Abstract
While surveying undiscovered fungal taxa in Korea, three rare zygomycetous fungal strains, CNUFC-PTF2-1, CNUFC-TF3-1, and CNUFC-ESAF3-1, were isolated from soil, leaf, and freshwater samples, respectively. The strains were analyzed morphologically as well as phylogenetically based on the internal transcribed spacer region and 28S rDNA sequences. Sequence analysis of the two loci revealed that the isolates, CNUFC-PTF2-1, CNUFC-TF3-1, and CNUFC-ESAF3-1, were identified as Backusella circina, Circinella muscae, and Mucor ramosissimus, respectively. These species have not yet been previously described in Korea.
KEYWORDS: Backusella circina; Circinella muscae; Mucor ramosissimus, taxonomy, zygomycetous fungus
1. Introduction
The Mucorales, which is classified into the subphylum Mucoromycotina [1], is the largest order of fungi. Members of this group are ubiquitous saprophytes in nature. They are commonly found in soil and decaying vegetation, and can also be found in grains [2–4]. Mucorales members grow and invade quickly on easily digestible substrates, such as those containing starches, sugars, and hemicelluloses [2]. Currently, 14 families are placed in this order based on the analysis of a multigene (act-1, EF-1α, 18S, and 28S rRNA) data set [3].
The genus Backusella, which belongs to the subphylum Mucoromycotina, order Mucorales, and family Backusellaceae, was established by Ellis & Hesseltine (1969) with the type species B. circina [5]. Species belonging to this genus are characterized by the production of both sporangia and sporangiola, as well as by the formation of transitorily curved sporangiophores [4–6]. They are typically isolated from soil, leaf litter, and other plant debris, as well as from dung samples, such as those from human, agouti, and insects [7–10]. For a long time, the genus Backusella contained only three species: B. circina, B. lamprospora, and B. ctenidia. However, Walther et al. [4] recently revised the order Mucorales based on internal transcribed spacer (ITS) and 28S rDNA sequence data and transferred some species of Mucor to the genus Backusella because they had transitorily recurved sporangiophores, while B. ctenidia was transferred to the genus Mucor. Based on these criteria, this genus now comprises 13 species, among which one species is registered in Korea (Source: www.indexfungorum.org as of April 2018).
The genus Circinella, which belongs to the subphylum Mucoromycotina, order Mucorales, and family Lichtheimiaceae, was established in 1873 by van Tieghem and Le Monnier [11]. It is closely related to Mucor, but differs in that it has sporangiophores with circinate branches bearing sporangia [12]. Members of this genus are characterized by the production of sporangiophores bearing circinate branches terminated by globose multispored sporangia with persistent sporangial walls [12]. After Hesseltine and Fennell [12] monographed this genus, several additional species were included [13–16]. To date, 10 species belonging to this genus are known according to the Index Fungorum (www.indexfungorum.org). They are commonly isolated from soil, dung, sand beach, and hydrocarbon-polluted sand [2,17,18].
The genus Mucor belongs to the subphylum Mucoromycotina, order Mucorales, and family Mucoraceae, and was described by Fresenius in 1850, comprising the largest number of species within the Mucorales [19]. Specimens of this genus are characterized by the formation of non-apophysate sporangia and production of simple or branched sporangiophores without basal rhizoids. Zygospores have opposed, non-appendaged suspensors [20]. Mucor species are easily isolated from soil, fruits, vegetables, stored grains, insect, and dung [2,21–24]. Several species of this genus are of great interest to the biotechnology industry due to their ability to produce enzymes such as proteases, amylase, lipases, phytase, and polygalacturonase [25–27], while some species are considered as the causal agents of cutaneous zygomycosis in humans [28]. The traditional taxonomic classification of Mucor species was determined based on morphological characteristics such as size and shape of the sporangia and mode of reproduction (sexual or asexual). Recently, molecular studies have been performed to evaluate mucoralean species [3,4]. These studies have indicated that Mucor is polyphyletic. Based on the phylogenetic analysis of ITS and large subunit rDNA regions of several mucoralean species, Walther et al. [4] observed that some Mucor species with curved sporangiophores were grouped with Backusella Hesselt. & J. J. Ellis. Therefore, these Mucor species were transferred to Backusella. Nine species have been recorded, including three new species from freshwater, tangerine fruit, and rat feces samples in Korea [10,23].
During an inventory of fungal species from soil, leaf, and freshwater samples, three interesting fungal strains belonging to the order Mucorales were assigned to the genera Backusella, Circinella, and Mucor.
The objective of this study was to morphologically and molecularly characterize three unrecorded species in Korea: B. circina, C. muscae, and M. ramosissimus.
2. Materials and methods
2.1. Isolation of fungal strains from leaf, soil, and freshwater samples
Leaves of Toxicodendron sylvestre were collected from Daegak Mountain, Sinsi Island, Gunsan, Korea. Collected samples were stored in sterile polyethylene bags. Samples were cleaned under running tap water to remove debris before use. Leaf tissue pieces were cut into small fragments, surface-disinfested with 2% NaOCl solution and 70% ethanol for 1 min each, washed three times with sterile distilled water, plated on potato dextrose agar (PDA; BD Biosciences, Franklin Lakes, NJ) supplemented with the antibiotic streptomycin sulfate (0.5 mg/mL, Sigma-Aldrich, St. Louis, MO), and incubated at 25 °C for 3–7 days.
Soil samples were collected from Geumgol Mountain, Jin Island (Jindo), Korea. Freshwater samples were collected from Eulsukdo, Busan, Korea. The samples were transported in sterile 50-mL Falcon tubes and stored at 4 °C until examination. Fungi were isolated using the serial dilution plating method. In this technique, 1 mL of water or 1 g of soil was mixed with 9 mL of sterile distilled water and shaken for 15 min at 25 °C; serial dilutions ranging from 10−1 to 10−4 were prepared. An aliquot of 0.1 mL from each dilution was transferred to PDA supplemented with the antibiotic streptomycin sulfate (0.5 mg/mL) and incubated at 25 °C for 3–7 days. To isolate pure cultures, individual colonies with various morphologies were picked, transferred to PDA, and subcultured until pure mycelia were obtained. All pure isolates, including those of B. circina, C. muscae, and M. ramosissimus, were stored in 20% glycerol at −80 °C at the Chonnam National University Fungal Collection (CNUFC), Gwangju, Korea. B. circina, C. muscae, and M. ramosissimus strains isolated in our study were designated CNUFC-PTF2-1 and CNUFC-PTF2-2, CNUFC-TF3-1 and CNUFC-TF3-2, CNUFC-ESAF3-1 and CNUFC-ESAF3-2, respectively. Strain CNUFC-PTF2-1 was also deposited at the Culture Collection of the National Institute of Biological Resources (NIBR, Incheon, Korea), strain CNUFC-TF3-1 was deposited at Korean Agricultural Culture Collection (Wanju, Korea), and strain CNUFC-ESAF3-1 was deposited at the Culture Collection of the Nakdonggang National Institute of Biological Resources (NNIBR, Sangju, Korea).
2.2. Morphological studies
Pure cultures of B. circina, C. muscae, and M. ramosissimus were cultured on synthetic mucor agar (SMA; 40 g dextrose, 2 g asparagine, 0.5 g KH2PO4, 0.25 g MgSO4·7H2O, 0.5 g thiamine chloride, and 15 g agar in 1 L of deionized water). The plates were incubated at 10, 15, 20, 25, 30, 35, and 40 °C in the dark for 5 days. Fragments of mycelia were removed from the cultures, placed on microscope slides with lactophenol solution (Junsei Chemical Co. Ltd., Tokyo, Japan) and observed under a light microscope (Olympus, Tokyo, Japan). The fine structure of C. muscae was observed using scanning electron microscopy (Hitachi S4700; Hitachi, Tokyo, Japan). The isolates were fixed in 2.5% paraformaldehyde-glutaraldehyde in 0.05 M phosphate buffer (pH 7.2) for 2 h and then washed with 0.05 M cacodylate buffer (Junsei Chemical Co. Ltd.). Cellular membranes were preserved by fixing the samples in 1% osmium tetroxide (Electron Microscopy Sciences, Hatfield, PA) diluted in 0.05 M cacodylate buffer for 1 h, washing again in 0.05 M cacodylate buffer, dehydrating in graded ethanol (Emsure, Darmstadt, Germany) and isoamyl acetate (Junsei Chemical Co. Ltd.), and drying in a fume hood. Finally, the samples were sputter-coated with gold and observed under a Hitachi S4700 field emission scanning electron microscope at the Korea Basic Science Institute (Gwangju, Korea).
2.3. DNA extraction, PCR, and sequencing
Genomic DNA was extracted directly from the mycelia of fungal isolates using the Solgent Genomic DNA prep Kit (Solgent Co. Ltd., Daejeon, South Korea). The ITS region and large subunit of 28S rDNA were amplified with the primer pairs ITS4 and ITS5 [29] and LROR and LR5F [30], respectively. The PCR mixture (total volume, 20 μL) contained fungal DNA template, 5 pmol/μL of each primer, and Accupower PCR Premix (Taq DNA polymerase, dNTPs, buffer, and tracking dye; Bioneer Corp., Daejeon, Korea). PCR products were purified using the Accuprep PCR Purification Kit (Bioneer Corp.) according to the manufacturer’s instructions. DNA sequencing was performed on an ABI 3700 Automated DNA sequencer (Applied Biosystems, Inc., Foster City, CA).
2.4. Phylogenetic analysis
The fungal sequences obtained from the GenBank database (Table 1) were aligned using Clustal_X v.1.83 [31] and edited with Bioedit v.5.0.9.1 [32]. Phylogenetic analyses were performed using MEGA 6 software [33] and maximum likelihood (ML) was constructed by Kimura’s two-parameter correction method. M. amphibiorum, M. indicus, and Rhizomucor pusillus were used as outgroups. The reliability of internal branches was assessed using the p-distance substitution model with 1000 bootstrap replicates. Sequence data were compared with similar sequences available in the GenBank databases using nucleotide Basic Local Alignment Search Tool (BLASTn).
Table 1.
Taxa, collection numbers, sequences, and GenBank accession numbers used in this study.
| Taxon name | Collection no. (Isolate no.) | GenBank accession no. |
|
|---|---|---|---|
| ITS | 28S | ||
| Backusella circina | CBS 128.70T | JN206258 | JN206529 |
| B. circina | KH1 | JX644544 | JX644491 |
| B. circina | CBS 323.69 | JN206259 | – |
| B. circina | KH9 | – | JX644492 |
| B. circina | CNUFC-PTF2-1 | MH262302 | MH262312 |
| B. circina | CNUFC-PTF2-2 | MH262303 | MH262313 |
| B. constricta | URM7322 | KT937159 | KT937156 |
| B. indica | CBS 786.70 | JN206255 | JN206526 |
| B. gigacellularis | CCIBt 3866 | KF742415 | – |
| B. grandis | CBS 186.87T | JN206252 | JN206527 |
| B. lamprospora | CBS 118.08T | JN206268 | JN206531 |
| B. lamprospora | CBS 195.28 | JN206271 | JN206530 |
| B. locustae | CNUFC-SFB2T | KY449291 | KY449290 |
| B. locustae | CNUFC-SFB4 | KY449293 | KY449292 |
| B. oblongielliptica | CBS 568.70 | JN206278 | JN206533 |
| B. oblongispora | CBS 569.70 | JN206251 | JN206407 |
| B. recurva | KH6 | – | JX644497 |
| B. recurva | KH7 | – | JX644498 |
| B. recurva | CBS 318.52 | JN206261 | – |
| B. recurva | CBS 673.75 | JN206264 | – |
| B. tuberculispora | CBS 562.66 | JN206267 | JN206525 |
| B. tuberculispora | CBS 570.70 | JN206266 | – |
| B. variabilis | CBS 564.66 | JN206254 | KC012658 |
| B. variabilis | CBS 564.66 | JN206253 | – |
| Circinella angarensis | CBS 173.62 | JN205849 | JN206551 |
| C. chinensis | CBS 140.28 | JN205855 | JN206549 |
| C. lacrymispora | CBS 101757 | JN206289 | JN206608 |
| C. minor | CBS 142.81 | JN205861 | JN206552 |
| C. mucoroides | CYD1000719 | KF805760 | KF805746 |
| C. muscae | CCD1000215 | – | KF805745 |
| C. muscae | CBS 141.28 | JN205853 | JN206548 |
| C. muscae | CBS 107.13 | JN205854 | – |
| C. muscae | CYR003 | – | KF805748 |
| C. muscae | D00122901 | KF805764 | KF805750 |
| C. muscae | CNUFC-TF3-1 | MH262304 | MH262314 |
| C. muscae | CNUFC-TF3-2 | MH262305 | MH262315 |
| C. simplex | CBS 428.80 | JN206213 | JN206445 |
| C. umbellata | CBS 160.49 | JN205858 | HM849722 |
| C. umbellata | CBS 101.16 | JN205857 | JN206553 |
| Mucor amphibiorum | CBS 763.74T | HM999957 | – |
| M. amphibiorum | NRRL28633 | – | AF113466 |
| M. circinelloides | CBS 338.71 | JN205998 | – |
| M. circinelloides | CBS 635.65 | JN205997 | – |
| M. circinelloides | UTHSC 04-1961 | – | FN650657 |
| M. circinelloides | UTHSC 06-1667 | – | FN650656 |
| M. circinelloides f. janssenii | CBS 232.29 | JN206007 | – |
| M. circinelloides f. janssenii | CBS 206.68 | JN206004 | – |
| M. circinelloides f. janssenii | CBS 205.68NT | HM999952 | – |
| M. circinelloides f. janssenii | CBS 526.68 | – | JN206426 |
| M. circinelloides f. circinelloides | CBS 195.68 | – | NG_055735 |
| M. circinelloides f. circinelloides | Kw1378 | – | FM246460 |
| M. circinelloides f. lusitanicus | CBS 108.17 | JN205980 | FN650665 |
| M. circinelloides f. lusitanicus | CBS 851.71 | JN205982 | – |
| M. circinelloides f. lusitanicus | CBS 111228 | JN205989 | – |
| M. circinelloides f. lusitanicus | CBS 242.33 | JN205987 | – |
| M. circinelloides f. lusitanicus | CBS 968.68 | HM999953 | – |
| M. circinelloides f. lusitanicus | UTHSC 03-1823 | – | FN650662 |
| M. circinelloides f. lusitanicus | NRRL 3631 | – | AF113467 |
| M. circinelloides f. lusitanicus | CBS 236.35 | JN205979 | – |
| M. fragilis | CTSP F1 | EU862184 | EU862173 |
| M. fragilis | FSU 6164 | EU484238 | – |
| M. plumbeus | CBS 634.74 | HM999955 | – |
| M. plumbeus | CBS 226.32 | JN205916 | – |
| M. indicus | CBS 226.29 | HM999956 | HM849690 |
| M. sinensis | CBS 204.74 | JN205899 | – |
| M. racemosus | UWFP 788 | – | AY213712 |
| M. racemosus f. chibinensis | CBS 636.67 | JN205904 | – |
| M. racemosus f. racemosus | CBS 260.68T | – | NG_055727 |
| M. ramosissimus | CBS 135.65NT | NR_103627 | FN650666 |
| M. ramosissimus | ATCC 28933 | – | AY213715 |
| M. ramosissimus | CNUFC-ESAF3-1 | MH262306 | MH262316 |
| M. ramosissimus | CNUFC-ESAF3-2 | MH262307 | MH262317 |
| Phascolomyces articulosus | CBS 113.76 | JX665039 | JN206547 |
| Rhizomucor pusillus | CBS 354.68 | AF461764 | HM849716 |
| Zychaea mexicana | CBS 441.76 | JN205845 | JN206545 |
Bold letters indicate isolates and accession numbers determined in our study.
ATCC: American Type Culture Collection, Manassas, VA, USA; CBS: Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; CNUFC: Chonnam National University Fungal Collection, Gwangju, South Korea; ITS: internal transcribed spacer; NRRL (ARS Culture Collection, Peoria, Illinois); UTHSC, Fungal Testing Laboratory, Department of Pathology at the University of Texas Health Science Center, San Antonio, Texas, USA; T and NT: ex-type and ex-neotype strains.
3. Results
3.1. Phylogenetic analysis
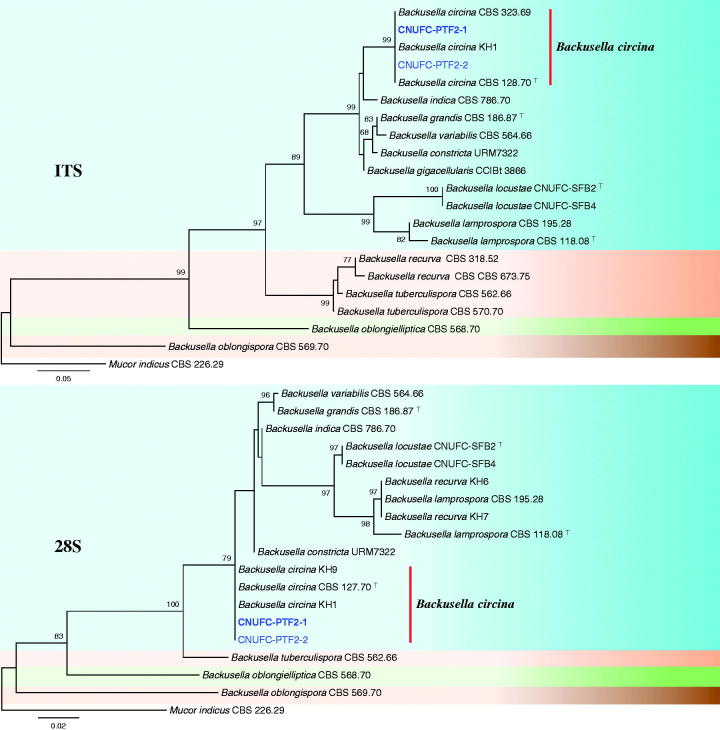
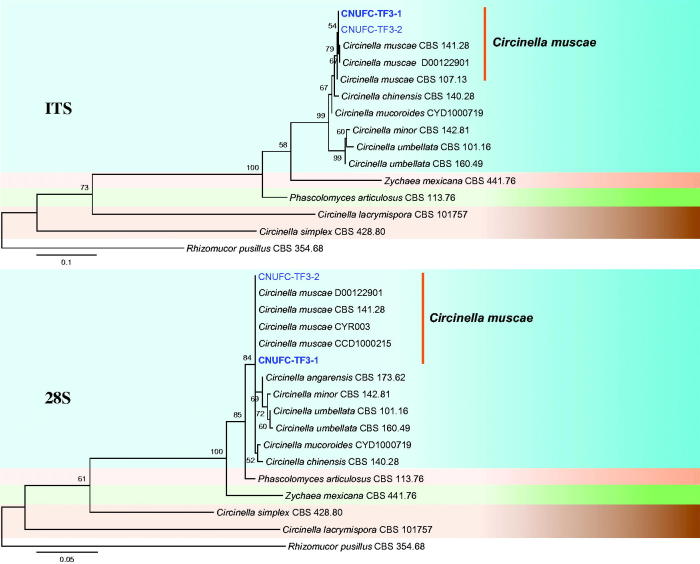
A BLAST search of ITS sequences via the NCBI database indicated that the isolates CNUFC-PTF2-1, CNUFC-TF3-1, and CNUFC-ESAF3-1 most closely resembled B. circina (GenBank accession no. JX644544), C. muscae (GenBank accession no. KF805764), and M. ramosissimus (GenBank accession no. NR_103627) with 100% (637/637 bp), 99.8% (601/602 bp), and 99.8% (566/567 bp) homology, respectively. The 28S rDNA sequences of B. circina (GenBank accession no. JN206529), C. muscae (GenBank accession no. KF805750), and M. ramosissimus (GenBank accession no. FN650666) showed 100% (674/674 bp), 98.9% (550/556 bp), and 100% (694/694 bp) homology with the 28S rDNA sequences of the isolates CNUFC-PTF2-1, CNUFC-TF3-1, and CNUFC-ESAF3-1, respectively. Based on the ITS and 28S rDNA trees, the isolates CNUFC-PTF2-1, CNUFC-TF3-1, and CNUFC-ESAF3-1 were identical to B. circina, C. muscae, and M. ramosissimus, respectively (Figures 1–3).
Figure 1.
Phylogenetic tree based on maximum likelihood analysis of internal transcribed spacers (ITS) and 28S rDNA sequences for Backusella circina CNUFC-PTF2-1 and B. circina CNUFC-PTF2-2. Mucor indicus was used as the outgroup. Bootstrap support values ≥50% are indicated at the nodes. The bar indicates the number of substitutions per position.
Figure 2.
Phylogenetic tree based on maximum likelihood analysis of internal transcribed spacers (ITS) and 28S rDNA sequences for Circinella muscae CNUFC-TF3-1 and C. muscae CNUFC-TF3-2. Rhizomucor pusillus was used as the outgroup. Bootstrap support values ≥50% are indicated at the nodes. The bar indicates the number of substitutions per position.
Figure 3.
Phylogenetic tree based on maximum likelihood analysis of internal transcribed spacers (ITS) and 28S rDNA sequences for Mucor ramosissimus CNUFC-ESAF3-1 and M. ramosissimus CNUFC-ESAF3-2. Mucor amphibiorum was used as the outgroup. Bootstrap support values ≥50% are indicated at the nodes. The bar indicates the number of substitutions per position.
3.2. Taxonomy
3.2.1. Taxonomy of CNUFC-PTF2-1
Backusella circina J.J. Ellis & Hesselt., Mycologia 61: 865 (1969) (Table 2, Figure 4).
Table 2.
Morphological characteristics of CNUFC-PTF2-1 compared to Backusella circina reference strain.
| Character | CNUFC-PTF2-1 | Backusella circinaa |
|---|---|---|
| Colony color | Rapid-growing, first white then light gray, reverse light gray | Rapid-growing, first white then light olive-gray |
| Sporangiophores | 6.0–20.0 µm in width, variable in length | Up to 9–16 µm in width, variable in length |
| Sporangia | Globose to subglobose, multispored, 27.5–66.5 × 27.0–63.5 µm | Globose and subglobose, 35–100 µm |
| Columellae | Subglobose to oblong, 18.1–30.4 × 20.3–34.6 µm | Subglobose to oblong, 11–35 × 11–30 μm |
| Unispored sporangiola | Globose to subglobose 10.3–19.5 × 10.0–19.1 µm | Globose to subglobose, (4.5–)6–16(–26) μm in diam., wall spinulose |
| Sporangiospores | Subglobose to ovoid, 6.5–11.0 × 6.0–10.2 μm | Subglobose to ovoid, (6.4–)7.2–10 (–12.8) × (5.6–)6.4–9.2 (–10) μm |
| Zygospores | Absent | Globose to subglobose, (35–)40–70(–80) μm in diam. |
From the description by Ellis and Hesseltine [5].
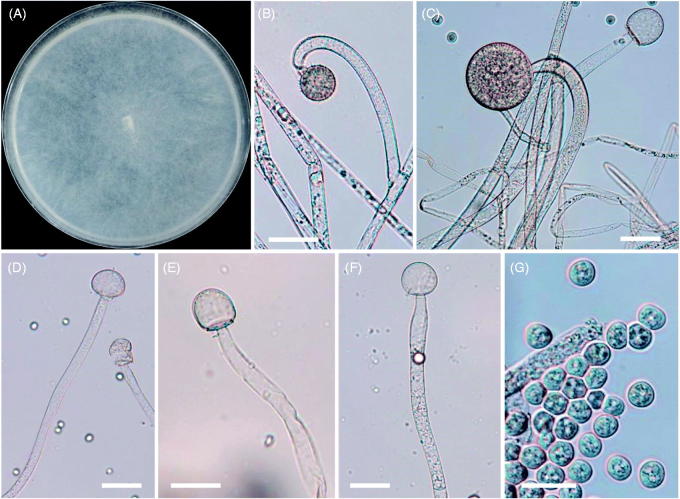
Figure 4.
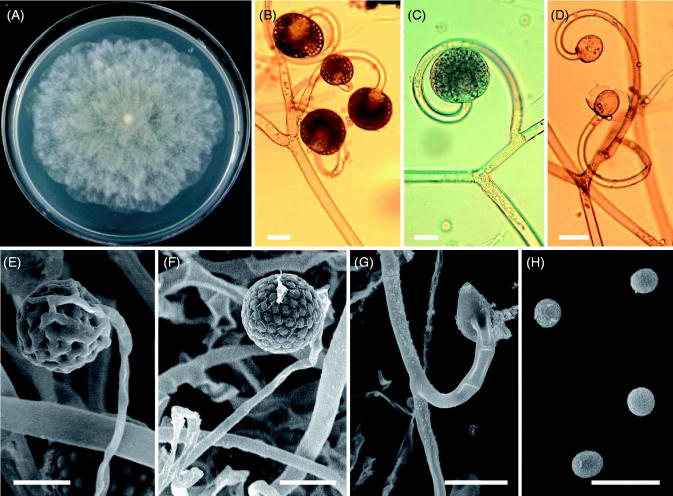
Morphology of Backusella circina CNUFC-PTF2-1. (A) Colony in synthetic mucor agar after 5 days at 25 °C; (B) Forming unispored sporangiola on curved short sporangiophores; (C) Sporangium on curved simple sporangiophores; (D–F) Columellae; (G) Sporangiospores (scale bars: B–G = 20 μm).
=Mucor pseudolamprosporus H. Nagan. & Hirahara, Hiroshima Jogakuin College Bull.: 167 (1968)
Description: Colonies grew rapidly at 25 °C on SMA, filling the Petri plate (diameter, 90 mm) after 5 days of incubation. The colonies were initially white, but later turned light gray. The colony reverse was light gray. Sporangiophores were 6–20 μm wide, erect, branched, and irregular. Sporangia were globose to subglobose, multispored, and measured 27.5–66.5 μm × 27.0–63.5 μm. Columellae were subglobose to oblong, and measured 18.1–30.4 μm × 20.3–34.6 μm. Unispored sporangiola were abundant, globose to subglobose, wall spinulose, and measured 10.3–19.5 μm × 10.0–19.1 μm. Sporangiospores were subglobose to ovoid, and measured 6.5–11.0 μm × 6.0–10.2 μm. Zygospores were not observed on this medium. Optimal growth was observed at 25 °C, slow growth was observed at 10 and 35 °C, and no growth was observed at 37 °C.
3.2.2. Taxonomy of CNUFC-TF3-1
Circinella muscae (Sorokin) Berl. & De Toni, Sylloge Fungorum 7: 216 (1888) (Table 3, Figure 5).
Table 3.
Morphological characteristics of CNUFC-TF3-1 compared to Circinella muscae reference strain.
| Character | CNUFC-TF3-1 | Circinella muscaea |
|---|---|---|
| Colony color | Rapid-growing, first white then brown; reverse brown, irregularly zonate | Rapid-growing, first white then Saccardo’s Olive |
| Sporangia | Globose, multispored, yellow to dark gray with age, 31.9–70.2 × 31.5–69.2 µm | Globose, multispored, 30–62 µm, some up to 100 µm |
| Columellae | Pyriform, subglobose, oval, conical, 17.5–43.3 × 15.5–36.5 µm | Pyriform, oblong, or conical 15–20 × 20–36 µm |
| Sporangiospores | Globose, 4.1–9.5 µm | Globose, sometimes short oval, 3–7 µm, mostly 5–6 µm |
| Zygospores | Absent | Globose, golden brown to reddish brown, 30–65 µm |
From the description by Hesseltine and Fennell [12].
Figure 5.
Morphology of Circinella muscae CNUFC-TF3-1. (A) Colony in synthetic mucor agar after 5 days at 25 °C; (B–D, G) Sporangia and columellae borne on circinate sporangiophores; (E, F) Young and mature multispored sporangia; (H) Sporangiospores. (B–D: observed under light microscope; E–H: observed by scanning electron microscopy) (scale bars: B–F, G = 20 μm, H = 10 μm).
≡Helicostylum muscae Sorokin, Bull. Soc. Imp. Nat. Moscou: 256 (1870)
=Circinella spinosa Tiegh. & G. Le Monn., Annales des Sciences Naturelles Botanique 17: 305 (1873)
=Circinella sydowii Lendn., Bulletin de la Société Botanique de Genève 5: 29 (1913)
Description: Colonies grew rapidly at 25 °C on SMA, filling the Petri plate (diameter, 90 mm) after 7 days of incubation. The colonies were initially white, but later turned brown. The colony reverse was brown and irregularly zonate. Sporangiophores were 6.3–10.8 μm in width, variable in length, and often circinate below the sporangium. Sporangia were globose, yellow to dark gray, multispored, and measured 31.9–70.2 μm × 31.5–69.2 μm. Collumellae were diverse in shape, pyriform, subglobose, oval, conical, and measured 17.5–43.3 μm × 15.5–36.5 μm. Sporangiospores were globose and measured 4.1–9.5 μm. Zygospores were not observed on this medium. Optimal growth was observed at 25 °C, slow growth was observed at 10 and 35 °C, and no growth was observed at 40 °C.
3.2.3. Taxonomy of CNUFC-ESAF3-1
Mucor ramosissimus Samouts., Mater. Mikol. Fitopat. Ross.: 210 (1927) (Table 4, Figure 6)
Table 4.
Morphological characteristics of CNUFC-ESAF3-1 compared to Mucor ramosissimus reference strain.
| Character | CNUFC-ESAF3-1 | Mucor ramosissimusa |
|---|---|---|
| Colony color | Rapid-growing, gray | Deep olive gray to mouse gray |
| Sporangia | Globose to subglobose, 15.5–60.1 × 14.8–56.3 µm | Globose, up to 70 (80) µm |
| Columellae | Globose, cylindrical–ellipsoidal, 12.7–41.8 × 11.8–36.8 µm | Applanate, up to 40 × 50 µm |
| Sporangiospores | Subglobose, ellipsoidal, 4.3–8.5 × 3.9–6.8 µm | Subglobose to broadly ellipsoidal, 4–7(8) µm in diam. or 5–8 × 4.5–6 µm |
| Zygospores | Absent | Absent |
From the description by Schipper [21].
Figure 6.
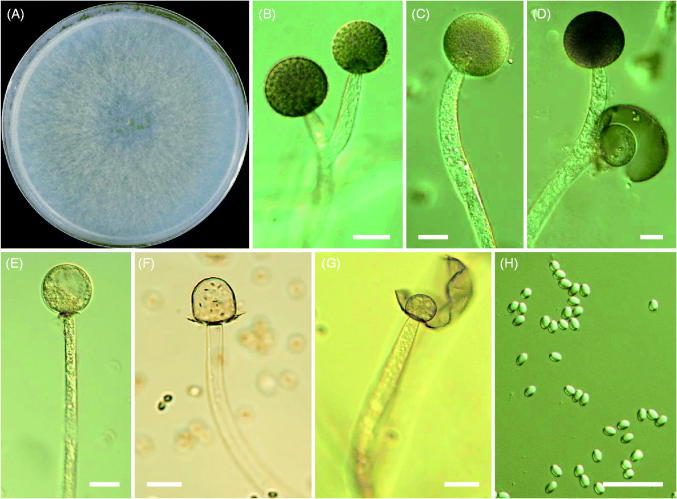
Morphology of Mucor ramosissimus CNUFC-ESAF3-1. (A) Colonies in synthetic mucor agar after 5 days at 25 °C; (B–D) Branched sporangiophores and sporangia; (E–G) Columella with clear collar present; (H) Sporangiospores (scale bars: B–H = 20 μm).
Description: Colonies grew rapidly at 25 °C on SMA, filling the Petri plate (diameter, 90 mm) after 5 days of incubation. The color of the colony was gray. Sporangia were globose to subglobose and measured 15.5–60.1 μm × 14.8–56.3 μm. Columellae were globose, cylindrical–ellipsoidal, and measured 12.7–41.8 μm × 11.8–36.8 μm. Sporangiospores were subglobose, ellipsoidal, and measured 4.3–8.5 μm × 3.9–6.8 μm. Zygospores were not observed on this medium. Optimal growth was observed at 25 °C, slow growth was observed at 10 °C, and no growth was observed at 35 °C.
4. Discussion
To date, few studies have reported new and undescribed zygomycetous fungi in Korea [10,23,34,35]. Particularly, species of Backusella and Circinella are rarely found in Korea. Thus, our finding of B. circina, C. muscae, and M. ramosissimus species not only establishes new records, but also provides knowledge regarding the occurrence and distribution of rare species within these genera.
Variability in nucleotide sequences in the ITS region has been reported by several authors as a critical barcode marker for identifying fungi at the species level, including in the order Mucorales [36,37]. In previous studies, we successfully identified species of Mucorales using this marker [10,22,23,38].
In the ITS and 28S phylogenetic trees, CNUFC-PTF2-1 and CNUFC-PTF2-2 isolated from soil samples clustered in the clade containing B. circina CBS 128.70 (type species) (Figure 1). Although the morphological features of our isolate were similar to those of B. circina described by Ellis and Hesseltine [5], there were differences in the diameter of sporangia and unispored sporangiola. Sporangia sizes reported in literature range from 35 to 100 μm [5], which are larger than our maximum measurements. The unispored sporangiola (4.5–)6–16(–26) μm [5] was larger than that of our isolate. The B. circina strain may exhibit morphological similarities to B. lamprospora, such as the production of subglobose sporangiospores [7]. However, B. circina differs from B. lamprospora in its production of large numbers of spiny-walled and unispored sporangiola. Furthermore, in the phylogenetic tree, the strain formed a separate branch from that of B. lamprospora. Molecular data confirmed the morphological identification of CNUFC-PTF2-1 as B. circina.
Isolates CNUFC-TF3-1 and CNUFC-TF3-2 formed a group with strains of C. muscae (Figure 2). The results of our analysis of molecular data were consistent with the phylogeny presented by Walther et al. [4]. Comparing the colony morphology and culture characteristics of the isolate on SMA medium with previous descriptions by Hesseltine and Fennell [12], the present isolate was generally similar to isolates of C. muscae. Gonzalez et al. [17] and Zheng et al. [18] isolated this species from sand beach and hydrocarbon-polluted sand, respectively. Accordingly, this is the first reported isolation of C. muscae from a leaf of T. sylvestre. C. muscae has been shown to transform androst-4-ene-3,17-dione and produce extracellular enzymes such as proteases [39,40]. This finding suggests that strain CNUFC-TF3-1 may be useful in biotechnological applications and requires further investigation.
Isolate CNUFC-ESAF3-1 was grouped with strains of M. ramosissimus CBS 135.65 (neo-type species) based on phylogenetic analyses (Figure 3). The morphological characteristics of the M. ramosissimus isolate in this study were similar to those previously described by Schipper [21]. However, sporangia were smaller than that of M. ramosissimus described by Schipper (up to 70–80 μm). The M. ramosissimus strain may be confused with M. circinelloides due to its production of subglobose sporangiospores and sympodially branched sporangiophores. However, there are clear genetic differences between these two species. The results of our analysis of molecular data of this species were consistent with the phylogeny presented by Álvarez et al. [41] and Walther et al. [4]. Species of M. ramosissimus produce extracellular enzymes, such as endopolygalacturonase and lipase, and secondary metabolites, such as phytoalexin elicitor [42–44]. Recently, several studies have focused on applying Mucorales members to produce ethanol and biomass by-product [45]. Particularly, M. ramosissimus has been reported as a potential ethanol-producing mold [46].
This is the first report of B. circina, C. muscae, and M. ramosissimus in Korea. Future studies should investigate their ability to produce extracellular enzymes and potential applications in biotechnology.
Funding Statement
This work was in part supported by the Graduate Program for the Undiscovered Taxa of Korea, and in part by the Project on Survey and Discovery of Indigenous Fungal Species of Korea funded by NIBR and Project on Discovery of Fungi from Freshwater and Collection of Fungarium funded by NNIBR of the Ministry of Environment (MOE), and in part carried out with the support of Cooperative Research Program for Agriculture Science and Technology Development [PJ013744], Rural Development Administration, Republic of Korea. This work was in part supported by the BK21 plus program through the National Research Foundation (NRF) funded by the Ministry of Education of Korea.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Hibbett DS, Binder M, Bischoff JF, et al. A higher-level phylogenetic classification of the fungi. Mycol Res. 2007;111:509–547. [DOI] [PubMed] [Google Scholar]
- 2.Benny GL. The methods used by Dr. R. K. Benjamin, and other mycologists, to isolate Zygomycetes. Aliso. 2008;26:37–61. [Google Scholar]
- 3.Hoffmann K, Pawłowska J, Walther G, et al. The family structure of the Mucorales: a synoptic revision based on comprehensive multigene-genealogies. Persoonia. 2013;30:57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walther G, Pawłowska J, Alastruey-Izquierdo A, et al. DNA barcoding in Mucorales: an inventory of biodiversity. Persoonia. 2013;30:11–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis JJ, Hesseltine CW. Two new members of the Mucorales. Mycologia. 1969;61:863–872. [Google Scholar]
- 6.Kirk PM. Nomenclatural novelties. Index Fungorum. 2012;11:1–1. [Google Scholar]
- 7.Benny GL, Benjamin RK. Observations on Thamnidiaceae (Mucorales). New taxa, new combinations, and notes on selected species. Aliso. 1975;8:301–351. [Google Scholar]
- 8.de Souza JI, Marano AV, Pires-Zottarelli CLA, et al. A new species of Backusella (Mucorales) from a Cerrado reserve in Southeast Brazil. Mycol Prog. 2014;13:975–980. [Google Scholar]
- 9.Lima DX, Voigt K, de Souza CAF, et al. Description of Backusella constricta sp. nov. (Mucorales, ex Zygomycota) from the Brazilian Atlantic Rainforest, including a key to species of Backusella. Phytotaxa. 2016;289:59–68. [Google Scholar]
- 10.Wanasinghe DN, Phukhamsakda C, Hyde KD, et al. Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018;89:1–236. [Google Scholar]
- 11.van Tieghem P, Le Monnier G. Recherches sur les Mucorinées. Ann Sci Nat. 1873;17:261–399. [Google Scholar]
- 12.Hesseltine CW, Fennell DI. The genus Circinella. Mycologia. 1955;47:193–212. [Google Scholar]
- 13.Hesseltine CW, Ellis JJ. Notes on Mucorales, especially Absidia. Mycologia. 1961;53:406–426. [Google Scholar]
- 14.Faurel L, Schotter G. Notes mycologiques VI. Sur quelques champignons coprophiles d'Afrique Equatoriale. Cah La Maboké. 1965;3:123–133. [Google Scholar]
- 15.Patil SD, Kale JC. A new species of Circinella van Tiegh. and Le Monn. Curr Sci. 1981;50:544–544. [Google Scholar]
- 16.Arambarri AM, Cabello MN. Circinella lacrymispora sp. nov., a new mucoral isolated from Argentine soils. Mycotaxon. 1996;57:145–149. [Google Scholar]
- 17.González MC, Murueta-Figueroa N, Medina-Ortiz C, et al. New record of Circinella muscae from a hydrocarbon polluted sand beach of Tabasco, Mexico. Mycotaxon. 2010;113:111–117. [Google Scholar]
- 18.Zheng RY, Liu XY, Wang YN. Circinella (Mucorales, Mucoromycotina) from China. Mycotaxon. 2017;132:43–62. [Google Scholar]
- 19.Spatafora JW, Benny GL, Lazarus K, et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia. 2016;108:1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benny GL, Humber RA, Voigt K. Zygomycetous fungi: Phylum Entomophthoromycota and subphyla Kickxellomycotina, Mortierellomycotina, Mucoromycotina, and Zoopagomycotina In: McLaughlin DJ, Spatafora JW, editors. Mycota VII, Part A, Systematics and evolution. New York (NY): Springer-Verlag; 2014. p. 209–250. [Google Scholar]
- 21.Schipper MAA. On Mucor circinelloides, Mucor racemosus and related species. Stud Mycol. 1976;12:1–40. [Google Scholar]
- 22.Nguyen TT, Duong TT, Lee HB. Characterization of two new records of Mucoralean species isolated from gut of soldier fly larva in Korea. Mycobiology. 2016;44:310–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen TT, Jung HY, Lee YS, et al. Phylogenetic status of two undescribed zygomycete species from Korea: Actinomucor elegans and Mucor minutus. Mycobiology. 2017;45:344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Souza CAFD, Lima DX, Gurgel LMS, et al. Coprophilous Mucorales (ex Zygomycota) from three areas in the semi-arid of Pernambuco, Brazil. Braz J Microbiol. 2017;48:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson DP, Eribo BE. Extracellular enzyme production by Rhizopus and Mucor species on solid media. Can J Microbiol. 1984;30:126–128. [DOI] [PubMed] [Google Scholar]
- 26.Alves MH, Campos-Takaki GM, Porto ALF, et al. Screening of Mucor spp. for the production of amylase, lipase, polygalacturonase and protease. Braz J Microbiol. 2002;33:325–330. [Google Scholar]
- 27.de Souza PM, Bittencourt MLA, Caprara CC, et al. A biotechnology perspective of fungal proteases. Braz J Microbiol. 2015;46:337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribes JA, Vanover-Sams CL, Baker DJ. Zygomycetes in human disease. Clin Microbiol Rev. 2000;13:236–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego, CA, USA: Academic Press; 1990. p. 315–322. [Google Scholar]
- 30.Lee HB. Molecular phylogenetic status of Korean strain of Podosphaera xanthii, a causal pathogen of powdery mildew on Japanese thistle (Cirsium japonicum) in Korea. J Microbiol. 2012;50:1075–1080. [DOI] [PubMed] [Google Scholar]
- 31.Thompson JD, Gibson TJ, Plewniak F, et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 33.Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YS, Jung HY, Lee HB, et al. National list of species of Korea Ascomycota, Glomeromycota, Zygomycota, Myxomycota, Oomycota.Korea: National Institute of Biological Resources Ministry of Environment; 2015. [Google Scholar]
- 35.Nguyen TT, Choi YJ, Lee HB. Isolation and characterization of three unrecorded Zygomycete fungi in Korea: Cunninghamella bertholletiae, Cunninghamella echinulata, and Cunninghamella elegans. Mycobiology. 2017;45:318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwarz P, Bretagne S, Gantier JC, et al. Molecular identification of Zygomycetes from culture and experimentally infected tissues. J Clin Microbiol. 2006;44:340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White MM, James TY, O'Donnell K, et al. Phylogeny of the Zygomycota based on nuclear ribosomal sequence data. Mycologia. 2006;98:872–884. [DOI] [PubMed] [Google Scholar]
- 38.Li GJ, Hyde HD, Zhao RL, et al. Fungal diversity notes 253-366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;78:1–237. [Google Scholar]
- 39.de Azevedo Santiago ALCM, de Souza-Motta CM. Isolation of Mucorales from processed maize (Zea mays L.) and screening for protease activity. Braz J Microbiol. 2008;39:698–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heidary M, Habibi Z. Microbial transformation of androst-4-ene-3,17-dione by three fungal species Absidia griseolla var. igachii, Circinella muscae and Trichoderma virens. J Mol Catalysis B Enzymatic. 2016;126:32–36. [Google Scholar]
- 41.Álvarez E, Cano J, Stchigel AM, et al. Two new species of Mucor from clinical samples. Med Mycol. 2011;49:62–72. [DOI] [PubMed] [Google Scholar]
- 42.Simões K, Dietrich SMC, Hahn MG, et al. Purification and characterization of a phytoalexin elicitor from spores of the saprobe Mucor ramosissimus. Rev Bras Bot. 2005;28:735–744. [Google Scholar]
- 43.Marques MR, Buckeridge MS, Braga MR, et al. Characterization of an extracellular endopolygalacturonase from the saprobe Mucor ramosissimus Samutsevitsch and its action as trigger of defensive response in tropical plants. Mycopathologia. 2006;162:337–346. [DOI] [PubMed] [Google Scholar]
- 44.Ko HS, Taguchi H, Takizawa K, et al. The enzymatic approach of zygomycosis – causing Mucorales. Kor J Med Mycol. 2007;12:9–17. [Google Scholar]
- 45.Satari B, Karimi K. Mucoralean fungi for sustainable production of bioethanol and biologically active molecules. Appl Microbiol Biotechnol. 2018;102:1097–1117. [DOI] [PubMed] [Google Scholar]
- 46.Schwarz P, Lortholary O, Dromer F, et al. Carbon assimilation profiles as a tool for identification of zygomycetes. J Clin Microbiol. 2007;45:1433–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]