Abstract
Designing self-assembling RNA ring structures based on known 3D structural elements connected via linker helices is a challenging task due to the immense number of motif combinations, many of which do not lead to ring-closure. We describe an in silico solution to this design problem by combinatorial assembly of RNA 3-way junctions, bulges, and kissing loops, and tabulating the cases that lead to ring formation. The solutions found are made available in the form of a web-accessible Ring Catalog. As an example of a potential use of this resource, we chose a predicted RNA square structure consisting of five RNA strands and demonstrate experimentally that the self-assembly of those five strands leads to the formation of a square-like complex. This is a demonstration of a novel “design by catalog” approach to RNA nano-structure generation. The URL https://rnajunction.ncifcrf.gov/ringdb can be used to access the resource.
Keywords: RNA motifs, 3D structural elements, Sequence design, Nanoconstruct, RNA nanoparticle, Self-assembly
1. Introduction
Self-assembling RNA nanostructures can be designed using the tectonics approach of utilizing known 3D structural motifs, and brought into a new molecular context by connecting them via linker helices [1]. Due to their geometry, modularity, and function, RNA motifs can be employed as building blocks towards designing new structures. Examples of such a building-block approach are the design of the RNA tecto-square [2], the antiprism [3] and the hexameric ring [4]. Moreover, some nanoconstructs were used as scaffolds to create RNA nanoparticles (NP) with different applications by attaching various RNA functional moieties to the scaffolds [5]. Such functional units included fluorescent tags, aptamers, and individual siRNAs targeting particular genes, or multiple siRNAs for combinatorial RNA interference and enhanced synergistic effect [6]. Designed RNA nanoparticles were successfully tested in vitro and in vivo, and proved to silence effectively in cell cultures and in vivo. The functionalized RNA nanoparticles have potential for targeted therapies and other biomedical applications [7–21].
This research highlights the question of what constitutes an “RNA motif”. Frequently, RNA structural elements are called motifs if they are modular and recurrent [22,23]. Note, however, that by connecting structural elements via relatively long linker helices, as is the case for many of the generated ring structures, the structural elements are stabilized, so that the requirement for modularity and re-occurrence is lower. Furthermore, some RNA nanoconstructs, such as the RNA cube [24,25], do not make use of known structural elements, but in a way can be considered as “RNA origami” that employ sequence complementarity for assuring self-assembly and tensegrity for ensuring structural rigidity [24]. It has been noted before, that designing RNA structures with geometries of rings or cages is far from trivial [26]. One can view it as the molecular equivalent of being asked to build a house using strangely shaped bricks containing odd angles. Most combinations of 3D motifs will not lead to closed rings. One variable that the designer has is to introduce connecting linker helices such that ring-closure is improved. The optimal length of such linker helices can be found using a combinatorial search that iterates through all linker helix length combinations. Frequently, even such a combinatorial search for the best linker helix length combinations will, for a given set of 3D motifs, not lead to ring closure. In other words, if one is being tasked to design an RNA ring structure using 3D motifs, it is not immediately obvious which combination of motifs and connecting linker helices will lead to the best ring closure. One approach to the “ring-closure problem” is to utilize idealized motifs that correspond to a closed ring-structure, perform molecular dynamics on motifs, and identify in the simulation trajectory structural candidates of motifs that are similar to the idealized motifs corresponding to ring-closure [27]. The success of that approach demonstrates that the structural flexibility inherent in some RNA motifs may allow for ring closure even if the static motif structures extracted from the experimentally determined atom coordinate data do not appear to correspond to precisely closed rings [27]. In the case of the phi29 DNA packaging RNA, Guo and coworkers demonstrated the use of different motif combinations that lead to the formation of different trimer, tetramer, pentamer, hexamer and heptamer RNA nanoparticles [17,28–31].
Here, we present a solution to this ring-design problem, by performing extensive combinatorial searches for ring structures among motif combinations and tabulating these found ring structures. This tabulation is made web-accessible in the form of the Ring Catalog. We show, using the example of an RNA square, how this catalog can be a useful starting point for identifying RNA scaffolds that form the foundation for designed ring-like RNA nanoconstructs.
NanoTiler is a Java-based multifunctional software capable of creating various such RNA-based nanoconstructs [32]. One of its algorithms (called JunctionScanner) can parse structures from the PDB database, extract RNA structural elements (such as junctions, kissing hairpin loops, internal loops/bulges) and employ them for creating new RNA structures. Applying JunctionScanner to the PDB database has led to the RNAJunction database, which to date comprises more than 13,000 multi-way junctions, internal loops/bulges, and kissing loops [33]. NanoTiler can perform combinatorial searches for ring-like constructs of various sizes employing the RNA motifs established in the RNAJunction Database.
2. Methods and materials
2.1. Generation of the Ring Catalog
2.1.1. Choice of motifs
In order to reduce the computational complexity, as well as the redundancy of the data, a subset of motifs was chosen. Chosen motifs have the property of being “cluster representatives” of the RNAJunction database. A motif cluster in the sense of the RNAJunction database is a set of motifs with the same sequence and NC-IUBMB nomenclature notation [34]. The database also provides a definition of a motif that is chosen to represent a cluster (called cluster representative). In other words, representatives of different clusters cannot have identical sequences and NC-IUBMB nomenclatures. Choosing a set of cluster representatives is thus a way to obtain a non-redundant subset of all motifs. The chosen motifs to generate ring-like structures were 3-way junctions, internal loops/bulges and kissing loops. This choice represents a trade-off of comprehensiveness while avoiding a “combinatorial explosion” of the motif combinations that have to be searched.
2.1.2. Generation of Ring structures
The NanoTiler program suite was used to collect the above-mentioned motifs and to perform a combinatorial search for ring-like structures. Its “growscan” command utilizes all combinations of the loaded motifs and iteratively places them with different lengths of connecting helices into different topologies. The potential ring structures are automatically recognized and written to the file system, provided that a computed quality score is below a defined threshold. The structures in the Ring Catalog were generated with the command growscan --ring-export-limit 15 --loaded 1.
For the case of constructs consisting of two different motifs, the additional option “--loaded 2” was used to ensure that the ring-search is only performed for cases where exactly two motifs have been successfully imported. Note that all combinations of connectivities are examined. In other words, if two motifs are loaded, the system will attempt to find rings utilizing the first motif alone, the second motif alone, and rings consisting of both motifs.
Each ring structure is scored by the quality of the ring closure. The details of the ring-closure score have been described previously. This score is zero for a perfectly closing ring structure and is greater than zero for imperfect ring-closure [32]. The chosen empirical value of 15 as the maximally allowed ring-closure score is a compromise between requiring steric feasibility and allowing for molecular flexibility in order to find viable solutions. The only inputs are RNA 3D motifs extracted from the PDB databank and idealized RNA helix parameters for interpolating linker helices.
2.1.3. Topology classification
The utilized ring-search approach of combinatorially “growing” structures, starting from one or two motifs until ring structures are attained, implies that the structures found can correspond to rings with different sizes and motif connectivities. This can lead to the potential challenge of categorizing the resulting structures in a meaningful way. Classifying structures is a non-trivial classical problem in the field of computational chemistry. We provide a solution to the nucleic acid nanostructure topology classification problem by utilizing existing computational chemistry software for classifying organic molecules. As depicted in Fig. 1, a nucleic acid nanostructure can be converted to a “pseudo-molecule” by identifying the structural motifs as “pseudo-atoms” and the connecting helices as “bonds”. The resulting pseudo-molecules can then be classified using existing computational chemistry approaches. NanoTiler utilizes the “tra” program developed by Faulon and co-workers, which computes molecular descriptors for a given chemical structure of an organic molecule [35–38]. Unlike other molecular descriptor systems described in the literature, these molecular descriptors are not “colliding”, in other words two different molecular structures do not lead to the same descriptor. This approach can thus be used for classifying potentially complicated nucleic acid nanostructures that consist of motifs connected by linker helices.
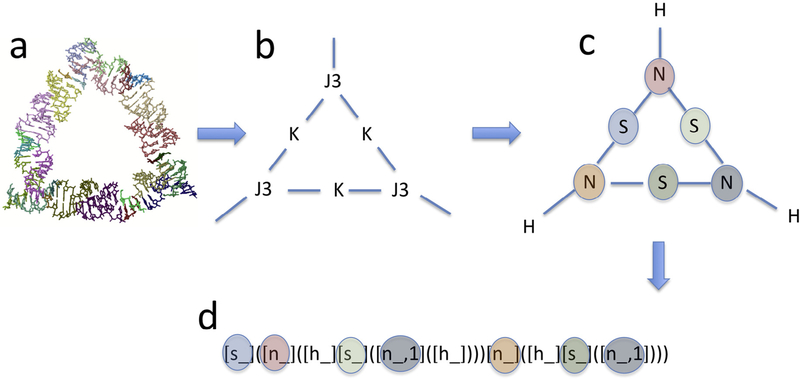
Fig. 1.
Creation of a topology descriptor for a given RNA structure. As an example, a ring structure consisting of three kissing loops connected by three 3-way junctions is shown (a). This structure is converted to a graph (b) with vertices representing motifs and edges representing helical connections between motifs. Next, the motifs are converted to pseudo-atoms such that the valency of the pseudo-atoms is equal to the number of helices emanating from each motif (c). The resulting pseudo-molecule is then converted into a molecular descriptor using computational chemistry software (d). This topology descriptor describes the molecular connectivity as a directed acyclic graph [38]. The parentheses correspond to the recursion inherent in the graph traversal. While the descriptor may not be optimal for human readability, it has the advantage that different topologies will lead to different descriptors and is thus well suited for grouping a set of molecular structures. The corresponding non-hydrogen pseudo-atoms in (c) and (d) are colored alike in order to show one possible way to traverse the connectivity graph in order to arrive at the molecular descriptor. In this example, the descriptor can be interpreted that the pseudo-molecule consists of a (pseudo) S atom (light-blue), that is connected to two N pseudo-atoms (red and orange), which are in turn each connected to H and S pseudo-atoms. The notation [n_,1] in the molecular descriptor (grey) indicates that one pseudo “N” atom (representing a 3-way junction) is referred to twice in the directed acyclic graph, thus indicating a cyclical structure.
2.2. Post-processing of Ring-structures
The structures generated by the combinatorial ring-search algorithms have to be post-processed in order to obtain the RNA sequences able to assemble experimentally. The NanoTiler program suite was used to perform the following steps (also shown schematically in Fig. 2):
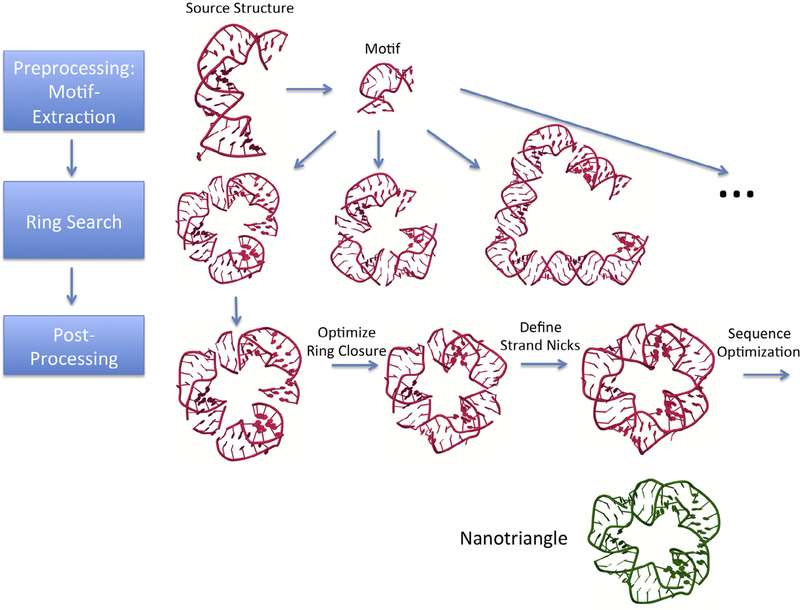
Fig. 2.
Depiction of the processing steps for generating an RNA ring structure. Preprocessing involves extracting from RNA structures those motifs that are intended to be used for the combinatorial search. The Search phase involves iterating through all motifs and motif connectivities, while storing found ring structures. The post-processing phase entails determining toeholds, start sequences, strand nicks and optimized sequences for the found ring structures, followed by a refinement of the 3D model. Depicted is the example of the recently published nanotriangle (shown in green, adapted from PDB structure 5CNR). The nanotriangle structure, as well as the shown motif search example, are based on the internal loop motif extracted from source structure 4PHY. A ring search results in structures connected by linker helices with lengths of 2, 3 and 13 base pairs. The ring structure with 3-base pair linker helix corresponds to the 5CNR published nanotriangle structure. The ring structures initially found by our algorithm contain a structural gap due to imperfect ring closure. In the post-processing stage, the gap is reduced by distributing the helical strain among all linker helices. The strand nicks are redefined as well. Sequence optimization needs to be performed to obtain improved RNA sequences for experimental testing.
Use a combination of fusing and splitting sequence fragments until a desired strand connectivity of the 3D molecular model is attained. The resulting molecular complexes should be connected via kissing loops or toeholds. If the structure assembles via loop-loop interactions, the sequence nicks should be placed preferably in positions that do not disturb the interacting motifs’ tertiary structures. Another consideration is to place nicks such that potential future extensions of strand ends at the nick site do not lead to steric clashes.
Subject the nucleic acid sequences to computational sequence optimization. We utilized the NanoFolder web server for this step [39]. This involves preparing a text description of a target secondary structure. The target secondary structure descriptor should take into consideration that linker helices are subject to the optimization process, while the sequence regions corresponding to the connected structural motifs remain unchanged.
Optionally, the obtained optimized sequences can be used to update the 3D molecular model by “mutating” (“remodeling”) the computational 3D structure accordingly. This 3D model can then be further refined; molecular mechanics minimization was performed with the Tinker 7.1 software and the Amber ff99 force field [40,41].
2.3. Motif used for experimentally verified structure
A square-like RNA ring structure was chosen from the Ring Catalog in order to verify its assembly process experimentally. This structure has been post-processed as described in Section 2.2. This ring structure consists of 4 copies of a 3-way junction motif. The 3-way junction was previously extracted using the JunctionScanner program from the crystal structure of the large ribosomal subunit (PDB ID 2OGM) and deposited in the RNAJunction database with the accession number 12316. The corresponding nucleotides are labeled in the PDB file as 2073–2076, 2179–2183, and 2202–2208.
2.4. Experimental assembly of the RNA nanoconstruct
In order to test that the predicted NanoTiler structures assemble in vitro, a square nanoconstruct was chosen to be investigated experimentally. Due to cost efficiency, the RNA strands were synthesized enzymatically from the corresponding PCR amplified DNA templates. The DNA templates were designed to have an additional 20 nucleotides corresponding to the T7 RNA polymerase promoter region [42]. The ssDNA template, the forward and the reverse primer sequences were automatically generated with the help of the publicly available website (http://rna.bgsu.edu/oldwebsite/rnatodna.html) from the input RNA sequences. The DNA oligos were purchased from IDT and PCR amplified on a T100 Thermal Cycler (Bio-Rad) using MyTaq. The amplified DNA was purified and concentrated with the DNA Clean & Concentrator Kits (Zymo Research). PCRed products were confirmed with 1.5% agarose gels containing Ethidium Bromide. The transcription of RNA strands was performed at 37° for 4 h with T7 RNA polymerase, followed by RNA purification on 8% polyacrylamide (Acrylamide: Bis-Acrylamide 37.5:1), 8 M urea denaturing gels. The RNA recovery from the gel slices was done overnight at 4 °C in 300 mM NaCl, 10 mM Tris, 0.5 mM EDTA (pH 7.5) elution buffer, followed by ethanol precipitation with 100% ethanol and two washes with 90% ethanol. The precipitated RNAs were vacuum-dried and dissolved in Tris-EDTA buffer or double-deionized water. Equimolar amounts of strands were assembled at 45 °C for 30 min, preceded by a 2 min at 95 step to allow any secondary structure to unfold. Assembly of the nanoconstruct was performed in 89 mM Trisborate buffer (pH 8.3) containing 2 mM Mg++ and 50 mM K+, according to protocols described previously [24]. Assembled constructs were analyzed with 8% polyacrylamide (Acrylamide/Bis-acrylamide 37.5:1) non-denaturing gels (native PAGE) containing either 2 mM or 4 mM Mg++. The samples were loaded with a loading buffer containing either 2 mM or 4 mM Mg++, 0.01% Xylene cyanol, 0.01% Bromphenol blue and 50% Glycerol. The formation of assembled constructs was visualized with Ethidium Bromide total-staining on FMBIO II Scanner (Hitachi) (For a schematic of the experimental procedure steps, see Fig. 3).
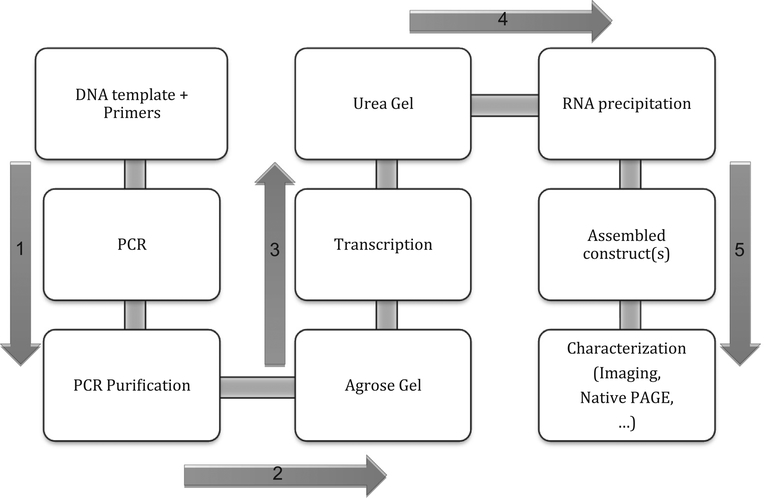
Fig. 3.
Workflow for experimental generation of self-assembling RNA nanostructures. The DNA templates are PCR-amplified and purified before transcription. These steps are validated on 1% agarose gels. Transcription of DNA into RNA strands is performed at 37 °C. Transcribed RNA strands are purified on 8% Acrylamide 8 M Urea gels, the RNAs eluted overnight from the gel bands, and alcohol-precipitated. The square constructs are assembled at 45 °C, according to the protocol described in “Methods and Materials”. Characterization of the nanoconstructs can be performed through various methods, such as Native PAGE or microscopy.
3. Results
3.1. Ring Catalog
The results of the combinatorial search for ring structures among combinations of kissing-loops, bulges and junctions are made available in the form of web-accessible HTML pages, with images and information about each potential ring structure. At the top level, structures are classified by the types of used 3D structural elements. The number of identified ring structures is shown in Table 1. Screenshots of the Ring Catalog can be seen in Fig. 4. It needs to be emphasized again that the provided structural models are not refined and their sequences are not optimized. Instead, the provided structures are intended to be a starting point for further optimization and refinement. The additional steps required to obtain RNA sequences that lead to self-assembling strands are described in the next section.
Table 1.
An overview of the number of identified ring structures. The type of motifs that are used as “input” for the structural search is shown in columns “Type I” and “Type II”. Shown in parenthesis is the number of utilized motifs of a certain type. “Structures”: number of identified ring structures. “Topologies”: number of different topologies.
| Type I | Type II | Structures | Topologies |
|---|---|---|---|
| Internal loop/bulges (1786) | – | 21 | 4 |
| 3-Way junction (129) | – | 4 | 4 |
| Kissing loop (21) | Internal loop/bulges (1786) | 302 | 2 |
| Kissing loop (21) | 3-Way junction (129) | 897 | 4 |
| Kissing loop (21) | Kissing loop (21) | 6 | 2 |
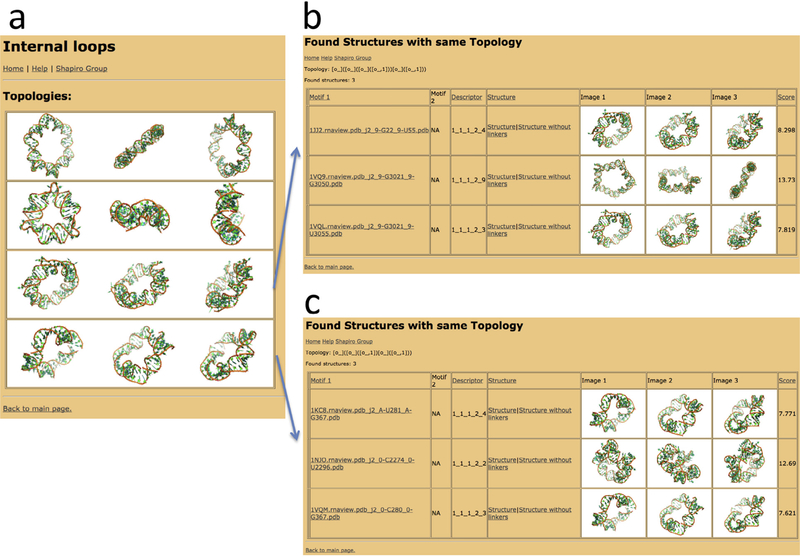
Fig. 4.
Screenshots of the Ring Catalog web resource. (a) Screenshot of the list of topologies found for rings consisting of one type of internal loop or bulge. (b) Screenshot of the list of found ring structures consisting of one type of internal loop/bulge forming a pentagon-like structure. (c) Screenshot of the list of found rings structures consisting of four copies of one type of internal loop/bulge. The tables shown in (b) and (c) provide for each ring structure information about the involved motifs (“Motif 1”, “Motif 2”), the utilized connectivity rule (“Descriptor”), a downloadable 3D structure model (link “Structure” for the unoptimized ring structure and “Structure without linkers” for a structure without linker helices), snapshots of the structures (“Image”), and a ring-closure quality score (“Score”). The “Descriptor” is a five-number encoding of the connectivity rule used for the ring-assembly algorithm: it contains information about the utilized motifs (first two numbers), motif helices (third and fourth number) and linker helix length (5th number).
3.2. Post-processing of Ring structures
The structures provided by the Ring Catalog consist of 3D RNA motifs linked by helices. The structures are, however, not “fused”, nor are their sequences optimized and additional processing of the sequences has to be performed. For the square structure tested experimentally (shown in Fig. 5), the following post-processing steps were performed:
Fig. 5.
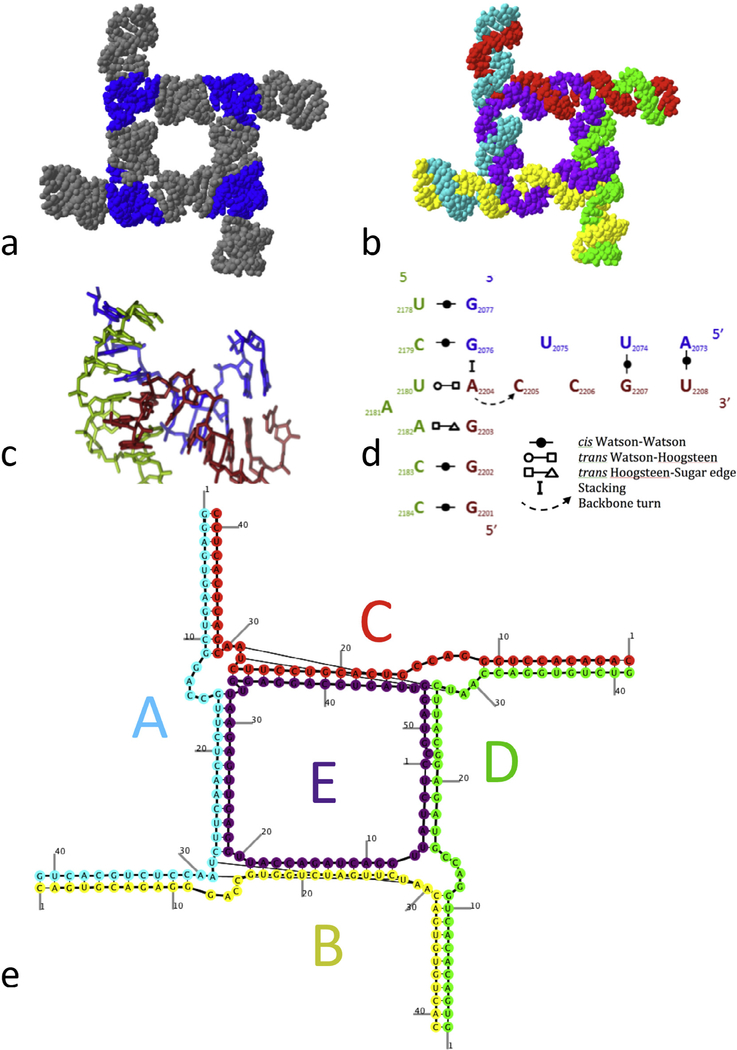
The planar square nanoconstruct. (a) The square colored by motifs (helices, 3-way junctions); Blue-3-way junctions; Grey: helical structural elements connecting the 3-way junctions, acting as struts to hold and position the structural elements. The length of the helix connectors was determined by a combinatorial search for ring-structures. (b) The squared colored by comprising RNA strands. A “side RNA strand” (shown in (b) in blue, red, yellow and green) interacts with 2 adjacent side strands and the “core” strand (shown in purple). The core strand interacts with all the 4 side strands, thus ensuring the consolidation of the complex. (c) 3D structure of the 3-way junction employed to design the square construct. The junction was extracted from a crystal structure of the large ribosomal subunit (PDB file 2OGM). (d) 2D schematic representation of the 3-way junction. The base pairing between nucleotides are depicted with symbols according to the Leontis-Westhof base pairing classification as shown in the legend [53]. (e) Secondary structure diagram (as predicted by the NanoFolder program) of the RNA square with a color-coding that is the same as in Table 2, Fig. 5b and Fig. 6 [39]. Not shown are 50 single-stranded start-sequence motifs (sequence GGGAAA, see Table 2).
Ring-closure of the found ring structures was optimized using the NanoTiler “ringfix” script. The underlying algorithm introduces slight twists and bends to the helices that are connecting the motifs. In other words, helical parameters are modified in order to optimize the ring-closure score;
Sequence fragments corresponding to 3D structural elements and linker helices were computationally concatenated using the NanoTiler “ringfuse” script. The algorithm generates a suggested set of strand nicks corresponding to toeholds lengths of 10 nucleotides [32];
Sequence “GGGAAA” or variants, required for the transcription initiation, were computationally concatenated to the 5′ end [43];
Sequences were optimized using the NanoFolder web server;
The obtained sequences were used to generate a new 3D model via in silico 3D “mutations” applied to the 3D model generated in step 1. Note that the “GGGAAA” sequence motifs at the 50 end of the RNA sequences are designed to be single-stranded and were not included in the 3D model;
The 3D model was further refined using energy minimization via the Tinker software.
The sequences of all RNA strands used for the experimentally tested square construct are provided in Table 2. We also developed a Galaxy front-end for these post-processing steps; however, this will be described in a separate publication.
Table 2.
Sequences of RNA strands for the planar square nanoconstruct. The “side” strands A-D consist of 47 nucleotide while the “core” strand E consists of 58 nucleotides. Each strand has the required “GGGAAA” sequence for transcription initiation concatenated at the 5′ end of the original RNA sequences. The colors of the shown sequences correspond to the colors of the molecular models shown in Fig. 5b and e.
| RNA | Sequence |
|---|---|
| A | GGGAAAGGAGUGAGUCGGACCGUUCUCAACUUCUAACCUCUGCACUG |
| B | GGGAAACAGUGCAGAGGGACCGUGGUCUAGUUCUAACAGUGUGUCAC |
| C | GGGAAACAGACACCUGGGACCGUCACGUCCUUCUAACGACUCACUCC |
| D | GGGAAAGUGACACACUGGACCGUAGAGGCAUUCUAACCAGGUGUCUG |
| E | GGGAAACUCUAUUGGACUAGACCAUUGGAGUUGAGAAUUGGAGGACGUGAUUGGAUGC |
3.3. Results of experimental assemblies
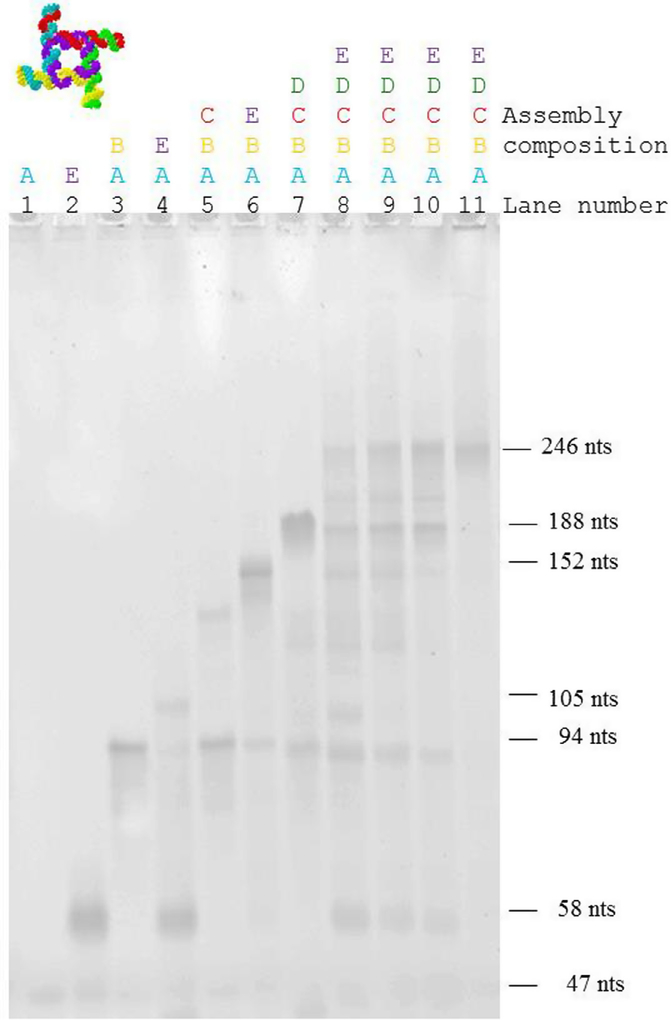
The strands required for the assembly of the square construct were produced enzymatically, assembled in Mg-containing Tris-Borate buffer, and visualized with native-PAGE (shown in Fig. 6). Note that the buffer Mg++ concentration is 2 mM for lanes 1–10 and 4 mM for lane 11. Increasing size complexes were observed as the number of strands added to the assembly was increased. Native-PAGE lanes 1 and 2 represent the band for a single strand, strand A (47 nucleotides), and strand E (58 nucleotides) respectively. Their electrophoretic mobilities reflect the difference in their sizes. Lanes 3–4 illustrate the formation of the dimer (strands A and B and strands A and E), trimer (strands A, B, and C, and strands A, B, and E in lanes 5–6) and tetramer (strands A, B, C, and D, lane 7). The formation of the pentamer, and thus of the entire square complex, is shown in lanes 8–11, for which all strands (A through E) were assembled in a single reaction (lane 8: 37 °C, lane 9: 45 °C, lane 10: 55 °C (2 mM Mg), lane 11: 55 °C (4 mM Mg)). At 55 °C and 4 mM Mg concentration (lane 11), the square assemble cooperatively, and no residual monomer, dimer, trimer or tetramer is observed.
Fig. 6.
Formation of molecular assemblies of the square nanoconstruct. Monomers (lane 1: strand A, 47 nts; lane 2: strand E, 58 nucleotides), dimers (lane 3: strands A + B, 94 nts; lane 4: strands A + E, 105 nts), trimers (lane 5: strands A + B + C; lane 6: strands A + B + E), tetramer (lane 7: strands A + B + C + D) and pentamers (strands: A + B + C + D + E, lanes 8–11). As expected, the dimer A + E (corresponding to 105 nts) displays a slower electrophoretic mobility compared with the dimer A + B (corresponding to 94 nucleotides). Only fractions of strands A and E assemble in a trimer, and the prominent band for the monomer E is observed for this assembly. The trimers formed by strands A, B, and C, and A, B, and E assemble more readily. This can be explained by the difference in the interaction strength between two strands in the dimer, and three strands in the trimer. The tetramer (strands A, B, C, and D,) seems to be thermodynamically less stable, and bands corresponding to the monomer, dimer, trimer and tetramer can be observed for the assembly. Lanes 1–10 correspond to 2 mM Mg++ concentration, while lane 11 corresponds to 4 mM Mg++ concentration. Lanes 8–11 correspond to the pentamer (strands A,B,C,D,E) with different assembly conditions (lane 8: 37 °C, lane 9: 45 °C, lane 10: 55 °C (2 mM Mg), lane 11: 55 °C (4 mM Mg)). The pentamer assembles as expected. At 55 °C and 4 mM Mg concentration, the square assembles without formation of lower molecular weight assemblies. The color-coding corresponds to the molecular models shown in Figs. 5b and e.
4. Discussion
Interestingly, the published self-assembling RNA ring structures are mostly designed synthetic RNA constructs [2,44,45]. Symmetric self-assembling RNA structures do not appear to be very common in nature. Indeed, Cayrol et al. state: “Natural RNAs, unlike many proteins, have never been reported to form extended nanostructures, despite their wide variety of cellular functions.”[46] An example of a natural self-assembling structure where RNA self-assembles in conjunction with a protein is the ToxIN type III system [47]. This is a symmetric trimeric bacterial system, where a potentially toxic protein is inhibited by an RNA antitoxin inhibitor. Another example of a naturally occurring RNA multimer is the hexameric phi29 DNA packaging motor [48].
A particularly interesting case that connects our approach to a published RNA ring structure is the RNA nanotriangle [45]. This RNA nanostructure was obtained by utilizing an internal loop motif that was part of a previously solved crystal structure [49]. By using the same RNA 3D motif as an input for our computational pipeline, a ring structure with 3-base pair long linker helices is generated (corresponding to 11 base pairs per side). The found structure has the same number of base pairs as the published nanotriangle. The involved computational processing steps are shown in Fig. 2. The output ring structure corresponds to the experimentally determined ring structure (shown in green in Fig. 2). Intriguingly, we find yet another ring structure where the linker helix consists of only two base pairs (corresponding to only 10 base pairs per side). This even smaller ring structure corresponds, according to the computer model, to a better ring closure compared to the slightly larger version. The published nanotriangle is not part of the Ring Catalog for two reasons: first, it is based on a motif that is part of a structure released in 2013, after the establishment of the underlying motif data set. Secondly, our default ring search score cutoff is more “restrictive”, thus requiring better ring closure of the initial solution, and less helical strain of the closed ring. It is encouraging that the nanotriangle structure corresponding to a more permissive ring closure was successfully demonstrated to form experimentally, revealing that the Ring Catalog can be expanded to allow for more permissive ring closures.
It has been noticed that to design ring-like structures from RNA motifs, for example a square, is not at all trivial, because many combinations of the chosen motif set do not correspond to closed ring structures [26]. Indeed, the ring design problem can be viewed as a form of the inverse problem: In an inverse problem – in the mathematical sense – the observations are given, but the causal factors that contribute to the observations are unknown and are yet to be determined. Similarly, for the ring-design problem, we may know what we want to design, i.e. a square-like structure, but do not know initially which combination of motifs lead to the target structure. Furthermore, the inverse problem is ill-posed, because it violates all three 3 conditions of a well-posed problem. According to Jacques Hadamard, a well-posed problem should have the properties that (i) a solution exists, (ii) the solution is unique, and (iii) the solution changes in a continuous fashion as a function of the initial conditions [50]. In contrast, for a given set of motifs and linker helices, there are target RNA structures not attainable, in other words no solution exists. As we show in the form of the Ring Catalog, there are many different motif combinations that lead to similar structures with the same topology – in other words condition (ii) of unique solutions of well-posed problems is violated. The choice of motifs and linker helices is also an inherently discrete optimization problem, thus violating condition (iii) of well-posed problems.
A general solution for the inverse “ring design” problem (what motifs are necessary to obtain a target structure) is to reverse the problem into a “forward” approach (“what nanostructures can be obtained given a set of motifs”). Solutions to the “forward” version of the design problem can be tabulated. These tabulated solutions can then be used as a look-up table for solving the more difficult original problem of creating a target 3D structure. Providing a catalog of “motif-feasible” RNA nanostructures in order to facilitate their design is a novel paradigm. The provided Ring Catalog is, to our knowledge, a first instance of such a tabulation of RNA nanostructures. It should be noted that such searches are never fully exhaustive because more motifs can be added, more combinations of motifs can be allowed, and the quality score cut-offs can be made more permissive. Also, the presented workflow is the reverse of the more common paradigm of starting with RNA sequences, then predicting their corresponding secondary and 3D structure. Instead, one starts with a chosen 3D scaffold from the Ring Catalog (which determines its secondary structure) and determines RNA sequences using sequence optimization. Note that the connecting helices contain placeholder sequences, consisting of consecutive Gs or Cs. Such consecutive repetitive nucleotide sequences may lead to transcription or folding ambiguities. In other words, before attempting to assemble the structures, one should apply sequence optimization.
By identifying motifs with “pseudo-atoms” and linker helices with “bonds”, we are able to classify these nanoconstructs in a consistent fashion using established methods of computational chemistry [32,38]. More recently, a similar strategy has been applied to the design of RNA and DNA super-lattices [32,51]. The illustrated example of a ring-like structure generated by NanoTiler and deposited into the Ring Catalog is of a planar square. This construct is composed of five chains which constitute 3-way junctions at the corners (Fig. 5a). This 3-way junction was extracted by JunctionScanner from the crystal structure of the large ribosomal subunit of Deinococcus radiodurans, PDB file 2OGM [52], and then deposited in the RNAJunction database. This junction (Fig. 5c) is composed of three strands: nucleotides 2073–2076, 2179–2183 and 2202–2208, including the flanking Watson-Crick base pairs. The junction gives rise to helical structural elements which form approximately 90° angles between their axes.
The four strands that constitute the sides of the square are 47 nucleotides in length (shown as yellow, blue, red, and green strands in Fig. 5b) and are here referred to as “side strands”. Each side-strand is forming intermolecular interactions with two other side strands. A 58-nucleotide long “core” strand (colored purple in Fig. 5b) ties the structure together by forming base pairs with all four side strands. We tested experimentally the assembly of one, two, three, four and all five RNA strands. Native PAGE of the in vitro assemblies clearly shows the formation of the monomer, dimer, trimer, tetramer and the pentamer – which is the entire construct (See Fig. 6). This demonstrates that the Ring Catalog can be used as a resource to design and characterize a plethora of RNA ring nanostructures. These nanoconstructs can be functionalized to generate RNA Nanoparticles (NPs) with a variety of therapeutic and biomedical applications.
5. Conclusion
In this work, we described a novel tabulation of computer-generated RNA nanoscale ring structures that utilize motifs connected by linker helices. Using the example of an RNA square consisting of 5 strands, we provide experimental results that show that this catalog of ring structures can be a starting point for RNA nanostructure designs and can be the foundation of a design-by-catalog approach.
Acknowledgements
We thank Jennifer Miller and Stuart Le Grice for providing the in-house T7 RNA polymerase. We thank Noah Hoffman for creating secondary structure diagrams. This work has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under Contract No. HHSN261200800001E. This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Abbreviations:
- 3D
three dimensional
- NP
nanoparticle
- PAGE
polyacrylamide gel electrophoresis
- nts
nucleotides
References
- [1].Jaeger L, Westhof E, Leontis NB, TectoRNA: modular assembly units for the construction of RNA nano-objects, Nucleic Acids Res. 29 (2) (2001) 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chworos A et al. , Building programmable jigsaw puzzles with RNA, Science 306 (5704) (2004) 2068–2072. [DOI] [PubMed] [Google Scholar]
- [3].Severcan I et al. , A polyhedron made of tRNAs, Nat. Chem 2 (9) (2010) 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yingling YG, Shapiro BA, Computational design of an RNA hexagonal nanoring and an RNA nanotube, Nano Lett 7 (8) (2007) 2328–2334. [DOI] [PubMed] [Google Scholar]
- [5].Afonin KA et al. , Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine, Nat. Protoc 6 (12) (2011) 2022–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Afonin KA et al. , Multifunctional RNA nanoparticles, Nano Lett 14 (10) (2014) 5662–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dao BN et al. , Triggering RNAi with multifunctional RNA nanoparticles and their delivery, DNA RNA Nanotechnol. 1 (1) (2015) 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Feng L et al. , Ocular delivery of pRNA nanoparticles: distribution and clearance after subconjunctival injection, Pharm. Res 31 (4) (2014) 1046–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Guo P, The emerging field of RNA nanotechnology, Nat. Nanotechnol 5 (12) (2010) 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Khaled A et al. , Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology, Nano Lett 5 (9) (2005) 1797–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Khisamutdinov EF, Jasinski DL, Guo P, RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry, ACS Nano 8 (5) (2014) 4771–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Khisamutdinov EF et al. , Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles, Nucleic Acids Res. 42 (15) (2014) 9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ohno H, Inoue T, Designed regular tetragon-shaped RNA-protein complexes with ribosomal protein L1 for bionanotechnology and synthetic biology , ACS Nano (2015). [DOI] [PubMed] [Google Scholar]
- [14].Ohno H et al. , Synthetic RNA-protein complex shaped like an equilateral triangle, Nat. Nanotechnol 6 (2) (2011) 116–120. [DOI] [PubMed] [Google Scholar]
- [15].Osada E et al. , Engineering RNA-protein complexes with different shapes for imaging and therapeutic applications, ACS Nano 8 (8) (2014) 8130–8140. [DOI] [PubMed] [Google Scholar]
- [16].Rychahou P et al. , Delivery of RNA nanoparticles into colorectal cancer metastases following systemic administration, ACS Nano 9 (2) (2015) 1108–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Shu D et al. , Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics, Nat. Nanotechnol 6 (10) (2011) 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shu Y et al. , Stable RNA nanoparticles as potential new generation drugs for cancer therapy, Adv. Drug Deliv. Rev 66 (2014) 74–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shu Y et al. , Fabrication of pRNA nanoparticles to deliver therapeutic RNAs and bioactive compounds into tumor cells, Nat. Protoc 8 (9) (2013) 1635–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shukla GC et al. , A boost for the emerging field of RNA nanotechnology, ACS Nano 5 (5) (2011) 3405–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Afonin KA et al. , In silico design and enzymatic synthesis of functional RNA nanoparticles, Acc. Chem. Res 47 (6) (2014) 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Leontis NB, Lescoute A, Westhof E, The building blocks and motifs of RNA architecture, Curr. Opin. Struct. Biol 16 (3) (2006) 279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hendrix DK, Brenner SE, Holbrook SR, RNA structural motifs: building blocks of a modular biomolecule, Q. Rev. Biophys 38 (3) (2005) 221–243. [DOI] [PubMed] [Google Scholar]
- [24].Afonin KA et al. , In vitro assembly of cubic RNA-based scaffolds designed in silico, Nat. Nanotechnol. 5 (9) (2010) 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Afonin KA et al. , Computational and experimental characterization of RNA cubic nanoscaffolds, Methods 67 (2) (2014) 256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bida JP, Das R, Squaring theory with practice in RNA design, Curr. Opin. Struct. Biol 22 (4) (2012) 457–466. [DOI] [PubMed] [Google Scholar]
- [27].Kasprzak W et al. , Use of RNA structure flexibility data in nanostructure modeling, Methods 54 (2) (2011) 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Guo P et al. , Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation, Mol. Cell 2 (1) (1998) 149–155. [DOI] [PubMed] [Google Scholar]
- [29].Shu D et al. , Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology, Nano Lett 4 (9) (2004) 1717–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shu Y et al. , Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs, RNA 19 (6) (2013) 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Haque F et al. , Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers, Nano Today 7 (4) (2012) 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bindewald E et al. , Computational strategies for the automated design of RNA nanoscale structures from building blocks using NanoTiler, J. Mol. Graph. Model 27 (3) (2008) 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bindewald E et al. , RNAJunction: a database of RNA junctions and kissing loops for three-dimensional structural analysis and nanodesign, Nucleic Acids Res 36 (Database issue) (2008) D392–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lilley DM, Structures of helical junctions in nucleic acids, Q. Rev. Biophys 33 (2) (2000) 109–159. [DOI] [PubMed] [Google Scholar]
- [35].Churchwell CJ et al. , The signature molecular descriptor. 3. Inverse-quantitative structure-activity relationship of ICAM-1 inhibitory peptides, J. Mol. Graph. Model 22 (4) (2004) 263–273. [DOI] [PubMed] [Google Scholar]
- [36].Faulon JL, Churchwell CJ, Visco DP Jr., The signature molecular descriptor. 2. Enumerating molecules from their extended valence sequences, J. Chem. Inf. Comput. Sci 43 (3) (2003) 721–734. [DOI] [PubMed] [Google Scholar]
- [37].Faulon JL, Collins MJ, Carr RD, The signature molecular descriptor. 4. Canonizing molecules using extended valence sequences, J. Chem. Inf. Comput. Sci. 44 (2) (2004) 427–436. [DOI] [PubMed] [Google Scholar]
- [38].Faulon JL, Visco DP Jr., Pophale RS, The signature molecular descriptor. 1. Using extended valence sequences in QSAR and QSPR studies, J. Chem. Inf. Comput. Sci 43 (3) (2003) 707–720. [DOI] [PubMed] [Google Scholar]
- [39].Bindewald E et al. , Multistrand RNA secondary structure prediction and nanostructure design including pseudoknots, ACS Nano 5 (12) (2011) 9542–9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ren P, Wu C, Ponder JW, Polarizable atomic multipole-based molecular mechanics for organic molecules, J. Chem. Theory Comput 7 (10) (2011) 3143–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Case DA et al. , AMBER 2015, University of California, San Francisco, 2015. [Google Scholar]
- [42].Rosa MD, DNA sequence for the T7 RNA polymerase promoter for T7 RNA species II, J. Mol. Biol 147 (1) (1981) 199–204. [DOI] [PubMed] [Google Scholar]
- [43].Weston BF, Kuzmine I, Martin CT, Positioning of the start site in the initiation of transcription by bacteriophage T7 RNA polymerase, J. Mol. Biol 272 (1) (1997) 21–30. [DOI] [PubMed] [Google Scholar]
- [44].Grabow WW et al. , Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes, Nano Lett. 11 (2) (2011) 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Boerneke MA, Dibrov SM, Hermann T, Crystal-structure-guided design of self-assembling RNA nanotriangles, Angew. Chem. Int. Ed. Engl 55 (12) (2016) 4097–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cayrol B et al. , A nanostructure made of a bacterial noncoding RNA, J. Am. Chem. Soc. 131 (47) (2009) 17270–17276. [DOI] [PubMed] [Google Scholar]
- [47].Short FL et al. , Selectivity and self-assembly in the control of a bacterial toxin by an antitoxic noncoding RNA pseudoknot, Proc. Natl. Acad. Sci. U.S.A 110 (3) (2013) E241–E249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chen C, Zhang C, Guo P, Sequence requirement for hand-in-hand interaction in formation of RNA dimers and hexamers to gear phi29 DNA translocation motor, RNA 5 (6) (1999) 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Boerneke MA et al. , Functional conservation despite structural divergence in ligand-responsive RNA switches, Proc. Natl. Acad. Sci. U.S.A 111 (45) (2014) 15952–15957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hadamard J, Sur les problèmes aux dérivées partielles et leur signification physique, Princeton Univ. Bull (1902) 49–52. [Google Scholar]
- [51].Barnaby SN et al. , Modular and chemically responsive oligonucleotide “bonds” in nanoparticle superlattices, J. Am. Chem. Soc 137 (42) (2015) 13566–13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Davidovich C et al. , Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity, Proc. Natl. Acad. Sci. U.S.A 104 (11) (2007) 4291–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Leontis NB, Westhof E, Geometric nomenclature and classification of RNA base pairs, RNA 7 (4) (2001) 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]