Abstract
Objective:
The diversity of tumour characteristics among glioma patients, even within same tumour grade, is a big challenge for disease outcome prediction. A possible approach for improved radiological imaging could come from combining information obtained at the molecular level. This review assembles recent evidence highlighting the value of using radiogenomic biomarkers to infer the underlying biology of gliomas and its correlation with imaging features.
Methods:
A literature search was done for articles published between 2002 and 2017 on Medline electronic databases. Of 249 titles identified, 38 fulfilled the inclusion criteria, with 14 articles related to quantifiable imaging parameters (heterogeneity, vascularity, diffusion, cell density, infiltrations, perfusion, and metabolite changes) and 24 articles relevant to molecular biomarkers linked to imaging.
Results:
Genes found to correlate with various imaging phenotypes were EGFR, MGMT, IDH1, VEGF, PDGF, TP53, and Ki-67. EGFR is the most studied gene related to imaging characteristics in the studies reviewed (41.7%), followed by MGMT (20.8%) and IDH1 (16.7%). A summary of the relationship amongst glioma morphology, gene expressions, imaging characteristics, prognosis and therapeutic response are presented.
Conclusion:
The use of radiogenomics can provide insights to understanding tumour biology and the underlying molecular pathways. Certain MRI characteristics that show strong correlations with EGFR, MGMT and IDH1 could be used as imaging biomarkers. Knowing the pathways involved in tumour progression and their associated imaging patterns may assist in diagnosis, prognosis and treatment management, while facilitating personalised medicine.
Advances in knowledge:
Radiogenomics can offer clinicians better insight into diagnosis, prognosis, and prediction of therapeutic responses of glioma.
Introduction
Gliomas, which comprise 27% of all brain tumours, are lethal primary malignant brain tumours originating from the interstitial tissue of the brain.1 Gliomas are categorised as diffuse astrocytic and oligodendroglial tumours, other astrocytic tumours, ependymal cell types and neuronal and mixed neuronal-glial tumours according to the World Health Organization (WHO) guidelines. A recent upgrade of the WHO guidelines feature integrated molecular parameters into histology that underlines the importance of radiogenomics in the classification of tumour entities.2, 3 The severity of the grade depends on tumour growth, localized invasion, cell pleomorphism, mitotic activity, vascular proliferation, necrosis, and resistance to therapy.
To date, MRI is the modality of choice as it offers valuable information on overall tumour structure, composition, physiology and function.4 Tumour characteristics examined such as intensity distribution, enhancement, size, shape, structure, location, volume, border, focality, subventricular zone involvement, cystic changes, the percentage of necrosis and tumour volume are often inadequate for clinical use because of the irregular shape and heterogeneous composition of the tumours.5–9 Histopathological grading serves as the gold-standard but suffers from several drawbacks such as intra- and interobserver variability, sampling error, tumour heterogeneities, and risk of surgical complications in patients.10 Quantitative imaging biomarkers derived from advanced MRI techniques, namely diffusion-weighted imaging, perfusion-weighted imaging, diffusion tensor imaging, diffusion kurtosis imaging and magnetic resonance spectroscopy are used to define tumour morphology and functionality.4, 11,12
Glioma detection and grading at its earliest stage is crucial for early intervention to improve prognosis and minimise neurocognitive risks. The problem of grading glioma accurately is not trivial. High diversity of tumour properties, even within a single tumour, is a big challenge to determine the grades and subtypes. The heterogeneous nature of the tumours further complicates histopathological observations and this can affect treatment decisions and management. To cap the complexity of the disease, different responses to treatments among patients are often seen due to the differences in the genetic profiles of the tumours.13, 14 Hence, the use of radiogenomic biomarkers may provide a holistic approach for the treatment of glioma.
Radiogenomics is an evolving new field that studies the link between gene expression patterns and imaging phenotypes for diagnosis, prognosis, and prediction of therapeutic responses in cancer.15, 16 The underlying inter- and intratumoral gene expression patterns that steer the unique characteristics and morphological manifestation of glioma can be captured by quantitative imaging.5,9,15–19 Radiogenomics holds the potential for targeted therapies, whereby therapeutic treatments are tailored to the individual tumour’s genetic profile based on indications from imaging features. There is a need to identify biomarkers that can reflect genetic profiles to better characterise the tumours, so that clinicians can make better decisions when administering treatment.
While there have been a number of studies looking at this aspect in glioma grading, it is still unclear which genes or pathways offer the most comprehensive personalised approach in practice.20, 21 This paper aims to provide a systematic review of these recent studies specifically looking at the use of MRI biomarkers in characterising glioma. We plan to stratify radiophenotypes that could serve as molecular surrogates to infer specific gene expression patterns from the review.
Methods and Materials
Eligibility criteria and search strategy
We performed a systematic review of imaging biomarkers (radiogenomics) of glioma literature according to the PRISMA (Preferred Reporting Items for Systemic Review and Meta-Analyses) guidelines.22, 23 Our review comprised of a detailed set of research questions and a search strategy that included screening criteria for titles and abstracts, followed by the selection of full-text articles. The detailed research questions were established using the patient, intervention, comparator, outcome and study design approach. The questions were devised as follows: what are the key genes associated with imaging characteristics of gliomas? What are the changes in gene expression of the tumours? Are gene expression patterns linked to specific MR imaging features? What are the correlations between the radiogenomic biomarkers associated with the tumours and the phenotypes reflected by MRI?
The inclusion criteria for full-text article assessment were randomised or cohort MRI studies of glioma patients. The exclusion criteria were studies on paediatric populations, radiotherapy or chemotherapy studies and drug studies such as clinical trials, animal experiments, biopsies or histopathological studies, cell culture, and toxicity tests. Pubmed and Google Scholar were used to search the Medline database. The keywords used in Medline included “glioma”, “magnetic resonance imaging”, “MRI”, “biomarkers” and “glioblastoma multiforme”. Full-article assessments were conducted to determine the compliance of the studies with the inclusion and exclusion criteria. The searches were done by PS and reviewed by NR, JHDW, and AAA respectively.
Study selection and data extraction
Only studies published in English after 2002 were selected and the last search was on 30 October 2017. Relevant data regarding imaging features and molecular profiles were extracted from each article. The data collected were categorised into gene groups, associated with different imaging characteristics of tumour.
Results
Study selection
The literature search and study selection showed 59 records were included in the final stage of the literature review where 38 full text articles investigated on quantifiable biomarkers (Figure 1). From the records, 14 articles were related to quantifiable imaging parameters (Table 1) while 24 articles investigated the relations between imaging biomarkers and genetic profiles (Table 2). There were overlaps in both of the tables as some of the studies investigated several parameters. The main findings of the studies were also recorded (Supplementary Material 1) while the PRISMA checklist was provided as Supplementary Material 2.
Figure 1.
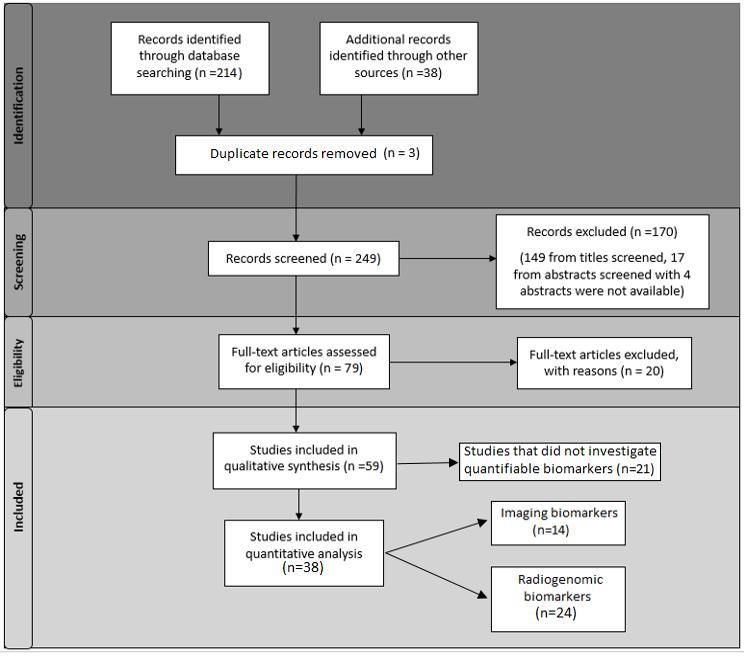
Literature assessment. Flow diagram of literature assessment.
Table 1.
Quantitative MRI biomarkers mentioned in the studies
| Characteristics | Imaging biomarkersa | Techniques | Number of studies | Ref |
| Heterogeneity | Enhancement and necrosis | MRI | 1 | 4 |
| Vascularity | Uncorrected CBV ratio and FPS ratio | MRI + PWI (DSC / DCE) | 6 | 24 |
| Min and max relative CBV and relative CBF | 11, 25,26 | |||
| Ktrans and Ve | 27 | |||
| Peak height in ET & non-ET | 28 | |||
| Non-Gaussian diffusion/Cell density/ cellularity | ADC, slow diffusion coefficient (Dslow), DDC and heterogeneity index (α) | MRI + DWI/IVIM | 3 | 4, 29,30 |
| Infiltrations along WM tracts/ micro-vascularity | FA, MD, and tensor decomposition p & q maps & fDM | MRI + DTI | 6 | 26, 31,32 |
| Relative anisotropy and radial diffusivity | 33 | |||
| Diffusion trace in ET | 28 | |||
| Metabolite changes | Lip/tCho | MRI + MRS | 3 | 11 |
| Cho/Cr, MI/Cr, Lac/Cr, NAA/Cr | 31 | |||
| Lipid quantification: Signal loss ratio in solid and cystic subregions | MRI + MRS+IOP | 34 | ||
| Kurtosis | Mean kurtosis | DKI | 1 | 11 |
ADC, apparent diffusion co-efficient; CBF, cerebral bloodflow; CBV, cerebral blood volume; Cho, choline; Cr, creatine; DDC, distributed diffusion coefficient; DWI, diffusion-weighted imaging; DTI, diffusion tensor imaging; ET, enhancing tumour; IVIM, intravoxel incoherent motion; FA, fractional anisotropy; fDM, functional diffusion map; FPS, first pass slope; Ktrans, volume transfer constant; IOP, in and opposed-MRI; Lip, lipid; MD, mean diffusivity; MI, myo-inositol; Lac, lactate; MRS, magnetic resonance spectroscopy; NAA, N-acetyl aspartate; PWI, perfusion-weighted imaging; Ve, volume of extravascular extracellular space per unit volume of tissue; WM, white matter.
MRI refers to structural MRI [T1-weighted, T2-weighted and Fluid Attenuation Inversion Recovery (FLAIR) sequences].
only biomarkers that are statistically significant (p < 0.05) are reported.
Table 2.
The radiogenomic biomarkers linking imaging features/phenotypes to gene expression patterns in the studies
| Genes/Molecular biomarkers | Characteristics | Imaging biomarkers | Number of studies | Ref |
| EGFR | Diffusion | relative CBV, PSR | 10 | 13 |
| Morphology | Anatomic location (radiogenomic maps) | 35, 36 | ||
| Percentage of CE, NE, necrosis & oedema and largest diameter on lesion | 7 | |||
| Morphology, diffusion & interaction with ECM | Border sharpness, restricted water diffusion, ADC | 37 | ||
| Gene expressions | CE, necrosis, mass effect, oedema, cortical involvement, CE:N volume ratio, T2heterogeneity | 38 | ||
| Infiltration, proliferation, neurogenesis and synaptic transmission | 39 | |||
| Perfusion | VP & Ktrans | 40 | ||
| Normalized CBV & CBF | 41 | |||
| Mean & relative TBF | 42 | |||
| MGMT methylation status | Perfusion | Ktrans | 5 | 43 |
| Normalized CBV | 44 | |||
| Morphology | Anatomic location | 35, 36 ] | ||
| Textures | Correlation, energy, entropy & local intensity |
45 | ||
| IDH1 | Morphology | Location | 4 | 35 |
| Metabolite changes | Percentage of CE, NE, necrosis & oedema and largest diameter on lesion 2-hydroxyglutarate (2HG) |
7 [46, 47 |
||
| Perfusion | TBF | 48 | ||
| TP53 | Morphology | Percentage of CE, NE, necrosis & oedema and largest diameter on lesion | 2 | 7 |
| Gene expressions | CE, necrosis, mass effect, oedema, cortical involvement, CE:N volume ratio, T2 heterogeneity | 38 | ||
| PTEN loss | Morphology | Anatomic location | 2 | 36 |
| Percentage of CE, NE, necrosis & oedema and largest diameter on lesion | 7 | |||
| Ki-67 index | Diffusion, perfusion, metabolite change & genomics | relative CBF, FA, ADC, Cho/Cr, NAA/Cho, NAA/Cr, Lac/Cr & MI | 3 | 49, 50 |
| Gene expressions | CE, necrosis, mass effect, oedema, cortical involvement, CE:N volume ratio, T2 heterogeneity | 38 | ||
| VEGFR | Morphology and textural | Shape, texture, edge sharpness of necrotic core and surrounding CE rim | 2 | 51 |
| Vascularization | CBV | 52 | ||
| 1p/19q codeletions | Vascularization | relative CBV | 1 | 53 |
| GAP4 and WWTR1 genes | Intensities (ROI), sharpness of lesion boundaries, boundary shapes | Edge sharpness of necrotic portion | 1 | 18 |
| HRAS copy number variation | Contrast enhancement and genetic expressions | Proportion of enhancing tumour & T1/FLAIR ratio | 1 | 8 |
| Periostin and miR-219 | Cellular invasion | Edema/invasion FLAIR volumes | 1 | 54 |
| Molecular subclasses of GBM | Hemodynamics | relative CBV | 1 | 52 |
ADC, apparent diffusion coefficient; CBF, cerebral blood flow;CBV, cerebral blood volume; CE, contrast enhancement; CE:N, contrast-enhancing volume to the necrotic tumour volume ratio; ECM, extra cellular matrix; EGFR,epidermal growth factor receptor; Cho, choline; Cr, creatine; FA, fractional anisotropy; IDH1,isocitrate dehydrogenase1; Ki-67antigen; Ktrans, volume transfer constant; Lac, lactate; MGMT, O6-methylguanine-DNA-methyltransferase; MI, myo-inositol; NAA, N-acetyl aspartate; NE, non-enhanced; PDGFA, platelet-derived growth factor; ROI, region of interest; PSR, percent signal recovery; PTEN, phosphatase and tensin homolog; TBF, tumour blood flow; TP53, tumour protein p53; VEGFR, vascular endothelial growth factor receptor; VP, plasma volume.
Findings
Table 1 lists the quantitative MRI biomarkers that are reported in the literature reviewed. Figures 2–6 show the structural and functional images of different glioma grades acquired from conventional and advanced MRI techniques, in relation to gene expression. We list the gene expression profiles linked to glioma characteristics in Table 2.
Figure 2.
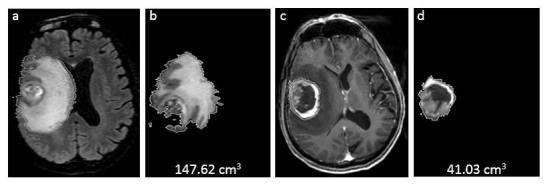
A case of grade IV GBM with EGFR amplification/overexpression. MRI features showing greater ratio of T2 bright volume to the enclosed T1 enhancing volume in GBM: (a) CUBE FLAIR images depicting perilesional oedema, (b) calculated 3D-T2 bright volume (147.62 cm3), (c) T1W post-contrast showing Rt parietal enhancing GBM with internal necrosis, and (d) calculated 3D-T1-enhancing volume including internal necrosis (41.03 cm3). EGFR, endothelial growth factor receptor; GBM, glioblastoma multiformes.
Figure 3.
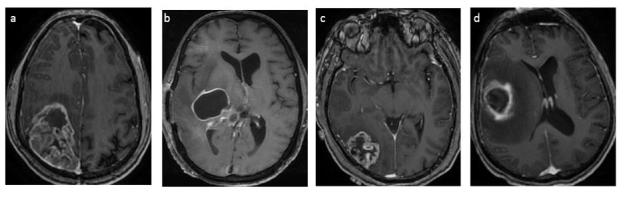
MRI post-gadolinium images of various grade IV GBM with hypermethylated and unmethylated MGMT. Imaging features showing: (a) mixed-nodular in a patient with hypermethylated MGMT and (b) ring enhancement in unmethylated MGMT. Another two cases demonstrating (c) preferential location of grade IV GBM with hypermethylation of the MGMT promoter located in parietal and occipital lobes, and (d) unmethylated of the MGMT promoter in the temporal lobes. GBM, glioblastoma multiformes; MGMT, O6-methylguanine-DNA-methyltransferase.
Figure 4.
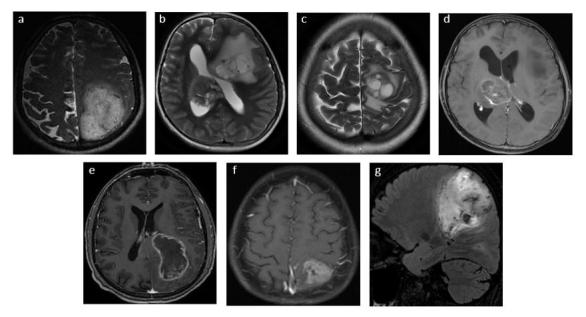
MRI features of various grade IV GBM patients with IDH1 mutation. The features included T2W images showing (a) large size at time of diagnosis (b) multifocality, and (c) cystic components. (d) Post-gadolinium T1W showing non-enhancing solid tumour component, (e) greater frequency of contact with the ventricles, and *(f) usually less necrotic (<50% of tumour volume, and (g) T2 FLAIR showing less perilesional oedema (<50% of tumour volume). GBM, glioblastoma multiformes; IDH1, isocitratedehydrogenase 1.
Figure 5.
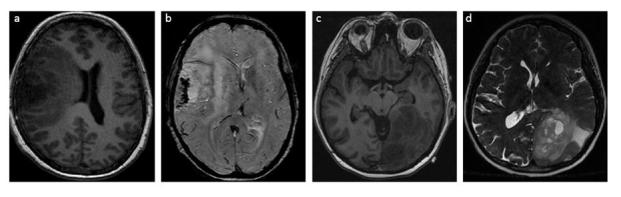
MRI features of grade III ODG patients with of 1p/19q co-deletion. Imaging findings showing: (a) indistinct borders on T1W, (b) GRE sequence with paramagnetic susceptibility and calcification and, (c-d) mixed signal intensities on T1W and T2W. GRE, gradient echo; ODG, oligodendroglioma.
Figure 6.
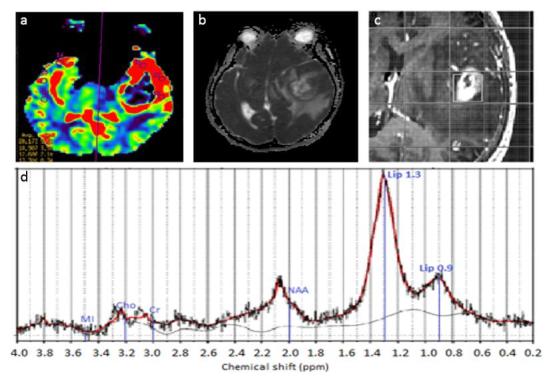
The MRI images of a grade IV GBM with prominent palisading necrosis, microvascular proliferation, Ki-67 index ~15–20% in a few cellular areas. Imaging findings showing: (a) relative CBV colour map where high blood volume was seen at the rim area, (b) decreased ADC shown as hypointense area compared to CSF in tumour region, (c) the voxel placement in SVS, and (d) the corresponding brain spectra acquired using LC Model where MI, Cho, Cr, NAA & Lip peaks are labelled. Elevated lipid peaks and Cho, with decreased NAA were apparent in the spectrum. ADC, apparent diffusion co-efficient; CBV, cerebral blood volume; Cho, choline; Cr, creatine; CSF, cerebrospinal fluid; GBM, glioblastoma multiformes; Lip, lipid; MI, myo-inositol; NAA, N-acetyl aspartate; SVS, single voxel spectroscopy.
The key genes
The gene expression profiles found to be associated with the imaging features are listed in the following sections. The order of the gene expression profiles discussed is according to numbers of studies done, rather than their interpretive significance. Figure 7 is a schematic diagram to summarise the relationship between glioma morphology, imaging features, and gene expression profiles, which can be inferred from MRI techniques. From the figure, a complex pattern of involvement is evident as a single gene may have roles in different tumour characteristics, meanwhile, a single tumour characteristic could be due to many different genes.
Figure 7.
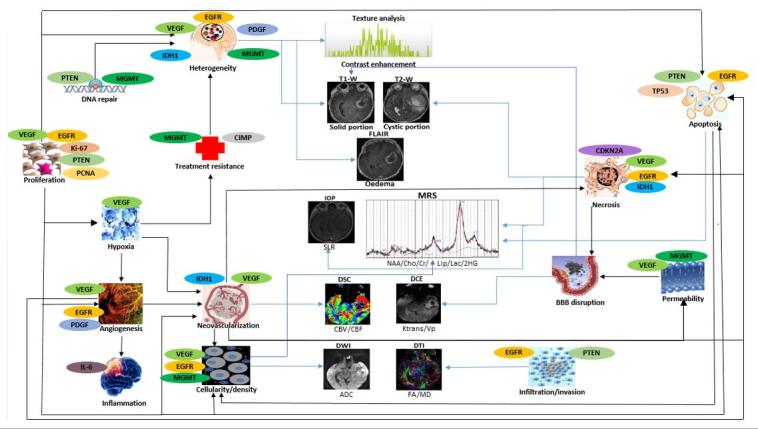
Radiogenomic approach for glioma characterisation. A schematic diagram to illustrate the relationship of glioma morphology with gene expressions and imaging characteristic. Black arrows indicate associations between different glioma morphology while blue arrows represent the linking between glioma morphology and MRI. The images displayed are for visual guide only. ADC, apparent diffusion co-efficient; BBB, blood-brain barrier; CBF, cerebral blood flow;CBV, cerebral blood volume; CDKN2A,cyclin-dependent kinase inhibitor; Cho, choline; Cr, creatine; DCE, dynamic contrast-enhanced; DSC, dynamic susceptibility contrast; DWI, diffusion-weighted imaging; DTI, diffusion tensor imaging; EGFR, endothelial growth factor receptor; FA, fractional anisotropy; IDH1, isocitratedehydrogenase 1; IOP, in and opposed-MRI; Ktrans, volume transfer constant; Lac, lactate; Lip, lipid; MD, mean diffusivity; MGMT,O6-methylguanine-DNA-methyltransferase; MRS, magnetic resonance spectroscopy; NAA, N-acetyl aspartate; PCNA, proliferating cell nuclear antigen; PDGF, platelet-derived growth factor; PTEN, phosphatase andtensin homolog; SLR, signal loss ratio; VEGF, vascularendothelial growth factor; VP, plasma volume.
Epidermal growth factor/receptor (EGFR)
EGFR is the receptor for epidermal growth factor, and amplification/overexpression of the EGFR locus is found in about 42% of primary glioblastoma multiformes (GBM).35 EGFR amplification in histologically pure anaplastic oligodendroglioma (ODG) is indicative of GBM. EFGR overexpression indicated poor outcome and correlated with decreased overall survival in GBM.7, 55 The stratification of GBM into four distinct molecular subtypes (classic, mesenchymal, neural and proneural) are differed by distinct prognoses and responses to therapy based on gene expression.56 The classic subtype has a strong association with astrocytic signature with EGFR amplification.
EGFR was identified as a significant glioma biomarker in 41.7% of the studies reviewed. The pathway activation of EGFR is associated with increased motility, invasion, angiogenesis, tumour cell proliferation, reprogramming of tumour metabolism, and inhibition of apoptosis.36, 37,57
Contrast enhancement of the solid region of tumour in T1W (T1 weighted) is often related to the aggressiveness of lesions,4, 6,9 however, many low-grade gliomas show enhancement and one-third of non-enhancing gliomas are malignant.6 The solid region of the tumour and its surrounding tissues are comprised of actively proliferating cells such as invasive tumour cells, microglial cells and reactive astrocytes.31 In terms of enhancement, EGFR amplification/overexpression was associated with higher T1 +C (post-contrast) and T2/FLAIR hyperintense volume, higher ratio of the contrast enhancing volume to the necrotic tumour volume and greater ratio of T2-bright volume to T1-enhancing volume (including internal necrosis) in GBM35, 36,38,39,51,58 (Figure 2). EGFR amplification/ overexpression/ mutation is related to angiogenesis, with a resultant increase in cerebral blood volume (CBV), cerebral blood flow (CBF), plasma volume and contrast transfer coefficient in MR perfusion.36, 40,41 Metabolite changes such as reduced N-acetyl-aspartate (NAA) levels, lower creatine (Cr) and lower myoinositol (MI) in high-grade gliomas (HGG) and increased lactate proportionally with volumes of necrosis lesion,11,31,49,59–62 and restricted water diffusion36, 37 are also related to EGFR amplification/ overexpression/ mutation.
O6-methylguanine-DNA-methyltransferase (MGMT)
The second gene that appears most frequently in the studies reviewed (20.8%) is the MGMT gene and has been reported for 30–60% in GBM. The MGMT gene encodes a DNA repair protein that is involved in cellular defense against mutagenesis and toxicity from alkylating agents.43 GBM with MGMT promoter methylation demonstrated more favourable prognosis in terms of longer median survival.6, 48,51,63,64 GBM with MGMT promoter methylation showed better treatment response6, 51 due to decreased MGMT protein expression that reduces DNA repair activity against temozolomide, a DNA alkylating agent. Thus, the sensitivity to therapy improves due to the increase in endothelial permeability that facilitates the penetration of drugs and their delivery.43, 63
Hypermethylated MGMT tumours tend to have mixed-nodular enhancement, non-temporal lobe lesions, and often show radiation or treatment-induced pseudo-progression.44 On the contrary, unmethylated MGMT tumours have high occurrences of temporal lobe lesions, ring enhancement, and true progression35 (Figure 3). Tumour characteristics such as cellular density, treatment response, and texture features are linked to MGMT methylation status.9, 43,45,53,59 Increased apparent diffusion coefficient (ADC) values derived from DWI implicate changes in tumoral water diffusion incited by necrosis or apoptosis,49 and a higher degree of spatial heterogeneity has been observed in contrast-enhancing unmethylated MGMT tumours.4, 9,29 Treatment responses were apparent in infiltrative low-grade gliomas (LGG) as reflected by changes in DTI metrics such as pure isotropic components of diffusion (p) and mean diffusivity (MD) at the tumour borders.32
Isocitrate dehydrogenase 1 (IDH1)
IDH1 encodes a metabolic enzyme known as IDH1, which catalyses the conversion of isocitrate to alpha-ketoglutarate. Mutations in IDH1 are frequently seen in diffuse LGG and secondary GBM.3,61–64 IDH1 mutations are also one of the genetic features related to the proneural subtype of GBM51 that carry better clinical prognosis in terms of overall survival and progression-free survival,48 and bear favourable overall survival in diffuse astrocytomas and anaplastic astrocytoma.3, 61
GBM with IDH1 mutations tend to be in the left frontal lobe, larger at diagnosis, may be multifocal, have a high prevalence of non-enhancing tumours, cystic and diffuse components, greater frequency of contact with brain ventricles with less necrosis detection and extent of oedema, less frequent vascular abnormalities, increased oligodendroglial morphology and also metabolite changes35, 46,60,64 (Figure 4). Glioblastomas without IDH1 mutations showed larger volumes of contrast enhancement seen in T1W + C.21, 35 The conversion to alpha-ketoglutarate by IDH1 gene is observable using MRS as the elevation of 2-hydroxyglutarate (2HG) co-detected at 2.25ppm and 4.02 ppm that reflect changes in tumour cellularity.46, 47
1p/19q codeletion status
The combined loss of 1p and 19q chromosome arms is uncommon in glioma and is considered the earliest genetic hallmark of ODG, whereby it is seen in 50–70% of the neoplasms.65 20 The complete loss of both chromosomes is associated with good prognosis, longer progression-free survival and increased sensitivity to chemotherapy in ODG and oligoastrocytoma.66, 67
In conventional MRI studies, ODG with 1p/19q loss is more likely to have indistinct borders on T1W images, mixed-signal intensities on T1W and T2W, paramagnetic susceptibility effect, calcification and infiltrative growth patterns65, 68 (Figure 5). Elevated relative CBV with 1p/19q codeletions suggested increased neovascularity in glioma with oligodendroglial components.67 The increased ADC values in ODG and 1p/19q codeleted mixed oligoastrocytomas (OA) were associated with the fraction of the tumour cells (relative number of tumour cells per total cells) and degree of axonal disruption in tumour subregions.66
TP53
TP53 is a tumour suppressor gene, which encodes a tumour suppressor protein that responds to cellular stresses by inducing cell cycle arrest, apoptosis, senescence, DNA repair or metabolism changes.7, 51,69 TP53 mutations are mainly found in astrocytomas and are associated with poor survival.61 High incidence of IDH1 mutations are seen in TP53 mutations in early gliomagenesis of LGG.61 GBMs with TP53 mutations were reported to be smaller in size compared to the wild type, presented as areas that were hyperintense on T2W FLAIR images.7
Ki-67 protein
The Ki-67 antigen is a nuclear protein encoded by MKI67 gene, that is used as a histopathological indicator of cellular proliferation and growth.62, 63 Ki-67 is identified in paraffin-embedded sections made with the monoclonal antibody MIB-1.62, 63,70 The Ki-67 index is measured as the percentage of positively stained nuclei.71 A high Ki-67 index correlates positively with tumour grades and prognosis (overall survival).30
High proliferation activities suggested as the elevation of Ki-67 index are related to higher relative CBV in GBM.13 In linkage with water mobility heterogeneity, an inverse correlation is seen between Ki-67 index with ADC across glioma grades9, 30,70,71 (Figure 6). Positive correlations are also seen between metabolite alterations of choline (Cho/Cr), lactate over creatine ratio (Lac/Cr) and MI with Ki-67 index.49, 71,72 Elevated Cho with cell proliferation and malignancy was linked to oncogenic transformation triggered by hypoxia19, 31,49,55,62 while the decrease in Cho levels was related to necrosis. Lac is the product of anaerobic glycolysis while MI is a marker for glial cells.
Other candidate genes as radiogenomic markers
Although less significantly associated, other genes have also been linked as potential radiogenomic markers and are discussed below.
Vascular endothelial growth factor (VEGF) gene, encodes the vascular endothelial growth factor, promotes endothelial proliferation, new blood vessel formation and growth of the new vessels into interstitial tissues.9, 11,38,62 Overexpression of VEGF has been linked to ODG progression7 and associated with contrast-enhancing tumours, hypoxia, angiogenesis, and oedema in GBM.9, 33,73 Areas of non-enhancing tumour in GBM imply decreased vascular permeability corresponded with low VEGF levels.60, 74 Upregulated VEGF is also associated with malignancy and microvascular density75 although no direct approach to quantifiable parameters found.
Platelet-derived growth factor (PDGF) is a growth factor that regulates cellular differentiation and responses to tissue damage.76 PDGF overexpression has been reported for 11% in glioma of all grades76, 77 and indicates enriched oligodendrocytic signature in the proneural subtype of GBM.39, 51 In GBM, PDGF is linked to intratumoural heterogeneity evaluated using histogram and texture analysis by assessing the spread of the grey level values of image voxels and the spatial relationship of the pixels.45,78–81
PTEN (Phosphatase and tensin homolog) regulates cell proliferation, adhesion, invasion, apoptosis and DNA damage repair,7, 36,51 is downregulated in brain tumours. PTEN loss is frequently observed in the frontal lobe of the brain (86.3%), while PTEN deficiency is significantly higher in the left lateral ventricle (42.9%) of GBM patients.35
Cyclin-dependent kinase inhibitor (CDKN2A) codes for a protein that acts as a tumour suppressor by regulating the cell cycle.69 CDKN2A deletions were reported at 42.6% in necrotic tumour of GBM patients.7 The classic subtype of GBM also has a strong association with CDKN2A deletion (92%).51
Proliferating cell nuclear antigen (PCNA) codes a protein that aids leading strand synthesis during DNA replication. Overexpression of this gene has been implicated as an indicator of malignancy and poor prognosis in glioma.39, 49
Another gene of interest is Periostin, where its upregulation is correlated with cellular invasion and oedema in GBM.73 It induces invasion probably through epithelial-mesenchymal transformation, where high expression is observed in the mesenchymal GBM subtype that leads to poor survival. CpG island methylator phenotype (CIMP)-positive is also associated with poor prognosis and treatment response.53
Discussion
This review discusses the recent advances in correlating genomic changes with imaging phenotypes. This may help clinicians to further appreciate the use of genomic information for characterisation of glioma and discrimination of glioma grades in facilitating treatment planning and management. While more work is needed to explore the molecular pathways further so that better correlations can be established, together with validation by other studies, this approach serves as an important and emerging area for an applied clinical use.
Targeted therapy
Tumour molecular heterogeneity not only varies across patients but also throughout a single tumour, indicating broad genetic alterations and adaptation to the microenvironment.15 Genomic heterogeneity can cause treatment resistance and highly heterogeneous tumours have a higher tendency for tumour progression.5 The radiogenomic approach enables identification of genes that are directly involved in cell growth, infiltration, proliferation, differentiation, apoptosis, neurogenesis, and synaptic transmission.39 Activated oncogenic signalling pathway via genetic mutations in EGFR/P13K/Akt and Ras/RAF/MEK pathways are major drivers for tumorigenesis.52 Targeting signalling pathway with tyrosine kinase inhibitors and using bevacizumab as a VEGF inhibitor are the targeted therapies being studied in GBM.53, 82 Inhibition of genes that regulate lipid metabolism to induce cell death makes a promising molecular target in treating malignant glioma.82
This review provides insights into possible radiogenomic markers that could reliably link the imaging features to molecular signatures of the tumours. The imaging features are potentially useful markers as non-invasive molecular surrogates to infer genetic expression profiles of tumour. The restructuring of WHO guideline recognises the importance of incorporating genetic features (i.e., IDH1 status and 1p/19q codeletion status) into histology for classification of the diffuse glioma.2, 3
Current research indicates:
EGFR amplification/overexpression are associated with contrast enhancement in GBM, increase in perfusion metric, metabolite changes, and restricted water diffusion. High-grade gliomas, which are mostly heterogeneous with the presence of solid enhancing rim and cystic portion implies a higher possibility of EGFR amplification.
Hypermethylated MGMT tumours showed mixed-nodular enhancement, non-temporal lobe lesions, and often show radiation or treatment-induced pseudo-progression. Treatment management can be facilitated by assessment of MGMT methylation status of the patient to ensure effective treatment response in concomitant and adjuvant chemoradiotherapy with temozolomide.
Astrocytomas and ODG that harbour IDH1 mutation exhibit more favourable prognosis and response to chemotherapy compared to the wild types. Thus, patients that benefit from chemotherapy could be identified. GBM with IDH1 mutations are larger at diagnosis, may be multifocal with left frontal lobe predominance, may be non-enhancing, have cystic and diffuse components, have a greater frequency of contact with brain ventricles, infrequent vascular abnormalities, less extent of necrosis and oedema.
ODG with 1p/19q loss demonstrated indistinct borders on T1W images, mixed-signal intensities on T1W & T2W, paramagnetic susceptibility effect, calcification, elevated CBV, and infiltrative growth patterns.
Increased proliferation as indicated by elevated Cho/Cr ratio, restricted diffusion and increased lipid correspond with higher Ki-67 index in relation to increased proliferation activities.
Recommendations for future research
Integration of molecular imaging with MRI techniques offers insights into the genetics in glioma. Genetic changes lead to metabolic reprogramming of the biosynthesis of glucose, glutamine, lipids, protein, DNA, and RNA for rapid growth and cell division of the tumour.52 Metabolite characteristics of GBM include enhanced glycolysis, elevated glutaminolysis and exacerbated lipogenesis. Potential research includes inhibiting glucose metabolism as regulated by HK2, PKM2, and IDH; and lipid metabolism as regulated by sterol regulatory element binding protein, acetyl-CoA carboxylase, fatty acid synthase and low-density lipoprotein receptor52 as target for personalised treatment. The linkage between the genetic profile and imaging phenotype to implicate metabolite regulations is another potential radiogenomic study. The presence of lipid in brain tumours has sparked new interest in glioma lipidomics using lipid quantification.34, 82,83 Lipids have roles in necrosis, apoptosis,84 cellular membrane breakdown55 and signal transduction. The elevated lipid fractions quantified using MRS and in- and opposed-phase (IOP) are related to tumour aggressiveness.11, 31,34
Further research in linking tumour characteristics such as metabolite changes, DTI and DKI metrics with molecular signatures could add more values to the understanding of gliomagenesis.11, 24 Quantification of angiogenesis and neovascularisation biomarkers with VEGF expression using PWI (i.e. CBV & permeability maps), arterial spin labelling (i.e. tumour blood flow) and intravoxel incoherent motion (IVIM) (i.e. molecular diffusion coefficient) will be of interest.4,6,9,11,19,24–28,30,35,42,43,49,85 The association of VEGF and inflammatory marker, interleukin-6 (IL-6), is another potential research interest as angiogenesis is also highly related to inflammation.75 Future works in the area of radiogenomics should explore molecular imaging, nanoparticle imaging, computer-aided detection, and targeted therapies. Most of the studies reported the comparison between binary groups (HGG vs LGG, or GBM vs control). Multiple group analysis should be done to compare the glioma grades to provide a better evaluation of the tumour characteristics.34, 86 Variation in imaging acquisition protocol among institutions, tumour sampling, different region of interests, and difficulties in matching the imaging dimension with molecular profiles are the major challenges for integration of imaging and molecular genetic features.
Conclusion
Our review provides insights into possible “personalised” imaging biomarker for precision therapy in glioma based on molecular signatures that provide fundamental information to facilitate decision-making by clinicians in determining treatment and management of tumour that will most likely benefit the patient.
Footnotes
Funding: This work was supported by the Fundamental Research Grant Scheme (FP009-2016). The authors gratefully acknowledge the essential contributions of the research staff of University of Malaya Research Imaging Centre (UMRIC).
Contributor Information
Pohchoo Seow, Email: swpohchoo@gmail.com.
Jeannie Hsiu Ding Wong, Email: jeannie_wong80@um.edu.my.
Azlina Ahmad-Annuar, Email: azlina_aa@um.edu.my.
Abhishek Mahajan, Email: drabhishek.mahajan@yahoo.in.
Nor Aniza Abdullah, Email: noraniza@um.edu.my.
Norlisah Ramli, Email: norlisahramli@gmail.com.
REFERENCES
- 1.Ostrom QT, Gittleman H, de Blank PM, Finlay JL, Gurney JG, McKean-Cowdin R, et al. . American brain tumor association adolescent and young adult primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Neuro Oncol 2016; 18(suppl 1): i1–i50. doi: 10.1093/neuonc/nov297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleihues P, Louis DN, Scheithauer BW, Rorke LB, Reifenberger G, Burger PC, et al. . The WHO classification of tumors of the nervous system. J Neuropathol Exp Neurol 2002; 61: 215–25. doi: 10.1093/jnen/61.3.215 [DOI] [PubMed] [Google Scholar]
- 3.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. . The 2016 World Health Organization classification of tumors of the central Nervous system: a summary. Acta Neuropathol 2016; 131: 803–20. doi: 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 4.Guzmán-De-Villoria JA, Mateos-Pérez JM, Fernández-García P, Castro E, Desco M, et al. . Added value of advanced over conventional magnetic resonance imaging in grading gliomas and other primary brain tumors. Cancer Imaging 2014; 14: 35. doi: 10.1186/s40644-014-0035-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RGPM, Granton P, et al. . Radiomics: Extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012; 48: 441–6. doi: 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Upadhyay N, Waldman AD. Conventional MRI evaluation of gliomas. Br J Radiol 2011; 84: S107–S111. doi: 10.1259/bjr/65711810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutman DA, Cooper LA, Hwang SN, Holder CA, Gao J, Aurora TD, et al. . MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 2013; 267: 560–9. doi: 10.1148/radiol.13120118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicolasjilwan M, Hu Y, Yan C, Meerzaman D, Holder CA, Gutman D, et al. . Addition of MR imaging features and genetic biomarkers strengthens glioblastoma survival prediction in TCGA patients. Journal of Neuroradiology 2015; 42: 212–21. doi: 10.1016/j.neurad.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellingson BM. Radiogenomics and imaging phenotypes in glioblastoma: novel observations and correlation with molecular characteristics. Curr Neurol Neurosci Rep 2015; 15: 506. doi: 10.1007/s11910-014-0506-0 [DOI] [PubMed] [Google Scholar]
- 10.Jackson RJ, Fuller GN, Abi-Said D, Lang FF, Gokaslan ZL, Shi WM, et al. . Limitations of stereotactic biopsy in the initial management of gliomas. Neuro Oncol 2001; 3: 193–200. doi: 10.1093/neuonc/3.3.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Cauter S, De Keyzer F, Sima DM, Croitor Sava A, D'Arco F, Veraart J, et al. . Integrating diffusion kurtosis imaging, dynamic susceptibility-weighted contrast-enhanced MRI, and short echo time chemical shift imaging for grading gliomas. Neuro Oncol 2014; 16: 1010–21. doi: 10.1093/neuonc/not304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buckler AJ, Bresolin L, Dunnick NR, Sullivan DC, For the Group, et al. . A collaborative enterprise for multi-stakeholder participation in the advancement of quantitative imaging. Radiology 2011; 258: 906–14. doi: 10.1148/radiol.10100799 [DOI] [PubMed] [Google Scholar]
- 13.Gupta A, Young RJ, Shah AD, Schweitzer AD, Graber JJ, Shi W, et al. . Pretreatment dynamic susceptibility contrast MRI perfusion in glioblastoma: prediction of EGFR gene amplification. Clin Neuroradiol 2015; 25: 143–50. doi: 10.1007/s00062-014-0289-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pope WB, Kim HJ, Huo J, Alger J, Brown MS, Gjertson D, et al. . Recurrent glioblastoma multiforme: ADC histogram analysis predicts response to bevacizumab treatment. Radiology 2009; 252: 182–9. doi: 10.1148/radiol.2521081534 [DOI] [PubMed] [Google Scholar]
- 15.Rutman AM, Kuo MD. Radiogenomics: Creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol 2009; 70: 232–41. doi: 10.1016/j.ejrad.2009.01.050 [DOI] [PubMed] [Google Scholar]
- 16.Narang S, Lehrer M, Yang D, Lee J, Rao A, et al. . Radiomics in glioblastoma: current status, challenges and potential opportunities. Transl Cancer Res 2016; 5: 383–97. doi: 10.21037/tcr.2016.06.31 [DOI] [Google Scholar]
- 17.Jaffe CC. Imaging and genomics: is there a synergy? Radiology 2012; 264: 329–31. doi: 10.1148/radiol.12120871 [DOI] [PubMed] [Google Scholar]
- 18.Gevaert O, Mitchell LA, Achrol AS, Xu J, Echegaray S, Steinberg GK, et al. . Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology 2014; 273: 168–74. doi: 10.1148/radiol.14131731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glunde K, Pathak AP, Bhujwalla ZM. Molecular–functional imaging of cancer: to image and imagine. Trends Mol Med 2007; 13: 287–97. doi: 10.1016/j.molmed.2007.05.002 [DOI] [PubMed] [Google Scholar]
- 20.Louis DN, Perry A, Burger P, Ellison DW, Reifenberger G, von Deimling A, et al. . International society of neuropathology-haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol 2014; 24: 429–35. doi: 10.1111/bpa.12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delfanti RL, Piccioni DE, Handwerker J, Bahrami N, Krishnan A, Karunamuni R, et al. . Imaging correlates for the 2016 update on WHO classification of grade II/III gliomas: implications for IDH, 1p/19q and ATRX status. J Neurooncol 2017; 135: 601–9. doi: 10.1007/s11060-017-2613-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JPA, et al. . The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med 2009; 6: e1000100. doi: 10.1371/journal.pmed.1000100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutton B, Salanti G, Caldwell DM, Chaimani A, Schmid CH, Cameron C, et al. . The PRISMA extension statement for reporting of systematic reviews incorporating network meta-analyses of health care interventions: checklist and explanations. Ann Intern Med 2015; 162: 777–84. doi: 10.7326/M14-2385 [DOI] [PubMed] [Google Scholar]
- 24.Nakajima S, Okada T, Yamamoto A, Kanagaki M, Fushimi Y, Okada T, et al. . Differentiation between primary central nervous system lymphoma and glioblastoma: a comparative study of parameters derived from dynamic susceptibility contrast-enhanced perfusion-weighted MRI. Clin Radiol 2015; 70: 1393–9. doi: 10.1016/j.crad.2015.08.004 [DOI] [PubMed] [Google Scholar]
- 25.Friedman SN, Bambrough PJ, Kotsarini C, Khandanpour N, Hoggard N, et al. . Semi-automated and automated glioma grading using dynamic susceptibility-weighted contrast-enhanced perfusion MRI relative cerebral blood volume measurements. Br J Radiol 2012; 85: e1204–e1211. doi: 10.1259/bjr/13908936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bauer AH, Erly W, Moser FG, Maya M, Nael K, et al. . Differentiation of solitary brain metastasis from glioblastoma multiforme: a predictive multiparametric approach using combined MR diffusion and perfusion. Neuroradiology 2015; 57: 697–703. doi: 10.1007/s00234-015-1524-6 [DOI] [PubMed] [Google Scholar]
- 27.Jia Z, Geng D, Xie T, Zhang J, Liu Y, et al. . Quantitative analysis of neovascular permeability in glioma by dynamic contrast-enhanced MR imaging. J Clin Neurosci 2012; 19: 820–3. doi: 10.1016/j.jocn.2011.08.030 [DOI] [PubMed] [Google Scholar]
- 28.Macyszyn L, Akbari H, Pisapia JM, Da X, Attiah M, Pigrish V, et al. . Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro Oncol 2016; 18: 417–25. doi: 10.1093/neuonc/nov127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moffat BA, Chenevert TL, Lawrence TS, Meyer CR, Johnson TD, Dong Q, et al. . Functional diffusion map: A noninvasive MRI biomarker for early stratification of clinical brain tumor response. Proc Natl Acad Sci U S A 2005; 102: 5524–9. doi: 10.1073/pnas.0501532102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan R, Haopeng P, Xiaoyuan F, Jinsong W, Jiawen Z, Chengjun Y, et al. . Non-Gaussian diffusion MR imaging of glioma: comparisons of multiple diffusion parameters and correlation with histologic grade and MIB-1 (Ki-67 labeling) index. Neuroradiology 2016; 58: 121–32. doi: 10.1007/s00234-015-1606-5 [DOI] [PubMed] [Google Scholar]
- 31.Bieza A, Krumina G. The value of magnetic resonance spectroscopy and diffusion tensor imaging in characterization of gliomas growth patterns and treatment efficiency. J Biomed Sci Eng 2013; 06: 518–26. doi: 10.4236/jbise.2013.65066 [DOI] [Google Scholar]
- 32.Castellano A, Donativi M, Rudà R, De Nunzio G, Riva M, Iadanza A, et al. . Evaluation of low-grade glioma structural changes after chemotherapy using DTI-based histogram analysis and functional diffusion maps. Eur Radiol 2016; 26: 1263–73. doi: 10.1007/s00330-015-3934-6 [DOI] [PubMed] [Google Scholar]
- 33.Cortez-Conradis D, Favila R, Isaac-Olive K, Martinez-Lopez M, Rios C, Roldan-Valadez E, et al. . Diagnostic performance of regional DTI-derived tensor metrics in glioblastoma multiforme: simultaneous evaluation of p, q, L, Cl, Cp, Cs, RA, RD, AD, mean diffusivity and fractional anisotropy. Eur Radiol 2013; 23: 1112–21. doi: 10.1007/s00330-012-2688-7 [DOI] [PubMed] [Google Scholar]
- 34.Ramli N, Khairy AM, Seow P, et al. . Novel application of chemical shift gradient echo in- and opposed-phase sequences in 3 T MRI for the detection of H-MRS visible lipids and grading of glioma. European Radiology 2015; 26: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellingson BM, Lai A, Harris RJ, Selfridge JM, Yong WH, Das K, et al. . Probabilistic Radiographic Atlas of Glioblastoma Phenotypes. AJNR Am J Neuroradiol 2013; 34: 533–40. doi: 10.3174/ajnr.A3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Fan X, Zhang C, Zhang T, Peng X, Qian T, et al. . Identifying radiographic specificity for phosphatase and tensin homolog and epidermal growth factor receptor changes: a quantitative analysis of glioblastomas. Neuroradiology 2014; 56: 1113–20. doi: 10.1007/s00234-014-1427-y [DOI] [PubMed] [Google Scholar]
- 37.Young RJ, Gupta A, Shah AD, Graber JJ, Schweitzer AD, Prager A, et al. . Potential role of preoperative conventional MRI including diffusion measurements in assessing epidermal growth factor receptor gene amplification status in patients with glioblastoma. AJNR Am J Neuroradiol 2013; 34: 2271–7. doi: 10.3174/ajnr.A3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, et al. . Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A 2008; 105: 5213–8. doi: 10.1073/pnas.0801279105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, et al. . Gene Expression Profiling of Gliomas Strongly Predicts Survival. Cancer Res 2004; 64: 6503–10. doi: 10.1158/0008-5472.CAN-04-0452 [DOI] [PubMed] [Google Scholar]
- 40.Arevalo-Perez J, Thomas AA, Kaley T, Lyo J, Peck KK, Holodny AI, et al. . T1-weighted dynamic contrast-enhanced MRI as a noninvasive biomarker of epidermal growth factor receptor vIII status. American Journal of Neuroradiology 2015; 36: 2256–61. doi: 10.3174/ajnr.A4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kickingereder P, Bonekamp D, Nowosielski M, Kratz A, Sill M, Burth S, et al. . Radiogenomics of glioblastoma: machine learning–based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology 2016; 281: 907–18. doi: 10.1148/radiol.2016161382 [DOI] [PubMed] [Google Scholar]
- 42.Yoo R-E, Choi SH, Cho HR, Kim TM, Lee S-H, Park C-K, et al. . Tumor blood flow from arterial spin labeling perfusion MRI: A key parameter in distinguishing high-grade gliomas from primary cerebral lymphomas, and in predicting genetic biomarkers in high-grade gliomas. Journal of Magnetic Resonance Imaging 2013; 38: 852–60. doi: 10.1002/jmri.24026 [DOI] [PubMed] [Google Scholar]
- 43.Ahn SS, Shin N-Y, Chang JH, Kim SH, Kim EH, Kim DW, Na-Young S, Jong Hee C, et al. . Prediction of methylguanine methyltransferase promoter methylation in glioblastoma using dynamic contrast-enhanced magnetic resonance and diffusion tensor imaging. J Neurosurg 2014; 121: 367–73. doi: 10.3171/2014.5.JNS132279 [DOI] [PubMed] [Google Scholar]
- 44.Yoon RG, Kim HS, Paik W, Shim WH, Kim SJ, Kim JH, et al. . Different diagnostic values of imaging parameters to predict pseudoprogression in glioblastoma subgroups stratified by MGMT promoter methylation. Eur Radiol 2017; 27: 255–66. doi: 10.1007/s00330-016-4346-y [DOI] [PubMed] [Google Scholar]
- 45.Korfiatis P, Kline TL, Coufalova L, Lachance DH, Parney IF, Carter RE, et al. . MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med Phys 2016; 432835–44. doi: 10.1118/1.4948668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, et al. . 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med 2012; 18: 624–9. doi: 10.1038/nm.2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leather T, Jenkinson M, Das K, Poptani H, et al. . Magnetic resonance spectroscopy for detection of 2-hydroxyglutarate as a biomarker for IDH mutation in gliomas. Metabolites 2017; 7: 29. doi: 10.3390/metabo7020029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamashita K, Hiwatashi A, Togao O, Kikuchi K, Hatae R, Yoshimoto K, et al. . MR imaging–based analysis of glioblastoma multiforme: estimation of IDH1 mutation status. AJNR Am J Neuroradiol 2016; 37: 58–65. doi: 10.3174/ajnr.A4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fudaba H, Shimomura T, Abe T, Matsuta H, Momii Y, Sugita K, et al. . Comparison of multiple parameters obtained on 3T pulsed arterial spin-labeling, diffusion tensor imaging, and MRS and the Ki-67 labeling index in evaluating glioma grading. AJNR Am J Neuroradiol 2014; 35: 2091–8. doi: 10.3174/ajnr.A4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.E. Taylor T, B. Furnari F, K. Cavenee W. Targeting EGFR for treatment of glioblastoma: molecular basis to overcome resistance. Curr Cancer Drug Targets 2012; 12: 197–209. doi: 10.2174/156800912799277557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. . An integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010; 17: 98–110. doi: 10.1016/j.ccr.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ru P, Williams T, Chakravarti A, Guo D, et al. . Tumor metabolism of malignant gliomas. Cancers 2013; 5: 1469–84. doi: 10.3390/cancers5041469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Itakura H, Achrol AS, Mitchell LA, Loya JJ, Liu T, Westbroek EM, et al. . Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med 2015; 7: ra138. doi: 10.1126/scitranslmed.aaa7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aghi M, et al. Magnetic resonance imaging characteristics predict epidermal growth factor receptor amplification status in glioblastoma. Clinical Cancer Research 2005; 11: 8600–5. doi: 10.1158/1078-0432.CCR-05-0713 [DOI] [PubMed] [Google Scholar]
- 55.Li X, Lu Y, Pirzkall A, McKnight T, Nelson SJ, et al. . Analysis of the spatial characteristics of metabolic abnormalities in newly diagnosed glioma patients. Journal of Magnetic Resonance Imaging 2002; 16: 229–37. doi: 10.1002/jmri.10147 [DOI] [PubMed] [Google Scholar]
- 56.Park J-H, Lee H, Makaryus R, Yu M, Smith SD, Sayed K, et al. . Metabolic profiling of dividing cells in live rodent brain by proton magnetic resonance spectroscopy (1HMRS) and LCModel analysis. PLoS One 2014; 9: e94755. doi: 10.1371/journal.pone.0094755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, Jin H, Lu Y, Oh J, Chang S, Nelson SJ, et al. . Identification of MRI and1H MRSI parameters that may predict survival for patients with malignant gliomas. NMR Biomed 2004; 17: 10–20. doi: 10.1002/nbm.858 [DOI] [PubMed] [Google Scholar]
- 58.Lukas L, Devos A, Suykens JAK, Vanhamme L, Howe FA, Majós C, et al. . Brain tumor classification based on long echo proton MRS signals. Artif Intell Med 2004; 31: 73–89. doi: 10.1016/j.artmed.2004.01.001 [DOI] [PubMed] [Google Scholar]
- 59.Wang Y, Fan X, Zhang C, Zhang T, Peng X, Li S, et al. . Anatomical specificity of O6-methylguanine DNA methyltransferase protein expression in glioblastomas. J Neurooncol 2014; 120: 331–7. doi: 10.1007/s11060-014-1555-6 [DOI] [PubMed] [Google Scholar]
- 60.Carrillo JA, Lai A, Nghiemphu PL, Kim HJ, Phillips HS, Kharbanda S, et al. . Relationship between Tumor Enhancement, Edema, IDH1 Mutational Status, MGMT Promoter Methylation, and Survival in Glioblastoma. American Journal of Neuroradiology 2012; 33: 1349–55. doi: 10.3174/ajnr.A2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, Zhang T, Li S, Fan X, Ma J, Wang L, et al. . Anatomical localization of isocitrate dehydrogenase 1 mutation: a voxel-based radiographic study of 146 low-grade gliomas. Eur J Neurol 2015; 22: 348–54. doi: 10.1111/ene.12578 [DOI] [PubMed] [Google Scholar]
- 62.Mahajan A, Goh V, Basu S, Vaish R, Weeks AJ, Thakur MH, et al. . Bench to bedside molecular functional imaging in translational cancer medicine: to image or to imagine? Clin Radiol 2015; 70: 1060–82. doi: 10.1016/j.crad.2015.06.082 [DOI] [PubMed] [Google Scholar]
- 63.Mahajan A, Moiyadi AV, Jalali R, Sridhar E, et al. . Radiogenomics of glioblastoma: a window into its imaging and molecular variability. Cancer Imaging 2015; 151. doi: 10.1186/1470-7330-15-S1-P1425888983 [DOI] [Google Scholar]
- 64.Lai A, Kharbanda S, Pope WB, Tran A, Solis OE, Peale F, et al. . Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. Journal of Clinical Oncology 2011; 29: 4482–90. doi: 10.1200/JCO.2010.33.8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Megyesi JF, et al. Imaging correlates of molecular signatures in oligodendrogliomas. Clinical Cancer Research 2004; 10: 4303–6. doi: 10.1158/1078-0432.CCR-04-0209 [DOI] [PubMed] [Google Scholar]
- 66.Khayal IS, VandenBerg SR, Smith KJ, Cloyd CP, Chang SM, Cha S, et al. . MRI apparent diffusion coefficient reflects histopathologic subtype, axonal disruption, and tumor fraction in diffuse-type grade II gliomas. Neuro Oncol 2011; 13: 1192–201. doi: 10.1093/neuonc/nor122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Law M, Brodsky JE, Babb J, Rosenblum M, Miller DC, Zagzag D, et al. . High cerebral blood volume in human gliomas predicts deletion of chromosome 1p: Preliminary results of molecular studies in gliomas with elevated perfusion. J Magn Reson Imaging 2007; 25: 1113–9. doi: 10.1002/jmri.20920 [DOI] [PubMed] [Google Scholar]
- 68.Brown R, Zlatescu M, Sijben A, Roldan G, Easaw J, Forsyth P, et al. . The use of magnetic resonance imaging to noninvasively detect genetic signatures in oligodendroglioma. Clin Cancer Res 2008; 14: 2357–62. doi: 10.1158/1078-0432.CCR-07-1964 [DOI] [PubMed] [Google Scholar]
- 69. CDKN2A gene (cyclin dependent kinase inhibitor 2A): US National Library of Medicine (NIH).cited 2017 15 Nov.
- 70.Calvar JA, Meli FJ, Romero C, Yánez MLCP, Martinez AR, Lambre H, et al. . Characterization of brain tumors by MRS, DWI and Ki-67 labeling index. J Neurooncol 2005; 72: 273–80. doi: 10.1007/s11060-004-3342-2 [DOI] [PubMed] [Google Scholar]
- 71.Khayal IS, Crawford FW, Saraswathy S, Lamborn KR, Chang SM, Cha S, et al. . Relationship between choline and apparent diffusion coefficient in patients with gliomas. Journal of Magnetic Resonance Imaging 2008; 27: 718–25. doi: 10.1002/jmri.21288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Demerath T, Simon-Gabriel CP, Kellner E, Schwarzwald R, Lange T, Heiland DH, et al. . Mesoscopic imaging of glioblastomas: are diffusion, perfusion and spectroscopic measures influenced by the radiogenetic phenotype? Neuroradiol J 2017; 30: 36–47. doi: 10.1177/1971400916678225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zinn PO, Majadan B, Sathyan P, Singh SK, Majumder S, Jolesz FA, et al. . Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS One 2011; 6: e25451. doi: 10.1371/journal.pone.0025451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y, Wang K, Li H, Wang J, Wang L, Dai J, et al. . Identifying the association of contrast enhancement with vascular endothelia growth factor expression in anaplastic gliomas: a volumetric magnetic resonance imaging analysis. PLoS One 2015; 10: e0121380. doi: 10.1371/journal.pone.0121380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reynés G, Vila V, Martín M, Parada A, Fleitas T, Reganon E, et al. . Circulating markers of angiogenesis, inflammation, and coagulation in patients with glioblastoma. J Neurooncol 2011; 102: 35–41. doi: 10.1007/s11060-010-0290-x [DOI] [PubMed] [Google Scholar]
- 76.Nazarenko I, Hede S-M, He X, Hedrén A, Thompson J, Lindström MS, et al. . PDGF and PDGF receptors in glioma. Ups J Med Sci 2012; 117: 99–112. doi: 10.3109/03009734.2012.665097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fleming TP, Saxena A, Clark WC, et al. . Amplification and/or overexpression of platelet-derived growth factor receptors and rpidermal growth factor receptor in human glial tumors. Cancer Research 1992; 52: 4550. [PubMed] [Google Scholar]
- 78.Alic L, Niessen WJ, Veenland JF. Quantification of heterogeneity as a biomarker in tumor imaging: a systematic review. PLoS One 2014; 9: e110300. doi: 10.1371/journal.pone.0110300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Herlidou-Même S, Constans JM, Carsin B, Olivie D, Eliat PA, Nadal-Desbarats L, et al. . MRI texture analysis on texture test objects, normal brain and intracranial tumors. Magn Reson Imaging 2003; 21: 989–93. doi: 10.1016/S0730-725X(03)00212-1 [DOI] [PubMed] [Google Scholar]
- 80.Zacharaki EI, Wang S, Chawla S, Soo Yoo D, Wolf R, Melhem ER, et al. . Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 2009; 62: 1609–18. doi: 10.1002/mrm.22147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kickingereder P, Burth S, Wick A, Götz M, Eidel O, Schlemmer H-P, et al. . Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology 2016; 280: 880–9. doi: 10.1148/radiol.2016160845 [DOI] [PubMed] [Google Scholar]
- 82.Guo D, Bell EH, Chakravarti A. Lipid metabolism emerges as a promising target for malignant glioma therapy. CNS Oncol 2013; 2: 289–99. doi: 10.2217/cns.13.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lim CJ, Ng KH, Ramli N, Azman RR, et al. . Evaluation of the application of chemical shift for the detection of lipid in brain lesion. Radiography 2011; 17: 43–8. doi: 10.1016/j.radi.2010.10.003 [DOI] [Google Scholar]
- 84.Fan G. Magnetic resonance spectroscopy and gliomas. Cancer Imaging 2006; 6: 113–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jain R, Poisson L, Narang J, Gutman D, Scarpace L, Hwang SN, et al. . Genomic mapping and survival prediction in glioblastoma: molecular subclassification strengthened by hemodynamic imaging biomarkers. Radiology 2013; 267: 212–20. doi: 10.1148/radiol.12120846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jingqin L, Chengjie X. DiagTest3Grp: an R package for analyzing diagnostic tests with three ordinal groups. J Stat Softw 2012; 51: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]


