Abstract
Pathogenic bacteria are armed with potent effector proteins that subvert host signaling processes during infection1. The activities of bacterial effectors and their associated roles within the host cell are often poorly understood, particularly for Chlamydia trachomatis2, a WHO-designated neglected disease pathogen. We identify and explain remarkable dual Lys63-deubiquitinase (DUB) and Lys-acetyltransferase (AcT) activities in the Chlamydia effector ChlaDUB1. Crystal structures capturing intermediate stages of each reaction reveal how the same catalytic center of ChlaDUB1 can facilitate such distinct processes, and enable the generation of mutations that uncouple the two activities. Targeted Chlamydia mutant strains allow us to link the DUB activity of ChlaDUB1 and of the related, dedicated DUB ChlaDUB2 to fragmentation of the host Golgi apparatus, a key process in Chlamydia infection for which effectors have remained elusive. Our work illustrates the incredible versatility of bacterial effector proteins, and provides important insights toward understanding Chlamydia pathogenesis.
During infection, many Gram-negative pathogenic bacteria translocate effector proteins directly into host cells to modify signaling pathways important for invasion, survival, and replication. One particularly interesting family of effectors are those belonging to the CE clan of cysteine proteases. Members of this family were variously found to be proteases for ubiquitin-like (Ubl) modifiers, deubiquitinases (DUBs), or even Ser/Thr acetyltransferases (AcTs)3,4,5,6,7,8,9,10,11, which is striking considering that they all share a structurally similar Cys protease fold. Physiologically, the activities are used against host inflammatory pathways. Deubiquitinases in particular are used by a wide range of pathogens to switch off ubiquitin (Ub)-dependent inflammatory signaling processes12, or interfere with microbe-directed autophagy (xenophagy) pathways. CE family DUBs such as Legionella SidE, Salmonella SseL, or Chlamydia ChlaDUB1 have been shown to mediate inhibition of autophagy, NF-κB signaling or cell death, during infection10,13,14,15. Similarly, the AcT activities of Yersinia YopJ and Salmonella AvrA modify phosphorylation sites, and directly block MAP kinase activation required for inflammatory signaling and innate immunity7,8,9.
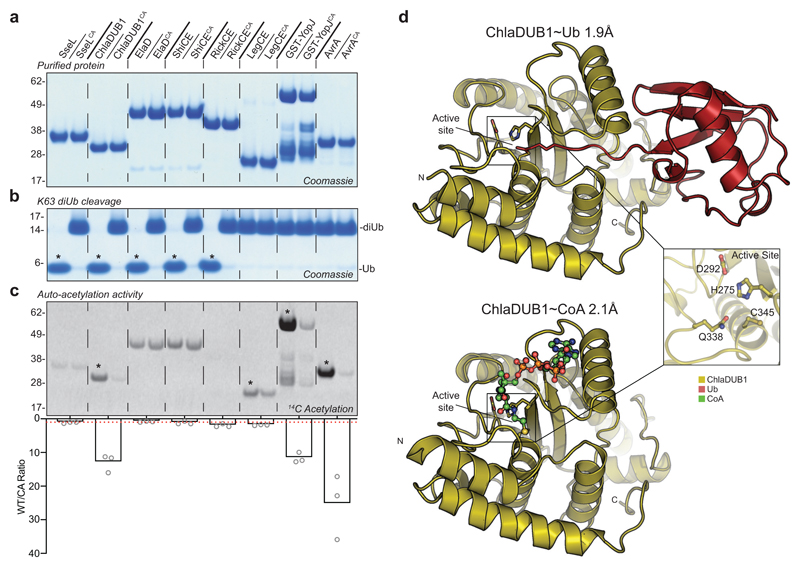
Recent phylogenetic analyses and crystal structures have started to explain the seemingly disconnected catalytic activities among CE family members, but the conundrum of the identical catalytic fold has remained intriguing. Indeed, a direct biochemical comparison of DUB and AcT activities in CE family proteins has not yet been performed. We used our panel of purified bacterial CE enzymes from a range of pathogens alongside their catalytically inactive variants (Fig. 1a) to test for DUB activity by monitoring cleavage of K63-linked diUb (Fig. 1b). In parallel, we tested for AcT activity by monitoring auto-acetylation via radioisotope incorporation following incubation with 14C Acetyl-Coenzyme A (Fig. 1c). This analysis revealed Salmonella SseL, Escherichia ElaD, Shigella ShiCE, and Rickettsia RickCE to be dedicated DUBs, and identified Legionella LegCE, Yersinia YopJ, and Salmonella AvrA to be dedicated AcTs.
Figure 1. Identification of specialized and dual-function CE-clan enzymes.
a) Panel of purified bacterial CE-clan enzymes and their catalytically inactive Cys-to-Ala mutants. b) Deubiquitinase assay monitoring cleavage of K63-linked diUb following overnight incubation. c) Acetyltransferase assay monitoring 14C incorporation following a 2 h incubation of each protein with 14C-labeled Acetyl-CoA. Below, histogram representation of the WT/CA 14C incorporation ratio following normalization of the 14C autoradiography signal to the Coomassie stain. The average of three replicate experiments is plotted. A WT/CA ratio of one indicates no AcT activity, and is denoted by a red dashed line. Gels in a, b, and c are representative of triplicate experiments. All uncropped gels are shown in Supplementary Fig. 10. Asterisks indicate appreciable DUB (b) or AcT (c) activity. d) ChlaDUB1 complex crystal structures that capture intermediate stages of deubiquitinase (top) and acetyltransferase (bottom) activities. Inlay, a representative view of the ChlaDUB1 active site showing the Cys-His-Asp catalytic triad and the Gln oxyanion hole.
Remarkably, Chlamydia ChlaDUB1 could perform both DUB and AcT reactions, seemingly using the same catalytic Cys residue (compare Fig. 1, b and c). ChlaDUB1 is phylogenetically distinct from the YopJ-like family11, but showed similar rates of auto-acetylation compared to YopJ and AvrA (although YopJ acetylation of its substrate MEK2 (ref. 8) is markedly faster, see Supplementary Fig. 1a,b,c). Importantly, ChlaDUB1 auto-acetylation occurs at Lys residues (Supplementary Fig. 1d,e) whereas YopJ-like family members predominantly target Ser/Thr residues7,8,9. Furthermore, ChlaDUB1 AcT activity is not regulated by phytic acid (inositol hexakisphosphate, IP6) (Supplementary Fig. 1f,g), in contrast to YopJ-like enzymes16,17. This identified ChlaDUB1 as a bona-fide Lys-AcT in addition to being a Lys63-specific DUB.
To explain how ChlaDUB1 could perform two seemingly disparate chemical reactions, namely deubiquitination – a hydrolysis reaction, and acetylation – a condensation reaction, we determined crystal structures of the enzyme bound to Ub, and bound to Coenzyme A (CoA) at 1.9 Å and 2.1 Å resolution, respectively (Fig. 1d and Supplementary Table 1). The structures showed hardly any conformational changes between each other, or in comparison to previous apo structures (pdb-id 5HAG11, 5B5Q15) with overall RMSDs <1 Å (Fig. 1e and Supplementary Fig. 2a), but revealed distinct binding sites for Ub and CoA.
The ChlaDUB1~Ub structure (Supplementary Fig. 2b) was obtained using the Ub activity based probe Ub-propargylamide (UbPA), which covalently links one Ub molecule into the enzymatic S1 site (Supplementary Fig. 3). ChlaDUB1 forms similar interactions with Ub as compared to other CE proteases10,11,18,19,20, involving both the Ile44 and Ile36 hydrophobic patches of Ub (Supplementary Fig. 2c,d).
The ChlaDUB1~CoA structure (Supplementary Fig. 2e) revealed a disulfide bridge between the cofactor’s cysteamine and the catalytic Cys, and identified a charge-complementary binding site for CoA near the active site (Supplementary Fig. 2f). The ChlaDUB1 CoA binding site is distinct from the CoA binding sites of the YopJ-like effector HopZ1a16 and arylamine N-acetyltransferases (NATs)21 (Supplementary Fig. 2g,h), and also removed from the Ub binding site (Supplementary Fig. 3).
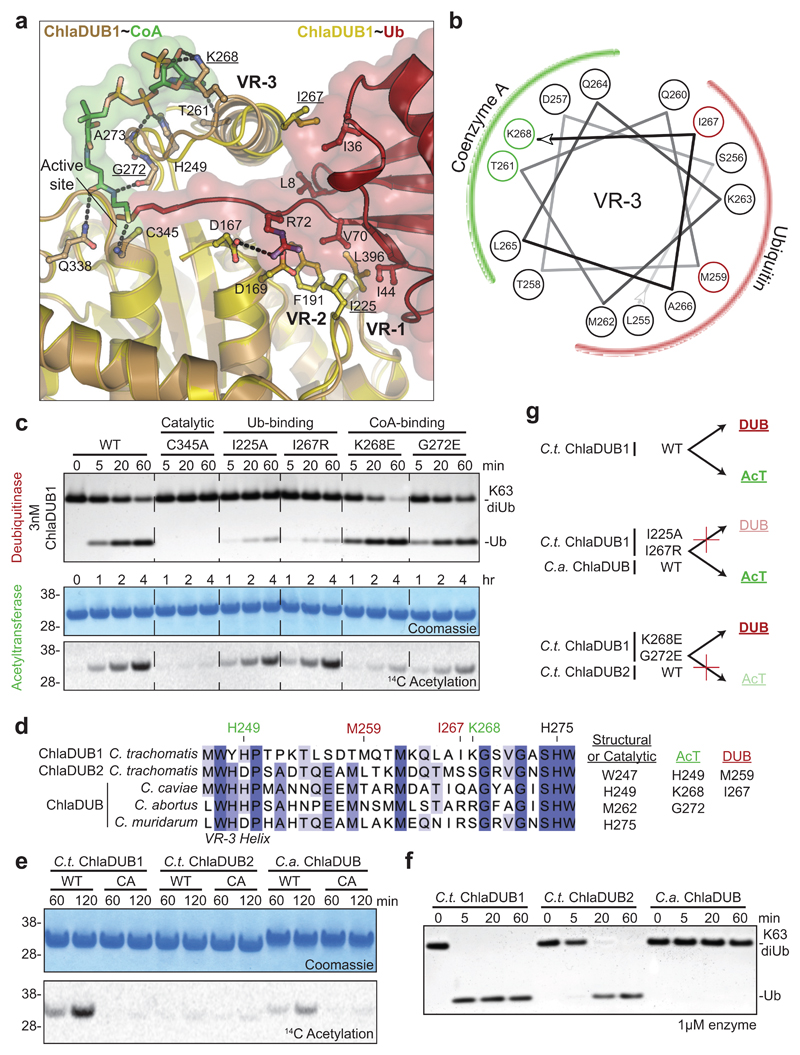
Both structures together reveal the importance of an inserted helix that is unique to ChlaDUB1 and not present in other CE enzymes from bacteria, viruses, or eukaryotes; we had previously annotated this element as Variable Region 3 (VR-3)11. One face of this VR-3 helix contacts the adenosine and phosphate groups of the CoA molecule (Fig. 2a,b). Remarkably, the opposite face of the VR-3 helix binds the Ile36-patch of Ub (Fig. 2a,b, Supplementary Fig. 2i). This arrangement enables both DUB and AcT activities to utilize the same active site (Fig. 1d and Supplementary Fig. 4a,b), via spatially separated, independent binding sites for Ub and CoA. Consistently, Ub compromises AcT activity, but when the Ub C-terminus is missing, auto-acetylation is restored (Supplementary Fig. 4c). Separate binding sites for Ub and CoA further enabled us to uncouple DUB and AcT activity. ChlaDUB1 AcT activity was strongly diminished by mutation of K268E (VR-3), or G272E without affecting DUB activity. In contrast, DUB activity was abrogated by I267R (VR-3) or I225A mutation, yet these mutants did not affect auto-acetylation (Fig. 2a,c, and Supplementary Fig. 5a).
Figure 2. Molecular dissection of dual deubiquitinase/acetyltransferase activities.
a) Close-up of the ChlaDUB1:CoA (brown:green) and ChlaDUB1:Ub (tan:red) interfaces with key interacting residues shown in ball-and-stick. Hydrogen bonds are shown as dashed lines. b) Helical wheel diagram illustrating the amphipathic nature of the ChlaDUB1 VR-3 helix, and its interactions with Ub or CoA (colored in red and green, respectively). c) Deubiquitinase (top) and acetyltransferase (bottom) assays illustrating that while both activities require the catalytic Cys residue, mutations in the Ub-binding and CoA-binding regions separate the two functions. A representative gel is shown of triplicate experiments. d) Sequence alignment of the VR-3 helix for orthologous Chlamydia ChlaDUB enzymes. The catalytic His, CoA-binding (green) and Ub-binding (red) residues are marked, and additional contacts are listed. e) 14C acetylation assay with the ChlaDUB orthologues from C. trachomatis (C.t. ChlaDUB2) and C. abortus (C.a. ChlaDUB). f) Deubiquitinase assay monitoring K63-linked diUb cleavage by the Chlamydia ChlaDUB orthologues. Gels in e and f are representative of triplicate experiments. All uncropped gels are shown in Supplementary Fig. 10. g) Schematic depicting how deubiquitinase and acetyltransferase functions can be separated either by structure-guided mutation or evolution as represented by the ChlaDUB orthologues.
The VR-3 helix is central to dual activities in C. trachomatis (C.t.) ChlaDUB1 and present in all Chlamydia ChlaDUB homologues, such as ChlaDUB of C. abortus (C.a.), a cattle pathogen that is transmissible to humans. A 1.5 Å crystal structure of C.a. ChlaDUB confirmed the register of the predicted VR-3 helix as shown in the sequence alignment (Fig. 2d, Supplementary Fig. 5d,e, and Supplementary Table 1). Importantly, the Ub- and CoA-coordinating residues within VR-3 are not jointly conserved (Fig. 2d), and we hypothesized that C.t. ChlaDUB2 should be a dedicated DUB, while C.a. ChlaDUB should be a dedicated AcT. Indeed, these predictions could be confirmed biochemically (Fig. 2e,f and Supplementary Fig. 5b,c). Together, our data strongly suggested that Chlamydia species evolved ChlaDUB effectors with dual activities and potentially multiple functions.
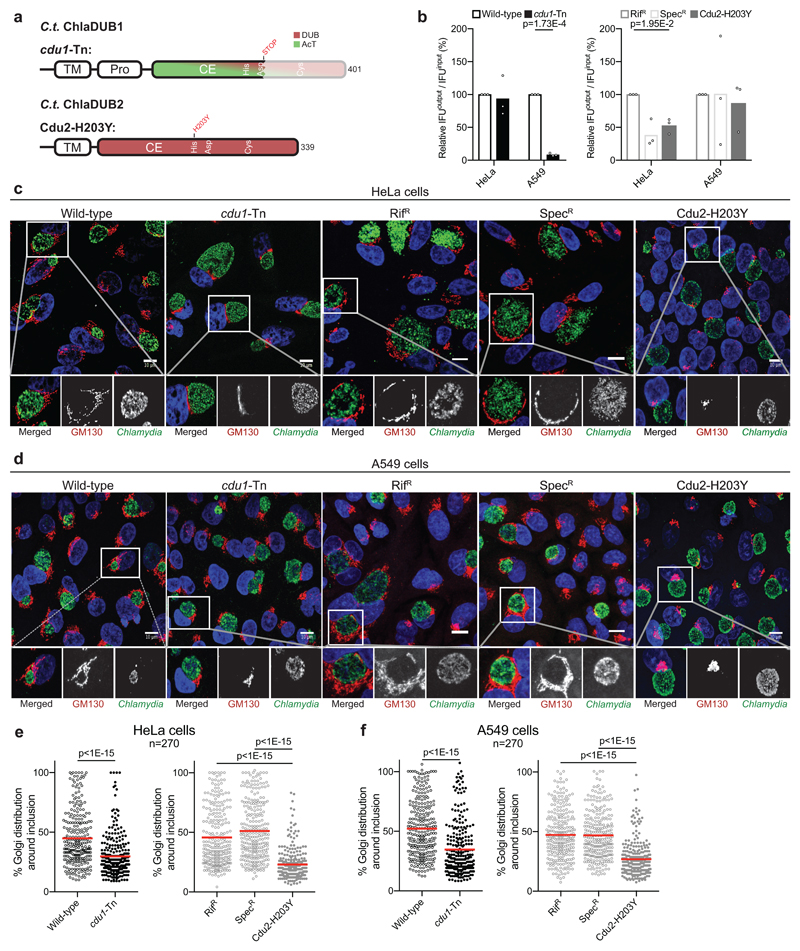
Functional characterization of Chlamydia effectors has remained challenging, mostly due to the rudimentary tools available for genetic manipulation of Chlamydia22. Nonetheless, we set out to uncover roles for the DUB/AcT ChlaDUB1 and the dedicated DUB ChlaDUB2 utilizing mutant strains harboring catalytically inactive ChlaDUB1 and ChlaDUB2 variants. One strain, containing a mutation leading to an amino acid substitution in the ChlaDUB2 catalytic His residue (H203Y) that inactivates the enzyme (Supplementary Fig. 6a), was identified from a collection of chemically mutagenized C. trachomatis strains23. This strain was back-crossed to wild-type C. trachomatis and a clean recombinant strain harboring only the ChlaDUB2 H203Y variant was isolated (Cdu2-H203Y) (Fig. 3a, Supplementary Table 2, 3). For ChlaDUB1, we obtained a recently characterized ChlaDUB1 mutant strain15 generated by transposon mutagenesis that introduced an early stop codon before the catalytic Cys residue (cdu1-Tn) (Fig. 3a, Supplementary Table 2).
Figure 3. ChlaDUB function is required for C. trachomatis Golgi fragmentation.
a) Topology diagram illustrating C.t. ChlaDUB1 and C.t. ChlaDUB2 domain architecture, with active site residues annotated within the catalytic domains. Changes present in the cdu1-Tn and Cdu2-H203Y defective strains are shown above. b) C. trachomatis growth assay measured as inclusion forming units (IFU) output per IFU input following a 48 h infection in either HeLa or A549 cells. Values were normalized to 100% for wild-type. Statistical significance compared to parental controls was measured using a two-tailed Welch’s t-test. HeLa: Wild-type – cdu1-Tn, p=0.768; RifR – Cdu2-H203Y, p=0.0195; SpecR – Cdu2-H203Y, p=0.392. A549: Wild-type – cdu1-Tn, p=0.000173; RifR – Cdu2-H203Y, p=0.615; SpecR – Cdu2-H203Y, p=0.791. n=3. c) Representative confocal images showing Golgi fragmentation and redistribution around the Chlamydia inclusion following a 26 h infection of HeLa cells. Samples were immunostained with anti-GM130 (cis-Golgi, red) and anti-Slc1 (Chlamydia, green) antibodies, and Hoechst stained (DNA, blue). Isolated channels for the boxed region are shown below, and full versions are shown in Supplementary Fig. 6b. Scale bar corresponds to 10 µm. d) As in c) for A549 cells. Full versions are shown in Supplementary Fig. 6c. e) Quantification of c) following measurement of Golgi distribution around the circumference of the Chlamydia inclusion in 90 cells for each of three independent replicates. Mean values are shown as a red bar with individual data points overlaid. Statistical significance compared to parental was measured using a two-tailed Mann-Whitney test. Wild-type – cdu1-Tn, p<1E-15; RifR – Cdu2-H203Y, p<1E-15; SpecR – Cdu2-H203Y, p<1E-15. Separated plots for each replicate are shown in Supplementary Fig. 6d. f) As in e) for A549 cells. Wild-type –cdu1-Tn, p<1E-15; RifR – Cdu2-H203Y, p<1E-15; SpecR – Cdu2-H203Y, p<1E-15. Separated plots for each replicate are shown in Supplementary Fig. 6e.
The ChlaDUB1 and ChlaDUB2 loss of function strains left us in the privileged position to assess the effects of either enzyme on host biology, and on contributions to Chlamydia infection. The cdu1-Tn mutant strain did not significantly reduce the number of infectious progeny in HeLa cells as compared to a wild-type strain. This was markedly different in A549 cells, a human adenocarcinomic lung epithelial cell line, in which infection with the cdu1-Tn mutant strain reduced progeny by 90% (Fig. 3b). The latter was comparable to the effect of this strain in vivo using a transcervical mouse model of infection, whereas primary human fimbriae cells showed a bacterial growth defect only after prior stimulation with interferon-γ15. Surprisingly, the Cdu2-H203Y strain showed little to no growth defect in either HeLa or A549 cell lines compared to its two parental strains (see Methods) (Fig. 3b), suggesting that ChlaDUB1, with its additional AcT activity, may play a unique role in Chlamydia infection.
Next, we inspected infected cells by confocal microscopy. A prominent feature of Chlamydia-infected cells is the fragmentation and subsequent redistribution of the Golgi apparatus into ministacks that surround the pathogen-containing vacuole (termed the inclusion) at approximately 20 hours post infection24,25. Because ChlaDUB1 and ChlaDUB2 are actively expressed and secreted at this time post infection and have been shown to localize to the outside of the inclusion membrane where they could interact with neighboring organelles15,28, we used our cdu1-Tn and Cdu2-H203Y mutant strains to test for a contribution to Golgi redistribution following infection. Remarkably, at 26-hours post infection, both the cdu1-Tn and Cdu2-H203Y mutant strains showed a dramatic impairment in redistribution of the Golgi apparatus (Fig. 3c-f and Supplementary Fig. 6b-e). Since both ChlaDUB1 and ChlaDUB2 mutant strains affected host Golgi redistribution, this strongly suggested that DUB activity is required in this process. Moreover, the comparable individual impact of each mutant strain on Golgi redistribution (Fig. 3e,f) indicates either non-redundant roles for each DUB, or a strict dose dependency on DUB-activity introduced by Chlamydia to invoke the observed cell biological effect. Finally, a similar extent of Golgi redistribution was seen in A549 but also HeLa cells, contrasting the different impact of ChlaDUB mutant strains on bacterial growth rates (compare Fig. 3c-f with 3b). This lack of correlation had been observed previously in HeLa cells, e.g. with InaC that regulates Golgi redistribution without impacting generation of bacterial progeny23.
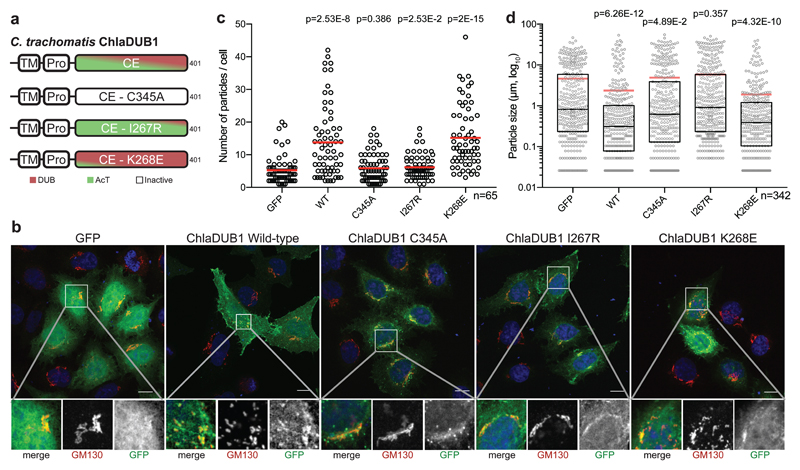
Our data suggested that ChlaDUB1 and ChlaDUB2 may have unrecognized roles in manipulating Golgi morphology and dynamics, which was corroborated in a simplified system (Fig. 4a). Strikingly, we found that Golgi fragmentation was readily induced by sole expression of either ChlaDUB1 or ChlaDUB2 in HeLa cells (Fig. 4b, Supplementary Fig. 7a,b, and Supplementary Fig. 8a,b). Expression of wild-type ChlaDUB1 and (to a lesser extent) ChlaDUB2 resulted in significant Golgi fragmentation as measured either by the number or the size of Golgi-stained puncta. Importantly, active site mutations eliminated this effect, which was again more pronounced for ChlaDUB1 (Fig. 4c,d, Supplementary Fig. 7c,d, and Supplementary Fig. 8c,d). All ChlaDUB1 constructs showed an enriched localization to the Golgi apparatus, indicating that the introduced mutations exclusively affect activity (Fig 4b, Supplementary Fig. 7b). ChlaDUB2 appeared to primarily localize to the endoplasmic reticulum (Supplementary Fig. 8e), which could explain its reduced ability to induce Golgi fragmentation as compared to ChlaDUB1. Using our structure-guided mutations that separate ChlaDUB1 DUB and AcT function (Fig. 2g and 4a), the Golgi fragmentation could be assigned as a DUB-dependent effect: A DUB-deficient I267R mutant was as defective in Golgi fragmentation as a catalytically-inactive C345A construct, while an AcT-deficient ChlaDUB1 K268E mutant retained its Golgi fragmenting capabilities (Fig. 4b-d and Supplementary Fig. 7a-d).
Figure 4. ChlaDUB deubiquitinase activity is required for C. trachomatis Golgi fragmentation.
a) Topology diagram illustrating the constructs used to characterize activity dependence of Golgi fragmentation following expression of ChlaDUB1 in mammalian cells. Separation-of-function mutations were selected from structural and biochemical work discussed in Fig. 2. b) Representative confocal images showing Golgi fragmentation in HeLa cells following expression of GFP-tagged ChlaDUB1. Samples were immunostained with anti-GM130 (cis-Golgi, red) and DAPI stained (DNA, blue). GFP fluorescence is shown in green. Isolated channels for the boxed region are shown below, and full versions are shown in Supplementary Fig. 7b. Scale bar corresponds to 10 µm. c) Quantification of cis-Golgi-stained puncta from b) for ~65 cells in each of three independent replicates (two remaining replicates are plotted in Supplementary Fig. 7c). Mean values are shown as red bars with individual data points overlaid. Statistical significance compared to GFP control was measured using a two-tailed Mann-Whitney test. GFP – WT, p=2.53E-8; GFP – C345A, p=0.386; GFP – I267R, p=0.0253; GFP – K268E, p=2E-15. d) Measurement of cis-Golgi-stained puncta size from b) for ~65 cells in each of three independent replicates (two remaining replicates are plotted in Supplementary Fig. 7d). Mean values are shown as red bars, median values are shown as black bars inside a quartile box plot, with individual data points overlaid. Statistical significance compared to GFP control was measured using a two-tailed Mann-Whitney test. GFP – WT, p=6.26E-12; GFP – C345A, p=0.0489; GFP – I267R, p=0.357; GFP – K268E, p=4.32E-10.
Together, we here unveil a remarkable case of protein moonlighting29, wherein a bacterial effector, C. trachomatis ChlaDUB1, performs two distinct enzymatic activities within its catalytic site, leading to separable cellular functions. Single amino acid substitutions can toggle between the activities, and this is used by closely related orthologues and paralogues in this enzyme family to modulate function (Fig. 2g). We further establish that the DUB activities present in ChlaDUB1 and also in the dedicated DUB paralogue ChlaDUB2, are necessary and sufficient for the fragmentation of the host Golgi apparatus, a prerequisite to Golgi redistribution around the Chlamydia inclusion. This adds ChlaDUB1 and ChlaDUB2 to the limited list of effectors and host factors implicated in this striking cell biological phenomenon23–27 (see Supplementary Fig. 9). In overexpression studies, ChlaDUB1 has also been implicated with inhibition of NF-κB signaling13 and cell death15, and it is tempting to speculate that some of these effects are conferred by the AcT activity of ChlaDUB1. While this requires further study, the importance of ChlaDUB1 for Chlamydia infectivity15 and its unique enzymatic nature make it an interesting candidate for future Chlamydia-targeted therapeutics.
Methods
Cloning and molecular biology
Generation of Salmonella Typhimurium SseL, Chlamydia trachomatis ChlaDUB1, Escherichia coli ElaD, Shigella flexneri ShiCE, Rickettsia bellii RickCE, Legionella pneumophila LegCE, Yersinia pestis YopJ, and Salmonella Typhimurium AvrA constructs was described previously11. Sequences for Chlamydia trachomatis ChlaDUB2 and Chlamydia abortus ChlaDUB were obtained via gene synthesis (Life Technologies). Following amplification with KOD polymerase (EMD Millipore), the genes were inserted into the pOPIN-B or pOPIN-GFP vector30 with the In-Fusion cloning method (Takara Bio USA). All mutagenesis was performed using the Quikchange method (Agilent).
Protein expression and purification
Expression and purification of SseL (24-340), ElaD (2-407), ShiCE (2-405), RickCE (378-691), LegCE (141-360), YopJ (1-288), and AvrA (1-288) has been described previously11. C.t. ChlaDUB1 (130-401), C.t. ChlaDUB2 (80-339), and C.a. ChlaDUB were expressed in E. coli Rosetta2 pLacI cells (Novagen) at 18°C for 20 h following induction with 0.2 mM IPTG an OD600 of 0.8-1.0. Cells were harvested in 25 mM Tris (pH 7.4), 200 mM NaCl, 2 mM ß-mercaptoethanol (Buffer A) and subjected to one freeze-thaw cycle. EDTA-free Complete protease inhibitor tablets (Roche), DNase, and Lysozyme were added prior to lysis by sonication. The resulting lysate was centrifuged at 35000 x g for 25 min, and applied to Talon resin (Takara Bio USA). The resin was washed with Buffer A thoroughly prior to elution with Buffer A containing 250 mM imidazole. During overnight dialysis back to Buffer A at 4°C, the His-tag was cleaved with His-3C protease. Following a reverse affinity step over regenerated Talon resin, the resulting protein was then concentrated (10,000 MWCO, EMD Millipore) and applied to a gel filtration column (Superdex75, GE Healthcare) equilibrated in 25 mM HEPES (pH 8.0), 150 mM NaCl, 5 mM DTT. Pure protein-containing fractions were concentrated, aliquoted, and flash-frozen for storage at -80°C.
Deubiquitinase assays
All enzymes were diluted to a ‘2x’ concentration in 25 mM Tris (pH 7.4), 150 mM NaCl, 10 mM DTT and allowed to fully reduce for 20 min at room temperature. 6 µM diUb stocks were prepared in 100 mM Tris (pH 7.4), 100 mM NaCl, 10 mM DTT and mixed 1:1 with 2x enzyme prior to incubation at 37°C. Samples were quenched in reducing LDS sample buffer (ThermoFisher), resolved by SDS-PAGE, and visualized using silver stain (BioRad).
Ub/Ubl KG-TAMRA protease assays were performed as described previously11.
Acetylation assays
All enzymes were diluted to 5 µM in 25 mM HEPES (pH 8.0), 50 mM NaCl, 0.5 mM DTT and incubated with 60 µM [1-14C] Acetyl-CoA (60 mCi/mmol, PerkinElmer) at 37°C for the indicated time. The panel shown in Figure 1 and all subsequent assays with YopJ or AvrA additionally included 200 nM inositol hexakisphosphate (IP6). Reactions were quenched with reducing LDS sample buffer (ThermoFisher) and resolved by SDS-PAGE prior to staining with Coomassie. Gels were then dried and exposed to a Phosphor screen for several days prior to imaging on a Typhoon scanner (GE Healthcare). 14C autoradiography intensity was quantified using ImageJ31 and normalized to the Coomassie stain signal.
Protein crystallization
The ChlaDUB1 (130-401)~Ub complex was purified by gel filtration following an overnight reaction at room temperature with 2-fold molar excess Ub-PA suicide probe32. Native ChlaDUB1~Ub crystals were obtained using protein prepared in 25 mM Tris (pH 7.4), 125 mM NaCl, 4 mM DTT and crystallized at 10 mg/mL in 0.1 M MES (pH 6), 20% PEG 6000, with a 400 nL sitting drop at 1:1 protein:precipitant ratio. SeMet ChlaDUB1~Ub crystals were obtained using protein at 7 mg/mL in 0.1 M HEPES (pH 7.1), 18% PEG 8K, with a 200 nL sitting drop at 1:1 protein:precipitant ratio. The ChlaDUB1~CoA complex was crystallized by addition of 2 mM CoA to 12 mg/mL ChlaDUB1, and mixing with 0.1 M HEPES (pH 7.2), 20% PEG 8000 at a 1:1 protein:precipitant ratio in a 400 nL sitting drop. C. a. ChlaDUB (108-377) was prepared in 25 mM HEPES (pH 8.0), 150 mM NaCl, 5 mM DTT and crystallized in 0.1 M Tris (pH 7), 0.2 M calcium acetate, 20% PEG 3000 with a 400 nL sitting drop at 1:1 protein:precipitant ratio. All crystals were cryoprotected with mother liquor containing 25% glycerol. Cryoprotectant for ChlaDUB1~CoA crystals also contained 5 mM CoA.
Data collection, structure determination, and refinement
Data were collected at 100K at the Diamond Light Source (DLS) beam lines I02, I03, and I04 (see Supplementary Table 1). Data collections were performed at 0.9798, 0.9795, and 0.9794 Å wavelength for the ChlaDUB1~Ub, ChlaDUB1~CoA, and C.a. ChlaDUB structures, respectively. Integration and scaling were performed using XDS33 and Aimless34, respectively. The ChlaDUB1~Ub structure was solved experimentally using a SeMet SAD dataset with PHENIX AutoSol and AutoBuild35.36,37. ChlaDUB1~CoA and C.a. ChlaDUB structures were solved using molecular replacement in Phaser38 using the apo ChlaDUB1 structure (pdb id 5HAG). Iterative rounds of model building and refinement were performed using COOT39 and PHENIX35, respectively. Ramachandran statistics (favored/allowed/outliers) for the ChlaDUB1~Ub, ChlaDUB1~CoA, and C.a. ChlaDUB structures were 97.2/2.8/0,97.4/2.6/0, and 98.0/2.0/0, respectively. All figures were generated using PyMOL (www.pymol.org).
Cell lines
HeLa, A549, and Vero cell lines were obtained from ATCC, where they were authenticated by morphology, karyotyping, and STR analyses. Stocks were routinely tested and confirmed negative for mycoplasma contamination.
Chlamydia growth conditions and infections
HeLa, A549, and Vero cells were grown in high glucose DMEM supplemented with L-glutamine, sodium pyruvate (Gibco, Life Technologies) and 10% FBS (Mediatech, CellGro), at 37 °C in a 5% CO2 humidified incubator. All cells infected with Chlamydia were centrifuged at 3,500 rpm for half an hour at 10°C immediately upon infection.
Chlamydia strains
All Chlamydia strains (Supplementary Table 2) were derived from C. trachomatis LGV biovar L2 434/Bu (wild-type). The cdu1-Tn strain was generously provided by Scott Hefty (The University of Kansas) and described previously15. The cdu2-G607A (Cdu2-H203Y) allele was identified in a collection of chemically mutagenized C. trachomatis L2 434/Bu strains by whole genome sequencing of a collection of pooled mutant strains23. Strain CTL2M467 was identified to harbor the cdu2-G607A single nucleotide variant (SNV) by Sanger sequencing of the cdu2 (CTL0246) locus. Vero cells seeded in a 6 well plate were infected with CTL2M467. At 48 hours post infection (hpi), cell monolayers were lysed by hypotonic lysis, lysates sonicated, and bacterial cells collected by centrifugation at 14,000 rpm for 15 minutes at 4 °C. Bacterial cell pellets were resuspended in 1X DNAse I buffer (New England Biolabs) and treated with 4 Units of DNAse 1 (New England Biolabs) for 1 hour at 37 °C to deplete co-purifying Vero DNA. Following a wash with PBS buffer, total DNA was isolated with a DNA isolation kit (DNeasy tissue and blood kit, Qiagen, Valencia, CA) following the manufacturer’s instructions. One μg of CTL2M467 enriched DNA was fragmented with NEBNext dsDNA Fragmentase (New England Biolabs) and DNA sequencing libraries prepared with a NEBnext DNA Library Prep Kit for Illumina according to manufacturer’s instructions. Libraries were sequenced with the MiSeq DNA Sequencing Platform (Illumina, Inc. San Diego, CA) at the Duke University IGSP sequencing facility. Genome assembly and single nucleotide variant (SNV) identification was performed with Geneious version 6 (Biomatters, http://www.geneious.com/). The C. trachomatis L2 434/Bu genome (GenBank no. NC_010287) was used as reference sequence. All SNV’s identified (Supplementary Table 3) were independently verified by Sanger sequencing. M467 rs22 was isolated from a backcross of parental strain CTL2M467 (rifampin resistant-RifR) with a spectinomycin resistant L2 434/Bu strain (SpecR) as described previously40. Vero cells were co-infected with strains CTL2M467 (RifR) and L2 434/Bu (SpecR) at an MOI of 3 and a ratio of 1:1. At 48 hpi, crude cell lysates prepared in SPG buffer (0.25 M sucrose, 10 mM sodium phosphate, 5 nM glutamic acid) were used to infect Vero cells seeded in a 6 well plate. At 2 hpi, an agarose/DMEM overlay supplemented with rifampin (200 ng/mL) and spectinomycin (200 μg/mL) was added to infected cells as previously described40 and cells incubated for 14 days. Recombinant strains were isolated from twenty-four individual plaques and expanded in Vero cells. All recombinant strains were genotyped by PCR for presence of CTL2M467 parental non-synonymous SNVs (Supplementary Table 3). Recombinant strain number 22 (M467 rs22) was found to harbor only the parental cdu2-G607A mutation (Cdu2-H203Y). Chlamydia strains were maintained as frozen stocks in SPG buffer.
Chlamydia growth assays
HeLa and A549 cells were seeded in wells of two 96 well plates (input and output plates). Cells were infected with Chlamydia strains at an MOI of 0.6. At 24 hpi, infected cells in input plate were fixed with ice cold Methanol and stored in PBS. At 48 hpi, crude lysates in SPG were prepared from infected cells in output plates and a series of 1/10 dilutions were used to immediately infect corresponding HeLa or A549 cells seeded in wells of a 96 well plate. At 24 hpi, cells were fixed with ice cold Methanol. Fixed cells were stained with rabbit anti-Slc141 and Hoechst. Images from stained cells were captured on an EVOS cell imaging system (ThermoFisher scientific) with a 20X objective. Inclusion forming units (IFUs) were quantified using Image J (NIH). Output IFU’s from each Chlamydia inoculum was normalized to their respective input IFU’s. Chlamydia IFU production was measured from 3 independent biological replicates.
Visual and quantitative analysis of Golgi redistribution around Chlamydia inclusions
Imaging
HeLa and A549 cells grown on glass coverslips were infected with Chlamydia strains at MOIs of 0.8. At 26 hpi, cells were fixed with pre-warmed (37°C) 3% formaldehyde in PBS for 20 minutes at room temperature. All washes and antibody staining’s were performed with pre-warmed (37°C) PBS and antibody solutions respectively. Fixed cells were stained with rabbit anti-Slc141 and mouse anti-GM130 antibodies (BD Biosciences), and Hoechst. Z-stacks of stained cells were captured on a Zeiss 880 inverted fluorescence microscope with a 63X objective (Zeiss).
Golgi redistribution quantification
For each Z-stack, maximum intensity Z-projections were generated with Image J (NIH). The length of Golgi (defined by GM130 staining) distributed around Chlamydia inclusions and the length of each inclusion perimeter (defined by Slc1 staining) were measured using the line tool from Image J (NIH). The ratio of Golgi length to inclusion perimeter length was determined, and values are expressed as a percent. Three independent experiments were performed to assess Golgi distribution. Golgi distribution was assessed from 6 fields for a total of 90 cells per independent experiment.
High-resolution imaging
Standard deviation Z-projections of captured images (see above) were generated and images minimally processed with Image J (NIH).
ChlaDUB expression in mammalian cells
Imaging
HeLa cells grown on glass coverslips were transfected with 1 µg of plasmid using Genejuice (EMD Millipore). At 23-hours post-transfection, cells were fixed with 4% paraformaldehyde, immunostained for GM130 (BD Biosciences 610822). Nucleic acids were stained with DAPI. Images were collected on a Nikon Eclipse Ti microscope with a Super Plan Fluor ELWD 40XC objective or a 3i Marianas spinning disk inverted confocal microscope with a 63X oil objective and a CMOS camera (Hamatsu). The images were then processed using SlideBook software and Photoshop CS4 Version 11.0 (Adobe).
Quantification of Golgi fragmentation
Images were converted from 16-bit to 8-bit binary using Fiji. Following selection of the region of interest, the number and surface area of Golgi-stained particles were quantified using the Analyze Particles tool of Fiji. A minimum of three independent experiments were performed to assess Golgi fragmentation, each consisting of ~65 counted cells. Multinucleated cells, as well as cells that were cycling through mitosis, were excluded from the analysis.
Western blotting
HeLa cells were transfected with 1 µg of plasmid using Genejuice (EMD Millipore). At 23-hours post-transfection, cells were lysed in RIPA buffer and the protein amount was assessed using the Pierce BCA Protein Assay Kit (Thermo). 20 µg of lysates were resolved by SDS-PAGE and transferred onto nitrocellulose. Membranes were blocked in 5% milk/TBST for 1 h and probed for GFP (sheep, 1:1000, made in-house) and actin (rabbit, 1:10,000, Sigma A2266) for 1 h at room temperature. The IRDye 680LT Donkey anti-Rabbit IgG (LI-COR 926-68023) and IRDye 800CW Donkey anti-Goat IgG (cross-reacts with sheep IgG, LI-COR 926-32214) secondary antibodies were used at a concentration of 1:10,000 in 5% milk. Membranes were scanned using a LI-COR CLx Odyssey system and the Image Studio software, and minimally processed in Photoshop CS4 Version 11.0 (Adobe).
Statistics
All statistical analyses were performed using GraphPad Prism 7.0. Chlamydia growth assays were analyzed using a two-tailed Welch’s t-test, all other analyses used a two-tailed Mann-Whitney test. All experiments contained three biological replicates. Data from these replicates is either combined or plotted separately, as described in the figure legends.
Supplementary Material
Acknowledgements
We thank members of our laboratories for reagents and advice, particularly Dr. Lee Dolat (Duke University) for his contribution to some preliminary Chlamydia infection work. Access to DLS was supported in part by the EU FP7 infrastructure grant BIOSTRUCT-X (contract no. 283570). Work in the D.K. lab was funded by the Medical Research Council (U105192732), the European Research Council (724804), and the Lister Institute for Preventive Medicine. J.N.P. was supported on an EMBO Long-Term Fellowship. Work in the R.H.V. lab was funded by the National Institute of Health (R01AI100759 to R.H.V.) and the National Institute of Allergy and Infectious Diseases (STI CRC U19 AI084044 to R.J.B. and R.H.V.). E.B. was supported by North West Cancer Research.
Footnotes
Data availability. The data that support the findings in this study are available from the corresponding author upon request. Coordinates and structure factors for the ChlaDUB1~Ub, ChlaDUB1~CoA, and C.a. ChlaDUB structures have been deposited with the protein data bank accession codes 6GZS, 6GZT, and 6GZU respectively.
Author Contributions
Conceptualization, J.N.P. and D.K.; Investigation, J.N.P., R.J.B., E.B., K.N.S., L.D. and B.S.; Methodology, R.J.B., R.H.V., M.J.C., and S.U.; Writing, J.N.P. and D.K.; Funding Acquisition, D.K., R.H.V., R.J.B., S.U., and M.J.C.
Competing Interests statement
The authors declare no competing interests.
References
- 1.Lin YH, Machner MP. Exploitation of the host cell ubiquitin machinery by microbial effector proteins. J Cell Sci. 2017;130:1985–1996. doi: 10.1242/jcs.188482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastidas RJ, Valdivia RH. Emancipating Chlamydia: Advances in the genetic manipulation of a recalcitrant pathogen. Microbiol Mol Biol Rev. 2016;80:411–427. doi: 10.1128/MMBR.00071-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rytkönen A, et al. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc Natl Acad Sci USA. 2007;104:3502–3507. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misaghi S, et al. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol Microbiol. 2006;61:142–150. doi: 10.1111/j.1365-2958.2006.05199.x. [DOI] [PubMed] [Google Scholar]
- 5.Catic A, Misaghi S, Korbel GA, Ploegh HL. ElaD, a deubiquitinating protease expressed by E. coli. PLoS One. 2007;2:e381. doi: 10.1371/journal.pone.0000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chosed R, et al. Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases. J Biol Chem. 2007;282:6773–6782. doi: 10.1074/jbc.M608730200. [DOI] [PubMed] [Google Scholar]
- 7.Mukherjee S, et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 8.Mittal R, Peak-Chew SY, McMahon HT. Acetylation of MEK2 and IκB kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc Natl Acad Sci USA. 2006;103:18574–18579. doi: 10.1073/pnas.0608995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones RM, et al. Salmonella AvrA coordinates suppression of host immune and apoptotic defenses via JNK pathway blockade. Cell Host Microbe. 2008;3:233–244. doi: 10.1016/j.chom.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 10.Sheedlo MJ, et al. Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination. Proc Natl Acad Sci USA. 2015;112:15090–15095. doi: 10.1073/pnas.1514568112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pruneda JN, et al. Molecular basis for ubiquitin and ubiquitin-like specificities in bacterial effector proteases. Mol Cell. 2016;63:261–276. doi: 10.1016/j.molcel.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corn JE, Vucic D. Ubiquitin in inflammation: the right linkage makes all the difference. Nat Struct Mol Biol. 2014;21:297–300. doi: 10.1038/nsmb.2808. [DOI] [PubMed] [Google Scholar]
- 13.Le Negrate G, et al. ChlaDub1 of Chlamydia trachomatis suppresses NF-kappaB activation and inhibitis IkappaBalpha ubiquitination and degradation. Cell Microbiol. 2008;10:1879–1892. doi: 10.1111/j.1462-5822.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- 14.Mesquita FS, et al. The Salmonella deubiquitinase SseL inhibits selective autophagy of cytosolic aggregates. PLoS Pathog. 2012;8:e1002743. doi: 10.1371/journal.ppat.1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer A, et al. Chlamydia trachomatis-containing vacuole serves as deubiquitination platform to stabilize Mcl-1 and to interfere with host defense. eLife. 2017;6:e21465. doi: 10.7554/eLife.21465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang ZM, et al. Structure of a pathogen effector reveals the enzymatic mechanism of a novel acetyltransferase family. Nat Struct Mol Biol. 2016;23:847–852. doi: 10.1038/nsmb.3279. [DOI] [PubMed] [Google Scholar]
- 17.Mittal R, Peak-Chew SY, Sade RS, Vallis Y, McMahon HT. The acetyltransferase activity of the bacterial toxin YopJ of Yersinia is activated by eukaryotic host cell inositol hexakisphosphate. J Biol Chem. 2010;285:19927–19934. doi: 10.1074/jbc.M110.126581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reverter D, Lima CD. A basis for SUMO protease specificity provided by analysis of human senp2 and a senp2-SUMO complex. Structure. 2004;12:1519–1531. doi: 10.1016/j.str.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 19.Reverter D, et al. Structure of a complex between NEDD8 and the Ulp/Senp protease family member Den1. J Mol Biol. 2005;345:141–151. doi: 10.1016/j.jmb.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 20.Shen L, et al. Structural basis of NEDD8 ubiquitin discrimination by the deNEDDylating enzyme NEDP1. EMBO J. 2005;24:1341–1351. doi: 10.1038/sj.emboj.7600628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fullam E, et al. Divergence of cofactor recognition across evolution: coenzyme A binding in a prokaryotic arylamine N-acetyltransferase. J Mol Biol. 2008;375:178–191. doi: 10.1016/j.jmb.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 22.Sixt BS, Valdivia RH. Molecular genetic analysis of Chlamydia species. Annu Rev Microbiol. 2016;70:179–198. doi: 10.1146/annurev-micro-102215-095539. [DOI] [PubMed] [Google Scholar]
- 23.Kokes M, et al. Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of Chlamydia. Cell Host Microbe. 2015;17:716–725. doi: 10.1016/j.chom.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heuer D, et al. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature. 2009;457:731–735. doi: 10.1038/nature07578. [DOI] [PubMed] [Google Scholar]
- 25.Dumoux M, Hayward RD. Membrane contact sites between pathogen-containing compartments and host organelles. Biochim Biophys Acta Mol Cell Biol Lipids. 2016;1861:895–899. doi: 10.1016/j.bbalip.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 26.Wesolowski J, et al. Chlamydia hijacks ARF GTPases to coordinate microtubule posttranslational modifications and Golgi complex repositioning. mBio. 2017;8:e02280–16. doi: 10.1128/mBio.02280-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rejman Lipinski A, et al. Rab6 and Rab11 regulate Chlamydia trachomatis development and golgin-84-dependent Golgi fragmentation. PLoS Pathog. 2009;5:e1000615. doi: 10.1371/journal.ppat.1000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Hybiske K, Stephens RS. Direct visualization of the expression and localization of chlamydial effector proteins within infected host cells. Pathog Dis. 2018;76 doi: 10.1093/femspd/fty011. fty011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson B. An overview of protein moonlighting in bacterial infection. Biochem Soc Trans. 2014;42:1720–1727. doi: 10.1042/BST20140236. [DOI] [PubMed] [Google Scholar]
- 30.Berrow NS, et al. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucl Acids Res. 2007;35:e45. doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ekkebus R, et al. On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J Am Chem Soc. 2013;135:2867–2870. doi: 10.1021/ja309802n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W. XDS. Acta Crytallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crytallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crytallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crytallogr D Biol Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crytallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crytallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen BD, Valdivia RH. Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc Natl Acad Sci USA. 2012;109:1263–1268. doi: 10.1073/pnas.1117884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen YS, et al. The Chlamydia trachomatis type III secretion chaperone Slc1 engages multiple early effectors, including TepP, a tyrosinephosphorylated protein required for the recruitment of CrkI-II to nascent inclusions and innate immune signaling. PLoS Pathog. 2014;10:e1003954. doi: 10.1371/journal.ppat.1003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.