Abstract
The well-known age-related mitochondrial dysfunction deeply affects heart because of the tissue’s large dependence on mitochondrial ATP provision. Our study revealed in aged rat heart a significant 25% decrease in mtDNA relative content, a significant 29% increase in the 4.8 Kb mtDNA deletion relative content, and a significant inverse correlation between such contents as well as a significant 38% decrease in TFAM protein amount. The TFAM-binding activity to specific mtDNA regions increased at those encompassing the mtDNA replication origins, D-loop and Ori-L The same mtDNA regions were screened for different kinds of oxidative damage, namely Single Strand Breaks (SSBs), Double Strand Breaks (DSBs), abasic sites (AP sites) and oxidized bases as 7,8-dihydro-8-oxoguaninc (8oxoG). A marked increase in the relative content of mtDNA strand damage (SSBs, DSBs and AP sites) was found in the D-loop and Ori-L regions in the aged animals, unveiling for the first time in vivo an age-related, non-stochastic accumulation of oxidative lesions in these two regions that appear as hot spots of mtDNA damage. The use of Formamidopyrimidinc glycosylase (Fpg) demonstrated also a significant age-related accumulation of oxidized purines particularly in the D-loop and Ori-L regions. The detected increased binding of TFAM to the mtDNA damage hot spots in aged heart suggests a link between TFAM binding to mtDNA and loss of mitochondrial genome likely through hindrance of repair processes.
Keywords: Aging, Rat heart, MtDNA, Damage hot spots, TFAM binding
1. Introduction
Mitochondria are fundamental for cell homeostasis being the major producers of AΤΡ through oxidative phosphorylation and the hub of various metabolic processes [1]. The mitochondrial involvement in aging is well acknowledged and it cooperates, with other causes, to induce the progressive decline of functions characterizing the aging process [2]. The age-related mitochondrial dysfunction involves the organelle’s maintenance (mitochondrial biogenesis, dynamics and turnover) and its metabolic functionality, often eliciting tissue-specific alterations [3,4]. Particularly affected by the mitochondrial dysfunction with aging are those tissues largely dependent on the ΛΤΡ provided by oxidative phosphorylation, such as skeletal muscle, central nervous system, and heart. Indeed, since 90% of the AΤΡ required by cardio- myocytes derives from organelle’s bioenergetics [5,6], the relevance of mitochondrial dysfunction to heart function can easily be surmised. In the recent up-rise of research interest for the mitochondrial role in heart aging, different quality control processes of the organelle, including mitophagy, unfolded protein response and dynamics balance, have begun to be finely dissected [7,8], Yet, relatively little is known about the relevance of mitochondrial biogenesis to cardiac senescence. Our previous works analyzed several age-related alterations in mitochondrial biogenesis in the frontal cortex [9], cerebral hemispheres and cerebellum [10], liver [11,12] and soleus muscle [13] from rats, supporting the idea that some mitochondrial changes occurred with aging in all tissues, whereas others were tissue-specific. In particular, with aging a shared loss of the mitochondrial DNA (mtDNA) content was reported, whereas changes in the amount of mitochondrial transcription factor A (TFAM) [14–17], deeply involved in mtDNA transcription and copy number regulation, were found to be tissue-specific. Furthermore, in vivo TFAM binding to specific mtDNA regions, assayed with quantitative or semi-quantitative techniques, showed increased binding to the regions encompassing the origins of mtDNA replication (D-loop and Ori-L regions) in aged rat liver [11] and muscle [13], but not in the frontal cortex [13]. It was thus suggested that such increased TFAM binding might have induced mtDNA loss through hindering mtDNA repair and/or replication processes [11,12]. This idea was also supported by the possibility, raised according to in vitro findings, that TFAM may also be involved in mtDNA repair [18,19]. Furthermore, human TFAM is able to bind with great affinity to G-quadruplex structures, naturally present in the D-loop region, which might be implicated in mtDNA replication and maintenance [20]. It was therefore interesting to verify if the age-related increase in TFAM binding to the regions encompassing the origins of mtDNA replication might be common to other tissues and might be linked with increased DNA damage in the same regions. In order to address this research question, we decided to analyze mtDNA content and integrity as well as TFAM amount and functionality in young and old rat hearts.
2. Material and methods
2.1. Animals
For the present study, 2-month-old Fischer 344 × Brown Norway (F344BNF1) male rats were obtained and housed until they became 6- month-old (young, n = 4), and 27-month-old (old, n = 5). Rats were obtained from the National Institute of Aging colony (Indianapolis, IN, USA) and housed at the Department of Aging and Geriatric Research, Division of Biology of Aging, College of Medicine, University of Florida, Gainesville, FL, USA, in a specific pathogen-free facility accredited by the American Association for Accreditation of Laboratory Animal Care. Health status, body weight (BW), and food intake were monitored daily. The study was approved by the Institutional Animal Care and Use Committee at the University of Florida. All procedures were performed in accordance with the National Institutes of Health guidelines for the care and use of laboratory animals. Rats were sacrificed by rapid decapitation and the heart was immediately removed. After removal of the atria and the right ventricle, samples were snap-frozen in iso-pentane cooled by liquid nitrogen and stored in liquid nitrogen until use.
2.2. Measurement of mtDNA and mtDNA 4.8 Kb deletion content
The relative content of mtDNA and mtDNA 4834 bp (4.8 Kb) deletion was measured using quantitative real time polymerase chain reaction (qPCR). Reactions were performed via SYBR Green chemistry on a QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Fluorescence spectra were monitored by the QuantStudio Real-Time PCR Software vl.3 (Applied Biosystems, Foster City, CA, USA). Each sample was analyzed in triplicate in 10pL final volume containing iTaq SYBR Green Supermix PCR 1 × Master Mix (Bio-Rad Laboratories Inc., Hercules, CA, USA), 0.5 μΜ forward and reverse primers, and DNA template (3 ng). After 10 min of denaturation at 95 ˚C, amplification proceeded for 40 cycles, each consisting of denaturation at 95 ˚C for 1 s, annealing and extension at 60 ˚C for 20 s (Fast block). The quantification of the mtDNA content (mtDNA primer set) relative to nuclear DNA (ß-actin primer set) was determined as previously reported [11]. The relative abundance of the 4.8 Kb deletion (4.8 Del primer set) normalized to mtDNA content was determined according to the formula 2ΔCT× — 2ΔCTb, where ΔCT× is the difference between the CT values obtained using the 4.8 Del primer set and the mtDNA primer set, and ΔcTb is the difference between the CT values obtained using the mtDNA primer set and the ß-actin primer set Primer sequences are reported in Table 1.
Table 1.
Oligonucleotide primer sequences.
| Primer set | Forward primer | Reverse primer | (nps) | (nps) |
|---|---|---|---|---|
| mtDNA | 5’GGTTCTTACTTCAGGGCCATCA3’ | 5’TGATTAGACCCGTTACCATCGA3’ | 15,785–15,806 | 15,868–15,847 |
| β-actin | 5’CCCAGCCATGTACGTAGCCA3’ | 5’CGTCTCCGGAGTCCATCAC3’ | 2181–2200 | 2266–2248 |
| 4.8 Del | 5’AAGGACGAACCTGAGCCCTAATA3’ | 5’CGAAGTAGATGATGCGTATACTGTA3’ | 8109–8131 | 13,020–12,996 |
| RT D-loop | 5’CACCCCCTACACCTGAAACTT3’ | 5’TTTGTGTCGGGAAATTTTACCAAT3’ | 16,092–16,112 | 16,250–16,227 |
| RT Ori-L | 5’CAGCTAAATACCCTACTTACTGG3’ | 5’GCCCCCTTTTTACCAAAAAGCC3’ | 5120–5142 | 5270–5249 |
| RT ND1 | 5’AACGCCCTAACATCAATTGTATTCC3’ | 5’TGGTCATATCGAAAACGGGGG3’ | 3442–3466 | 3590–3570 |
| D-loop long | 5’TCTGGTCTTGTAAACCAAAAATGA3’ | 5’TGGAATTTTCTGAGGGTAGGC3’ | 15,302–15,325 | 16,302–16,282 |
| D-loop short | 5’TCTGGTCTTGTAAACCAAAAATGA3’ | 5’TGATGGTGGGGAGGTAGTTC3’ | 15,302–15,325 | 15,378–15,359 |
| Ori-L long | 5’AACCAGACCCAAACACGAAA3’ | 5’CTATTCCTGCTCAGGCTCCA3’ | 4414–4433 | 5407–5388 |
| Ori-L short | 5’CGTAAACCGTTGACTCTTTTCA3’ | 5’CTATTCCTGCTCAGGCTCCA3’ | 5328–5349 | 5407–5388 |
| ND1 long | 5’AGGACCATTCGCCCTATTCT3’ | 5’CGCCAACAAAGACTGATGAA3’ | 3390–3409 | 4399–4380 |
| ND1 short | 5’AGGACCATTCGCCCTATTCT3’ | 5’TGATGTTAGGGCGTTTATTAGGA3’ | 3390–3409 | 3456–3434 |
2.3. Western blotting
Whole-tissue extracts were obtained from 50 mg of left ventricle. Samples were pulverized under liquid nitrogen with a porcelain mortar and pestle. The powder was suspended in ice-cold lysis buffer [220 mM mannitol, 70 mM sucrose, 20 mM Tris-HCl pH 7.4, 5 mM MgCl2, 5 mM ethylene glycol tetraacetic acid (EGTA)], and 1 mM ethylenediaminete- traacetic acid (EDTA)]. Samples were homogenized, precleared by centrifugation and the supernatant fraction was recovered. Ten μg total proteins, quantified with the Bradford method (Bio-Rad Laboratories Inc., Hercules, CA, USA), were separated by gel electrophoresis on 4–12% Bis-Tris Criterion XT precast gels (Bio-Rad Laboratories Inc., Hercules, CA, USA) and electroblotted onto polyvinylidene fluoride membranes (Amersham Pharmacia Biotech Inc., Piscata way, NJ, USA). Immunoblotting was performed with a rabbit anti-TFAM antibody (1:30,000), a rabbit anti-GAPDH antibody (1:100,000; Cell Signaling Technology, Beverly, ΜΛ, USA, cat. n. 2118), and a peroxidase-conjugated anti-rabbit secondary antibody at a dilution of 1:10,000 (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA). The antibody against TFAM was custom-made as previously described [11]. Bands were revealed by enhanced chemiluminescence (ECL). Signals were analyzed by laser densitometry with the Chemi Doc System and Image Lab software (Bio-Rad Laboratories Inc., Hercules, CA, USA). The densito- metric value (OD units) of each TFAM band was then related to that of the respective band of GAPDH.
2.4. Mitochondrial DNA immunoprecipitation
The binding of TFAM to specific regions of mtDNA was analyzed using mtDNA immunoprecipitation (mIP) following the procedure described elsewhere [9]. Due to the small size of the tissue samples the mIP analysis could be performed only in the old rats and in no young animals. Briefly, frozen samples of 60 mg each were submitted, in sequential steps, to cross-linking, termination of cross-linking, brief homogenization and centrifugation. Each obtained pellet was washed in PBS, suspended with homogenization buffer, manually homogenized, briefly centrifuged, and incubated in lysis buffer. The obtained DNA was subjected to shearing by sonicadon and checking of the size range (between 500 and 900 bp) of the fragments by electrophoresis on a 1.2% agarose gel. Each sample was diluted with 3 volumes of FSB buffer (5 mM EDTA, 20 mM Tris HCl pH 7.5, 50 mM NaCl), pre-deared with 75 μL of protein A-agarose/Salmon sperm 50% DNA (Upstate, distributed by Millipore Corporate Headquarters, Billerica, ΜA, USA) for 2 mL of sample on a rotator at 4 °C for 30 min and centrifuged at lOOOrpm for 1 min. The resuldng pellet was suspended in 300 μL of FSB and divided into three aliquots: the input (100 μL), without im- munopreripitarion, was stored at — 80 ˚C until the cross-linking reversal; the other two aliquots (lOOμL each) were incubated overnight at 4°C with the above described rabbit and-TFAM antibody (1:50 dilution) and without antibody (-Ab), respectively. The next day 15 μL of Protein A-agarose/Salmon sperm 50% DNA (Upstate, distributed by Millipore Corporate Headquarters, Billerica, ΜA, USA) were added to each sample for 2 h at 4 ˚C to isolate protein-DNA complexes. After a brief centrifugation, the pellets were washed three times with 1 mL of RIPA buffer (140 mM NaCl), three times with 1 mL of RIPA buffer (500 mM NaCl), three times with 1 mL of LiCl buffer and twice with 1 mL of TE. The final pellets, suspended in TE containing 0.5% SDS (200 μL), together with the original input, were incubated at 65 ˚C for 6 h for the thermal reversal of the cross-linking. All DNA samples (treated with specific antibody, without Ab, and the input) were ethanol-precipitated overnight. The resulting pellets, after being washed with cold 70% ethanol, dried, and suspended in 200 μL of sterile ultrapure H20, were treated with 10 μg of RNase A (50μg/μL) for one hour at 37 ˚C and incubated with 20 μg of Proteinase K (100 μg/μL) and SDS (0.25%) at 37 ˚C overnight After a first extraction with phenol/ chloroform/isoamyl alcohol (25:24:1) and a second extraction with chloroform/isoamyl alcohol (24:1), all DNAs were ethanol-precipitated overnight, centrifuged, washed with cold 70% ethanol, dried and suspended in 60 μL of 10 mM Tris-HCl pH 8.0. Input and mIP mtDNAs were subjected to qPCR analysis as described in the following section.
2.5. Quantitative PCR of mIP DNA
The measurement of the relative content of mtDNA im- munoprecipitated by ΤFAΜ was carried out by qPCR. To analyze the D- loop, Ori-L, and ND1 mtDNA regions bound by TFAM, rt D-loop, rt Ori- L, and rt ND1 primer sets were used (Table 1). qPCR reactions were performed via SYBR Green chemistry on a QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) and fluorescence spectra were monitored by the QuantStudio Real-Time PCR Software vl.3 (Applied Biosystems, Foster City, CA, USA). The reaction mixture (total volume 20 μL) consisted of iTaq SYBR Green Supermix PCR 1 × Master Mix (BioRad Laboratories Inc., Hercules, CA, USA), 0.2 μM forward and reverse primers, and 2.5 μL of the input or of the immunoprecipitated with anti-TFAM or without antibody DNA aliquots. After 10 min of denaturation at 95 ˚C, amplification proceeded for 40 cycles, each consisting of denaturation at 95 ˚C for 1 s, annealing and extension at 60 ˚C for 20 s (Fast block). The calculation of the relative content of TFAM-bound mtDNA was performed according to the formula 2ΔCT× — 2ΔCTb, where ΔcT× is the difference between the CT values of the input and the immunoprecipitated sample and ΔCTb is the CT difference between the CT values of the input and the no-antibody sample [21], respectively for each analyzed region.
2.6. MtDNA damage analyses
2.6.1. Strand damage analysis
The relative content of strand damage in the D-loop, Ori-L, and ND1 mtDNA regions was determined according to [22], with modifications, by qPCR amplification of fragments of different lengths: long (long primer set) and short (short primer set) amplicons for each analyzed region. The method is based on the interference, due to SSBs, DSBs, and AΡ sites, on the polymerase-based DNA amplification. The primers for each region (listed in Table 1) were designed with the Primer Express software (Applied Biosystems, Foster City, CA, USA). The method was validated by primer-limiting experiments to determine the proper primer concentrations (0.5 μΜ for each primer pair), by evaluating the reaction efficiencies of the long and short amplicons. Amplification specificity was controlled by melting curve analysis and gel electrophoresis. qPCR amplification reactions were performed via SYBR Green chemistry on a QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) and fluorescence spectra were monitored by the QuantStudio Real-Time PCR Software vl.3 (Applied Biosystems, Foster City, CA, USA). The reaction mix (total volume 10μL) consisted of iTaq SYBR Green Supermix PCR l × Master Mix (BioRad Laboratories Inc., Hercules, CA, USA), 0.5 μΜ each forward and reverse primer (specific for the long or the short amplicon, respectively), and template DNA (3 ng of total DNA). The cycling conditions included a pre-incubation phase of 10 min at 95 ˚C followed by 40 cycles of 10 s 95 ˚C, 10 s 58 ˚C, and 10 s 72 ˚C (short fragments) or 50 s 72 ˚C (long fragments). Each sample was analyzed in triplicate. The calculation of the relative content of mtDNA strand lesions in old vs. young rats was performed using the formula 2−(ACT1ong − ΔCTshort), adapted from [23]. Briefly, the difference in the CT values between old and young rats, used as the reference, for the long and the short fragments, respectively, was used to determine the relative content of DNA strand damage in each tested region.
2.6.2. Modified purines analysis
Formamidopyrimidine glycosylase (Fpg) (New England Biolabs, Beverly, ΜA, USA) digestion of total DNA was used to detect oxidized purines, according to [24]. The method is based on the selective cleavage by Fpg at sites of oxidized purines, introducing SSBs that block the following amplification. The PCR amplification of the D-loop, Ori-L and ND1 mtDNA regions was conducted using the respective long primer sets (Table 1) on FPG-treated and untreated templates. The reaction mix (total volume 20 μL) consisted of DreamTaq Green PCR 1 × Master Mix (Thermo Fisher Scientific Inc, Waltham, ΜA, USA), 0.5 μΜ each forward and reverse primer, and template DNA (15 and 7.5 ng of Fpg- treated or untreated total DNA). The cycling conditions included a preincubation phase of 10 min at 95 ˚C followed by 18 cycles of 15 s 95 ˚C, 15 s 58 ˚C, and 1 min 72 ˚C. An aliquot (10 μL) of each PCR amplification was loaded on 1.3% agarose gel. Ethidium bromide-stained bands were visualized using Gel Doc XR documentation system (BioRad Laboratories Inc., Hercules, CA, USA). Band intensity was analyzed by Image Lab Software (BioRad Laboratories Inc., Hercules, CA, USA). Data are presented as the ratio between Fpg-treated and untreated band intensities, expressed as percentage.
2.7. Statistics
Data are expressed as mean ± SEM. The Student’s t-test or the oneway ANOVA test with the Tukey’s Multiple Comparison Test were used when appropriate to compare groups. Correlation between analyzed parameters was determined by the Pearson’s test All differences were considered significant at a 5% level. A specific statistical package was used (Stata Corp. 2005. Stata Statistical Software: Release. College Station, TX, USA).
3. Results
3.1. Age-related effect on content and integrity of mtDNA in rat heart
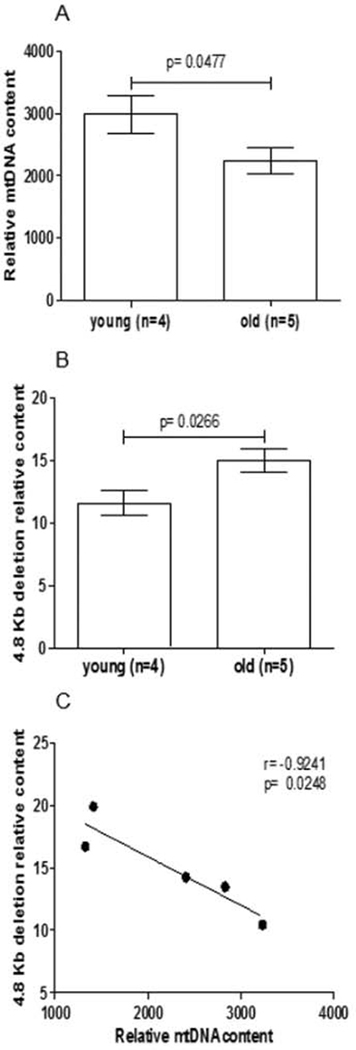
As mtDNA content was reported to be affected by aging in various rat organs [9–11,13], but only indirect indications were available for the heart [25], we determined the relative mtDNA content by qPCR in heart from young and old rats. A significant 25% reduction was observed in old compared with young rats (p = 0.0477, t-test; Fig. 1A) demonstrating the age-related loss of mtDNA also in the rat heart. We then verified the effect of age on mtDNA quality by determining, through qPCR, the relative content of the 4834 bp (4.8 Kb) deletion, used as a marker of the structural damage of the genome. A significant 29% increase in the abundance of such deletion was found in old relative to young rats (p = 0.0266, t-test; Fig. 1B), suggesting increased damage to mtDNA with aging. A statistically significant inverse correlation was found between mtDNA and 4.8 Kb deletion contents (r = — 0.9241, p = 0.0248, Pearson’s test; Fig. 1C), indicating that rats with a more pronounced mtDNA loss also showed an increased proportion of deleted species.
Fig. 1.
Age-related changes of the relative content of mlDNA and of the 4.8 Kb deletion and correlation between individual values in old rats. Bars represent the mean value ( ± SEM) of triplicates. A: relative mlDNA content in young (n = 4) and old rats (n = 5). p = t-test B: 4.8 Kb deletion relative content in young (n = 4) and old (n = 5) rats, p = t-l C: correlation between mlDNA and 4.8 Kb relative content in old rats (n 5). Dots arc the mean values of triplicates, r, p = Pearson’s test.
3.2. Effect of age on die amount and binding activity of TEAM in rat heart
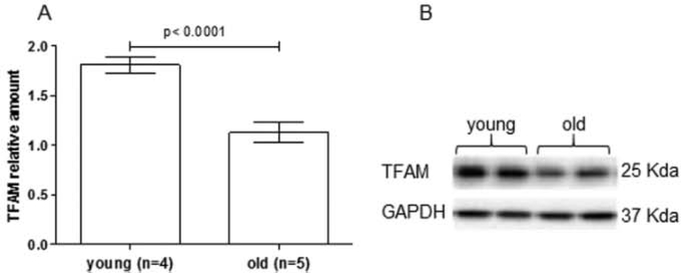
The relative amount of TFAM in the heart of young and old rats was determined by Western immunoblotting. A 38% decrease was found in old rats relative to their younger counterparts (p < 0.0001, t-test) (Fig. 2A).
Fig. 2.
Agc-related change of relative TFAM amount. A: the histogram shows the relative amount of TFAM normalized to GAPDH in young (n = 4) and old rats (n = 5). Bars represent the mean ( ± SEM) of triplicates, p t-test. B: representative Western blot carried out in two young and two old animals.
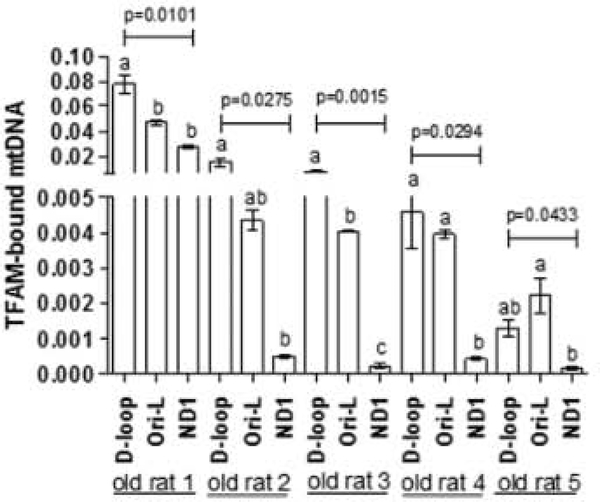
The age-related decrease in TFAM amount was consistent with the above reported mtDNA loss and questioned us about the activities performed by the factor that might have been also affected by aging. Therefore, mIP was carried out in samples from five old rats to verify an eventual differential binding of TFAM along the mtDNA molecule. The screened regions were the two encompassing the mtDNA origins of replication, namely D-loop and Ori-L, and a third one, included in the ND1 gene, that was chosen as a control, having no other functional role besides the coding one. The relative amounts of TFAM-bound mtDNA were determined by qPCR and were very different because of marked individual variability among old animals. As shown in Fig. 3, the difference in the relative amounts of TFAM-bound mtDNA among the three analyzed regions was statistically significant (p = 0.0101, p = 0.0275, p = 0.0015, p = 0.0294, p = 0.0433, respectively; one-way ANOVA). On overall the relative amounts of TFAM-bound mtDNA were higher at the D-loop and Ori-L regions than at ND1 (p < 0.05, Tukey’s Multiple Comparison Test).
Fig. 3.
Relative amounts of TFAM-bound mtDNA at the D-loop, Ori-L, and ND1 mtDNA regions determined by qRT-PCR in five old rats. Bars represent the mean value ( ± SEM) of the results obtained in each animal, p = one-way ANOVA Bars not sharing a common superscript differ significantly (p < 0.05, Tukey’s Multiple Comparison Test).
3.3. Effect of age on mtDNA damage in rat heart
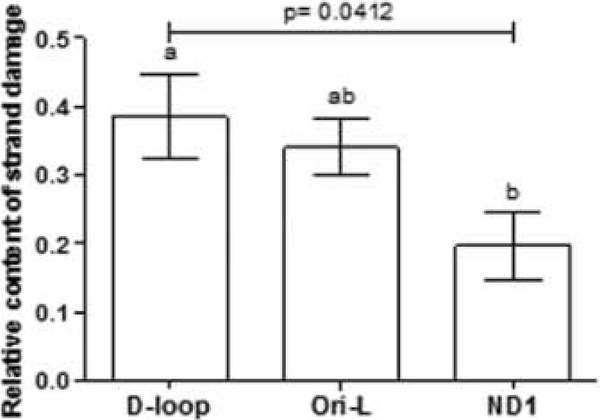
The overall tendency to an increased binding of TFAM to the D-loop and Ori-L regions in old rats prompted us to verify if there was an increased DNA damage at the same regions. Therefore, we set up an assay to evaluate, by means of an appropriate qPCR method, mtDNA strand damage (SSBs, DSBs, and AP sites) at specific mtDNA regions. Using the young rats value as the reference in the calculation, it was possible to determine the relative content of mtDNA strand damage in old animals at the same regions screened for TFAM binding. Such content was significantly different among the three regions (p = 0.0412, one-way ANOVA) (Fig. 4). In particular, the D-loop region of old rats showed a two-fold greater abundance of strand lesions relative to that of the ND1 region, whereas the Ori-L region was characterized by a 72% higher relative content of damage than that of ND1.
Fig. 4.
Relative content of mtDNA strand damage in old vs. young rats at the D- loop, Ori-L, and ND1 regions. Bars represent the mean value ( ± SEM) of triplicates. p = one-way ANOVA Bars not sharing a common superscript differ significantly (p < 0.05, Tukey’s Multiple Comparison Test).
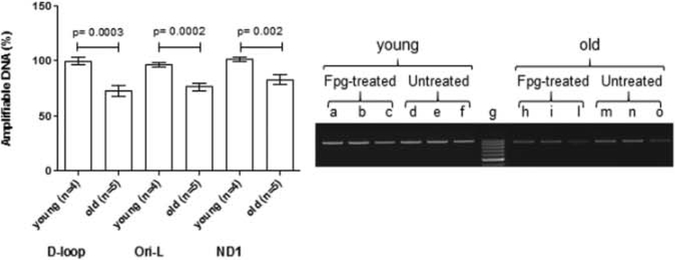
Previous in vitro studies found that TFAM preferentially bound to oxidized guanosine (8o×oG) [18.19]. We therefore decided to screen the same mtDNA regions for the presence of oxidized purines by using the oxidized purines-sensitive enzyme Fpg and comparing the amount of PCR-amplifiable template between Fpg-treated and untreated DNA. Expressing the result as a percentage, the closer to 100% was the value, the lower was the presence of oxidized purines and the resulting inhibition to the polymerase activity. In old rats the percentages of am- plifiable DNA were 72.7% in the D-loop region, 76.2% in the Ori-L region, and 86.1% in the ND1 region, respectively. The difference among the three regions approached the statistical significance (p = 0.0584, one-way ANOVA). In young rats, the percentage of ampli fiable DNA was close to 100% in all the analyzed regions (99.6%, 96.5%, and 100% in the D-loop, Ori-L and ND1 region, respectively). The amount of amplifiable DNA in Fpg-treated samples was sig- nificandy lower in old than in young rats in all the analyzed regions (p = 0.0003, p = 0.0002, and p = 0.002 for D-loop, Ori-L and ND1, respectively; t-test) (Fig. 5Λ), indicating that oxidized purines accumulated in old age.
Fig. 5.
Age-related increase in purine-specific inlDNA damage at the D-loop, Ori-L, and ND1 regions. A: bars represent the mean ( ± SEM) ratio of Fpg-lrcaled and untreated band intensities, expressed as percentage, n = number of analyzed animals, p = t-test B: representative gel analysis of Fpg-trealed and untreated total DNA from one young and one old raL 15 and 7.5 ng of total DNA were amplified using the D-loop long primer set. An aliquot of each PCR amplification was loaded onto agarose elhidium bromide-stained gel and analyzed for band intensities, a: molecular weight marker; a, b, d, e, h, i, m, and n: 15ng total DNA; c, f, 1, and o- 7.5 ng total DNA; g: molecular weight marker (Gene ruler 100 bp,Thermo Fisher Scientific Inc, Waltham,MA,USA).
4. Discussion
The heart is highly dependent on mitochondrial oxidative phosphorylation for energy supply. Hence, cardiomyocyte function and viability are deeply affected by mitochondrial dysfunction [26,27]. Though, how cardiomyocyte mtDNA maintenance is altered by aging is not yet fully understood. To gain insights into the age-related changes in mitochondrial biogenesis, we conducted a comprehensive investigation on mtDNA quantity and quality, and TFAM binding in the heart of old rats. In line with previous findings by our group in liver, frontal cortex and skeletal muscle [19,11,13], we detected a 25% decrease in the relative mtDNA content in the heart of old versus young rats. Furthermore, the relative content of the 4.8 Kb mtDNA deletion the so-called common deletion, [9,28], was determined as a marker of structural damage. We found a 29% increase in old animals in comparison to their younger counterparts. Notably, in old rats total mtDNA abundance was inversely correlated with the common deletion content, consistent with the suggested replicative advantage of the shorter (i.e., deleted) molecules versus the longer wild-type mtDNA [29]. Since TFAM amount was reported to be closely related to the mtDNA content [15,30], we then measured TFAM protein expression and found a 38% age-related decrease. This observation is in agreement with the results obtained in liver and soleus muscle of old rats [11,13] and might be due to Lon protease activity as it was reported not to decrease in aged rat heart [31]. Also, the binding activity of TFAM to specific regions of mtDNA was altered in the aged heart, being increased at the regions encompassing the mtDNA replication origins (D-loop and Ori-L) with respect to the control region ND1. Similar results were obtained in liver [11] and soleus muscle [ 13], but not in the frontal cortex [9]. A possible involvement of TFAM in mtDNA repair was suggested by previous in vitro studies demonstrating that TFAM preferentially bound to 8oxoG [18,19]. Such binding hindered the base excision repair (BER) mechanisms by inhibition of 8-oxoguanine DNA glycosylase (OGGI), ur- acil-DNA glycosylase (UDG), apurinic endonuclease 1 (ΑΡΕΙ), and DNA polymerase γ (pol γ) [19]. Therefore, the D-loop, Ori-L and ND1 regions were screened for different kinds of oxidative damage (i.e., SSBs, DSBs, ΛΡ sites, and 8oxoG). The relative content of mtDNA strand damage (SSBs, DSBs, and ΛΡ sites) was markedly increased in the regions encompassing the origins of replication relative to the ND1 region in old rats. In particular, the mtDNA damage was 95% higher in the D-loop and 72% higher in the Ori-L regions than in the ND1 counterpart. This suggests an age-related, non-stochastic accumulation of oxidative lesions in two functionally relevant regions that may represent hot spots of mtDNA damage. The qPCR method used to evaluate the relative content of strand damage, however, was not highly sensitive in the detection of oxidized purines, such as 8oxoG, because these adducts did not completely prevent the amplification process [22]. Therefore, we assessed, in the same three mtDNA regions, purine-specific mtDNA lesions by using the oxidized purines-sensitive enzyme Fpg. In young rats, the percentage of amplifiable DNA was almost equal to 100% in all the analyzed regions, whereas in old rats this percentage was 72.7% in the D-loop, 76.2% in the Ori-L, and 86.1% in the ND1 regions. This clearly demonstrates an age-related, significant accumulation of oxidized purines that was more relevant in the regions encompassing the origins of replication. These findings are in keeping with the age-related increased presence of oxidized purines, mosdy 8oxoG, in mtDNA from various rat tissues [32–38]. The results obtained through the determination of the mtDNA 4.8 Kb deletion content and the analysis of strand damage and of oxidized purines allowed us to report for the first time the age-related increase in different kinds of mtDNA oxidative damage. What is particularly intriguing in our study is the in vivo finding of two hot spot regions of mtDNA damage, D-loop and Ori-L, that are also characterized by increased binding of TEAM. It has recently been reported that a short hypoxia in pulmonary artery endothelial cells induced only oxidative modifications of bases and no alkali-detectable lesions, restricted to the D-loop region, which appeared extremely sensitive to oxidative damage. Furthermore, hypoxia also elicited an increased binding of TFAM to the D-loop region, likely because of the induced oxidative modifications since it was attenuated by Oggl overexpression. The authors suggested that the D-loop-specific increase in oxidative damage and subsequent enhanced TFAM-binding might have triggered the mtDNA replication verified in hypoxia since this condition, particularly in the vasculature, is a strong inducer of cell proliferation and vasculogenesis through up-regulation of VEGF expression [24]. According to our model, the presendy reported increased binding of TFAM to the damage hot spots might have prevented the normal repair process by hindering the repairing enzymes accessibility to the lesions and/or by trapping the accessed enzymes and preventing their activities. The unrepaired, persistent damage (especially in the form of DSBs, SS gaps or ΛΡ sites) might then have induced the degradation of the damaged molecules and the loss of mtDNA. In fact, persistent mtDNA damage may induce the degradation of the genome through different mechanisms verified in vitro: unrepaired SSBs in mtDNA were shown to trigger apoptotic signaling [39], and a persistent, although smaU, amount of AΡ sites and single-strand gaps was reported to lead to mtDNA degradation [40]. Also, some in vivo studies, about persistent ΛΡ sites induced in the mtDNA of mouse brain neurons [41] and mouse cardiomyocytes [42], demonstrated that such damage elicited a relevant mtDNA loss. Recently, the regulated induction of DSBs in mtDNA of a stable human cell line has been reported to drive the loss of mtDNA, likely through degradation of the damaged molecules [43]. Furthermore, the possible effect of increased TFAM-binding to the damage hot spots on the maintenance of mtDNA is supported by the recent paper by [44]. These authors reported how DNA repair attempts may sometime cause a more serious damage leading to the formation of a DNA-protein crosslink that might trap the repair enzyme on the DNA. Preliminary evidences suggest that this might occur for pol γ acting on ΛΡ sites oxidized at the 1-carbon of 2-deoxyribose [45]. Such DNA-protein crosslinks should probably be solved to restore the repair activity, but the increased TFAM-binding to the damage hot spots might enhance rather than reduce the persistence of crosslinks eventually formed, and thus might prevent effective repair processes. Therefore, accumulating evidences support the possibility that the age- related increased binding of TFAM to the damage hot spots might have prevented the normal repair process. Effectively, a recent and comprehensive review has indicated the failure of the repair mechanisms among the major causes of the age-related mtDNA mutations [46], strengthening our interpretation of data. Furthermore, the increased binding of TFAM to the damaged regions encompassing the origins of mtDNA replication might have also hindered a regular replication process, thus concurring to the age-related mtDNA loss. It is firmly established that ΛΡ sites can induce a stall of the pol γ activity during mtDNA replication [47] and that, as later described in 3T3 cells, the efficiency of pol γ to perform translesion synthesis through AP sites in vivo is low and when the molecule is too severely damaged usual BER mechanisms or even mtDNA degradation become the major pathways to process AP sites [48]. Further studies are necessary to verify the consistency of such mechanism in other tissues or model systems. However, the present findings offer a mechanistic framework that explains and links the age-related increased binding of TFAM to the damage hot spots of mtDNA with the loss of mitochondrial genome.
Acknowledgements
This research was supported by grants to AMSL (University of Bari- Progetti di Ateneo 2012; Istituto Banco di Napoli-Fondazione 2015), to CL (National Institute on Aging (NIA) ROI AGI7994), and to RB, RC, and EM: Fondazione Roma (NCDs Call for Proposal 2013), nonprofit research foundation “Achille and Linda Lorenzon”.
Footnotes
Conflict of interest
None declared.
References
- [1].Galluzzi L, Kcpp O, Trojel-Hansen C, Kroemer G, Mitochondrial control of cellular life, stress, and death, Circ. Res Ill (9) (2012) 1198–1207, 10.1161/ORCRESAHA.112.268946. [DOI] [PubMed] [Google Scholar]
- [2].Lopez Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G, The hallmarks of aging. Cell 153 (6) (2013) 1194–1217, 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Held NM, Houtkooper RH, Mitochondrial quality control pathways as determinants of metabolic health, Bioessays 37 (8) (2015) 867–876, 10.1002/bies.201500013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schcibye Knudsen M, Fang ELF, Croteau DL, Wilson DM III, Bohr VA, Protecting the mitochondrial powerhouse. Trends Cell Biol. 25 (3) (2015) 158–170, 10.1016/j.tcb.2014.ll.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lesncfsky EJ, Chen Q, Hoppcl CL, Mitochondrial metabolism in aging heart, Qrc. Res 118 (10) (2016) 1593–1611, 10.1161/CIRCReSAHA.116.307505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Martín-Fernández B, Gredilla R, Mitochondria and oxidative stress in heart aging, Age 38 (2016) 225–238, https://doi.org/l0.1007/sll357-016-9933-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shi R, Guberman M, Kirshenbaum LA, Mitochondrial quality control: the role of mitophagy in aging, Trends Cardiovasc. Med 28 (4) (2017) 246–260, 10.1016/j.tcm.2017.11.008. [DOI] [PubMed] [Google Scholar]
- [8].Tocchi A, Quarles FK, Basisty N, Gitari L, Rabinovitch PS, Mitochondrial dysfunction in cardiac aging, Biochim. Biophys. Acta 1847 (11) (2015) 1424–1433, 10.1016/jbbabio.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Picea A, Fracasso F. Pesce V, Cantatore P, Joseph AM, Leeuwenburgh C, Gadaleta MN, Lczza AMS, Age- and calorie restriction-related changes in rat brain mitochondrial DNA and TFAM binding, Age 35 (2013) 1607–1620, 10.1007/sl1357-012-9465-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nicassio L, Fracasso F, Sirago G, Musicco C, Picea A, Marzetti E, Calvani R, Cantatore P, Gadaleta MN, Pesce V, Dietary supplementation with acetyl-l-car- nitine counteracts age-related alterations of mitochondrial biogenesis, dynamics and antioxidant defenses in brain of old rats, Exp. Gerontol 98 (2017) 99–109, 10.1016/j.exger.2017.08.017. [DOI] [PubMed] [Google Scholar]
- [11].Picea A, Pesce V, Fracasso F, Joseph AM, Leeuwenburgh C, Lczza AMS, Aging and calorie restriction oppositely affect mitochondrial biogenesis through TFAM binding at both origins of mitochondrial DNA replication in rat liver, PLoS One 8 (2013) e74644, 10.1371/joumal.pone.0074644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Picea A, Pesce V, Sirago G, Fracasso F. Leeuwenburgh C, Lczza AMS, “What makes some rats live so long?” The mitochondrial contribution to longevity through balance of mitochondrial dynamics and mtDNA content, Exp. Gerontol 85 (2016) 33–40, https://doi.org/l0.1016/j.exger.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Picea A, Pesce V, Fracasso F, Joseph AM, Leeuwenburgh C, Lczza AMS. A comparison among the tissue -specific effects of aging and caloric restriction on TFAM amount and TFAM-binding activity to mtDNA in rat, Biochim. Biophys. Acta 1840 (7) (2014) 2184–2191, 10.1016/j.bbagen.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fisher RP, Clayton DA, Purification and characterization of human mitochondrial transcription factor 1, Mol. Cell. Biol 8 (1988) 3496–3509, 10.1128/MCB.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Larsson NG, Wang J, Wilhelmsson IL, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA, Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenes is in mice, NaL Genet 18 (3) (1998) 231–236, 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- [16].L Bestwick M, Shadel GS, Accessorizing the human mitochondrial transcription machinery, Trends Biochem. Sei 38 (6) (2013) 283–291, https://doi.org/10.10l6/j.tibs.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Picea A, Lezza AM, Regulation of mitochondrial biogenesis through TFAM-mi- tochondrial DNA interactions: useful insights from aging and calorie restriction studies, Mitochondrion 25 (2015) 67–75, 10.1016/j.mito.2015.10.001. [DOI] [PubMed] [Google Scholar]
- [18].Yoshida Y, Izumi II, Ise T, Uramoto IL, Torigoc T, Ishiguchi IL, Murakami T, Tanabc M, Nayakama Y, Itoh II, Kasai IL, Kohno K, Human mitochondrial transcription factor A binds preferentially to oxidatively damaged DNA, Biochcm. Biophys. Res. Commun 295 (2002) 945–951, 10.1016/S0006-291X(02)00757-X. [DOI] [PubMed] [Google Scholar]
- [19].Canugovi C, Maynard S, Bayne ACV, Sykora P, Tian J, de Souza Pinto NC. Croteau D, Bohr VA, The mitochondrial transcription factor A functions in mitochondrial base excision repair, DNA Repair 9 (10) (2010) 1080–1089, 10.1016/j.dnarep.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lyonnais S, Tarros Solé A, Rubio Cosíais A. Cuppari A, Brito R, Jaumot J, Gargallo R, Vilaseca M, Silva C, Granzhan A, Teuladc Fichou MP, ritja RF, Sola M, The human mitochondrial transcription factor A is a versatile G-quadruplex binding protein, Sd. Rep 7 (2017) 43992, 10.1038/srep4399Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Vercautcren JK, Pasko RA, Gleyzer N, Marino VM, Scarpulla RC, PGC-1-related coactivator immediate early expression and characterization of a CREB/NRF-1 binding domain associated with cytochrome c promoter occupancy and respiratory growth, Mol. Cell. Biol 26 (2006) 7409–7419, 10.1128/MCB.00585-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rothfuss O, Gasser T, Patcngc N, Analysis of differential DNA damage in the mitochondrial genome employing a semi-long run real-time PCR approach. Nucleic Acids Res. 38 (4) (2010) e24, 10.1093/nar/gkpl082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Livak KJ, Schmittgcn TD, Analysis of relative gene expression data using RealTime quantitative PCR and the 2 AACT method. Methods 25 (4) (2001) 402–408, 10.1006/mcth.2001.1262. [DOI] [PubMed] [Google Scholar]
- [24].Pastukh VM, Gorodnya OM, Gillespie MN, Ruchko MV, Regulation of mi tochondrial genome replication by hypoxia: the role of DNA oxidation in D-loop region, Free Radie. Biol. Med 96 (2016) 78–88, 10.1016/j.freeradbiomed.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ljubicic V, Joseph AM, Saleem A, Uguccioni G, Collu-Marchese M, Lai RYJ, Nguyen LMD, Hood DA, Transcriptional and post-transcriptional regulation of mitochondrial biogenesis in skeletal muscle: effects of exercise and aging, Biochim. Biophys. Acta 1800 (3) (2010) 223–234, 10.1016/j.bbagcn.2009.07.031. [DOI] [PubMed] [Google Scholar]
- [26].Kopsidas G, Kovalenko SA, Heffcman DR, Yarovaya N, Kramarova L, Stojanovski D, Borg J, Islam MM, Caragounis A, Linnane AW, Tissue mitochondrial DNA changes. A stochastic system, Ann. N. Y. Acad. Sei 908 (2000) 226–243, 10.1111/j.1749-6632.2000.tb06650.x. [DOI] [PubMed] [Google Scholar]
- [27].Hoppel CL, Lesncfsky EJ, Chen Q, Tandler B, Mitochondrial dysfunction in cardiovascular aging, Adv. Exp. Med. Biol 982 (2017) 451–464, 10.1007/978-3-319-55330_6.24. [DOI] [PubMed] [Google Scholar]
- [28].Gadaleta MN, Rainaldi G, Lczza AM, Milella F, Fracasso F, Cantatore P, Mitochondrial, DNA copy number and mitochondrial DNA deletion in adult and senescent rats, MutaL Res. 275 (1992) 181–193, 10.1016/0921-8734(92)90022-11. [DOI] [PubMed] [Google Scholar]
- [29].Fukui H, Moraes CT, Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons, Hum. Mol. Genet 18 (6) (2009) 1028–1036, 10.1093/hmg/ddn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ekstrand ML, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rusten P, Gustafsson CM, Larsson NG, Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet 13 (9) (2004) 935–944, 10.1093/hmg/ddhl09. [DOI] [PubMed] [Google Scholar]
- [31].Bota DA, Davies KJ, Mitochondrial Lon protease in human disease and aging: including an etiologic classification of Lon-related diseases and disorders, Free Radie. Biol. Med 100 (2016) 188–198, 10.1016/j.freeradbiomcd.2016.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hudson EK, Hogue BA, C Souza Pinto N, L Croteau D, Anson RM, Bohr VA, Hansford RG, Age associated change in mitochondrial DNA damage. Free Radie. Res 29 (6) (1998) 573–579, 10.1080/10715769800300611. [DOI] [PubMed] [Google Scholar]
- [33].Herrero JA, Barja G, Effect of aging on mitochondrial and nuclear DNA oxidative damage in the heart and brain throughout the life-span of the rat, J. Am. Aging Assoc 24 (2) (2001) 45–50, 10.1007/s11357-0010006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].López-Torres M, Gredilla R, Sanz A, Barja G, Influence of aging and long-term caloric restriction on oxygen radical generation and oxidative DNA damage in rat liver mitochondria. Free Radie. Biol. Med 32 (2002) 882–889, 10.1016/S0891.5849(02)007736. [DOI] [PubMed] [Google Scholar]
- [35].Van Remmcn H, L Hamilton M, Richardson A, Oxidative damage to DNA and aging, Exerc. Sport Sei. Rev 31 (3) (2003) 149–153. [DOI] [PubMed] [Google Scholar]
- [36].Haripriva D, Sangeetha P, Kanchana A, Balu M, Panneerselvam C, Modulation of age-associated oxidative DNA damage in rat brain cerebral cortex, striatum and hippocampus by L-camitinc, Exp. Gerontol. 40 (3) (2005) 129–135, 10.1016/j.exger.2004.10.006. [DOI] [PubMed] [Google Scholar]
- [37].Nakamoto H, Kaneko T, Tahara S, layashi EI, Naito 11, Radak Z, Goto S, Regular exercise reduces 8-oxodG in the nuclear and mitochondrial DNA and modulates the DNA repair activity in the liver of old rats, Exp. Gerontol 42 (4) (2007) 287–295, 10.1016/j.exger.2006.11.006. [DOI] [PubMed] [Google Scholar]
- [38].Savitha S, Panneerselvam C, Mitigation of age-dependent oxidative damage to DNA in rat heart by carnitine and lipoic acid, Mech. Ageing Dev 128 (2) (2007) 206–212, 10.1016/j.mad.2006.11.029. [DOI] [PubMed] [Google Scholar]
- [39].Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S, Szczcsny B, Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role of EXOG (5’EXO/endonudease) in their repair, J. Biol. Chem 286 (2011) 31975–31983, 10.1074/jbc.M110.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shokolcnko IN, L Wilson G, Alexcyev MF, Persistent damage induces mi tochondrial DNA degradation, DNA Repair 12 (7) (2013) 488–499, 10.1016/j.dnarcp.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lauritzen KH, Cheng C, Wiksen H, Bergersen LIL, Klungland A, Mitochondrial DNA toxicity compromises mitochondrial dynamics and induces hippocampal an tioxidant defenses, DNA Repair 10 (6) (2011) 639–653, https://doi.org/l0.1016/j.dnarep.2011.04.011. [DOI] [PubMed] [Google Scholar]
- [42].Lauritzen KH, L Klcppa, Aronscn JM, Eide L, Carlsen II, Haugen OP, Sjaastad L, Klungland A, Rasmussen LJ, Attramadal H, Storm-Mathisen J, Bergersen LH, Impaired dynamics and function of mitochondria caused by mtDNA toxicity leads to heart failure, Am. J. Physiol. Heart Qrc. Physiol 309 (3) (2015) 11434–11449, 10.1152/ajpheart.00253.2014. [DOI] [PubMed] [Google Scholar]
- [43].Moretton A, Morel F, Macao B, Lachaume P, Ishak L, Lcfebvre M, Garreau Balandicr I, Falkcnberg M, Farge G, Selective mitochondrial DNA degradation fol lowing double-strand breaks, PLoS One 12 (4) (2017) e0176795, 10.1371/joumal.pone.0176795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gaston RA, Demple B, Risky repair. DNA-protein crosslinks formed by mitochondrial base excision DNA repair enzymes acting on free radical lesions, Free Radie. Biol. Med 107 (2017) 146–150, 10.1016/j.freeradbiomed.2016.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, Shen B, Demple B, Removal of oxidative DNA damage via FEN 1 dependent long-patch base excision repair in human cell mitochondria, Mol. Cell. Biol 28 (2008) 4975–4987, 10.11128/MCB.004574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Pinto M, Moraes CT, Mechanisms linking mtDNA damage and aging, Free Radie. Biol. Med 85 (2015) 250–258, 10.1016/j.freeradbiomed.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pinz KG, Shibutani S, Bogenhagen DF, Action of mitochondrial DNA polymerase gamma at sites of base loss or oxidative damage, J. Biol. Chem 270 (1995) 9202–9206, 10.1074/jbc.270.16.9202. [DOI] [PubMed] [Google Scholar]
- [48].Kozhukhar N, Spadafora D, Fayzulin R, Shokolcnko IN, Alexeyev M, The efficiency of the translesion synthesis across abasic sites by mitochondrial DNA polymerase is low in mitochondria of 3T3 cells, Mitochondrial DNA A DNA Mapp. Seq. Anal 27 (6) (2016) 4390–4396, 10.3109/19401736.2015.1089539. [DOI] [PMC free article] [PubMed] [Google Scholar]