Abstract
Computer simulations are widely used to study molecular systems, especially in biology. As simulations have greatly increased in scale reaching cellular levels there are now significant challenges in managing, analyzing, and interpreting such data in comparison with experiments that are being discussed. Management challenges revolve around storing and sharing terabyte to petabyte scale data sets whereas the analysis of simulations of highly complex systems will increasingly require automated machine learning and artificial intelligence approaches. The comparison between simulations and experiments is furthermore complicated not just by the complexity of the data but also by difficulties in interpreting experiments for highly heterogeneous systems. As an example, the interpretation of NMR relaxation measurements and comparison with simulations for highly crowded systems is discussed.
1. Introduction
Computer simulations have become a central element in modern science as a bridge between experiments and theory [1]. Simulations are commonly applied to describe the evolution of complex systems in time and space based on theoretical models but in regimes where direct analytical or even numerical solutions are not feasible. The level of realism that is achieved by such simulations depends on the nature of the underlying models and may range from idealized conceptual views to physically highly 1accurate descriptions of the systems that are being studied. The most sophisticated simulations can, at least in principle, rival experiments and provide complete spatio-temporal information although the availability of computer resources limits the scales that can be accessed.
A key advantage of simulations is that essentially any question can be interrogated irrespective of practical limitations that may hinder experiments, including hypothetical scenarios that could not even be realized experimentally under any circumstances; however, computer simulations remain a fundamentally theoretical approach that is most valuable when new hypotheses or predictions are generated that can subsequently be subjected to experimental validation. A common strategy is thus an iterative approach where simulations and experiments rely on each other in the development of new mechanistic insights. While computer simulations are widely used today in all fields of science, they are especially valuable in biology where a high degree of system complexity challenges both experiments and theory [2, 3]. In fact, molecular dynamics simulations of biological macromolecules such as proteins and nucleic acids have become a staple in modern biological science having contributed much to our detailed mechanistic understanding of biomolecular processes [4]. Very recently, such simulations have been extended to cellular scales and simulations of entire cells in molecular detail will soon become reality [5].
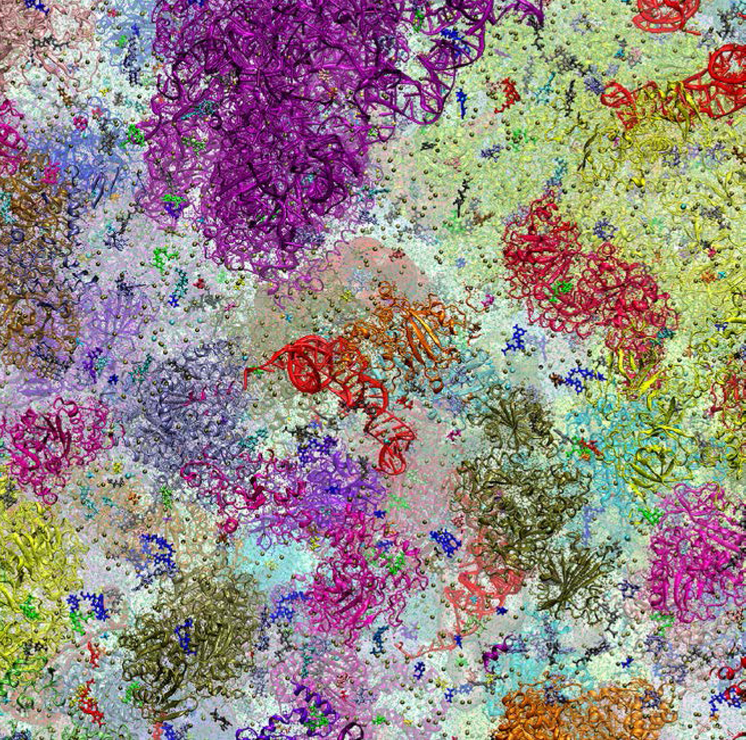
The main results of computer simulations are three-dimensional coordinate trajectories over time for a given system. Molecular systems are often described at an atomistic level of detail although higher (quantum-mechanical) or lower (coarse-grained) resolutions are possible. In addition to the system of immediate interest, the environment often also needs to be considered. For many molecular systems, especially in biology, this involves aqueous solvent so that water, ions, and co-solvents are part of the system, typically also in atomistic detail; therefore, system sizes between 50,000 and 1 M atoms are common nowadays for simulations of single macromolecules or macromolecular complexes [6], but much larger systems with as many as 100 M atoms have been reported for studies of many interacting molecules for the cytoplasm of a bacterial cell [7] (see figure 1). The time scales covered by such simulations are now routinely reaching 1 µs and in exceptional cases as much as 1 ms [8]. Depending on how often coordinates are saved, this means that a single simulation may generate data on terabyte to petabyte scales. The large amounts of data coupled with the high degree of complexity in many systems presents formidable data management and analysis challenges and it is also becoming increasingly difficult to compare with experiments. A further discussion of these challenges is the topic of this article.
Figure 1.
Model of bacterial cytoplasm in atomistic detail.
2. Data size challenges
The large amount of data generated by computer simulations intrinsically presents big data challenges. At the logistical level, the storage, management, and dissemination of terabyte-scale trajectory data is still not trivial even as the performance and capacity of storage resources continues to increase [9]. While it has become common at least in most areas of biological science that primary data is made publicly available when research findings are published, this is generally not the case for simulation studies [10]; furthermore, while there are public databases for essentially every kind of biological data generated by experiments or from computational analysis, there is no widely used resource for molecular dynamics simulations [11] despite many efforts to develop such databases [10, 12–16]. The main reason is that a database where the original trajectories are collected and simply made available for download is not practical because of network bandwidth limitations. This is especially true for the large cellular-scale simulations that are beginning to emerge, as it will likely remain impossible in the foreseeable future to efficiently transfer petabyte-scale data sets over the Internet.
One way to overcome such limitations is to reduce the data. This can be done by using only snapshots at infrequent time points and/or by removing less important parts of the system such as solvent. It is also possible to store the system of interest at a coarse-grained level even though the original simulations were carried out in atomistic detail with the idea that an atomistic level of resolution could be reconstructed on the fly if needed [17]. A different strategy is to maintain the full data sets but develop tools that allow remote analysis so that only the results of such analysis have to be transmitted instead of the actual data [10, 12]; however, this requires significant software and hardware infrastructure and may limit the flexibility in terms of what analysis can be carried out. The same challenges also apply even within a computational laboratory when there is not enough storage space to maintain all of the generated simulations available for direct access so that re-analysis and comparative studies of large sets of simulations become difficult tasks although it would be prudent to do so given the high computational costs to generate the data in the first place.
3. Information wealth challenges
A different challenge is how to fully capitalize on the wealth of information that is provided by simulations where every molecule is represented in atomistic detail. Early molecular dynamics simulations of biological macromolecules, where a single small protein or piece of DNA was studied over pico-to nanosecond time scales, could often be understood qualitatively by inspecting molecular movies generated from the trajectories but nowadays this is rarely a productive approach for analyzing simulation trajectories. In fact, in the largest cellular-scale simulations published recently that contain thousands of proteins [7, 18] it would take days just to look at every molecule for a minute each. Simulations are often carried out with a specific scientific question in mind and one way to navigate the large amount of information is to only focus on that question during the analysis in a strictly hypothesis-driven fashion; however, large-scale simulations of complex biological systems allow scientific discovery beyond the motivating question(s) that led to the simulations to be carried out initially.
One approach to attempt such discovery in a system where one cannot simply ‘look’ at what is happening is to carry out a battery of standard analyses to characterize structural and dynamic features followed by automated feature analysis to identify, for example, if the secondary structure in a given protein is lost compared to an experimental reference structure. It is more difficult, however, to recognize causal relationships whereby the loss of secondary structure in this example may be related to interactions with other molecules or locally altered solvent properties. Not knowing what to focus on a priori and being faced with too many possibilities when considering first correlations and then causal connections between different components in a system with as many as 100 M atoms presents a classical data science challenge. There is an ideal opportunity for the application of machine learning and artificial intelligence techniques to interpret the increasingly rich information that is being generated nowadays via simulation. While machine learning has seen some applications in the analysis of molecular dynamics data [19, 20], the unsupervised analysis of complex biomolecular simulations remains a key challenge that needs to be addressed. Molecular dynamics simulations of entire cells with billions of atoms and tens of thousands of macromolecules are on the horizon and the traditional manual analyses will become entirely inadequate for such data sets.
4. Connections between simulation and experiment
As computer simulations are based on theoretical models, the connection with experiments is crucial. At the onset, experiments are needed to define the composition of a given system and resolve molecular structures to provide initial coordinates from which simulations can be started. Experimental findings also usually generate the initial motivation to carry out simulations with the goal of developing a deeper understanding based on details extracted from the simulations that are not accessible experimentally. Once simulations have been carried out, comparisons with experiment are needed for validation and to follow up on predictions to further advance knowledge. As important as the connection between simulations and experiments is, there are many challenges: experimental conditions and scales are often quite different from what is being simulated and experiments cannot typically provide full atomistic resolution and picosecond time resolution at the same time as in the simulations.
Atomistic resolution in experiments is often obtained via extensive time-and ensemble averaging, as in X-ray crystallography, nuclear magnetic resonance spectroscopy, or cryo-electron microscopy. Simulations often rely on the ergodic hypothesis that states that the long-time average of a single system is equivalent to the ensemble average of the same system [21]; however, since the accessible time scales in the simulations are still relatively short, a single simulation of a single system rarely provides fully-converged conformational space averages. The situation can be improved by running multiple replicates of a given system, or by employing enhanced sampling strategies to accelerate the exploration of the conformational energy landscape with given computational resources [22–24]. Simulations of multiple components such as in simulations of cellular environments, on the other hand, do allow ensemble averaging if there are multiple copies of the same molecule present [7]. Because different copies in such systems experience different local environments and sample different regions of phase space, extensive conformational averaging may be achieved with such simulations even if the overall simulation lengths for such large systems are much more limited than for smaller, single-molecule systems.
Different copies of the same molecule experiencing different environments may also increase the complexity of the conformational energy landscape when interactions with the environment modulate biomolecular structure. There is increasing evidence that non-specific protein-protein interactions modulate biomolecular structure [25], but simulations suggest that such effects may be limited to only a small subset of molecules [7, 26]; for example, in a simulation of a bacterial cytoplasm, a few copies of the molecule pyruvate dehydrogenase, subunit A, were seen to unfold due to protein-protein interactions, whereas the majority of copies remained stably folded in their native state [7]. While such rare events can be easily discerned in simulations, experiments that rely on averages often cannot detect events that occur for only a small percentage of molecules making it difficult to compare simulations and experiments in such cases.
The comparison of kinetic and diffusive properties between simulation and experiments requires that both approaches cover the same time scales. Simulations are so far limited to millisecond time scales for single molecules and to the microsecond range for the largest cellular systems. Experiments may describe dynamics from femtoseconds to hours or longer depending on the method that is being used. NMR experiments are especially suitable to cover a wide range of time scales [27, 28]. NMR data is often compared with molecular dynamics simulations because processes occurring on different time scales can be isolated and matched to the simulation time scales [29–32]; however, the standard interpretation of NMR data usually depends on certain assumptions that can become problematic in highly crowded cellular systems. Using dynamic relaxation of protein backbone amide N-H vectors as an example (see figure 2), the resulting dynamics is a result of combining internal dynamics with rotational and translational diffusion. All three processes can be easily separated in the analysis of simulation data. NMR on the other hand measures the overall relaxation rates that combine fluctuations due to internal motions with rotational tumbling. In interpreting the experimental data, the usual assumption is that both the solvent environment and the rotational tumbling are isotropic; furthermore, a separation of time scales between internal and rotational motions is assumed. It is then possible to determine the extent of internal motions on short and long time scales as well as rotational diffusion times via so-called model-free analysis [33]; however, none of these assumptions may be true in highly crowded heterogeneous cellular systems because of anisotropy and coupling of internal and diffusional motions. Although one can formally carry through with the standard analysis of the experimental data, the results are likely going to become problematic in such a scenario. A better strategy would be the direct calculation of relaxation times from the simulation as it avoids the assumptions made in the experimental analysis [34]; however, this is also not without challenges, the simulations would then be required to not just reach sufficiently long time scales for convergence but also they also have to accurately capture the entire dynamic spectrum as a result of different processes, each with their respective time scales estimated correctly. Because of methodological limitations this is often not the case; for example, while the time scale of internal protein motions are perhaps accurate with current force fields, the diffusional motions that depend on the water model used in the simulations would be accelerated if the popular TIP3P model is used [35]. While a separate analysis of diffusion and internal dynamics could correct such artifacts, this is not easily done when calculating spectra for the overall dynamics. While this example focuses on NMR data, similar arguments can be made for comparisons with single-molecule fluorescence data, electron paramagnetic (EPR) spectroscopy, or other types of spectroscopic measurements.
Figure 2.
Dynamics of N-H vectors: internal motion (blue), rotational diffusion (green), translational diffusion (purple)
Beyond structural and dynamic information, simulations also allow the estimation of various energetic terms; for example, it is possible to calculate free energies of crowding, the free energy for transferring a given molecule from dilute solvent to a crowded environment, or interaction free energies within cellular environments [26, 36]. There are so far no good examples for experimental measurements of such quantities although this may not be impossible with innovative experimental setups.
Finally, it goes almost without saying that meaningful comparisons between experiments and simulations require that the same systems are being studied in silico and in vitro or in vivo. For traditional studies of a single molecule under dilute conditions that is relatively easy to accomplish because the goal of both experiments and simulations would be to study pure systems with a minimum of contaminants; however, cellular-scale biological systems are highly complex and while the simulated systems are well-defined, corresponding experimental systems are less controlled which makes it difficult to follow up on predictions made in simulation studies for such systems. For example, it would be next to impossible to carry out experiments on the same exact model cytoplasm that is shown in figure 1. Even although it may be possible to match the same exact initial molecular composition, active metabolism, protein and nucleic acid synthesis and degradation, and molecular diffusion and osmotic effects would likely lead to significant fluctuations in concentrations and molecular composition. On the other hand, simulations of complete cells with all of the biological function intact are not going to be feasible in the foreseeable future, therefore, matching simulated and experimentally studied cellular systems is a major challenge that will remain difficult to overcome even as the scale and complexity further increase.
5. Conclusions
The extension of computer simulations to cellular-scale system is exciting but also presents significant challenges for fully taking advantage of the vast data generated in such efforts. Efficient management and mechanisms of public sharing of the resulting large data sets is the first issue that needs to be addressed but the bigger issues are how to analyze and interpret such data sets effectively. As traditional approaches to the analysis of simulations do not scale well to systems with thousands of macromolecules, a greater emphasis on machine learning and artificial intelligence will be required in the future. The comparison with experimental data, a vital component in any simulation study, is furthermore complicated by a variety of factors, even though time and spatial scales are increasingly overlapping between simulations and experiments. A major issue is the complexity of cellular-scale systems where the interpretation of experimental data becomes more difficult and the increasing importance of rare events that can be observed in simulations but are difficult to see experimentally. There are also challenges with exactly matching systems between simulations and experiments, which will make it increasingly difficult to follow-up experimentally on predictions made by simulations of cellular environments. Understanding and addressing these challenges will be essential in maintaining the synergy between simulations and experiments in studying biological systems at increasingly larger scales.
Acknowledgments
Funding was provided by the National Institute of Health Grants R01 GM084953 and GM103695 (to MF), from the National Science Foundation Grant MCB 1330560 (to MF), the Fund from the High Performance Computing Infrastructure (HPCI) Strategic Program (hp120309, hp130003, hp140229, hp150233) and HPCI general trial use project (hp150145, hp160120) and FLAGSHIP 2020 project focused area 1 “Innovative drug discovery infrastructure through functional control of biomolecular systems” (hp160207, hp170254) of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (to YS), a Grant-in-Aid for Scientific Research on Innovative Areas “Novel measurement techniques for visualizing ‘live’ protein molecules at work” (No. 26119006) (to YS), a grant from JST CREST on “Structural Life Science and Advanced Core Technologies for Innovative Life Science Research” (to YS), RIKEN QBiC, iTHES, and Dynamic Structural Biology (to YS), a Grant-in-Aid for Scientific Research (C) from MEXT (No. 25410025) (to IY).
6. References
- [1].van Gunsteren WF, Berendsen HJC. 1990. Angew. Chemie 29 992–1023 [Google Scholar]
- [2].Karplus M, Petsko GA. 1990. Nature 347 631–9 [DOI] [PubMed] [Google Scholar]
- [3].Hospital A, Goñi JR, Orozco M, Gelpi JL. 2015. Adv. Appl. Bioinform. Chem 8 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Karplus M, McCammon JA. 2002. Nat. Struct. Biol 9 646–52 [DOI] [PubMed] [Google Scholar]
- [5].Feig M, Yu I, Wang P-h, Nawrocki G, Sugita Y. 2017. J. Phys. Chem. B [DOI] [PMC free article] [PubMed]
- [6].Perilla JR, Goh BC, Cassidy CK, Liu B, Bernardi RC, Rudack T, Yu H, Wu Z, Schulten K. 2015. Curr. Opin. Struct. Biol 31 64–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yu I, Mori T, Ando T, Harada R, Jung J, Sugita Y, Feig M. 2016. eLife 5 e19274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan YB, et al. 2010. Science 330 341–6 [DOI] [PubMed] [Google Scholar]
- [9].Kumar A, Grupcev V, Berrada M, Fogarty JC, Tu Y-C, Zhu X, Pandit SA, Xia Y. 2014. J. Big Data 2 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hospital A, Andrio P, Cugnasco C, Codo L, Becerra Y, Dans PD, Battistini F, Torres J, Goñi R, Orozco M, et al. 2016. Nucleic Acids Res 44 D272–D8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Thibault JC, Roe DR, Facelli JC, Cheatham TE. 2014. J. Cheminf 6 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Feig M, Abdullah M, Johnsson L, Pettitt BM. 1999. Future Gener. Comput. Syst 16 101–10 [Google Scholar]
- [13].Tai K, Murdock S, Wu B, Ng M H, Johnston S, Fangohr H, Cox SJ, Jeffreys P, Essex JW, P. Sansom MS. 2004. Org. Biomol. Chem 2 3219–21 [DOI] [PubMed] [Google Scholar]
- [14].Meyer T, D’Abramo M, Hospital A, Rueda M, Ferrer-Costa C, Pérez A, Carrillo O, Camps J, Fenollosa C, Repchevsky D, et al. 2010. Structure 18 1399–409 [DOI] [PubMed] [Google Scholar]
- [15].van der Kamp MW, Schaeffer RD, Jonsson AL, Scouras AD, Simms AM, Toofanny RD, Benson NC, Anderson PC, Merkley ED, Rysavy S, et al. 2010. Structure 18 423–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Thibault JC, Facelli JC, Cheatham TE. 2013. J. Chem. Inf. Model 53 726–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cheng Y-M, Gopal SM, Law SM, Feig M. 2012. IEEE Trans. Comput. Biol. Bioinf 6 476–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Feig M, Harada R, Mori T, Yu I, Takahashi K, Sugita Y. 2015. J. Mol. Graph. Modell 58 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Glazer DS, Radmer RJ, Altman RB. 2008. Pac. Symp. Biocomput 332–43 [PMC free article] [PubMed]
- [20].Rydzewski J, Nowak W. 2016. J. Chem. Theory Comput 12 2110–20 [DOI] [PubMed] [Google Scholar]
- [21].Feig M Molecular simulation methods. Computational modeling in lignocellulosic biofuel production Acs symposium series. 1052: American Chemical Society; 2010. p. 155–78. [Google Scholar]
- [22].Paul W, Muller M. 2002. Comput. Phys. Commun 146 113–7 [Google Scholar]
- [23].Okamoto Y 2004. J. Mol. Graph. Modell 22 425–39 [DOI] [PubMed] [Google Scholar]
- [24].Abrams C, Bussi G. 2014. Entropy 16 163–99 [Google Scholar]
- [25].Charlton L M, Barnes C O, Li C, Orans J, Young G B, Pielak G J. 2008. J. Am. Chem. Soc 130 6826–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Harada R, Tochio N, Kigawa T, Sugita Y, Feig M. 2013. J. Am. Chem. Soc 135 3696–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kay LE 1998. Biochem. Cell Biol 76 145–52 [DOI] [PubMed] [Google Scholar]
- [28].Kay LE 2016. J. Mol. Biol 428 323–31 [DOI] [PubMed] [Google Scholar]
- [29].Virk AS, Stait-Gardner T, Willis SA, Torres AM, Price WS. 2015. Front. Phys 3 1 [Google Scholar]
- [30].Graf J, Nguyen PH, Stock G, Schwalbe H. 2007. J. Am. Chem. Soc 129 1179–89 [DOI] [PubMed] [Google Scholar]
- [31].Roll C, Ketterle C, Faibis V, Fazakerley GV, Boulard Y. 1998. Biochemistry 37 4059–70 [DOI] [PubMed] [Google Scholar]
- [32].Chen JH, Brooks CL, Wright PE. 2004. J. Biomol. NMR 29 243–57 [DOI] [PubMed] [Google Scholar]
- [33].Lipari G, Szabo A. 1982. J. Am. Chem. Soc 104 4546–59 [Google Scholar]
- [34].Peter C, Daura X, van Gunsteren WF. 2001. J. Biomol. NMR 20 297–310 [DOI] [PubMed] [Google Scholar]
- [35].Yeh IC, Hummer G. 2004. J. Phys. Chem. B 108 15873–9 [Google Scholar]
- [36].Feig M, Sugita Y. 2012. J. Phys. Chem. B 116 599–605 [DOI] [PMC free article] [PubMed] [Google Scholar]