
CAZyme distribution predicted from F. ononidis genome. The number after each CAZyme means family. (a) GT families; (b) GH families; (c) AA, CBM, CE, and PL families. AA, auxiliary activities; GH, glycoside hydrolase; GT, glycosyltransferase; CBM, carbohydrates- binding module; PL, polysaccharide lyase.
Figure 3.
Venn diagrams of CAZymes predicted in F. ononidis by three different database searches including protein family database (Pfam), CAZyme database (dbCAN), and National Center for Biotechnology Information (NCBI) non-redundant database (NCBI-NR). (a) GT families; (b) GH families; (c) PL families; (d) CBM families; (e) CE families; and (f) AA families.
GTs have been classified into families based on amino acid sequence similarities [40,41]. Likewise, 100 GTs were identified in the three databases with amino acid sequences of predicted genes in the F. ononidis genome. However, many GTs showed different activity, even though the members of GT are closely related to their sequences [38]. It has been reported that polyspecific families including GT2 or GT4 have amino acid sequence similarities within only a small portion of the catalytic domain [38]. Therefore, Mukai et al. [42] suggested that a method that does not rely on sequence similarity is required for GT identification. The authors demonstrated that a computational method for determining the transmembrane region of Golgi-localized signal-anchor-type GTs and for discovering novel GTs [42]. Furthermore, additional studies based on structural, modeling, and mutational analysis are needed to identify and elucidate the enzymatic characteristics of these enzymes.
3.3. GHs in F. ononidis genome
GHs (EC 3.2.1.x) are key enzymes of carbohydrate metabolism that catalyze the hydrolysis of glycosidic bonds of carbohydrate substrates. GHs are involved in the degradation of the most biomass such as cellulose, hemicellulose, and starch [43,44]. GHs are classified into various families based on sequence similarity. At the time of writing (July 2018), the database comprises more than 507,392 classified and 8,914 non-classified GH sequences that have been divided into 153 families (CAZy database; http://www.cazy.org/). Presently, a total of 228 GHs were classified into 52 families predicted in the F. onionidis genome based on a dbCAN database search (Figure 2(b) and Supplementary data S3). GH family classification revealed 17 families that consisted of only 1 GH. GH16 was prominently present, with 31 genes (Figure 2(b) and Supplementary data S3). A total of 249 GHs were identified from the dbCAN, BLASTP (NCBI-NR), and protein family (Pfam 31.0) databases (Figure 3(b) and Supplementary data S5). Among them, 134 and 166 GHs were identified by NCBI-NR and Pfam 31.0 search, respectively, and 90 GHs were commonly identified among the three different databases (Figure 3(b) and Supplementary data S5).
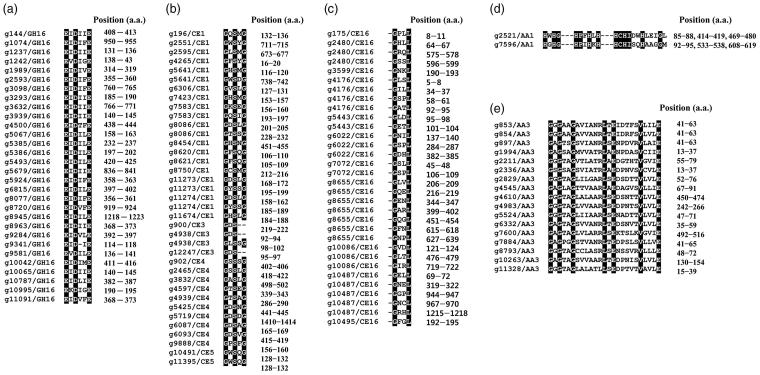
In previous studies, most of the enzymes belonging to the GH16 family were found to contain a conserved motif, EXDX(X)E, in their amino acid sequences [45,46]. Our results also showed the conserved motifs within amino acid sequences of genes predicted to encode GH16 families in F. ononidis, except one GH16 family (Figure 4(a) and Supplementary data S3). Among them, 8, 6, 5, and 2 GH16 families possessed a catalytic motif identical to EIDIIE, EIDILE, EIDIFE, and EIDVFE, respectively (Figure 4(a)). Kotake et al. [47] indicated that the first and last glutamic acid (E) residues are important for the catalytic activity of the GH16 family enzyme. Although further analysis is required to characterize their biochemical activities, these results suggest that these genes predicted to encode GH16 families might act as GHs, such as lichenase (EC 3.2.1.73), xyloglucan xyloglucosyltransferase (EC 2.4.1.207), agarase (EC 3.2.1.81), kappa-carrageenase (EC 3.2.1.83), endo-beta-1,3-glucanase (EC 3.2.1.39), endo-beta-1,3-1,4-glucanase (EC 3.2.1.6), and endo-beta-galactosidase (EC 3.2.1.103).
Figure 4.
Conserved motifs within (a) glycoside hydrolase family 16, (b) carbohydrate esterase family 1, (c) carbohydrate esterase family 16, (d) auxiliary activity family 1 (laccase), and (e) auxiliary activity family 3 (GMC oxidoreductase) amino acids of F. ononidis. Position (a.a.) refers to the start and end positions of the motif in the amino acid sequence.
GHs are essential for the processing of polysaccharides including cellulose and xylan of various plants and chitin, which are important carbon and nitrogen sources in nature [15]. The enzymes belonging to GH5–GH9, GH12, GH44, GH45, and GH48 families act as cellulases; those belonging to GH10, GH11, and GH30 families act as xylanases; and those belonging to GH18, GH19, and GH85 families act as xylanase chitinases [15,48]. Our results also reveal that F. ononidis contains genes associated with cellulase (18 GH5, 2 GH6, 3 GH7, 1 GH9, and 2 GH12 families), xylanase (3 GH10, 1 GH11, and 1 GH30), and chitinases (16 GH18 and 1 GH85) in its genome sequence (Figure 2(b) and Supplementary data S3). β-Glucosidases (EC 3.2.1.21), which are also members of GH families such as GH1 and GH3, convert cellobiose into glucose. CAZyme annotation also revealed F. ononidis contained these GH families with 1 GH1 and 12 GH3 families in its genome (Figure 2(b) and Supplementary data S3).
Plant cell walls often form complex polysaccharides structure with cellulose and xylan. Therefore, the synergistic activities of various GH families are required to degrade these complexes efficiently. Among various organisms in nature, fungi play a key role in the degradation of these complex polysaccharide structures and, therefore, are likely to be applied to the industrial and biotechnological applications, such as food, paper, textile, animal feed, and other chemicals, including biofuels [14]. Thus, our results suggest that F. ononidis has a strong potential for industrial and biotechnological applications, because of the over 200 genes predicted to encode various GHs in its genome.
3.4. PLs in F. ononidis genome
Polysaccharides are essential cellular components of all living organisms [49]. PLs (EC 4.2.2.-) act on polysaccharides to produce an unsaturated polysaccharide via a β-elimination mechanism [50].
To date, PLs have been classified into 28 families with more than 14,200 classified and 1,300 non-classified PL sequences in the CAZy database (http://www.cazy.org/). Pectate or pectin is less prominently present in plants than cellulose and hemicellulose [51,52]. The enzymes that catalyze the formation of polygalacturonan are designated pectate or pectin lyases [52]. Several fungi produce both pectin lyases and pectate lyases, which are often accompanied by other hydrolases to degrade pectin and/or pectate [53]. Pectin and pectate lyases have been classified into 6 PL families in the CAZy database: PL1, PL2, PL3, PL9, PL10, and PL22 (http://www.cazy.org/). All characterized pectin lyases (EC 4.2.2.10) are classified in the PL1 family. In addition, pectate lyases (EC 4.2.2.2 and EC 4.2.2.9) of fungal species belong to PL1, PL3, and PL9 (CAZy database; http://www.cazy.org/).
In F. ononidis, a total of 21 PLs classified in 9 families were identified in the genome according to a dbCAN database search (Figure 2(c) and Supplementary data S3). Among them, the PL1 and PL3 families were prominently present in the F. ononidis genome, with 5 genes each (Figure 2(c) and Supplementary data S3). Furthermore, 11 and 24 PLs were identified by NCBI-NR and Pfam 31.0 database searches, respectively, and 7 PLs were commonly identified among the three different databases (Figure 3(c) and Supplementary data S6).
Recently, the determination of the sequences of a variety of basidiomycetes has revealed information about genes encoding PLs. This information has the potential to be used in biotechnological applications. S. commune is one of the most efficient (hemi)cellulose degrading-basidiomycete because of the wealth of putative pectin-degrading lyases in its genome and, therefore, produces a high level of pectinase [34,54]. Similarly, diverse members of PL families including PL1, PL3, and PL9 in the F. ononidis genome suggest the potential of this fungus for various applications involved in the processing of polysaccharides in the future.
3.5. CBMs in F. ononidis genome
CBMs have carbohydrate-binding activity. They bind to carbohydrate ligands and enhance the catalytic efficiency of CAZymes [55]. Although CBMs have no hydrolytic activity, the enzymatic complex including GHs, GTs, and PLs, which contain CBM, more efficiently degrade substrate [56]. CBMs have been classified into 83 families with more than 133,400 classified in the CAZy database (CAZy database; http://www.cazy.org/).
In the present study, a total of 40 CBMs classified into 14 families with 17 CBM1 families were identified in the F. ononidis genome based on a dbCAN database search (Figure 2(c) and Supplementary data S3). Among them, 9 families including CBM5, CBM12, CBM20, CBM32, CBM35, CBM43, CBM48, CBM63, and CBM67 consisted of only one CBM (Figure 2(c) and Supplementary data S3). Moreover, 14 and 21 CBMs were identified by NCBI-NR and Pfam 31.0 database searches, respectively (Figure 3(d) and Supplementary data S7), and 3 CBMs (CBM1/5 and CBM21) were commonly identified among the three different databases (Figure 3(d) and Supplementary data S7). CBM is considered as an essential module in cellulase and cellobiohydrolases, and belongs to the GH6 and GH7 families [57]. In the present study, each GH6 and GH7 of the F. ononidis genome contained a CBM1 family (Supplementary data S3) and CBMs families were also identified in several CAZymes including 11 GHs, 6 CEs, and 1 AA, suggesting that those CAZymes may require CBM for their activities to efficiently degrade substrates.
3.6. CEs in F. ononidis genome
Esterases act on ester bonds and are widely used in industrial processes [58,59]. CEs are a class of esterases and catalyze the O-de- or N-deacylation to remove the ester of substituted saccharides [60].
To date, CEs have been classified into 16 families with more than 53,400 classified and 930 non-classified CE sequences in the current CAZy database (http://www.cazy.org/). CEs act on diverse substrates including xylan (acetylxylan esterases, EC 3.1.1.72), feruloyl-polysaccharide (feruloyl esterases, EC 3.1.1.73), acetic ester (acetyl esterases, EC 3.1.1.6), peptidoglycan (poly-N-acetylglucosamine deacetylases, EC 3.5.1.104), chitin (chitin deacetylases, EC 3.5.1.41), and pectin (pectinesterase, EC 3.1.1.11) among others [61]. In the present study, a total of 61 CEs classified into 10 families were identified in F. ononidis genome based on a dbCAN database search (Figure 2(c) and Supplementary data S3). CE1 families were prominently present with 19 and CE4 family was the second largest family with 11 in the F. ononidis genome (Figure 2(c)). However, a relatively low number of CEs were identified by an NCBI-NR search with 23 and 11 CEs commonly identified from the three different databases (Figure 3(e) and Supplementary data S8).
Several characteristic features in the amino acid sequences of CE families have been identified [62]. Some CE1, CE4, CE5, and CE7 families contain the GXSXG (Gly-Xaa-Ser-Xaa-Gly) conserved motif in their amino acid sequences [62]. However, CE2 and CE3 families contain the Gly-Asp-Ser-(Leu) (GDS[L]) motif in their amino acid sequences [62].
Our results also revealed those conserved motifs in the amino acid sequences of some CE families including CE1, CE3, CE4, and CE5 with GXSXG (Gly-Xaa-Ser-Xaa-Gly) and GDS (Gly-Asp-Ser) (Figure 4(b) and Supplementary data S3). In addition, esterases involved with class C β-lactamases have a Gly-Xaa-Xaa-Leu (GXXL) motif in their amino acid sequences [63,64]. Indeed, all members of the CE16 family identified in the F. ononidis genome contained multiple copies of these conserved motifs in their amino acid sequences (Figure 4(c) and Supplementary data S3).
CEs generally facilitate the access of GHs to accelerate the degradation of substrates, thereby leading to saccharification [65]. Therefore, our results suggest that F. ononidis having various CE families may be useful for industrial applications that require saccharification, such as food, animal feed, and biofuel production.
3.7. AAs in F. ononidis genome
The lignin degradation enzymes, such as lytic polysaccharide monooxygenases (LMPOs), are classified into AA families in the CAZy database (http://www.cazy.org/). Levasseur et al. [66] reported that the GH61 and CBM33 families were characterized as LPMOs, and these families were reclassified into a new category as AA. These AA families catalyze non-carbohydrate structural components, such as lignin, which is the primary cell wall component of plants [14].
These AAs are classified into 15 families with more than 10,300 classified families based on mainly amino acid sequence similarities in the CAZy database (http://www.cazy.org/). The AA members are grouped into eight families of ligninolytic enzymes and three families of LPMOs. F. ononidis contains a total of 11 AA families with 87 AAs in its genome (Figure 2(c) and Supplementary data S3). In addition, our results also show that F. ononidis contains the majority members of AA3 (GMC oxidoreductase; alcohol oxidase, aryl-alcohol oxidase/glucose oxidase, cellobiose dehydrogenase, pyranose oxidase), AA7 (glucooligosaccharide oxidase), and AA9 (lytic polysaccharide monooxygenase; GH61) family in its genome (Figure 2(c) and Supplementary data S3). Moreover, 54 and 102 associated with AAs were identified by NCBI-NR and Pfam 31.0 database search, respectively (Figure 3(f) and Supplementary data S9) and 33 AAs were commonly identified from the three different databases (Figure 3(f) and Supplementary data S9).
In previous studies, several AA families have been identified to contain the conserved motifs that are required for the interaction with substrates [67–70]. Laccases (EC 1.10.3.2) in the AA1 family (multi-copper oxidase) contain copper-binding motifs of His-Xaa-His-Gly (HXHG), His-Xaa-Xaa-His-Xaa-His (HXXHXH), and His-Cys-His-Xaa(3)-His-Xaa(4)-Met/Leu/Phe (HCHXXXHXXXXM/L/F) within their amino acid sequences [67]. It has also been reported that GMC oxidoreductase of the AA3 family possesses the β-α-β dinucleotide binding-motif, Gly-Xaa-Gly-Xaa-Xaa-Gly-Xaa18-Glu (GXGXXGX18E), which interacts with the flavin adenine dinucleotide cofactor [68–70]. Our results also demonstrate that laccases belonging to the AA1 family and most GMC oxidoreductases belonging to the AA3 family contain the copper-binding motif (Figure 4(d) and Supplementary data S3) and the β-α-β dinucleotide binding motif (Figure 4(e) and Supplementary data S3) within their amino acid sequences, respectively, suggesting that the AA1 and AA3 families may act as laccase and GMC oxidoreductase on non-carbohydrate structural components.
Plants cell walls often contain a significant amount of lignin (non-carbohydrate structural component), which is an obstacle to the conversion of biomass (mainly carbohydrates) into bioenergy, such as bioethanol. Several recent studies suggested that the use of microbial enzymes belong to AA members might be useful to degrade the recalcitrant lignin matrix for bioethanol production [71–73]. In the present study, a vast array of genes associated with AA family was identified found in the F. ononidis genome. These results suggest the strong potential of F. ononidis as a biomaterial and for bioenergy production through the degradation of non-carbohydrate structural components such as the recalcitrant lignin matrix.
3.8. CAZyme repertoire of F. ononidis and other fungal species
In the present study, analysis of the genome sequence of F. ononidis revealed a series of genes associated with GT, GH, PL, CE, AA, and CBM. For genome-wide comparisons, CAZymes of 7 fungal species were further identified by a dbCAN CAZyme database search [37]. In addition, CAZymes of 8 other fungal species were obtained from the CAZy database (http://www.cazy.org/) and the JGI Fungi Portal database (https://genome.jgi.doe.gov/programs/fungi/index.jsf). Table 4 shows the distribution of the total CAZymes of F. ononidis and 15 other fungal species (see also Supplementary data S10).
Table 4.
CAZymes of F. ononidis and other fungal species.
| Taxon | Species | AA | GH | GT | CE | CBM | PL | Total | Annotation |
|---|---|---|---|---|---|---|---|---|---|
| Basidiomycota | F. ononidis | 87 | 228 | 87 | 61 | 40 | 21 | 524 | dbCAN (this study) |
| F. velutipes | 85 | 239 | 84 | 63 | 44 | 25 | 540 | dbCAN (this study) | |
| A. bisporus | 81 | 174 | 54 | 33 | 44 | 9 | 395 | JGI | |
| C. cinerea | 111 | 195 | 83 | 60 | 105 | 16 | 570 | dbCAN (this study) | |
| L. bicolor | 55 | 170 | 96 | 18 | 31 | 7 | 377 | JGI | |
| L. edodes | 82 | 254 | 85 | 44 | 61 | 11 | 537 | dbCAN (this study) | |
| P. chrysosporium | 85 | 175 | 65 | 16 | 62 | 4 | 407 | JGI | |
| S. commune | 78 | 241 | 85 | 57 | 37 | 18 | 516 | dbCAN (this study) | |
| U. maydis | 28 | 113 | 61 | 29 | 10 | 2 | 243 | dbCAN (this study) | |
| C. neoformans | 14 | 97 | 70 | 5 | 12 | 4 | 202 | CAZy | |
| Ascomycota | C. militaris | 54 | 165 | 91 | 34 | 39 | 5 | 388 | dbCAN (this study) |
| T. reesei | 59 | 210 | 90 | 32 | 44 | 5 | 440 | dbCAN (this study) | |
| S. cerevisiae | 5 | 57 | 68 | 2 | 12 | 0 | 144 | CAZy | |
| A. nudulans | 33 | 267 | 91 | 30 | 46 | 23 | 490 | CAZy | |
| N. crassa | 35 | 177 | 80 | 21 | 42 | 4 | 359 | CAZy | |
| B. cinerea | 77 | 287 | 119 | 37 | 89 | 10 | 619 | CAZy |
In CAZyme identification based on the dbCAN database search, 87 GTs were identified from the F. ononidis genome and the GT2 family was prominently present, with 11 genes (Figure 2(a) and Supplementary data S3). More than 427,000 classified GT sequences have been divided into 105 families in the CAZy database (http://www.cazy.org/), of which more than 132,000 are GT2 family members. Breton et al. [38] reported that not all GTs from various organisms were present in the database, and the number of families will be gradually increased as new GTs are discovered. In the present study, the GT2 family was also prominently present in F. ononidis as well as other fungal species, implying that this family is a major component of the GT family of various organisms (Supplementary data S10).
Various members of the GH families were identified in most fungal species including F. ononidis. The GH16 family was most abundant, except for some ascomycetes including C. militaris, T. ressei, S. cerevisiae, and A. nudulans (Supplementary data S10). In addition, multiple copies of the GH5 and GH18 families in F. ononidis were similar to those in some other basidiomycetes. GH5 (formerly cellulase family A) is one of the largest of the GH families and utilizes a variety of substrates, such as cellulase, endomannanase, exoglucanases, exomannanases, β-glucosidase, and β-mannosidase [74]. GH18 family includes chitinase (EC 3.2.1.14), lysozyme (EC 3.2.1.17), and endo-β-N-acetylglucosaminidase (EC 3.2.1.96). Chitinases break down all forms of chitin at varying rates depending on the enzyme and the substrate [75]. Recently, various genes related to the degradation of cellulose, chitin, and xylan have been identified in the bacterial genome, suggesting industrial applicability, such as biopolymer degradation [76–78]. In addition to bacteria, several fungi also show high hydrolytic activity toward polysaccharides due to their degradation machinery [48,79,80]. Thus, the various GHs revealed in the F. ononidis genome as well as the various fungi suggests the potential for biotechnology applications.
Zhao et al. [80] reported that some PL families are phylum-specific, among which PL10 and PL11 are ascomycetes-specific, with PL15 only in basidiomycetes. In the present study, PL family 11 and 15 were found only in A. nidulans (ascomycete) and C. cinerea (basidiomycete), respectively. However, most of the other fungal species have no PL family 11 or 15. Moreover, our results revealed that PL5, -14, and -24 families are basidiomycetes-specific (Supplementary data S10). To date, all characterized pectin lyases belong to the PL1 family, and the fungal pectate lyases belong to families PL1, PL3, and PL9 (CAZy database). Our results showed that PL family 1 and 3 comprised the majority of PLs of F. ononidis as well as that of other fungal species. However, most fungal species did not have the PL9 family. This family was only found in F. ononidis, F. velutipes [16], S. commune [34], C. militaris [27], and A. nidulans [23] (Supplementary data S10). Xavier-Santos et al. [54] reported that S. commune (basidiomycete) produced higher polygalacturonase than A. niger in wheat bran cultures. Thus, because of their diverse roles in the environment, there is a compelling reason to discover new PLs with unique properties from the basidiomycetes.
F. ononidis does not have a unique CBM in its genome, but the distribution of multiple copies of the CBM1, CBM13, and CBM50 CBM is similar to some other fungal species. However, the abundance of some families differed between basidiomycetes and ascomycetes. Ascomycetes had more CBM18 family members than other basidiomycetes and the CBM5 and CBM12 families were not observed in all ascomycetes (Supplementary data S10). Zhao et al. [80] also demonstrated that ascomycetes have more CBM18 family members than basidiomycetes, while CBM5 and CBM12 have less. Interestingly, the distribution of CBM in fungal species showed that the largest number of CBMs, including 52 CBM family 1, were found in the coprophilic fungus C. cinerea genome. Fernandez-Fueyo et al. [71] also observed a vast array of genes associated with CBMs including CBM family 1 in C. cinerea genome.
The total number of CEs in F. ononidis was similar with that of F. velutipes, and the CE1, CE4, and CE16 families were prominently present in F. ononidis and other basidiomycetes. Presently, the abundance of the CE family also varied in some basidiomycetes and ascomycetes. For example, only 5 CEs, including 4 CE4 and 1 CE9 families, and 2 CE4 family members have been found in C. neoformans and S. cerevisiae (Supplementary data S10). In the CAZyme prediction against the dbCAN database, various CE classified in the CE10 family were identified in the F. ononidis genome (data not shown). However, all of the CE10 family members were not included in the study because they were found to act on non-carbohydrate substrates [15].
The total number of AAs in F. ononidis was similar to that of other white rot fungus, such as F. velutipes (white rot) [16], L. edodes (white rot) [30], S. commune (white rot-like) [34], and P. chrysosporium (white rot) [32], but not C. cinerea (white rot) [26] (Table 4 and Supplementary data S10). In addition, fungal species including L. bicolor (ectomycorrhizal fungus) [29], U. maydis (plant pathogen) [36], and C. neoformans (yeast) [28] have fewer AAs than other basidiomycetes. Unicellular and lipophilic fungi, such as C. neoformans, Rhodotorula glutinis, and Wallemia sebi, possess very few genes encoding polysaccharide degrading enzymes [14]. It has also been suggested that U. maydis possesses at least a gene encoding a polysaccharide degrading enzyme due to plant defense responses [32,36].
As described above, various members of CAZyme were identified in the F. ononidis genome including 228 GHs, 21 PLs, 61 CEs, and 87 AAs associated with polysaccharide and lignin degradation (Table 4). Our results reveal that the distribution of this CAZyme repertoire in F. ononidis is comparable to those of other basidiomycetes. Although further studies of these CAZymes are needed, the present study suggests that F. ononidis degradation machinery has great potential for industrial and biotechnological applications, such as food, paper, textile, animal feed, and other chemicals, including biofuels.
Supplementary Material
Funding Statement
This research was supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2016R1D1A1B03930808).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Arnolds E. Einige Pilze eines Halbtrockenrasen bei Detmold (Westfalen). Westfäl Pilzbriefe. 1977;11:29–39. [Google Scholar]
- 2.Ripková S, Adamčík S, Kučera V. Flammulina ononidis – a new species for Slovakia. Czech Mycol. 2008;60:221–230. [Google Scholar]
- 3.Łuszczyński J, Łuszczyńska B, Tomaszewska A. Flammulina ononidis – first record in Poland. Acta Mycol. 2014;49:79–85. [Google Scholar]
- 4.Ripková S, Hughes K, Adamčík S, et al. The delimitation of Flammulina fennae. Mycol Progress. 2010;9:469–484. [Google Scholar]
- 5.Hughes KW, McGhee LL, Methven AS, et al. Patterns of geographic speciation in the genus Flammulina based on sequences of the ribosomal ITS1-5.8S-ITS2 area. Mycologia. 1999;91:978–986. [Google Scholar]
- 6.Psurtseva NV. Culture of Flammulina velutipes (Fr.) P.Karst. (biology and economic importance. Mikol. i Fitopatol. l987;21:477–486. [Google Scholar]
- 7.Klan J, Baudisova D. Cultural, enzyme and genetic studies in the genus Flammulina Karst. Mycotaxon. 1992;43:341–350. [Google Scholar]
- 8.Nobles MJ. Identification of cultures of wood inhabiting Hymenomycetes. Can J Bot. 1965;26:281–413. [Google Scholar]
- 9.Marr CD. Laccase and peroxidase oxidation of spot test reagents. Mycotaxon. 1979;9:244–276. [Google Scholar]
- 10.Alekhina I, Psurtseva N, Yli-Mattila T. Isozyme analysis of LE(BIN) Collection Flammulina strains. Karstenia. 2001;41:55–63. [Google Scholar]
- 11.Psurtseva NY, Mnoukhina AY. Morphological, physiological and enzyme variability of Flammulina P. Karst. cultures. Mikol i Fitopatol. 1998;32:49–54. [Google Scholar]
- 12.Pérez-Butrón JL, Ferdnández-Vicente J. Una nueva especie de Flammulina P. Karsten, F. cephalariae (Agaricales) encontrada en España. Rev Catalana Micol. 2007;29:81–91. [Google Scholar]
- 13.Eriksson K, Blanchette RA, Ander P. Microbial and enzymatic degradation of wood and wood components. Berlin, Germany: Springer-Verlag; 1990. [Google Scholar]
- 14.Rytioja J, Hildén K, Yuzon J, et al. Plant-polysaccharide-degrading enzymes from basidiomycetes. Microbiol Mol Biol Rev. 2014;78:614–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lombard V, Golaconda Ramulu H, Drula E, et al. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park YJ, Baek JH, Lee S, et al. Whole genome and global gene expression analyses of the model mushroom Flammulina velutipes reveal a high capacity for lignocellulose degradation. PLoS One. 2014;9:e93560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanke M, Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–W467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchfink B, Xie C, Huson D. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. [DOI] [PubMed] [Google Scholar]
- 21.Finn RD, Coggill P, Eberhardt R, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emms D, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galagan JE, Calvo SE, Cuomo C, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. [DOI] [PubMed] [Google Scholar]
- 24.Staats M, van Kan JA. Genome update of Botrytis cinerea strains B05.10 and T4. Eukaryot Cell. 2012;11:1413–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morin E, Kohler A, Baker AR, et al. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc Natl Acad Sci USA. 2012;109:17501–17506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stajich JE, Wilke SK, Ahrén D, et al. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc Natl Acad Sci USA. 2010;107:11889–11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng P, Xia Y, Xiao G, et al. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol. 2011;12:R116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janbon G, Ormerod K, Paulet D, et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLOS Genet. 2014;10:e1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin F, Aerts A, Ahrén D, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. [DOI] [PubMed] [Google Scholar]
- 30.Chen L, Gong Y, Cai Y, et al. Genome sequence of the edible cultivated mushroom Lentinula edodes (Shiitake) reveals insights into lignocellulose degradation. PloS One. 2016;11:e0160336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galagan JE, Calvo SE, Borkovich KA, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. [DOI] [PubMed] [Google Scholar]
- 32.Martinez D, Larrondo LF, Putnam N, et al. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol. 2004;22:695–700. [DOI] [PubMed] [Google Scholar]
- 33.Fisk DG, Ball CA, Dolinski K, et al. Saccharomyces cerevisiae S288C genome annotation: a working hypothesis. Yeast. 2006;23:857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohm RA, De Jong JF, Lugones L, et al. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol. 2010;28:957–963. [DOI] [PubMed] [Google Scholar]
- 35.Li WC, Huang CH, Chen CL, et al. Trichoderma reesei complete genome sequence, repeat-induced point mutation, and partitioning of CAZyme gene clusters. Biotechnol Biofuels. 2017;10:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kämper J, Kahmann R, Bölker M, et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 2006;444:97–101. [DOI] [PubMed] [Google Scholar]
- 37.Yin Y, Mao X, Yang JC, et al. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Breton C, Šnajdrová L, Jeanneau C, et al. Structures and mechanisms of glycosyltransferases. Glycobiology. 2006;16:29R–37R. [DOI] [PubMed] [Google Scholar]
- 39.Lairson LL, Henrissat B, Davies GJ, et al. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. [DOI] [PubMed] [Google Scholar]
- 40.Coutinho PM, Deleury E, Davies GJ, et al. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. [DOI] [PubMed] [Google Scholar]
- 41.Campbell JA, Davies GJ, Bulone V, et al. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326:929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukai Y, Hirokawa T, Tomii K, et al. Identification of glycosyltransferases focusing on Golgi transmembrane region. Tigg. 2007;19:41–47. [Google Scholar]
- 43.Berlemont R, Martiny AC. Glycoside hydrolases across environmental microbial communities. PLoS Comput Biol. 2016;12:e1005300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hahn M, Olsen O, Politz O, et al. Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-beta-glucanase. J Biol Chem. 1995;270:3081–3088. [DOI] [PubMed] [Google Scholar]
- 46.Masuda S, Endo K, Koizumi N, et al. Molecular identification of a novel beta-1,3-glucanase from alkaliphilic Nocardiopsis sp. strain F96. Extremophiles. 2006;10:251–255. [DOI] [PubMed] [Google Scholar]
- 47.Kotake T, Hirata N, Degi Y, et al. Endo-beta-1,3-galactanase from winter mushroom Flammulina velutipes. J Biol Chem. 2011;286:27848–27854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berlemont R. Distribution and diversity of enzymes for polysaccharide degradation in fungi. Sci Rep. 2017;7:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sutherland IW. Polysaccharide lyases. FEMS Microbiol Rev. 1995;16:323–347. [DOI] [PubMed] [Google Scholar]
- 50.Yip VL, Withers SG. Breakdown of oligosaccharides by the process of elimination. Curr Opin Chem Biol. 2006;10:147–155. [DOI] [PubMed] [Google Scholar]
- 51.van den Brink J, de Vries RP. Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol. 2011;91:1477–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garron ML, Cygler M. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology. 2010;20:1547–1573. [DOI] [PubMed] [Google Scholar]
- 53.Linhardt RJ, Galliher PM, Cooney CL. Polysaccharide lyases. Appl Biochem Biotechnol. 1986;12:135–176. [DOI] [PubMed] [Google Scholar]
- 54.Xavier-Santos S, Carvalho CC, Bonfá M, et al. Screening for pectinolytic activity of wood-rotting basidiomycetes and characterization of the enzymes. Folia Microbiol. 2004;49:46–52. [DOI] [PubMed] [Google Scholar]
- 55.Boraston AB, Bolam DN, Gilbert HJ, et al. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382:769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shoseyov O, Shani Z, Levy I. Carbohydrate binding modules: biochemical properties and novel applications. Microbiol Mol Biol Rev. 2006;70:283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Várnai A, Mäkelä MR, Djajadi DT, et al. Carbohydrate-binding modules of fungal cellulases: occurrence in nature, function, and relevance in industrial biomass conversion. Adv Appl Microbiol. 2014;88:103–165. [DOI] [PubMed] [Google Scholar]
- 58.Bornscheuer UT. Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol Rev. 2002;26:73–81. [DOI] [PubMed] [Google Scholar]
- 59.Jaeger KE, Eggert T. Lipases for biotechnology. Curr Opin Biotechnol. 2002;13:390–397. [DOI] [PubMed] [Google Scholar]
- 60.Cantarel BL, Coutinho PM, Rancurel C, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Biely P. Microbial carbohydrate esterases deacetylating plant polysaccharides. Biotechnol Adv. 2012;30:1575–1588. [DOI] [PubMed] [Google Scholar]
- 62.Adesioye FA, Makhalanyane TP, Biely P, et al. Phylogeny, classification and metagenomic bioprospecting of microbial acetyl xylan esterases. Enzyme Microb Technol. 2016;93–94:79–91. [DOI] [PubMed] [Google Scholar]
- 63.Wei Y, Schottel JL, Derewenda U, et al. A novel variant of the catalytic triad in the Streptomyces scabies esterase. Nat Struct Biol. 1995;2:218–223. [DOI] [PubMed] [Google Scholar]
- 64.Petersen EI, Valinger G, Sölkner B, et al. A novel esterase from Burkholderia gladioli which shows high deacetylation activity on cephalosporins is related to beta-lactamases and DD-peptidases. J Biotechnol. 2001;89:11–25. [DOI] [PubMed] [Google Scholar]
- 65.Christov LP, Prior BA. Esterases of xylan-degrading microorganisms: production, properties, and significance. Enzyme Microb Technol. 1993;15:460–475. [DOI] [PubMed] [Google Scholar]
- 66.Levasseur A, Drula E, Lombard V, et al. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels. 2013;6:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reiss R, Ihssen J, Richter M, et al. Laccase versus laccase-like multi-copper oxidase: a comparative study of similar enzymes with diverse substrate spectra. PloS One. 2013;8:e65633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fernandez IS, Ruiz-Duenas FJ, Santillana E, et al. Novel structural features in the GMC family of oxidoreductases revealed by the crystal structure of fungal aryl-alcohol oxidase. Acta Crystallogr D Biol Crystallogr. 2009;65:1196–1205. [DOI] [PubMed] [Google Scholar]
- 69.Varela E, Jesús Martínez M, Martínez AT, Arylalcohol oxidase protein sequence: a comparison with glucose oxidase and other FAD oxidoreductases. Biochem Biophys Acta Protein Struct Mol Enzymol. 2000;1481:202–208. [DOI] [PubMed] [Google Scholar]
- 70.Wierenga RK, Drenth J, Schulz GE, et al. Comparison of the 3-dimensional protein and nucleotide structure of the FAD-binding domain of parahydroxybenzoate hydroxylase with the FAD-binding as well as NADPH-binding domains of glutathionereductase. J Mol Biol. 1983;167:725–739. [DOI] [PubMed] [Google Scholar]
- 71.Fernandez-Fueyo E, Ruiz-Duenas FJ, Ferreira P, et al. Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. Proc Natl Acad Sci USA. 2012;109:5458–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martínez AT, Ruiz-Dueñas FJ, Martínez MJ, et al. Enzymatic delignification of plant cell wall: from nature to mill. Curr Opin Biotechnol. 2009;20:348–357. [DOI] [PubMed] [Google Scholar]
- 73.Ruiz-Dueñas FJ, Martínez AT. Microbial degradation of lignin: how a bulky recalcitrant polymer is efficiently recycled in nature and how we can take advantage of this. Microb Biotechnol. 2009;2:164–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Henrissat B, Claeyssens M, Tomme P, et al. Cellulase families revealed by hydrophobic cluster analysis. Gene. 1989;81:83–95. [DOI] [PubMed] [Google Scholar]
- 75.The CAZypedia Consortium. Ten years of CAZypedia: a living encyclopedia of carbohydrate-active enzymes. Glycobiology. 2018;28:3–8. [DOI] [PubMed] [Google Scholar]
- 76.Berlemont R, Martiny AC. Phylogenetic distribution of potential cellulases in bacteria. Appl Environ Microbiol. 2013;79:1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Berlemont R, Martiny AC. Genomic potential for polysaccharides deconstruction in bacteria. Appl Environ Microbiol. 2015;81:1513–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Talamantes D, Biabini N, Dang H, et al. Natural diversity of cellulases, xylanases, and chitinases in bacteria. Biotechnol Biofuels. 2016;9:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eichlerová I, Homolka L, Žifčáková L, et al. Enzymatic systems involved in decomposition reflects the ecology and taxonomy of saprotrophic fungi. Fungal Ecol. 2015;13:10–22. [Google Scholar]
- 80.Treseder KK, Lennon JT. Fungal traits that drive ecosystem dynamics on land. Microbiol Mol Biol Rev. 2015;79:243–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao Z, Liu H, Wang C, et al. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics. 2013;14:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.