Abstract
Abstract: Epigenetic regulation of the chromatin landscape is often orchestrated through modulation of nucleosomes. Nucleosomes are composed of two copies each of the four core histones, H2A, H2B, H3, and H4, wrapped in ~150 bp of DNA. We focus this review on recent structural studies that further elucidate the mechanisms used by macromolecular complexes to mediate histone modification and nucleosome assembly. Nucleosome assembly, spacing, and variant histone incorporation are coordinated by chromatin remodeler and histone chaperone complexes. Several recent structural studies highlight how disparate families of histone chaperones and chromatin remodelers share similar features that underlie how they interact with their respective histone or nucleosome substrates. Post‐translational modification of histone residues is mediated by enzymatic subunits within large complexes. Until recently, relatively little was known about how association with auxiliary subunits serves to modulate the activity and specificity of the enzymatic subunit. Analysis of several recent structures highlights the different modes that auxiliary subunits use to influence enzymatic activity or direct specificity toward individual histone residues.
Keywords: Histone chaperones, chromatin regulation, chromatin remodeling, epigenetic mechanisms, histones, histone modification complexes
Introduction
Eukaryotic nucleosomes are built from an octamer of core histones containing two copies of each H3, H4, H2A, and H2B wrapped in roughly 150 base pairs of double stranded DNA, while additional DNA can associate with linker histone H1 to aid in chromatin compaction.1, 2 In the years following the first high‐resolution crystal structure of a nucleosome, the protocols for assembly and study of histone subcomplexes and nucleosomes in vitro have become widespread.3 Many researchers have worked to determine X‐ray crystal structures of a variety of nucleosomes containing different variant histones, histone modifications, and DNA sequences.3, 4 While the nucleosome is a substrate for a vast variety of enzymes and chromatin associated protein complexes, comparatively few structures of these additional cellular factors bound to nucleosomes have been determined.5, 6, 7, 8, 9, 10, 11 While X‐ray crystallography has been used to determine the majority of nucleosome and nucleosome‐bound structures,4, 6, 7, 8, 9 the development of single particle cryo–electron microscopy (cryo‐EM) has lead to a number of recent structures of cellular factors bound to nucleosomes. Notably, structures have been reported for the nucleosome in complex with chromatin remodelers Snf2,12 Chd1,13, 14 and INO80.15, 16 Chromatin remodelers contribute to proper nucleosome assembly/eviction, histone variant incorporation, and regulation of proper nucleosome positioning along DNA.17 Comparison of these structures reveals that separate families of chromatin remodelers use similar mechanisms of nucleosome association.
Nucleosome assembly is orchestrated by histone chaperone proteins in concert with enzymatic chromatin remodelers.18, 19 Histone chaperones are a diverse group of eukaryotic proteins that, unlike chromatin remodelers, bear little sequence homology and serve a variety chromatin maintenance functions. Histone chaperones are involved in transporting histones from the cytosol to the nucleus, properly poising histones for post‐translational modifications, and depositing histones onto DNA for nucleosome assembly.18, 20, 21 A variety of histone chaperones have evolved to associate with histone complexes harboring different histone variants to carry out their specific cellular functions.22 A detailed analysis of several recent structures of histone chaperones in complex with their histone substrates reveals that many different histone chaperones, despite having no sequence conservation, share structural and mechanistic similarities in how they associate with the conserved histone surface.
Post‐translational modifications (PTM) such as acetylation, methylation, phosphorylation, ubiquitination, and sumoylation regulate gene expression through covalently modifying specific residues of histones as well as other proteins. PTMs to histones can be introduced on free histones or histones within an assembled nucleosome and can effect changes in gene expression in two major ways. First, different classes of PTMs control the distribution of the heterochromatin and euchromatin by altering the electrostatic interaction between histone proteins and surrounding DNA. Second, the chemical moieties attached by histone‐modifying enzymes can be specifically recognized by histone‐binding reader proteins that help to recruit a variety of nuclear factors to further modulate the surrounding chromatin.23, 24, 25, 26 Many of these histone‐modifying enzymes function in multiprotein complexes. In recent years various structural and biochemical analyses have revealed the importance of noncatalytic subunits for their ability to modulate substrate specificity through differential substrate recruitment and allosteric regulation of the active site.27, 28, 29, 30, 31, 32, 33
In this review, we cover recent structural studies focusing on different nuclear complexes, which contribute to epigenetic regulation of chromatin through modulation of nucleosome assembly and post‐translational modification. First, we highlight several recent cryo‐EM studies that provide insight into the interaction between chromatin remodeler complexes and their nucleosome substrates. The second section focuses on X‐ray crystallographic analysis of histone chaperone proteins in complex with different histone subcomplexes. Comparison of several recent histone chaperone structures reveals previously uncharacterized structural similarity between different histone chaperones and provides insight into mechanisms of nucleosome assembly. In the final section, we cover recent cryo‐EM and X‐ray crystallographic structures of histone modifying complexes that are responsible for regulating the overall architecture of chromatin through acetylation and methylation. Analysis of these structures reveals that nonenzymatic auxiliary subunits within these complexes strongly contribute to modulation of the activity and substrate specificity of the catalytic subunit.
Structural analysis of nucleosome/chromatin remodeler complexes
Chromatin remodelers, which function individually or as members of larger multiprotein complexes to maintain proper chromatin structure, can be divided into four subfamilies: SWI/SNF, CHD, ISWI, and INO80.17 The SWI/SNF subfamily regulates chromatin access by evicting nucleosomes or sliding nucleosomes along DNA to open up specific areas of DNA, which can then be accessed by additional nuclear factors.34, 35 CHD and ISWI chromatin remodelers function to aid histones, initially deposited by histone chaperones, to develop into mature nucleosomes.36, 37 Additionally CHD and ISWI regulate nucleosome spacing by a nucleosome sliding mechanism.36, 37 Modulation of H2A/H2B and variant H2A.Z/H2B incorporation into nucleosomes is conducted by the INO80 subfamily.38, 39 Although the functions of chromatin remodelers are quite diverse, all chromatin remodelers share a greater affinity for nucleosomes over free DNA and use a conserved Swi2/Snf2 ATPase domain split into two RecA‐like lobes to conduct ATP‐dependent translocation of DNA from a fixed point on the nucleosome.17, 19 On binding to the nucleosome, the two RecA‐like lobes engage the same strand of DNA and hydrolyze ATP to catalyze the sequential binding and release of each individual lobe to propel DNA around the nucleosome.17, 19 The disruption of the DNA/histone contacts by remodeler‐catalyzed DNA translocation allows for remodelers to propel DNA around the nucleosome to conduct various nucleosome assembly/disassembly and positioning functions. All remodelers have been shown to engage the nucleosome with primary DNA contacts coming from the ATPase domain and additional contacts from ancillary domains or subunits depending on the subfamily.17 DNA footprinting experiments have revealed that remodelers associate with fixed regions of nucleosomal DNA; these regions are generally reported as specific SuperHelix Locations (SHL) that represents the number of double helical turns away from the nucleosome dyad at SHL 0.4 The major groove faces in toward the histones at full integer SHL values and the minor groove faces inward at one‐half SHL values, while ± values denote symmetric locations relative to the twofold symmetry axis through the dyad.4 Footprinting data indicate that SWI/SNF, CHD, and ISWI remodelers associate with nucleosomal DNA at SHL ±2,40, 41, 42 while INO80 has been reported to associate at the SHL +3, +5, and + 6 positions.38 Additional contacts between the ATPase motor domain and the N‐terminal tail of H4 have been reported to modulate the activity of ISWI chromatin remodelers but had not been observed for other families of remodelers to this point.43 Here, we discuss the recently reported cryo‐EM structures of Saccharomyces cerevisiae Snf2 bound to a nucleosome,12 Saccharomyces cerevisiae Chd1 bound to a nucleosome,13, 14 and a human INO80 subcomplex bound to a nucleosome16 (Fig. 1). Another cryo‐EM structure of an INO80 subcomplex from Chaetomium thermophilum bound to a nucleosome has also been reported;15 as this structure is comparable with the human complex, we will focus our discussions largely on the nucleosome‐bound structure of human INO80 for simplicity.
Figure 1.
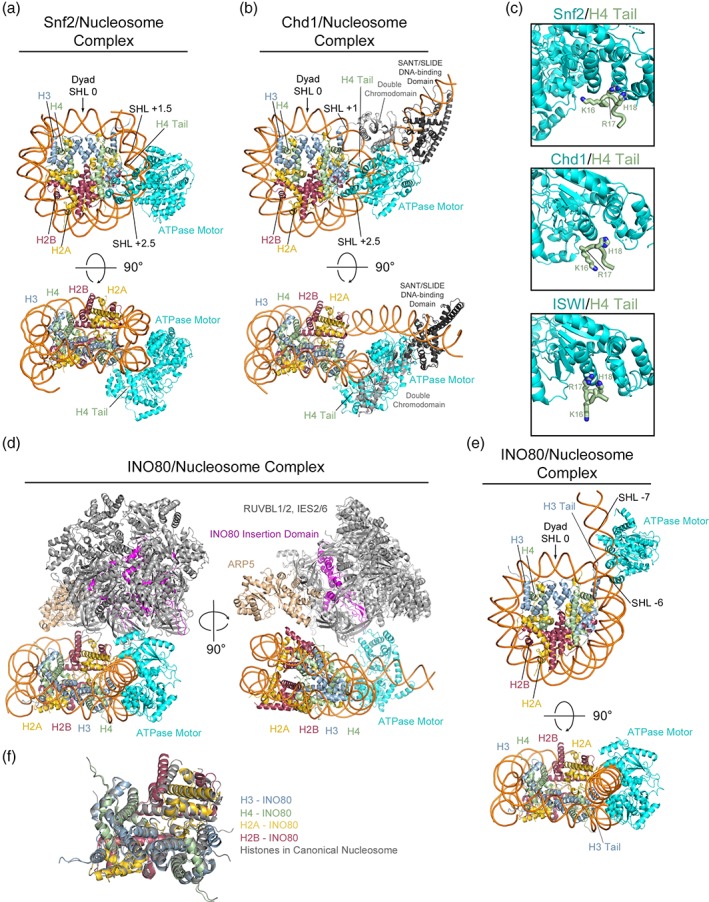
Cryo‐EM structures of Snf2, Chd1, and INO80 chromatin remodelers bound to nucleosomes. (a) Front and top view of 4.69 Å structure for S. cerevisiae Snf2 bound to a nucleosome, details of the interactions with nucleosomal DNA and the H4 tail are highlighted (PDB: 5X0Y). (b) Front and top view of 4.80 Å structure for S. cerevisiae Chd1 bound to a nucleosome, details of the interactions with nucleosomal DNA and the H4 tail are highlighted (PDB: 5O9G). (c) Comparison of the interaction between H4 N‐terminal tail residues 15–18 and the ATPase motor domains from Snf2 (top), Chd1 (middle), and ISWI (bottom) (PDB: 5JXT). (d) 4.8 Å structure of the human INO80 complex bound to a nucleosome (PDB: 6ETX). A rotated view details the assembly of the RUVBL1/2 heterohexamer and additional subunits on the INO80 insertion domain and highlights the locations of the INO80 ATPase motor domain and ARP5 subunit. (e) Front and top view of the INO80 ATPase motor bound to a nucleosome with additional domains and subunits removed for simplicity. Details of the interactions with nucleosomal DNA and the H3 tail are highlighted. (f) Alignment of the histone octamer from the INO80 complex structure (PDB: 6ETX) with the histone octamer from the crystal structure of the nucleosome alone (PDB: 1AOI) detailing the rearrangement on INO80 binding.
Snf2/nucleosome complex
The 4.69 Å structure for the Snf2/nucleosome complex reveals that the ATPase motor domain associates with the nucleosome at the SHL +2 position, in good agreement with previous biochemical studies [Fig. 1(a), top/bottom]. Interestingly two additional cryo‐EM classes were observed. One depicts a separate binding site for Snf2 at SHL +6, and a fractionally smaller class shows the nucleosome simultaneously engaged by two molecules of Snf2 at both SHL +2 and SHL+ 6.12 While the study did not discuss either of these additional binding sites at length, it is interesting that a small fraction of the sample had a nucleosome simultaneously engaged by two enzymes. This may have implications for the activity of the enzyme as a dimer, as there have been reports of other chromatin remodelers functioning as dimers engaged with a single nucleosome.42, 44, 45 Comparison with a recently reported 2.33 Å crystal structure46 of the ATPase motor domain of Snf2 in the absence of the nucleosome reveals that binding to nucleosomal DNA allows Snf2 to adopt an active conformation by stimulating rotation of the two RecA‐like lobes of the ATPase motor. This nucleosome‐induced rotation brings the catalytic “arginine fingers” into a conformation poised for ATP hydrolysis. Additionally, it is observed that binding of the ATPase motor domain to the nucleosome induces the formation of additional brace helices that help to stabilize this active conformation of the ATPase domain.
Chd1/nucleosome complex
The 4.8 Å structure of Chd1 bound to a nucleosome also strongly reflects the reported biochemical characterization of the interaction [Fig. 1(b), top/bottom]. Crosslinking and low‐resolution negative stain EM experiments suggest that the Chd1 ATPase motor and chromodomain likely associate with nucleosomal DNA at SHL +2 and SHL +1 locations, respectively, while the C‐terminal DNA‐binding SANT/SLIDE domain associates with and influences the partial unwrapping of exit DNA on the opposite gyre of the nucleosome.42 The Chd1/nucleosome structure reveals that the ATPase motor associates with nucleosomal DNA at SHL +2 and the chromodomain at SHL +1. The SANT/SLIDE DNA‐binding domain is associated with partially unwrapped exit DNA at the SHL −7 position on the opposite gyre. Similar to observations from the Snf2/nucleosome structure, comparison with the 3.7 Å crystal structure of the Chd1 ATPase motor domain alone reveals that binding to nucleosomal DNA induces rotation of the two RecA‐like lobes into an active conformation that is favorable for ATP hydrolysis.47 Another cryo‐EM analysis of S. cerevisiae Chd1 bound to the nucleosome identified an EM class in which one nucleosome is simultaneously engaged by two molecules of Chd1.14 The 4.5 Å structure demonstrates that the respective ATPase motor, chromodomain, and SANT/SLIDE domains of each Chd1 molecule occupy the same SHL positions on the opposing ± sides of the nucleosome, further verifying data that suggest chromatin remodelers may function as dimers engaged with a single nucleosome.42
H4 tail interaction is a common feature of chromatin remodelers
Interestingly, the structures of Snf2 and Chd1 bound to the nucleosome reveal a previously uncharacterized interaction between the H4 N‐terminal tail and the C‐terminal RecA‐like lobe of the ATPase motor domain. Binding of the H4 tail to the ATPase domain has been reported to modulate activity for the ISWI family of chromatin remodelers through competition with an inhibitory loop from the ISWI AutoN regulatory domain43, 48 [Fig. 1(c), top]. It has been postulated that the H4 tail may serve to anchor the ATPase motor domain to the correct location on the nucleosome given the proximity to the SHL ±2 site.48 The nucleosome bound structures for Snf2 and Chd1 show that H4 tail residues 15–18 occupy the same binding pocket as in ISWI [Fig. 1(c), middle/bottom], although Snf2 and Chd1 lack the AutoN regulatory domain with which these H4 residues compete for binding. H4 tail binding is important for proper ISWI activity, as mutation of H4R17 or acetylation of H4K16 have been shown to negatively impact ISWI remodeling activity.48 While these structures reveal that binding to the H4 N‐terminal tail is a shared feature of several families of chromatin remodelers, further study is necessary to verify the biological role of this interaction and explore the effects that any H4 PTMs have on Chd1 and Snf2 chromatin remodeling activity.
INO80/nucleosome complex
The structures of Snf2 and Chd1 bound to the nucleosome contain only domains from the enzymatic chromatin remodeler in complex with a nucleosome. The 4.8 Å structure of human INO80 in complex with a nucleosome contains a large INO80 sub‐complex composed of the INO80 enzymatic subunit and additional RUVBL1/2, IES2, IES6, and ARP5 subunits [Fig. 1(d), left/right]. Association of these subunits with the catalytic INO80 subunit is necessary for proper chromatin remodeling activity.49, 50 Assembly of the INO80 subcomplex is mediated by a large insertion domain between the N and C‐terminal lobes of the INO80 ATPase motor that associates with the RUVBL1/2 AAA+ heterohexamer, onto which the other subunits assemble [Fig. 1(d), left/right]. Differing from Snf2 and Chd1 that bind to the nucleosome at SHL +2, the structure reveals that the INO80 ATPase motor binds to a region of DNA by the entry point spanning SHL −6 to SHL‐7 [Fig. 1(e), top/bottom]. This is in relatively good agreement with in vitro footprinting experiments that report S. cerevisiae INO80 protects nucleosomal DNA at SHL −6, −5, and −3.38 The positioning of the ARP5 module proximal to the SHL −3 site helps to account for the reported protection of nucleosomal DNA in this region. While it is in close proximity, the ARP5 subunit does not appear to be in direct contact with nucleosomal DNA at the SHL −3 site in the human structure [Fig. 1(d), right]. However, analysis of the 4.3 Å structure of the Chaetomium thermophilum INO80 complex bound to a nucleosome reveals that the ARP5/IES6 module is in a direct interaction with the DNA at the SHL −2 and SHL −3 and makes additional contacts with H2B. It has been reported that ARP5 is essential for nucleosome sliding activity of the INO80 complex.49 Comparison of the DNA bound and unbound states of ARP5 in the two structures indicates that ARP5 may contribute to sliding through a process of dynamic binding and release of nucleosomal DNA at the SHL −3 position to aid in translocation of DNA around the histone octamer.
The structure of human INO80 bound the nucleosome additionally reveals that the N‐terminal H3 tail is proximal to the N‐terminal RecA‐like lobe of the INO80 ATPase motor domain [Fig. 1(e), top/bottom]. Although there is no electron density for the H3 tail residues directly binding to the domain, the authors postulated that the H3 tail could possibly bind and modulate ATPase activity of the INO80 complex similar to the H4 tail interaction with the C‐terminal RecA‐like lobes of ISWI, Snf2, and Chd1. Deletion experiments identify a region spanning residues 30–39 of H3, removal of which results in a significant decrease in the Hill coefficient for nucleosome binding as well as nucleosome sliding activity. As INO80 has been shown to function as a cooperative dimer in solution45 this decrease in Hill coefficient indicates that H3 binding to the INO80 ATPase motor plays a role in regulating cooperativity between the INO80 subunits within a functional dimer.
Comparison of the arrangement of the histone octamer between the INO80/nucleosome structure to the histones in the crystal structure of the nucleosome alone reveals that there are minor conformational rearrangements in H2B and H3 [Fig. 1(f)]. This histone rearrangement is likely due to INO80 binding to the nucleosome and causing a conformational distortion of the DNA wrap where one of the DNA gyres is slightly moved away from the other, pulling portions of the histones along with it. This rearrangement of the histones reflects the results of a recent study that reports that rearrangements within the histone octamer are necessary for proper nucleosome sliding activity by SNF2.51 Additionally, a recent cryo‐EM study identified several classes of partially unwrapped nucleosomes that have minor rearrangements in the histone octamer as a result of the DNA unwrapping.52 These rearrangements of histone positions are not evident in the structure of Chaetomium thermophilum INO80 bound to a nucleosome, further indicating that these two structures may represent two conformations of a dynamic complex.
Structural analysis of histone chaperone mediated nucleosome assembly
The tyrosine key to H3/H4
Although histone chaperones and histone binding proteins do not generally share sequence similarity, structural analysis reveals that many of these proteins have convergently evolved structurally similar features for histone interaction, likely due to the strong evolutionary conservation of histones. For example, the arginine anchor,5 an interaction between an arginine residue or residues from a histone binding protein and the acidic patch on H2A, has been observed as a common feature of several nucleosome‐binding proteins such as viral proteins,53 guanine exchange factors,6 and histone modifying enzymes.8, 54 Despite widely varying sequence and overall structure, the insertion of an arginine residue into the deep, acidic pocket formed by H2A residues E61, D90, and E92 is seen in many nucleosome‐bound structures. This phenomenon establishes a platform for competitive binding that allows for the association of a maximum of two factors per nucleosome at a time to help ensure orderly chromatin dynamics. This being the case, it follows that a similar mechanism may help provide for the specificity of different histone chaperones competing for the same pool of available histone proteins.
Through comparison of several structures of H3/H4 bound to different histone chaperones, we have identified a structurally similar feature where a tyrosine residue extends from the histone chaperone into a deep surface pocket on histones H3/H4; we name this interaction the “tyrosine key.” Comparison of the structures of UBN1,55 a subunit of the replication‐independent H3.3/H4‐specific HIRA‐containing histone chaperone complex, DAXX,56 an H3.3/H4‐specific chaperone associated with chromatin compaction at pericentromeres and telomeres, and MCM2,57 an H3/H4 chaperone that is part of the replicative helicase complex, bound to H3.3/H4 reveals the presence of a tyrosine residue in the same H3/H4 surface binding pocket. Although UBN1, DAXX, and MCM2 share no sequence conservation and are overall structurally dissimilar, Y132 of UBN1, Y222 of DAXX, and Y90 of MCM2 each insert into the deep surface pocket formed by H3 residues K64, Q68, S86, I89, and Q93 [Fig. 2(a)]. Here, the hydroxyl group of the tyrosine residue forms a hydrogen bond with H3 Q93 and the tyrosine aromatic ring establishes van der Waals interactions with the aliphatic side chain of H3 K64. Comparable with the arginine anchor, the tyrosine key likely serves to aid in recognition of the conserved histone surface and may help regulate H3/H4 binding and deposition through competition among multiple different histone chaperones for the same binding site.
Figure 2.
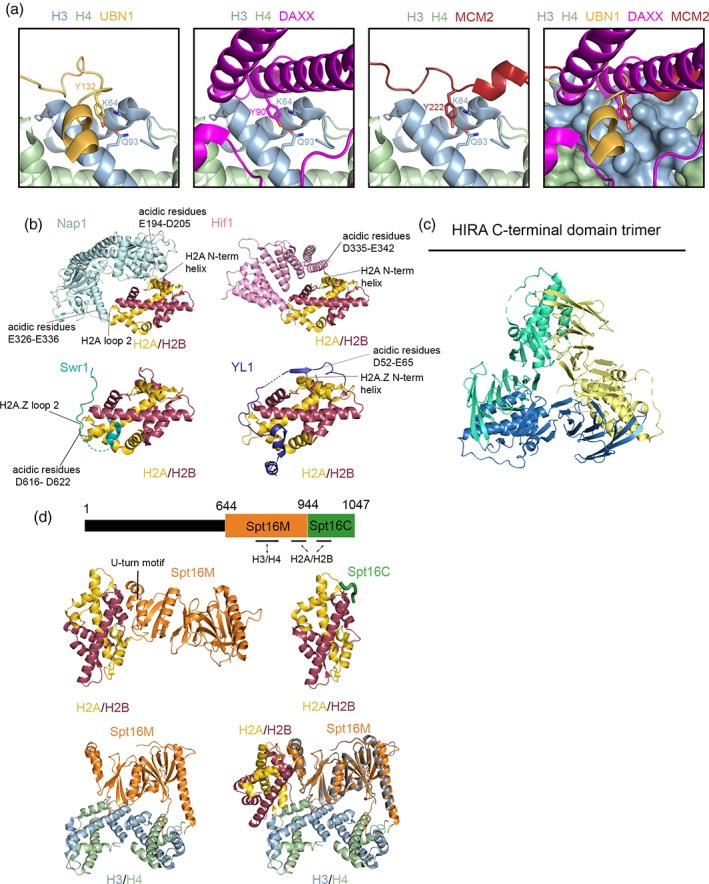
Molecular comparison of different histone chaperone proteins. (a) UBN1 (PDB: 4ZBJ), DAXX (PDB: 4HGA), and MCM2 (PDB: 5BNV) in complex with H3/H4. UBN1 Y132, MCM2 Y90, or DAXX Y222 fit into a surface pocket on H3 where it forms a hydrogen bond with H3Q93 and van der Waals interactions with H3K64. This deep association likely drives competition for H3/H4 binding to ensure orderly histone trafficking in a crowded cellular environment. (b) At least four uniquely functioning H2A/H2B chaperones, Nap1 (PDB: 5G2E), Hif1 (PDB: 5BT1), YL1 (PDB: 5FUG), and Swr1 (PDB: 4M6B), utilize patches of acidic residues to interact with the same basic residues on the negatively charged DNA‐binding surface of an H2A/H2B dimer to sequester the histones from DNA until the final step of nucleosome formation. (c) 2.45 Å crystal structure of the C‐terminal domain of HIRA (residues 644–1017; PDB: 5YJE) reveals homotrimer formation. (d) Top: Spt16 with identified H2A/H2B‐ and H3/H4‐binding domains. Middle: 2.35 Å resolution structure of Spt16M in complex with H2A/H2B (PDB: 4KHA) versus 1.8 Å resolution structure of Spt16C in complex with H2A/H2B (PDB: 4WNN) reveals that these two domains compete for some of the same H2B residues and may represent two unique interactions with H2A/H2B relevant to the FACT mechanism. Bottom: 2.98 Å resolution structure of Spt16M in complex with an H3/H4 tetramer (left; PDB: 4Z2M) overlayed with the Spt16M/H2A/H2B structure (right; H2A/H2B‐bound Spt16M in gray) demonstrates an additional, distinct histone binding domain associated with H3/H4, a feature potentially useful in H3/H4 tetramer stabilization and H2A/H2B dimer shuttling during chromatin reorganization.
DNA‐competitive binding of H2A/H2B chaperones
A number of structures featuring H2A/H2B dimers bound to different histone chaperones demonstrate that a wide variety of proteins which make multiple contacts with different surfaces of H2A/H2B have also evolved some common binding features. Comparison of the structures for H2A/H2B bound to Nap1, H2A/H2B bound to Hif1, H2A.Z/H2B bound to YL1, and H2A.Z/H2B bound to Swr1 reveals one common binding feature is a cluster of acidic residues that interact with the basic, DNA‐binding surface of H2A/H2B, presumably to compete for DNA binding and promote histone sequestration until nucleosome assembly.
Nap1, a histone chaperone tasked with escorting H2A/H2B dimers to the nucleus, shields two of the three DNA‐binding surfaces of the H2A/H2B dimer to prevent aberrant DNA association.58 These interactions are mediated by Nap1 C‐terminal residues 194–205, which associate with residues 16–21 of the H2A N‐terminal alpha helix, and E328‐E336 of Nap1's α8 helix that bind to H2A loop 2 [Fig. 2(b), top left]. This second interaction region mimics an acidic strand of DNA and thereby binds mutually exclusively with DNA by engaging basic lysine and arginine residues on the histone surface.
Hif1 is a histone chaperone that can interact with both H3/H4 and H2A/H2B. The structure of Hif1 has been determined in complex with H2A/H2B and reveals that Hif1 associates with the H2A/H2B DNA‐binding surface to mask it from unfavorable interactions, similarly to Nap1.59 Acidic residues of Hif1 interact with H2A and H2B residues on the same face as Nap1 [Fig. 2(b), top right]. Hif1 can bind H3/H4 tetramers in addition to H2A/H2B dimers, but biochemical studies suggest that interaction is largely mediated by residues outside of the acidic stretch used for binding the H2A/H2B DNA‐binding surface.59 Interestingly, however, H2A/H2B dimers are displaced by H3/H4 tetramers in an in vitro competition‐binding assay,59 suggesting that Hif1 plays a pivotal scaffold role in chromatin dynamics, as may other histone chaperones capable of binding to multiple core histones.
The catalytic Swr1 subunit of the SWR1 nucleosome‐remodeling complex, which exchanges H2A/H2B dimers for variant H2A.Z/H2B dimers, contains a C‐terminal Z domain that functions as an H2A.Z/H2B‐specific histone chaperone and binds to the same H2A/H2B face60 [Fig. 2(b), bottom left]. The 1.7 Å crystal structure of the Swr1‐Z domain bound to H2A.Z/H2B reveals that the Swr1‐Z domain binds to both the DNA‐binding and H3‐binding sites of the dimer, suggesting that it may serve the role of a histone chaperone in the H2A.Z/H2B exchange process catalyzed by Swr1. The crystal structure reveals that a stretch of acidic amino acids, D614‐D622, parallels the interactions seen across the other H2A/H2B chaperones. In vitro biochemical studies reveal that the Swr1‐Z domain facilitates H3/H4 tetrasome to nucleosome formation and its absence impairs nucleosome formation. This demonstrates the essential role of the histone chaperone function of this domain in the remodeling complex.60
YL1, an H2A.Z/H2B‐specific histone chaperone, also uses similar engagement with the DNA binding surface of H2A.Z‐H2B [Fig. 2(b), bottom right]. Residues D55‐E65 of its C terminal region interacts intimately with almost the entire DNA binding surface of H2A.Z.61 These residues act largely as a steric hindrance to DNA binding and echo the utility of H2A/H2B chaperones harboring acidic stretches that inhibit unfavorable chromatin interactions prior to nucleosome assembly.
Multimerization of H3/H4‐depositing chaperones
The HIRA histone chaperone complex, composed of the proteins HIRA, UBN1, CABIN1, and transiently ASF1a, drives replication‐independent deposition of H3.3/H4 onto DNA.62 A structure of a fragment of UBN1 bound H3.3/H4/Asf1 has been determined and demonstrates, together with previous Asf1/H3/H4 crystal structures, that Asf1 binds to the tetramerization interface of the H3/H4 dimer.55 Asf1 must ultimately be released prior to deposition, as it is preventing assembly of the H3/H4 heterotetramer that is likely formed prior to deposition onto DNA and nucleosome formation.63 To this end, it has been previously suggested that multimerization of the HIRA complex could aid in proper H3/H4 tetramer formation prior to deposition. Recent structural, biophysical, and cellular evidence demonstrate that trimerization of the HIRA subunit [Fig. 2(c)] is essential for proper H3.3/H4 deposition function of the complex.64 Mutations that abolish HIRA trimerization impair association with CABIN1 as well as H3.3/H4 deposition activity of the complex. Although further study is necessary to fully understand the biological role of HIRA trimer formation, this multimerization may play a role in regulating the formation of the H3.3/H4 tetramer.
While structural information is not yet available, biochemical data for CAF1, a replication‐dependent H3/H4 chaperone, has demonstrated that a 1:1:1:1:1 complex is formed between its three subunits (Cac1, Cac2, Cac3) and histones H3 and H4.65 This stoichiometry implies that this chaperone sequesters H3/H4 dimers until tetramerization is induced for DNA deposition. Two distinct DNA binding domains on Cac1 have been identified, and binding to DNA allows two CAF1/H3/H4 complexes to come into close proximity and induce deposition of a newly formed H3/H4 tetramer from the chaperone complex onto DNA.65 This mechanistic insight implies that multimerization or at least close colocalization of multiple CAF1 chaperones binding to DNA must be necessary for H3/H4 tetramer deposition into chromatin.
These recent studies echo the trend demonstrated by the histone chaperone HJURP, which has been shown to dimerize to assemble and deposit histone variant CENP‐A/H4 tetramers at the centromere66 as well as the chaperone Vps75, the structure of which has been determined as both a dimer67 and as a tetramer.68 Interestingly, it has been shown that, in equimolar concentrations of Vps75 and H3/H4, Vps75 forms tetramers that associate with histone dimers, while, in the presence of excess histones, Vps75 tetramers associate with histone tetramers. This observation suggests that Vps75 may have self‐chaperoning activity that enables it to multimerize and drive H3/H4 tetramer formation on DNA.68
Multiple histone‐binding domains within the FACT complex
FACT is a heterodimeric histone chaperone complex containing two subunits, Spt16 and SSRP1(human)/Pob3(yeast), that play a role in chromatin remodeling during transcription, replication, and DNA repair.63 FACT has been shown to interact with both H2A/H2B dimers and H3/H4 tetramers,21 suggesting it likely functions in multiple steps of nucleosome reorganization during these processes. Two different x‐ray crystal structures of FACT in complex with H2A/H2B have been determined: one in which the middle domain (Spt16M) from Chaetomium thermophilum Spt16 is in complex with H2A/H2B,69 and one in which a peptide from the C‐terminal domain of Saccharomyces cerevisiae Spt16 (Spt16C) is in complex with H2A/H2B.70 A third structure contains the human Spt16M in complex with human H3/H4.71
Hondele et al. first biochemically identified Spt16M as the minimum domain necessary for Spt16‐H2A/H2B interaction, but also reported the presence of another electrostatic interaction between H2A/H2B and Spt16C identified using Isothermal Titration Calorimetry (ITC)69 [Fig. 2(d), top]. Histone‐DNA aggregation, however, is only prevented in the presence of Spt16M, suggesting that this domain is relevant to chaperone function. The 2.35 Å structure of Spt16M in complex with H2A/H2B reveals that a U‐turn motif at the C‐terminal end of the Spt16M domain engages helix 1 of H2B. Specifically, H2B residues I36 and Y39 form hydrophobic interactions with the surrounding hydrophobic residues of Spt16M [Fig. 2(d), middle left]. Cell‐based deletion and mutation studies indicate that both Spt16M and Spt16C are essential for FACT function.
Kemble et al., however, report that only Spt16C confers histone binding and determined a 1.8 Å crystal structure of a peptide from this domain in complex with H2A/H2B.70 The structure reveals major contacts between three Spt16C residues, the N‐terminus of H2B helix 2, and one residue on H2A [Fig. 2(d), middle right]. Biochemical studies from this study fail to reproduce any contribution of Spt16M to relevant FACT/H2A/H2B binding. A full‐length structure of Spt16 is not available, but a comparison of the two structures suggests that interactions of both domains with an H2A/H2B dimer could not coexist [Fig. 2(d), middle]. Further biochemical analysis is needed to settle the binding capabilities of these two domains to H2A/H2B dimers. It is possible that both domains could be in competition for H2A/H2B binding, which may be relevant in the mechanism of nucleosome assembly mediated by FACT.
A 3.0 Å structure reveals that Spt16M also interacts with an H3/H4 tetramer utilizing two different contact sites: one where a short antiparallel beta sheet is formed between residues 745–750 of Spt16, residues 44–48 of H4, and residues 117–119 of H371 [Fig. 2(d), bottom left]. The second site of contact includes the other H3/H4 dimer of the complex, where Spt16 residues R847‐D856 form a combination of hydrophobic and hydrophilic interactions with residues on both H3 and H4 [Fig. 2(d), bottom left]. This second site falls on the H2A binding surface of the H3/H4 tetramer. Alignment of the different H2A/H2B and H3/H4 bound Spt16 structures reveals that Spt16 can potentially bind both H2A/H2B and H3/H4 simultaneously with different interaction surfaces [Fig. 2(d), bottom right]. This structural overlay may represent a mechanistic intermediate where Spt16 can bind to and stabilize H3/H4 tetramers as H2A/H2B dimers are deposited or shuttled out through interactions with Spt16M and/or Spt16C.
Auxiliary subunits serve a variety of molecular functions within enzymatic histone modifying complexes
Role of auxiliary subunits within histone acetyltransferase complexes
Lysine acetylation is one of the most extensively studied histone modifications, and its function in chromatin accessibility has been well established.72 The histone acetyltransferases (HAT) enzymes that mediate this modification can be classified into four major families based on sequence conservation; Gcn5/PCAF, MYST, p300/CBP, and Rtt109. HATs typically function as members of multi‐protein complexes where catalytic activation and substrate specificity can be modulated by other subunits within the complex.73, 74
The NuA4 complex, containing catalytic subunit Esa1 as well as additional subunits Epl1, Yng2, and Eaf6, is the main player in global H4 acetylation in S. cerevisiae, where Esa1 serves as the only HAT essential for cell cycle.75 Recent crystallographic and low‐resolution EM analyses of the yeast NuA4 complex suggest that Esa1 recognizes its target residues on the histone tail through a mechanism of substrate specificity that depends on the other subunits of the complex. The tetrameric core of the NuA4 complex consists of Esa1, Epl1, Yng2 and Eaf6, where the scaffold protein Epl1 serves as the platform for assembly of the complex [Fig. 3(a)]. Esa1 and the N‐terminus of Epl1 associate to form the catalytic core, while Yng2, Eaf6, and the C‐terminus of Epl1 form a helical bundle. The catalytic core and the helical bundle regions are connected by a loop in Epl1 referred to as the dual function loop (DFL), as it plays a key role in both active site rearrangement and in nucleosome binding. The 2.0 Å crystal structure of Esa1 in the absence of Epl1 suggests that a α2–β7 loop in the active site is highly conserved among MYST acetyltransferases and is important for catalysis and for substrate binding76 [Fig. 3(b)]. In the absence of Epl1, residues within the Esa1 α2–β7 loop interact with several α2 residues. The recent 3.5 Å crystal structure of the tetrameric NuA4 core complex reveals that α2–β7 loop residues R249, L264, and Y265 interact with a highly conserved FRR (F312, R313, R314) motif in Epl1 to reshape its substrate binding surface.30 This new binding surface has a preference for a substrate with a small residue (G or A) at the P‐1 position, allowing Esa1 to specifically target H4 (K5, K8, K12, K16). Additionally, low‐resolution cryo‐EM studies and biochemical assays reveal that substrate specificity of the Nua4 complex is further achieved through the N‐terminal portion of the Epl1 DFL, which associates with the nucleosome and helps position the Nua4 complex on the dish face of the nucleosome with the active site proximal to the H4 tail [Fig. 3(a)].
Figure 3.
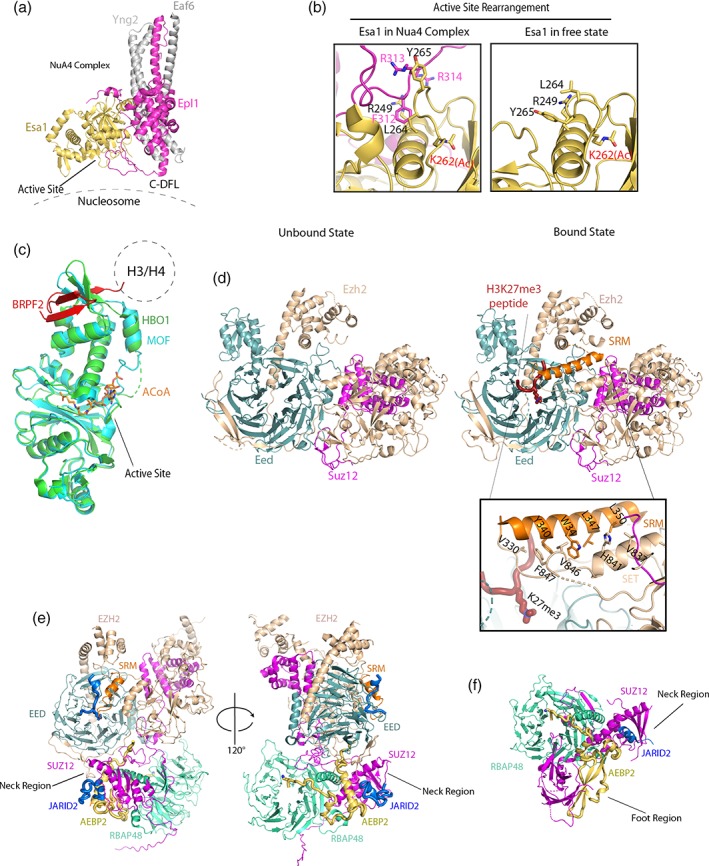
X‐ray crystallographic and cryo‐EM structures of enzymatic histone modifying complexes. (a) 2.0 Å crystal structure of the S. cerevisiae NuA4 complex containing Esa1, Epl1, Yng2 Eaf6. The active site of Esa1, DFL of Epl1 and the position of the NuA4 complex relative to nucleosome are indicated. (PDB: 5J9U). (b) Active site comparison of Esa1 in the NuA4 complex versus the free state. (PDB: 3TO9). (c) 2.4 Å crystal structure of the human HBO1 HAT domain in complex with an N‐terminal fragment of BRPF2 (PDB: 5GK9) overlayed with MOF (PDB: 2GIV) bound to A‐CoA with substrate binding site indicated. (d) The Chaetomium thermophilum PRC2 complex containing Eed, Suz12, Ezh2 in H3K27me3 peptide unbound (PDB: 5KJI) and bound state (PDB: 5KJH). (e) Cryo‐EM structure of the human PRC2 complex in complex with both auxiliary subunits, JARID2, and AEBP2 (PDB: 6C23). (f) Crystal structure of the human PRC2 complex in complex with both auxiliary subunits, JARID2 and AEBP2, featuring additional foot region (PDB: 5WAI).
The human MYST family of HATS, comprised of MOZ, Tip60, HBO1, MORF, and MOF, are evolutionarily conserved with S. cerevisiae Esa1 and characterized by a highly conserved MYST fold within the catalytic domain.73 The structure of the Gcn5 and PCAF catalytic HAT domain bound to an H3K14‐containing peptide reveals extensive backbone interaction with surrounding residues on the H3 tail, thus demonstrating that the catalytic Gcn5/PCAF HAT domain can bind its lysine substrate independently.77 In contrast to the Gcn5/PCAF HATs, MYST family HATs are thought to have lower affinity toward their substrates and are generally more promiscuous toward multiple lysine residues with in the same histone tails. MYST HATs often use other subunits of their respective complexes to provide substrate specificity.73, 77 For example, HBO1 can form two different tetrameric HAT complexes through association with JADE1/2/3, ING4/5, and hEaf6, or BRPF1/2/3, ING4/5, and hEaf6 to target H4 K5/K8/K12 or H3 K14/K23 for acetylation, respectively. The scaffold proteins BRPF and JADE serve as the platform for assembly and help determine substrate specificity of these different HBO1 complexes.31, 78, 79
A recent 2.4 Å crystal structure of the HBO1 HAT domain in complex with an N‐terminal fragment of BRPF2 reveals that BRPF2 binding modulates HBO1 substrate specificity through a mechanism differing from the Epl1/Esa1 interaction. The structure reveals that BRPF2 wraps around the C‐terminal part of the HBO1 HAT domain with the interaction stabilized by a network of hydrogen bonds and Van der Waals interactions between the two proteins [Fig. 3(c)]. Unlike Esa1 in the NuA4 complex, the active site of HBO1 does not undergo any conformational change when in complex with BRPF2. Further biochemical analysis highlights two different acidic patches within BRPF2 consisting of residue E41/E43/E45 and E62/D63/D64 that are responsible for binding and positioning the H3/H4 substrate proximal to the HBO1 active site.32 The crystal structure of the HBO1‐BRPF2 complex indicates that the histone binding site of BRPF2 is ~40 Å away from the active site. This suggests that BRPF2 may bind to the core of the H3/H4 histone, which positions the H3 tail in close proximity to the active site of HBO1.32 Supporting this observation is a recent biochemical analysis of the interaction between HBO1 and an N‐terminal fragment of JADE1, which highlights the importance of the N‐terminal fragment of JADE1 in recruiting the histone substrate to the catalytic domain.33 Biochemical studies also support the role of the histone core domain for JADE1‐mediated HBO1 recruitment for substrate‐specific acetylation. A more detailed molecular mechanism likely must await a structure of a more substantial HBO1 complex.
Roles of auxiliary subunits within of the PRC2 histone methyltransferase complex
Polycomb repressive complex 2 (PRC2) promotes the propagation and maintenance of heterochromatin formation through incorporating and promoting local spreading of the transcriptionally repressive H3K27me3 mark.27, 28 The PRC2 complex consists of four core subunits: EZH2, EED, SUZ12, and RBAP44/48. EZH2 serves as the catalytic subunit of the PRC2 complex, although the isolated catalytic SET domain from EZH2 remains in an inhibited state where both the substrate‐binding groove and cofactor‐binding pocket are inaccessible. Association with SUZ12 and EED bound to a pre‐existing H3K27me3 mark mediates allosteric rearrangement of the EZH2 SET domain for activity toward substrate H3K27 lysine residues.80, 81, 82, 83 In this way, the PRC2 complex functions through a positive feedback mechanism where H3K27me3 is the enzymatic product, while also serving to activate and recruit additional PRC2 complexes to the local area of chromatin through specific interaction of H3K27me3 (with an aromatic cage containing residues F97, Y148, Y365 in EED).84 This feedback mechanism contributes to the local spreading of heterochromatin. The RBAP48 subunit has a similar structural motif to that of EED and can further activate the PRC2 complex by binding to a different region of the H3 tail, through specific contacts with H3R2 and H3K4. H3K4 must be unmodified for association with RBAP48 as H3K4me3, which is associated with transcriptionally active chromatin, inhibits substrate binding through steric hindrance with the H3K4 binding pocket of RBAP48.85 These two lysine‐binding pockets in the PRC2 complex have a synergistic effect to ensure that the PRC2 complex functions in the correct chromatin region.
Recently several high‐resolution crystal structures of the PRC2 core complex containing EZH2, EED, and SUZ12 from Chaetomium thermophilum 29 and human86 bound or unbound to various H3 or JARID substrate peptides have been reported. Analysis of these structures reveals how substrate binding to the aromatic cage in EED and the stimulation‐responsive motif (SRM) in EZH2 activates the PRC2 complex through allosteric rearrangement of the active site of EZH2 [Fig. 3(d)]. These crystal structures reveal that in the absence of substrate peptide the SRM of EZH2 is in an unstructured form that cannot be traced in the electron density map. Binding of H3K27me3 or JARID2 K116me3 to the aromatic cage (F97, Y148, Y365) of EED and the SRM of EZH2 stimulates a rearrangement within the SRM, leading to formation of a new helix that interacts with the catalytic EZH2 SET domain to influence rearrangement of the catalytic pocket for activity toward unmodified H3K27 residue [Fig. 3(d)].29
In addition to the core subunits, the human PRC2 complex contains additional auxiliary subunits JARID2 and AEBP2 that function to localize the complex to target genes as well as to modulate PRC2 complex activity.87, 88 Both JARID2 and AEBP2 have DNA binding properties, which may help the PRC2 complex to anchor itself on the correct region of chromatin. JARID2 helps recruit the PRC2 complex to target genes with high GC content, as it contains a DNA binding domain with specificity toward these sequences.89, 90 JARID2 K116me3 structurally mimics H3K27me3 and has been shown to bind to the aromatic cage of EED and stimulate the PRC2 complex in the same manner as H3K27me3. AEBP2, which was initially identified as a transcriptional repressor, is a zinc‐finger protein with a nonspecific DNA binding activity that may also be involved in recruiting PRC2 to chromatin.91 AEBP2 also assists in overall structural stability to the PRC2 complex by making contacts with several subunits within the complex.80, 92 Similar to JARID2 association with EED, AEBP2 can also bind to the H3 binding pocket of RBAP48, using residues R295 and K294 to mimic the interactions from R2 and K4 in the H3 tail.85
A recent 3.9 Å cryo‐EM structure of the human PRC2 complex with JARID2 and AEBP2 and a larger fragment of SUZ12 containing both the VEFS‐box and a newly identified neck region (NR) reveals the role of SUZ12 in overall architecture and stability of the PRC2 complex [Fig. 3(e)]. SUZ12 makes contacts with every subunit and serves as a scaffold for assembly of PRC2 and its auxiliary subunits. In this structure, the K116me3 of JARID2 and K294 of AEBP2 mimic the binding of H3K27me3 and H3K4 at their respective binding sites. SUZ12 contains a zinc‐finger domain within the NR at the bottom of the PRC2 complex that plays an important role in an overall structural stability of the PRC2 complex by providing a platform for JARID2 and AEBP2 to bind and properly fold. The interaction between SUZ12 NR, helical segment, and K and R rich region of AEBP2 allows AEBP2 to function as the bridge between RBAP48 and the SET domain of EZH2 adding extra stability of the PRC2 complex. A recent 2.9 Å crystal structure of a smaller PRC2 sub‐complex containing SUZ12, RBBP4, JARID2, AEBP293 also reveals a previously uncharacterized foot region with additional contacts between SUZ12, JARID2, and AEBP2 [Fig. 3(f)]. Biochemical data demonstrate competitive binding of additional auxiliary subunits EPOP and PHF19 with JARID2 and AEBP2, respectively, within the foot region. This binding competition may have a role in modulating complex activity and specificity. Together, these structural analyses suggest that the catalytic lobe containing, EZH2, EED, and VEFS of SUZ12 is sufficient for catalysis. Altogether the neck and foot regions of the PRC2 complex containing SUZ12 and RBAP48 further assists the recruitment and activation of the PRC2 complex through interaction with JARID2, AEBP2, and other auxiliary proteins.
Discussion and future directions
The cryo‐EM structures determined for Snf2, Chd1, and INO80 have significantly extended the findings of previous biochemical characterization of remodeler/nucleosome interactions. Each remodeler associates with the nucleosome at the SHL location predicted by prior footprinting analysis. Interestingly, the structures reveal previously uncharacterized interactions with histone tails that may regulate proper nucleosome binding and chromatin remodeler activity. It is possible that post‐translation modification of the histone tails may influence remodeler binding and activity, as has been shown for ISWI, but further experimentation is necessary to explore the role of histone PTMs in regulation of Snf2, Chd1, and INO80.
Comparison of several structures of histone chaperones bound to their histone substrates exemplifies the evolutionary power of the conserved histone surface. Several different H3/H4 and H2A/H2B chaperones have converged on structurally homologous methods of histone binding, although these histone chaperones share little sequence conservation. We introduce the recent crystal structure of the HIRA C‐terminal domain trimer and discuss a possible role for HIRA trimerization in the formation of an H3.3/H4 tetramer prior to deposition onto DNA. Although further exploration is necessary to fully characterize the biological role of HIRA trimer formation. Analysis of several structures of the FACT subunit Spt16 bound to either H2A/H2B or H3/H4 reveals a possible mechanism of histone chaperone function in which Spt16 can associate with and stabilize an H3/H4 tetramer while a separate domain can simultaneously bind to a molecule of H2A/H2B for nucleosome assembly or disassembly function.
Analysis of several structures of enzymatic histone modifying complexes has revealed differing mechanisms through which nonenzymatic auxiliary subunits modulate the assembly and activity of the complex. The Epl1 subunit in the NuA4 HAT complex serves as a scaffold for assembly of other subunits and rearranges the active site of the catalytic Esa1 subunit to enhance activity toward specific lysine residues in the H4 tail. Conversely, association of the auxiliary subunit BRPF with the catalytic HBO1 HAT subunit does not mediate an active site rearrangement, but instead appears to recruit the histone substrate to the complex through binding to the H3/H4 core domain. Although further structural studies are necessary to reveal the molecular basis for BRPF binding to the H3/H4 core. PRC2 is a large histone methyltransferase complex comprised of the enzymatic EZH2 subunit, the scaffold protein SUZ12, and several different auxiliary subunits that modulate the activity of the complex. Several recent cryo‐EM and crystal structures have detailed a mechanism through which the EED and RBAP48 subunits associate with different regions of the H3 tail to induce a conformational rearrangement of the EZH2 active site for specific methylation of H3K27. Recent cryo‐EM and crystal structures have revealed the molecular details of how additional subunits, JARID2 and AEBP2, associate with PRC2 and mimic regions of H3 to activate the complex in the absence of H3. Interestingly binding of EED to an existing H3 K27me3 or JARID2 K116me3 is essential for proper activation of the complex, although both of these modified residues are also products of the complex and further experimentation is necessary to fully understand how the complex is initially activated.
Analysis of several recent structures of proteins and protein complexes that modulate the epigenetic regulation of chromatin, we have illuminated structural similarities in how different families of chromatin remodelers and histone chaperones associate with their respective nucleosome and histone substrates. We have also reviewed several mechanisms through which auxiliary subunits can modulate the specificity and activity of enzymatic chromatin modifying complexes. Nonetheless, this is only the tip of the iceberg of what structures of multiprotein chromatin regulatory complexes, still to come, will teach us about the molecular basis for chromatin assembly and modification.
References
- 1. Thoma F, Koller T (1977) Influence of histone H1 on chromatin structure. Cell 12:101–107. [DOI] [PubMed] [Google Scholar]
- 2. Kornberg RD (1974) Chromatin structure: a repeating unit of histones and DNA. Science 184:868–871. [DOI] [PubMed] [Google Scholar]
- 3. Luger K, Rechsteiner TJ, Richmond TJ (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol 304:3–19. [DOI] [PubMed] [Google Scholar]
- 4. Tan S, Davey CA (2011) Nucleosome structural studies. Curr Opin Struct Biol 21:128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McGinty RK, Tan S (2015) Nucleosome structure and function. Chem Rev 115:2255–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Makde RD, England JR, Yennawar HP, Tan S (2010) Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467:562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Girish TS, McGinty RK, Tan S (2016) Multivalent interactions by the Set8 histone methyltransferase with its nucleosome substrate. J Mol Biol 428:1531–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McGinty RK, Henrici RC, Tan S (2014) Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514:591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Morgan MT, Haj‐Yahya M, Ringel AE, Bandi P, Brik A, Wolberger C (2016) Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science 351:725–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chittori S, Hong J, Saunders H, Feng H, Ghirlando R, Kelly AE, Bai Y, Subramaniam S (2018) Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP‐N. Science 359:339–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y (2013) A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP‐C. Science 340:1110–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu X, Li M, Xia X, Li X, Chen Z (2017) Mechanism of chromatin remodelling revealed by the Snf2‐nucleosome structure. Nature 544:440–445. [DOI] [PubMed] [Google Scholar]
- 13. Farnung L, Vos SM, Wigge C, Cramer P (2017) Nucleosome‐Chd1 structure and implications for chromatin remodelling. Nature 550:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sundaramoorthy R, Hughes AL, El‐Mkami H, Norman DG, Ferreira H, Owen‐Hughes T (2018) Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome. eLife 7:e35720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eustermann S, Schall K, Kostrewa D, Lakomek K, Strauss M, Moldt M, Hopfner KP (2018) Structural basis for ATP‐dependent chromatin remodelling by the INO80 complex. Nature 556:386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ayala R, Willhoft O, Aramayo RJ, Wilkinson M, McCormack EA, Ocloo L, Wigley DB, Zhang X (2018) Structure and regulation of the human INO80‐nucleosome complex. Nature 556:391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clapier CR, Iwasa J, Cairns BR, Peterson CL (2017) Mechanisms of action and regulation of ATP‐dependent chromatin‐remodelling complexes. Nat Rev Mol Cell Biol 18:407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gurard‐Levin ZA, Quivy JP, Almouzni G (2014) Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu Rev Biochem 83:487–517. [DOI] [PubMed] [Google Scholar]
- 19. Narlikar GJ, Sundaramoorthy R, Owen‐Hughes T (2013) Mechanisms and functions of ATP‐dependent chromatin‐remodeling enzymes. Cell 154:490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang L, Serra‐Cardona A, Zhou H, Wang M, Yang N, Zhang Z, Xu RM (2018) Multisite substrate recognition in Asf1‐dependent acetylation of histone H3 K56 by Rtt109. Cell 174:818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hammond CM, Stromme CB, Huang H, Patel DJ, Groth A (2017) Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol 18:141–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mattiroli F, D'Arcy S, Luger K (2015) The right place at the right time: chaperoning core histone variants. EMBO Rep 16:1454–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447:407–412. [DOI] [PubMed] [Google Scholar]
- 24. Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403:41–45. [DOI] [PubMed] [Google Scholar]
- 25. Suganuma T, Workman JL (2011) Signals and combinatorial functions of histone modifications. Annu Rev Biochem 80:473–499. [DOI] [PubMed] [Google Scholar]
- 26. Bannister AJ, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ (2009) Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461:762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kasinath V, Faini M, Poepsel S, Reif D, Feng XA, Stjepanovic G, Aebersold R, Nogales E (2018) Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 359:940–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiao L, Liu X (2015) Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 350:aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu P, Li C, Chen Z, Jiang S, Fan S, Wang J, Dai J, Zhu P, Chen Z (2016) The NuA4 core complex acetylates nucleosomal histone H4 through a double recognition mechanism. Mol Cell 63:965–975. [DOI] [PubMed] [Google Scholar]
- 31. Lalonde ME, Avvakumov N, Glass KC, Joncas FH, Saksouk N, Holliday M, Paquet E, Yan K, Tong Q, Klein BJ, Tan S, Yang XJ, Kutateladze TG, Cote J (2013) Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev 27:2009–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tao Y, Zhong C, Zhu J, Xu S, Ding J (2017) Structural and mechanistic insights into regulation of HBO1 histone acetyltransferase activity by BRPF2. Nucleic Acids Res 45:5707–5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Han J, Lachance C, Ricketts MD, McCullough CE, Gerace M, Black BE, Cote J, Marmorstein R (2018) The scaffolding protein JADE1 physically links the acetyltransferase subunit HBO1 with its histone H3‐H4 substrate. J Biol Chem 293:4498–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen‐Hughes T (1999) Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400:784–787. [DOI] [PubMed] [Google Scholar]
- 35. Lorch Y, Zhang M, Kornberg RD (1999) Histone octamer transfer by a chromatin‐remodeling complex. Cell 96:389–392. [DOI] [PubMed] [Google Scholar]
- 36. Torigoe SE, Urwin DL, Ishii H, Smith DE, Kadonaga JT (2011) Identification of a rapidly formed nonnucleosomal histone‐DNA intermediate that is converted into chromatin by ACF. Mol Cell 43:638–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ocampo J, Chereji RV, Eriksson PR, Clark DJ (2016) The ISW1 and CHD1 ATP‐dependent chromatin remodelers compete to set nucleosome spacing in vivo. Nucleic Acids Res 44:4625–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brahma S, Udugama MI, Kim J, Hada A, Bhardwaj SK, Hailu SG, Lee TH, Bartholomew B (2017) INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat Commun 8:15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C (2004) ATP‐driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343–348. [DOI] [PubMed] [Google Scholar]
- 40. Zofall M, Persinger J, Kassabov SR, Bartholomew B (2006) Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol 13:339–346. [DOI] [PubMed] [Google Scholar]
- 41. Dang W, Bartholomew B (2007) Domain architecture of the catalytic subunit in the ISW2‐nucleosome complex. Mol Cell Biol 27:8306–8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nodelman IM, Bleichert F, Patel A, Ren R, Horvath KC, Berger JM, Bowman GD (2017) Interdomain communication of the Chd1 chromatin remodeler across the DNA gyres of the nucleosome. Mol Cell 65:447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clapier CR, Cairns BR (2012) Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature 492:280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Racki LR, Yang JG, Naber N, Partensky PD, Acevedo A, Purcell TJ, Cooke R, Cheng Y, Narlikar GJ (2009) The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature 462:1016–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Willhoft O, McCormack EA, Aramayo RJ, Bythell‐Douglas R, Ocloo L, Zhang X, Wigley DB (2017) Crosstalk within a functional INO80 complex dimer regulates nucleosome sliding. eLife 6:25782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xia X, Liu X, Li T, Fang X, Chen Z (2016) Structure of chromatin remodeler Swi2/Snf2 in the resting state. Nat Struct Mol Biol 23:722–729. [DOI] [PubMed] [Google Scholar]
- 47. Hauk G, McKnight JN, Nodelman IM, Bowman GD (2010) The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell 39:711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yan L, Wang L, Tian Y, Xia X, Chen Z (2016) Structure and regulation of the chromatin remodeller ISWI. Nature 540:466–469. [DOI] [PubMed] [Google Scholar]
- 49. Yao W, Beckwith SL, Zheng T, Young T, Dinh VT, Ranjan A, Morrison AJ (2015) Assembly of the Arp5 (actin‐related protein) subunit involved in distinct INO80 chromatin remodeling activities. J Biol Chem 290:25700–25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen L, Cai Y, Jin J, Florens L, Swanson SK, Washburn MP, Conaway JW, Conaway RC (2011) Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes ATP‐dependent nucleosome remodeling. J Biol Chem 286:11283–11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sinha KK, Gross JD, Narlikar GJ (2017) Distortion of histone octamer core promotes nucleosome mobilization by a chromatin remodeler. Science 355:aaa3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bilokapic S, Strauss M, Halic M (2018) Histone octamer rearranges to adapt to DNA unwrapping. Nat Struct Mol Biol 25:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barbera AJ, Chodaparambil JV, Kelley‐Clarke B, Joukov V, Walter JC, Luger K, Kaye KM (2006) The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311:856–861. [DOI] [PubMed] [Google Scholar]
- 54. Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE (2011) Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science 334:977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ricketts MD, Frederick B, Hoff H, Tang Y, Schultz DC, Singh Rai T, Grazia Vizioli M, Adams PD, Marmorstein R (2015) Ubinuclein‐1 confers histone H3.3‐specific‐binding by the HIRA histone chaperone complex. Nat Commun 6:7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu CP, Xiong C, Wang M, Yu Z, Yang N, Chen P, Zhang Z, Li G, Xu RM (2012) Structure of the variant histone H3.3‐H4 heterodimer in complex with its chaperone DAXX. Nat Struct Mol Biol 19:1287–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang H, Stromme CB, Saredi G, Hodl M, Strandsby A, Gonzalez‐Aguilera C, Chen S, Groth A, Patel DJ (2015) A unique binding mode enables MCM2 to chaperone histones H3‐H4 at replication forks. Nat Struct Mol Biol 22:618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Aguilar‐Gurrieri C, Larabi A, Vinayachandran V, Patel NA, Yen K, Reja R, Ebong IO, Schoehn G, Robinson CV, Pugh BF, Panne D (2016) Structural evidence for Nap1‐dependent H2A‐H2B deposition and nucleosome assembly. EMBO J 35:1465–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang M, Liu H, Gao Y, Zhu Z, Chen Z, Zheng P, Xue L, Li J, Teng M, Niu L (2016) Structural insights into the association of Hif1 with histones H2A‐H2B dimer and H3‐H4 tetramer. Structure 24:1810–1820. [DOI] [PubMed] [Google Scholar]
- 60. Hong J, Feng H, Wang F, Ranjan A, Chen J, Jiang J, Ghirlando R, Xiao TS, Wu C, Bai Y (2014) The catalytic subunit of the SWR1 remodeler is a histone chaperone for the H2A.Z‐H2B dimer. Mol Cell 53:498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Latrick CM, Marek M, Ouararhni K, Papin C, Stoll I, Ignatyeva M, Obri A, Ennifar E, Dimitrov S, Romier C, Hamiche A (2016) Molecular basis and specificity of H2A.Z‐H2B recognition and deposition by the histone chaperone YL1. Nat Struct Mol Biol 23:309–316. [DOI] [PubMed] [Google Scholar]
- 62. Ricketts MD, Marmorstein R (2017) A molecular prospective for HIRA complex assembly and H3.3‐specific histone chaperone function. J Mol Biol 429:1924–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Briana K, Dennehey JT, Histone chaperones in the assembly and disassembly of chromatin In: Abmayr S.M. Workman J.L., Eds. (2014) Fundamentals of Chromatin Ch. 2, New York: Springer‐Verlag, pp 29–67. [Google Scholar]
- 64. Ray‐Gallet D, Ricketts MD, Sato Y, Gupta K, Boyarchuk E, Senda T, Marmorstein R, Almouzni G (2018) Functional activity of the H3.3 histone chaperone complex HIRA requires trimerization of the HIRA subunit. Nat Commun 9:3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sauer PV, Timm J, Liu D, Sitbon D, Boeri‐Erba E, Velours C, Mucke N, Langowski J, Ochsenbein F, Almouzni G, Panne D (2017) Insights into the molecular architecture and histone H3‐H4 deposition mechanism of yeast Chromatin assembly factor 1. eLife 6:e23474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zasadzinska E, Barnhart‐Dailey MC, Kuich PH, Foltz DR (2013) Dimerization of the CENP‐A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J 32:2113–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tang Y, Meeth K, Jiang E, Luo C, Marmorstein R (2008) Structure of Vps75 and implications for histone chaperone function. Proc Natl Acad Sci USA 105:12206–12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hammond CM, Sundaramoorthy R, Larance M, Lamond A, Stevens MA, El‐Mkami H, Norman DG, Owen‐Hughes T (2016) The histone chaperone Vps75 forms multiple oligomeric assemblies capable of mediating exchange between histone H3‐H4 tetramers and Asf1‐H3‐H4 complexes. Nucleic Acids Res 44:6157–6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hondele M, Stuwe T, Hassler M, Halbach F, Bowman A, Zhang ET, Nijmeijer B, Kotthoff C, Rybin V, Amlacher S, Hurt E, Ladurner AG (2013) Structural basis of histone H2A‐H2B recognition by the essential chaperone FACT. Nature 499:111–114. [DOI] [PubMed] [Google Scholar]
- 70. Kemble DJ, McCullough LL, Whitby FG, Formosa T, Hill CP (2015) FACT disrupts nucleosome structure by binding H2A‐H2B with conserved peptide motifs. Mol Cell 60:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tsunaka Y, Fujiwara Y, Oyama T, Hirose S, Morikawa K (2016) Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev 30:673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sterner DE, Berger SL (2000) Acetylation of histones and transcription‐related factors. Microbiol Mol Biol Rev 64:435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Marmorstein R, Zhou MM (2014) Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol 6:a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Berndsen CE, Denu JM (2008) Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr Opin Struct Biol 18:682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook RG, Lucchesi JC, Allis CD (1998) ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci USA 95:3561–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yan Y, Barlev NA, Haley RH, Berger SL, Marmorstein R (2000) Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol Cell 6:1195–1205. [DOI] [PubMed] [Google Scholar]
- 77. Rojas JR, Trievel RC, Zhou J, Mo Y, Li X, Berger SL, Allis CD, Marmorstein R (1999) Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature 401:93–98. [DOI] [PubMed] [Google Scholar]
- 78. Saksouk N, Avvakumov N, Champagne KS, Hung T, Doyon Y, Cayrou C, Paquet E, Ullah M, Landry AJ, Cote V, Yang XJ, Gozani O, Kutateladze TG, Cote J (2009) HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol Cell 33:257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Avvakumov N, Lalonde ME, Saksouk N, Paquet E, Glass KC, Landry AJ, Doyon Y, Cayrou C, Robitaille GA, Richard DE, Yang XJ, Kutateladze TG, Cote J (2012) Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation. Mol Cell Biol 32:689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cao R, Wang L, Wang H, Xia L, Erdjument‐Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb‐group silencing. Science 298:1039–1043. [DOI] [PubMed] [Google Scholar]
- 81. Talbert PB, Henikoff S (2006) Spreading of silent chromatin: inaction at a distance. Nat Rev Genet 7:793–803. [DOI] [PubMed] [Google Scholar]
- 82. Margueron R, Reinberg D (2011) The Polycomb complex PRC2 and its mark in life. Nature 469:343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wu H, Zeng H, Dong A, Li F, He H, Senisterra G, Seitova A, Duan S, Brown PJ, Vedadi M, Arrowsmith CH, Schapira M (2013) Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations. PLoS One 8:e83737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K (2008) A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10:1291–1300. [DOI] [PubMed] [Google Scholar]
- 85. Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, Bunker RD, Wirth U, Bouwmeester T, Bauer A, Ly‐Hartig N, Zhao K, Chan H, Gu J, Gut H, Fischle W, Muller J, Thoma NH (2011) Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42:330–341. [DOI] [PubMed] [Google Scholar]
- 86. Justin N, Zhang Y, Tarricone C, Martin SR, Chen S, Underwood E, De Marco V, Haire LF, Walker PA, Reinberg D, Wilson JR, Gamblin SJ (2016) Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun 7:11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Son J, Shen SS, Margueron R, Reinberg D (2013) Nucleosome‐binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev 27:2663–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Grijzenhout A, Godwin J, Koseki H, Gdula MR, Szumska D, McGouran JF, Bhattacharya S, Kessler BM, Brockdorff N, Cooper S (2016) Functional analysis of AEBP2, a PRC2 Polycomb protein, reveals a Trithorax phenotype in embryonic development and in ESCs. Development 143:2716–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D (2010) Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24:368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE (2008) Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4:e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. He GP, Kim S, Ro HS (1999) Cloning and characterization of a novel zinc finger transcriptional repressor. A direct role of the zinc finger motif in repression. J Biol Chem 274:14678–14684. [DOI] [PubMed] [Google Scholar]
- 92. Kim H, Kang K, Kim J (2009) AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res 37:2940–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chen S, Jiao L, Shubbar M, Yang X, Liu X (2018) Unique structural platforms of Suz12 dictate distinct classes of PRC2 for chromatin binding. Mol Cell 69:840–852. [DOI] [PMC free article] [PubMed] [Google Scholar]


