Abstract
Accurate protein structure in the ligand-bound state is a prerequisite for successful structure-based virtual screening (SBVS). Therefore, applications of SBVS against targets for which only an apo structure is available may be severely limited. To address this constraint, we developed a computational strategy to explore the ligand-bound state of target protein, by combined use of molecular dynamics simulation, MM/GBSA binding energy calculation as well as fragment-centric topographical mapping. Our computational strategy is validated against low-molecular weight protein tyrosine phosphatase (LMW-PTP) and then successfully employed in the SBVS against protein tyrosine phosphatase receptor type O (PTPRO), a potential therapeutic target for various diseases. The most potent hit compound GP03 showed IC50 value of 2.89μM for PTPRO and possessed a certain degree of selectivity towards other protein phosphatases. Importantly, we also found that the neglection of ligand energy penalty upon binding partially account for the false positive SBVS hits. Preliminary structure-activity relationship of GP03 analogs is also reported.
Graphical Abstract

1. Introduction
Because of the increasing availability of three-dimensional structures of biological targets, structure-based ligand design is becoming more pervasive in current drug discovery1–3. Specifically, structure-based virtual screening (SBVS), which relies on molecular docking, is widely used in the early-stage of drug discovery to search a compound library for novel bioactive molecules against a certain drug target4–6. Although SBVS has successfully contributed to the discovery of many novel inhibitors, the method faces some limitations in its general applicability for diverse proteins targets. A significant complicating factor in SBVS is protein rearrangement upon ligand binding (induced-fit)7–9. Previous cross-docking studies have shown that docking a ligand to the non-native structure of a target protein leads to failure of docking in pose and affinity prediction10–12. These results imply that the use of apo crystal protein structures might lead to poor enrichment in virtual screening experiments. Thus, for cases in which only an unbound (apo) structure is available for the specific target protein, the SBVS method can be severely limited9, 13.
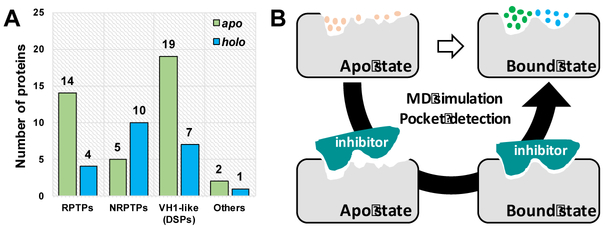
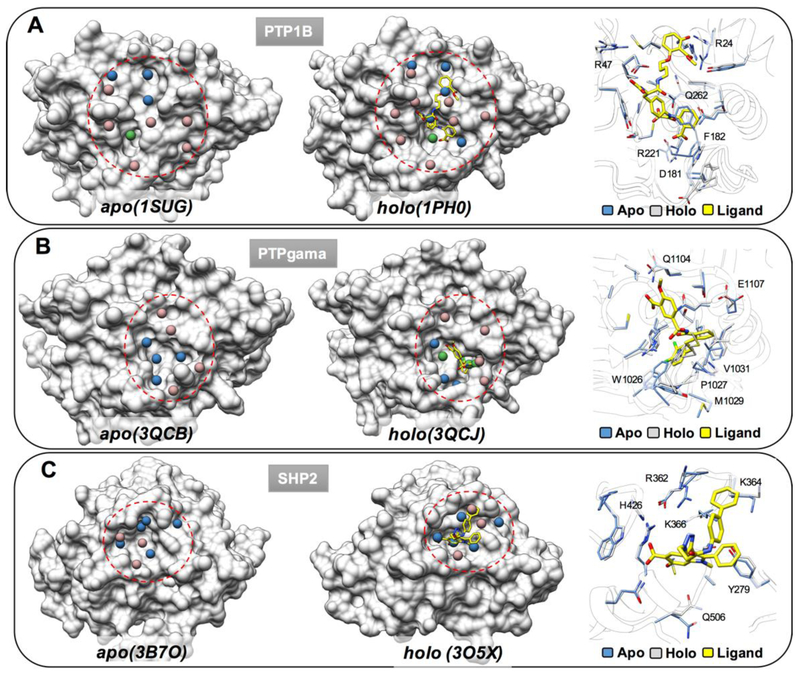
Protein tyrosine phosphatases (PTPs) play essential roles in cell signaling and have been recognized as underexploited targets for potential therapeutic intervention in many diseases, including cancer, diabetes, autoimmune disorders, and infectious diseases14–16. Although many structures have been determined for diverse members of the PTP family, often only the apo structure is available, especially for proteins that belong to receptor-type protein tyrosine phosphatases (RPTPs) and VH1-like PTPs (Figure 1A and Supporting Information Table S1). Furthermore, we compared the binding pockets between apo and holo crystal structures for three PTP family members (PTP1B, PTPgamma, SHP2 and LMW-PTP) and found ligand-induced conformation changes to be widely observable (Figure 2 and Figure 3A–C). Thus, the lack of bound state (holo) structures for many PTPs is likely to be a critical challenge to their reliable SBVS.
Figure 1.
(A) Analysis of apo and holo structures for different classes of PTPs in the RCSB Protein Data Bank17 (version June 2018). (B) Computational strategy to predict protein bound state from apo state.
Figure 2.
Comparison of the ligand binding pockets in apo (PDB: 1SUG18, 3QCB19 and 3B7O20) and holo (PDB: 1PH021, 3QCJ19 and 3O5X22) crystal structures of PTP1B (A), PTPgama (B) and SHP2 (C).
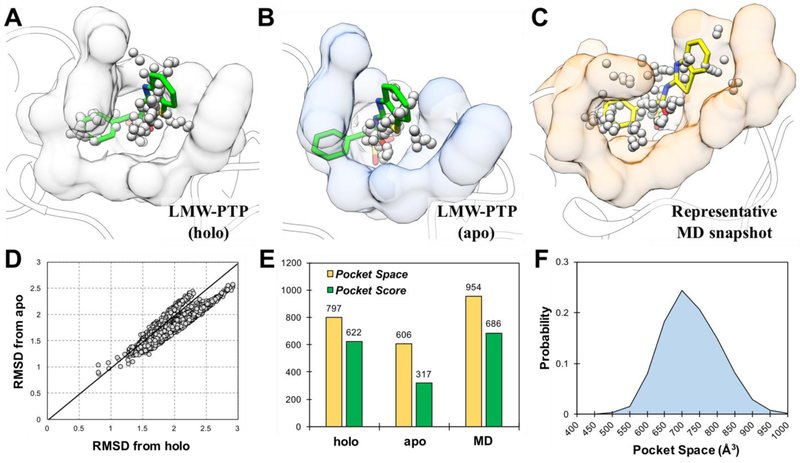
Figure 3.
Computational strategy validation using LMW-PTP. The binding pockets of LMW-PTP inhibitor in holo crystal structure (A), apo crystal structure (B) and representative MD snapshot (C) are calculated using AlphaSpace38, 39. (D) RMSD of binding site residues (within 5Å of LMW-PTP inhibitor) from apo and holo crystal structures. (E) Comparison of ligand binding pocket space and score in holo crystal structure, apo crystal structure and representative MD snapshot. (F) Probability of ligand binding pocket space during MD simulation.
Considering that experimental structure determination of protein-ligand complexes at atomic resolution can be time-consuming and costly, molecular dynamics (MD) simulation can serve as an alternative computational tool to generate multiple protein conformations23–25. In fact, previous studies suggest that certain snapshots from MD simulation can be more predictive in SBVS than experimental structures26–28. However, MD trajectories can include many poorly predictive structures as well, and how to select the most suitable structure(s) for SBVS remains elusive.
As a member of RPTPs, the protein tyrosine phosphatase receptor type O (PTPRO) has attracted significant attention for its essential roles in many diseases. For example, PTPRO has been recognized as a tumor suppressor, and hypermethylation and reduced expression of PTPRO has been observed in many kinds of cancer29–31. A recent study further suggested that PTPRO-mediated autophagy could prevent tumorigenesis32. PTPRO may also play roles in axon growth, vertebrate limb development, and regeneration33–35. In addition, inhibition of PTPRO using small molecules has reduced thioglycolate-induced peritoneal chemotaxis and improved ulcerative colitis in murine disease models36. Heretofore, few PTPRO inhibitors have been reported (Supporting Information Figure S1), thus there is a need to develop novel PTPRO inhibitors and to evaluate their therapeutic potential. Currently only two apo crystal structures (2G5937 and 2GJT20) are determined for PTPRO (Last visit of RCSB Protein Data Bank17: June 2018).
Herein, we designed an inexpensive computational workflow to search for a reliable bound state structure of PTPRO, starting from the apo structure (Figure 1B). First, a known ligand was used as a probe to induce conformational changes in the target protein during MD simulation. Second, an evaluation of MD snapshots was carried out on the basis of MM/GBSA binding energy calculation, structure clustering, and fragment-centric pocket analysis using AlphaSpace38, 39. As a new alpha sphere-based pocket detection tool, AlphaSpace is able to identify high-quality pockets at protein-ligand interfaces and has been successfully employed in the design of KIX/MLL inhibitor38. Finally, an MD snapshot exhibiting good ligand binding affinity as well as well-characterized, high-scoring binding pockets was selected as a favorable bound state structure and used in SBVS to identify novel PTPRO inhibitors. Our computational strategy was first validated using LMW-PTP, in which both apo and holo crystal structures are available, and then successfully employed in the SBVS of new inhibitors targeting PTPRO, where only apo crystal structure is available. Our prediction of a viable bound state structure to assist SBVS serves as a proof-of-concept study for our computational strategy, as well as for future in silico discovery of PTP inhibitors.
2. Results and Discussion
We began by validating our computational strategy (Figure 1B) using both apo (5KQP40) and holo (5KQG40) crystal structures of LMW-PTP. Pocket analysis of LMW-PTP crystal structures using AlphaSpace38, 39 revealed that the apo structure lacks proper binding pocket for benzene group of LMW-PTP inhibitor (Figure 3B). To test whether we could capture bound state structure of LMW-PTP using apo protein structure, the LMW-PTP inhibitor was docked to apo LMW-PTP structure and serve as a probe in MD simulation to induce conformation changes in LMW-PTP. As shown in Supporting Information Figure S2A, the binding of inhibitor in apo structure is very stable during 200-ns MD simulation (RMSD to crystal ligand structure = 0.71 ± 0.25 Å). Although it is very difficult to capture the same holo crystal structure during our MD simulation, the protein-ligand complex become closer to holo crystal structure than apo crystal structure in respect of binding site residues (Figure 3D). Then, clustering analysis was performed on the basis of RMSD values of ligand during MD simulation. Five clusters were generated with the cluster-1 represent 46% of all MD snapshots and the representative MD snapshot of cluster-1 exhibit the highest pocket space as well as pocket score among five representative MD snapshots (Supporting Information Figure S2B and S2C). Interestingly, the representative MD snapshot of cluster-1 possesses better binding pockets for LMW-PTP inhibitor than apo crystal structure and its pocket space and pocket score values are even higher than holo crystal structure (Figure 3E). As shown in Figure 3F, our MD simulation explored a large range of pocket spaces upon inhibitor binding and more than 95% MD snapshots possess a higher pocket space than apo crystal structure. Additionally, the Vina scores of LMW-PTP inhibitor in MD representative structure (−8.6 kcal/mol) is higher than that in apo crystal structure (−6.3 kcal/mol), indicating that the predicted bound state structure is more suitable for SBVS than apo crystal structure. Results above validated the feasibility of our computational strategy in predicting suitable bound state protein structure for SBVS using apo state structure.
We then compared two available apo crystal structures of PTPRO (PDB: 2G5937 and 2GJT20) and identified variation in their WPD-loop structure, which causes variation in the active site binding pockets (Supporting Information Figure S3). We docked the known PTPRO inhibitor (compound 1) into both PTPRO crystal structures, but only 2GJT was able to accommodate ligand binding, possibly due to the larger pockets observed in 2GJT. Figure 4A and 4C illustrates the initial binding mode of compound 1 predicted from molecular docking: (1) the 2-hydroxybenzoic acid group interacts with catalytic site residues in the P-loop, mimicking the cognate interaction between the phosphorylated PTP substrate and the P-loop; (2) the 1,2-diphenylethyne group occupies pockets located between the WPD-loop and the Q-loop; (3) the hexane group occupies a pocket beside the pTyr-loop.
Figure 4.
Prediction of the most favorable bound state structure of PTPRO. (A) Selected snapshots of compound 1 during 200ns MD simulation. (B) The RMSD values and calculated binding energies of compound 1 during MD simulation. (C) Binding pockets of compound 1 from initial docking result using crystal structure. (D) Binding pockets of compound 1 from representative MD snapshot. (E) Vina scores and occupied pocket space values of compound 1 in crystal structure and representative MD structure. Fragment-centric pocket analysis was performed using AlphaSpace38, 39. Pockets are represented by spheres, which are colored by pocket classification: core pockets (green), auxiliary pockets (blue), and minor pockets (rosy brown).
The docked model of PTPRO-inhibitor was then subjected to a 200ns MD simulation, during which we observed significant changes in the binding mode of compound 1. As shown in Figure 4A, the 2-hydroxybenzoic acid group remains in the catalytic P-loop, however, the 1,2-diphenylethyne group shifted gradually to the pTyr-loop (10–50ns) and finally binds to pockets in the Second-site loop (60–200ns). According to the ligand binding energies calculated using MM/GBSA, the binding mode of compound 1 during the MD simulation can be divided into three stages (Figure 4B). Calculated binding energies during Stage III (150–200ns) are approximately 5 kcal/mol lower than Stage I (0–50ns). Although the binding mode of compound 1 in Stage II is similar to that in Stage III, the calculated binding energies in Stage II are less stable, exhibiting greater fluctuation. Stage III represents the most stable binding model of the PTPRO-inhibitor complex.
In order to identify a representative bound state structure of PTPRO for SBVS, we performed clustering analysis using MD snapshots extracted from the last 50ns of the MD simulation (Stage III) and a representative structure from the most populated cluster, which possessed the highest pocket score as well as occupied space for compound 1 (Supporting Information Figure S4), was selected. Using AlphaSpace, we further analyzed the active site pockets from the apo crystal structure and the MD representative structure. Results indicated that both pocket space and pocket score of Site 1 (P-loop) and Site 2 (WPD-loop and Q-loop) are increased due to conformation changes induced by the ligand (Figure 5). Pockets located in Site 3 (Q-loop, pTyr-loop and Secondary-site loop), the initial binding site of the 1,2-diphenylethyne group from the docked compound 1, remain unchanged, but their interaction with the ligand has been lost (Figure 4A and Figure 5). The 2-hydroxybenzoic acid group in compound 1 translates from Site 3 to Site 2 during the MD simulation (Figure 4A and Figure 5). Interestingly, the induced-fit conformation changes are mainly observed in side-chain rearrangement, with the backbone remaining relatively fixed (Supporting Information Figure S5). These results highlight that AlphaSpace is able to reveal underutilized subpocket space and is sensitive to subtle conformation changes in the target protein. In comparison with the initial docking result using the crystal structure, both Vina score and occupied pocket space of compound 1 are increased in the representative MD snapshot (Figure 4E), which is in line with MM/GBSA results (Figure 4B). Moreover, the structural quality of our predicted bound state structure was verified using ProSA-web41. As shown in Figure S6 in Supporting Information, the Z-score of our predicted bound state structure is −6.99, which is comparable with crystal structure 2G59 (−7.54). The ProSA energy plot of our predicted bound state structure is also similar with crystal structure 2G59. This result supports the good quality of our predicted bound state structure for PTPRO. Taken together, we predicted a stable bound state complex for PTPRO using our computational strategy and the representative MD snapshot was carefully evaluated and finally selected as the most favorable bound state structure for PTPRO (Figure 4D). The workflow for MD snapshot selection is summarized in Supporting Information Figure S7.
Figure 5.
Comparison of the inhibitor binding sites in PTPRO. Panel A and B illustrate the pockets of three inhibitor binding sites (Site 1, Site 2 and Site 3) in crystal structure and MD representative structure. Pockets are represented by spheres, which are colored by pocket classification: core pockets (green), auxiliary pockets (blue), and minor pockets (rosy brown). Panel C and D present the total pocket score and pocket space for three inhibitor binding sites, comparing crystal structure and MD representative structure.
In order to further verify the reliability of our predicted bound state structure of PTPRO for SBVS (Figure 4D) and to identify new inhibitors, we performed docking-based virtual screening using the representative protein-ligand complex protein structure. According to results described above, the known PTPRO inhibitor mainly occupied pockets in Site 1 and Site 2 (Figure 6), so these two regions were targeted in our virtual screening. Although the limited number of known PTPRO inhibitors makes it impossible to reliably compare the performance of different docking programs, our previous study indicated that Gold and GoldScore achieve high accuracy in the virtual screening of another PTP family protein. Thus, in the current study, Gold and GoldScore were utilized in the virtual screening of PTPRO inhibitors.
Figure 6.
(A) Predicted bound state of PTPRO is illustrated with two major inhibitor binding sites on the left and the workflow for virtual screening on the right. (B) Chemical structures and predicted binding modes of compound GP03, GP07 and GP17. The inhibitory activities against PTPRO as well as occupied pocket space values are illustrated for each compound.
A commercial database containing more than 200,000 compounds was docked to PTPRO using Gold and ranked according to their GoldScore values. Autodock Vina was used to rescore the top 2,000 compounds. Then, 500 compounds with high GoldScore and Vina score were extracted for cluster analysis and visual inspection (Figure 6A). In each cluster, compounds that formed favorable interactions (e.g. multiple hydrogen-bonds) with PTPRO were prioritized. Using visualization, we also prioritized compounds binding Site 1 because (1) Site 1 is the major binding site for pTyr substrate; (2) AlphaSpace analysis revealed high-scoring, underutilized subpocket space in Site 1. At last, a total of 20 compounds were selected to purchase for biological evaluation (Supporting Information Table S2).
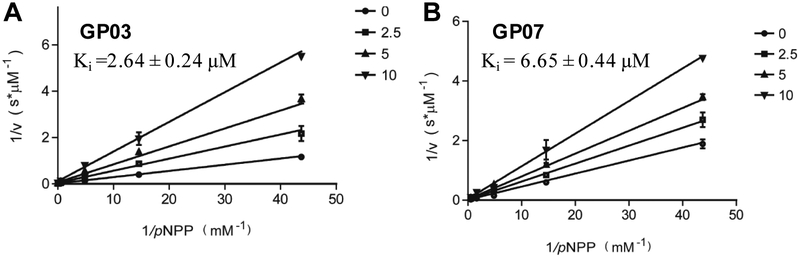
The initial screenings were conducted at a concentration of 100 μM, and compounds that exhibited more than 50% inhibition were further tested at different concentrations to calculate their IC50 values. Finally, three compounds were identified as novel PTPRO inhibitors with IC50 < 100 μM (Figure 6B). Specially, compound GP03 and GP07 possessed low micromolar IC50 values (2.89 μM and 6.08 μM, respectively) (Supporting Information Figure S8). Chemical structures of these new inhibitors were compared with the list of pan assay interference compounds (PAINs), and all of them passed the PAINs filter42. In addition, compound GP03 and GP07 showed structural novelty with respect to known PTPRO inhibitor compound 1, with ROCS Tanimoto score 0.53 and 0.48, respectively. The Lineweaver-Burk plots of the most potent inhibitors, shown in Figure 7, indicate that these compounds are competitive inhibitors for PTPRO with low micromolar Ki values (2.64 ± 0.24 μM for GP03 and 6.65 ± 0.44 μM for GP07).
Figure 7.
Kinetic analysis of PTPRO inhibition by GP03 (A)and GP07 (B). The Lineweaver-Burk plot displays a characteristic pattern of intersecting lines that indicates competitive inhibition.
As shown in Figure 6B, compound GP03 and GP07 bind only to pockets in Site 1 and their occupancy of Site 1 is increased compared to compound 1 (Figure 6A). Compound GP17 binds mainly to Site 1 pockets, but partially occupies a pocket in Site 2. However, the total pocket occupancy of compound GP17 is lower than that of GP03 and GP07, which might explain its weaker inhibitory activity (Figure 6B). Furthermore, we analyzed the pocket occupancy of GP03 throughout MD simulation of the complex (Supporting Information Figure S9) and detected significant unoccupied pocket space within Site 1 as well. Thus, the unoccupied pocket space, especially in Site 2, provides opportunities for structure-based optimization of these new PTPRO inhibitors. We further compared the docking scores of these new inhibitors between the crystal structure or the predicted bound state structure and found that the latter possesses a higher discriminatory power (Supporting Information Figure S10). Results above not only support the utility of our computational strategy (Figure 1B and Supporting Information Figure S7) in predicting a bound state structure for SBVS, but also provide several new chemotypes for PTPRO inhibition.
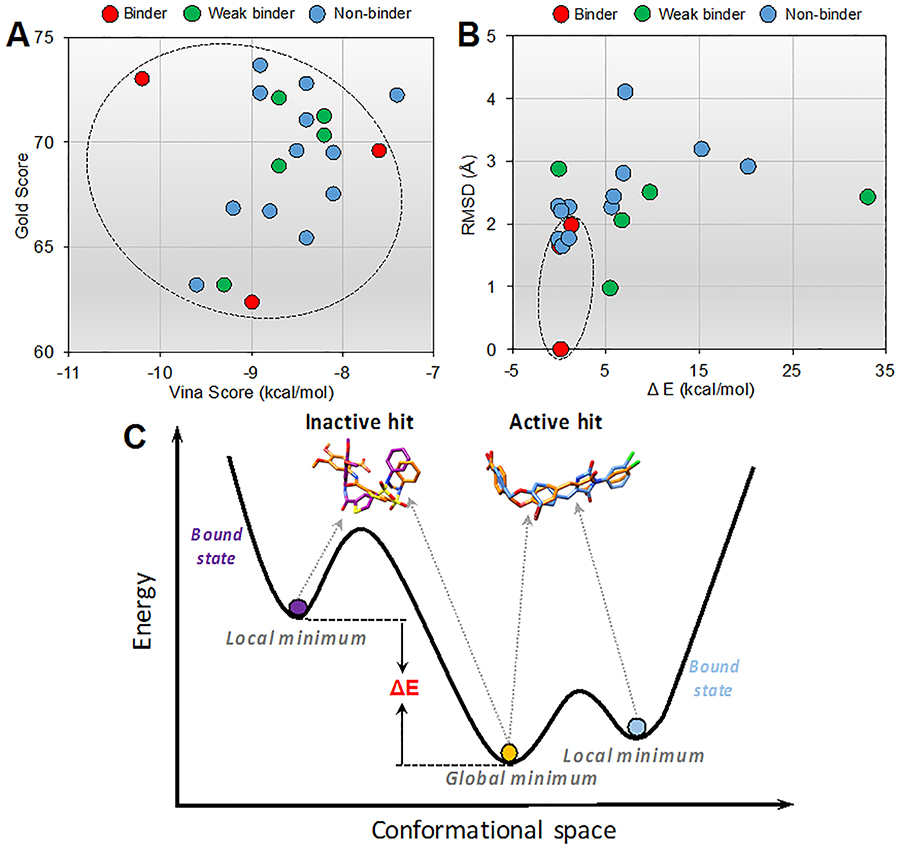
Although we have successfully identified three new PTPRO inhibitors from 20 virtual screening hits, docking scores failed to accurately distinguish three active hits (binders) from 17 inactive hits, including five weak binders (IC50 > 100μM) and 12 non-binders (no inhibition at 100μM) (Figure 8 and Supporting Information Table S2). In addition, the occupied pocket space values showed good correlation with the inhibitory activities of the three new PTPRO inhibitors, however, inactive hits also possess high occupied pocket space values, making it difficult to distinguish between active and inactive hits (Supporting Information Table S2).
Figure 8.
The ability to discriminate between binders and non-binders. (A) Docking scores of 20 virtual screening hits using Autodock Vina and Gold. (B) Energy difference and structure difference of each virtual screening hit between local minimum and global minimum. (C) Active and inactive hits are schematically represented by different energy wells on the ligand energy landscape, illustrating the magnitude of the conformational energy penalty upon binding.
In order to further understand the underlying reasons for false positive results in the current SBVS study and improve our ability to discriminate between active and inactive PTPRO inhibitors, we performed MD simulations as well as MM/GBSA binding energy calculations using the docking results of the 20 experimentally evaluated virtual screening hits. Results showed that general binding poses of the 20 hit compounds remained stable during MD simulations (Supporting Information Figure S11). Unfortunately, the MM/GBSA results still failed to discriminate between active and inactive PTPRO inhibitors (Supporting Information Figure S12). It should be mentioned that consideration of the ligand conformational energy penalty can be critical for accurate estimation of free energy of binding of inhibitors43, 44. However, due to the expensive computational cost and low predictive accuracy45–48, the energetics of the binding-induced conformation changes in the ligand are usually neglected in current docking scores and in the MM/GBSA method49–52. Thus, we sought to qualitatively evaluate the energy penalty for the binding-induced ligand conformation using an inexpensive computational approach.
Ligand strain energy, which can be defined as the potential energy difference between the bound state ligand conformation and the unbound ligand conformation, can serve as an approximation of the energy penalty associated with ligand binding53. However, positions of ligand atoms in the protein-ligand complex predicted by molecular docking contain significant uncertainties in bond lengths and angles. Small variations of bond lengths and angles in a ligand structure may result in an artificially large calculated energy penalty53. Taking these factors into consideration, we finally calculated ligand strain energy (ΔEstrain) using the following equation:
| (eq. 1) |
where Elocal is calculated by minimizing the docked ligand conformation to its closest local minimum (in the absence of the protein) and Eglobal is calculated using the global minimum ligand conformation. All ligand conformation energies were calculated using a fast and widely used force field, MMFF (Supporting Information Table S3). In addition, the structural differences (RMSD) between the local minimum and global minimum ligand conformations, which may also reflect the energetic penalty for the bound conformation, were measured (Figure 8B).
As expected, we found that inactive hits often possess higher energy barriers than active hits when adopting the bound conformer from the global minimum conformer (Figure 8B and 8C). The ligand strain energies of the three active PTPRO inhibitors (ΔEstrain < 1.5 kcal/mol) are lower than four weak binders and six non-binders (ΔEstrain > 5 kcal/mol) (Figure 8B). By considering the structural differences (RMSD < 2 Å), we could further distinguish the three active hits from one additional weak binder and three additional non-binders. Finally, we could qualitatively discriminate between the three active hits and 14 false positive inactive hits by combining the above criteria (ΔEstrain < 1.5 kcal/mol and RMSD < 2 Å). This treatment to approximate the conformational energy penalty of ligand binding, which is usually neglected in docking scores (and MM/GBSA calculation), may partially account for false positive results from the SBVS.
Because all classical PTPs possess conserved residues in the active site, the selectivity of current PTP inhibitors represents the major hurdle for their further development. To evaluate the selectivity of the three newly identified PTPRO inhibitors, we first tested their inhibition selectivity against PTP1B, VHR and STEP. As results show in Table 1, compound GP03 possessed 1.5–7 fold selectivity for PTP1B, VHR and STEP. Compound GP07 possessed good selectivity (8–12 fold) for VHR and STEP; however, it lacks selectivity for PTP1B. Compound GP17 exhibited the lowest selectivity with nearly equal inhibition activities for PTPTO, PTP1B, VHR, and STEP. We further tested the selectivity profile of compound GP03 against other protein phosphatases and observed 3–30 fold selectivity for PEST, LYP, PTPN18, Slingshot2, PPM1A, PPM1G and PP1 (Supporting Information Table S4). So, compound GP03 not only exhibits the highest potency from our SBVS study, but also exhibits varying degrees of selectivity over a panel of protein phosphatases.
Table 1.
Selectivity of compound GP03, GP07 and GP17 against PTP1B, VHR and STEP.
| PTP | PTP inhibition IC50 (μM) | ||
|---|---|---|---|
| GP03 | GP07 | GP17 | |
| PTPRO | 2.89 | 6.08 | 67.94 |
| PTP1B | 4.44 | 5.75 | 60.86 |
| VHR | 6.58 | 49.27 | 83.51 |
| STEP | 21.08 | 75.16 | 54.42 |
To further provide insight into the structure-activity relationship of compound GP03, we performed a hit-based substructure search using the Specs database. A total of eight analogues of GP03 were selected for biological evaluation. As shown in Table 2, analogues exhibited substitutions in the R2 position as well as in the R1 position. Introducing the ethoxy substituent in the benzyloxybenzene group gave a compound (GP03–1) with reduced potency (IC50 = 13.99 μM). On the other hand, moving the hydantoin scaffold to ortho-position of benzyloxybenzene led to an inactive compound GP03–2. Although moving the para-substituted carboxylic acid group in R2 position to the meta-position also led to inactive compounds (GP03–3 and GP03–5), the introduction of an amide group in R1 position slightly restored the inhibitory activity (GP03–4). This result suggests the importance of modifying positioning for the substituent in R1, at least for a compound that possesses 3-(phenoxymethyl) benzoic acid group in R2 position. Also, exchanging the carboxylic acid group for fluorine atom gave the inactive compound GP03–6. Thus, the benzoic acid group of GP03 is essential for its high inhibitory activity, possibly because of the benzoic acid forming hydrogen bonds with residues in the active site of PTPRO. Interestingly, exchanging the benzyloxybenzene group for the 2-phenylfuran group in R2 position gave compounds (GP03–7 and GP03–8) with reduced potencies (IC50= 26.09 and 50.06 μM, respectively). Although the 2-phenylfuran derivatives displayed lower inhibitory activity than compound GP03, these compounds still provide a new scaffold for PTPRO inhibition.
Table 2.
Structure-activity relationship of compound GP03 and derivatives.

| Compound | R1 | R2 | IC50 (μM) |
|---|---|---|---|
| GP03 |  |
 |
2.89 |
| GP03–1 |  |
 |
13.99 |
| GP03–2 |  |
 |
>100 |
| GP03–3 |  |
 |
>100 |
| GP03–4 |  |
 |
81.74 |
| GP03–5 |  |
 |
>100 |
| GP03–6 |  |
>100 | |
| GP03–7 |  |
 |
26.09 |
| GP03–8 |  |
 |
50.06 |
3. Conclusion
In summary, we predicted a favorable ligand-bound state for PTPRO by combined use of molecular dynamics simulation, MM/GBSA binding energy calculation, and AlphaSpace pocket analysis. By utilizing a selected representative bound state structure, docking-based virtual screening was performed and successfully identified several novel PTPRO inhibitors. Calculations of ligand strain energies revealed a potential underlying factor of false positive SBVS results. Moreover, the most potent new PTPRO inhibitor, compound GP03, also displayed certain degrees of selectivity over other protein phosphatases. Preliminary structure-activity relationships of analogs of GP03 were also explored. These newly identified inhibitors not only support that our predicted bound state structure (holo) of PTPRO is more robust as a predictive tool than the available crystal structures (apo), but also provide good starting points for the further development of PTPRO selective inhibitors.
4. Experimental Section
Materials.
The selected virtual screening hits were purchased from Specs database with purities confirmed by LC-MS and 1H-NMR (data available at http://www.specs.net/). The para-nitrophenyl phosphate (pNPP, CAS: 4264-83-9) was purchased from Sangon Biotech Co., Ltd. All other chemicals and reagents were purchased from Sigma.
Molecular Dynamics Simulation.
Molecular dynamics (MD) simulations were performed using Amber 14 package using AMBER14SB force field54 for protein and TIP3P model for water55. The starting conformation of each protein-inhibitor complex was taken from the molecular docking result of AutoDock Vina. The topology file for each ligand was generated using Antechamber56 with general AMBER force field (GAFF)57 and AM1-BCC charges58, 59. The Particle Mesh Ewald (PME)60 method with 12.0 Å cutoff was used to deal with all non-bonded interactions. The SHAKE algorithm61 was applied to constrain all bonds involving hydrogen atoms. After a series of minimizations and equilibrations, MD simulations were performed on GPUs using the CUDA version of PMEMD62 with periodic boundary condition. Berendsen thermostat method63 has been used to control the system temperature at 300 K. Other parameters were default values.
MM/GBSA Calculations.
Relative binding energies of ligands were calculated using molecular mechanics/generalized born solvent accessibility (MM/GBSA) methodology47, 49, 50. MM-GBSA calculations were performed by MM-PBSA.py module of Amber14. The binding energies (ΔGbind) are calculated as the sum of molecular mechanical and solvation energies as described by following equations:
| (eq. 2) |
where ΔEMM is the gas phase molecular mechanical energy; ΔGsol is the desolvation free energy; −TΔS represent the conformational entropy upon association of substrata at temperature T. Due to the expensive computational cost and low prediction accuracy45–48, entropies were not considered in current study.
For compound 1, all frames taken from the 200ns MD simulations were used in MM/GBSA calculation. The binding energy of each virtual screening hit was calculated using the snapshots taken from 2–20ns of each MD simulation.
Snapshot Selection.
All MD snapshots were analyzed using cpptraj module in AmberTools 15. Clustering analysis of LWM-PTP-inhibitor system and PTPRO-compound 1 system were performed using the hierarchical agglomerative approach as implemented in the AmberTools package. Protein-ligand interaction energies were calculated using MM/GBSA method. Binding pocket analysis was performed using AlphaSpace38, 39, a computational tool for fragment-centric topographical mapping of intermolecular interfaces.
AlphaSpace Pocket Analysis.
AlphaSpace38, 39 (www.nyu.edu/projects/yzhang/AlphaSpace/)) employs a geometric model based on Voronoi tessellation to identify and represent all concave interaction space across the protein surface as a set of alpha-atom/alpha-space pairs, which are then clustered into discrete fragment-centric pockets. The occupation status of each individual alpha-space within each pocket is evaluated based on the distance between its associated alpha-atom and the nearest atom from the ligand, using a 1.6 Å cutoff. The total pocket occupation by ligand is calculated by taking the sum of all occupied alpha-space volumes associated with ligand atoms. All detected pockets are classified as core (green sphere), auxiliary (blue sphere), or minor pockets (rosy brown sphere) by employing AlphaSpace pocket score as described before39.
Structure-based Virtual Screening.
Crystal structures of PTPRO were retrieved from the Protein Data Bank (PDB code: 2G59 and 2GJT) and prepared using the protein preparation workflow in Sybyl-x 1.1 (Tripos, Inc.). The protonation states of specific residues were calculated using the PDB2PQR server64. Molecular docking studies were carried out using the standard setting of Autodock Vina65 and Gold66. All ligands in the Specs database (www.specs.net) were prepared using Ligand Preparation module in Sybyl-x 1.1 with 3D structures generated by Concord. The predicted bound structure of PTPRO was used in virtual screening. Firstly, the database was screened using Gold program and top 2000 docking hits were selected according to the Gold Scores and then rescored using AutoDock Vina with local minimization. Then, the top ranking 500 compounds were clustered based on the FCFP_6 fingerprints calculation and then selected manually. Other parameters that are not mentioned were set at default values.
Enzyme Catalytic Assay.
The expression and purification of the PTPRO catalytic domain as well as other protein phosphatases were performed as described previously67–72. The effect of small molecule inhibitors on the PTP-catalyzed pNPP hydrolysis were determined at 25°C in 50 mM 3,3-dimethylglutarate buffer, and the ionic strength was adjusted to 0.15 M with NaCl (buffer A). The reaction was quenched at set time points using 1 M NaOH, and the ageneration of products was detected by monitoring the absorbance of pNP at 405 nm. The IC50 values were calculated by GraphPd Prism according to following equation:
Calculation of Ligand Strain Energy.
Ligand strain energy was defined as the energy difference between local minimum conformation of bound ligand and global minimum conformation of the unbound ligand. Docked conformations of each virtual screening hit was minimized to its closest local minimum using Sybyl-x 1.1 and MMFF force field. To identify the lowest energy conformation (global minimum), conformational analysis was performed for each ligand using the Random Search module in Sybyl-x 1.1. An energy cutoff of 3.0 kcal/mol and a RMS threshold of 0.2 Å above the global minimum were used, and the maximum number of conformations was set to 1000.
Supplementary Material
ACKNOWLEDGMENT
The authors gratefully acknowledge Professor Renxiao Wang at the Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, for providing the necessary software and NYU-ITS and NYUAD for providing computational resources.
Funding Sources
This work was supported by National Natural Science Foundation of China (No. 21672127 and 81874288), Key Research and Development Project of Shandong Province (Grant No. 2017CXGC1401), the Fundamental Research Funds of Shandong University (Grant No. 2016JC018). Y. Z. would acknowledge the support by NIH (R35-GM127040).
ABBREVIATIONS
- SBVS
structure-based virtual screening
- MD
molecular dynamics
- PTPRO
protein tyrosine phosphatase receptor type O
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Supporting figures and tables for additional results, 1H-NMR and HRMS spectral information for representative compounds (PDF)
Molecular formula strings (XLS)
Predicted bound state model of PTPRO (PDB)
The authors declare no competing financial interest.
Authors will release the atomic coordinates and experimental data upon article publication
REFERENCES
- 1.Kalyaanamoorthy S; Chen YP, Structure-Based Drug Design to Augment Hit Discovery. Drug Discov. Today 2011, 16, 831–839. [DOI] [PubMed] [Google Scholar]
- 2.Congreve M; Langmead CJ; Mason JS; Marshall FH, Progress in Structure Based Drug Design for G Protein-coupled Receptors. J. Med. Chem 2011, 54, 4283–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pei J; Yin N; Ma X; Lai L, Systems Biology Brings New Dimensions for Structure-based Drug Design. J. Am. Chem. Soc 2014, 136, 11556–11565. [DOI] [PubMed] [Google Scholar]
- 4.Cavasotto CN; Orry AJ, Ligand Docking and Structure-Based Virtual Screening in Drug Discovery. Curr. Top. Med. Chem 2007, 7, 1006–1014. [DOI] [PubMed] [Google Scholar]
- 5.Ripphausen P; Nisius B; Peltason L; Bajorath J, Quo Vadis, Virtual Screening? A Comprehensive Survey of Prospective Applications. J. Med. Chem 2010, 53, 8461–8467. [DOI] [PubMed] [Google Scholar]
- 6.Hou X; Li R; Li K; Yu X; Sun JP; Fang H, Fast Identification of Novel Lymphoid Tyrosine Phosphatase Inhibitors Using Target-Ligand Interaction-Based Virtual Screening. J. Med. Chem 2014, 57, 9309–9322. [DOI] [PubMed] [Google Scholar]
- 7.Hou X; Li K; Yu X; Sun JP; Fang H, Protein Flexibility in Docking-Based Virtual Screening: Discovery of Novel Lymphoid-Specific Tyrosine Phosphatase Inhibitors Using Multiple Crystal Structures. J. Chem. Inf. Model 2015, 55, 1973–1983. [DOI] [PubMed] [Google Scholar]
- 8.Therrien E; Weill N; Tomberg A; Corbeil CR; Lee D; Moitessier N, Docking Ligands into Flexible and Solvated Macromolecules. 7. Impact of Protein Flexibility and Water Molecules on Docking-Based Virtual Screening Accuracy. J. Chem. Inf. Model 2014, 54, 3198–3210. [DOI] [PubMed] [Google Scholar]
- 9.McGovern SL; Shoichet BK, Information Decay in Molecular Docking Screens against Holo, Apo, and Modeled Conformations of Enzymes. J. Med. Chem 2003, 46, 2895–2907. [DOI] [PubMed] [Google Scholar]
- 10.Verdonk ML; Mortenson PN; Hall RJ; Hartshorn MJ; Murray CW, Protein-Ligand Docking against Non-Native Protein Conformers. J. Chem. Inf. Model 2008, 48, 2214–2225. [DOI] [PubMed] [Google Scholar]
- 11.Seeliger D; de Groot BL, Conformational Transitions upon Ligand Binding: Holo-Structure Prediction from Apo Conformations. Plos Comput. Biol 2010, 6, e1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warren GL; Andrews CW; Capelli AM; Clarke B; LaLonde J; Lambert MH; Lindvall M; Nevins N; Semus SF; Senger S; Tedesco G; Wall ID; Woolven JM; Peishoff CE; Head MS, A Critical Assessment of Docking Programs and Scoring Functions. J. Med. Chem 2006, 49, 5912–5931. [DOI] [PubMed] [Google Scholar]
- 13.Teague SJ, Implications of Protein Flexibility for Drug Discovery. Nat. Rev. Drug Discov 2003, 2, 527–541. [DOI] [PubMed] [Google Scholar]
- 14.Yu ZH; Zhang ZY, Regulatory Mechanisms and Novel Therapeutic Targeting Strategies for Protein Tyrosine Phosphatases. Chem. Rev 2017, 118, 1069–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang ZY, Drugging the Undruggable: Therapeutic Potential of Targeting Protein Tyrosine Phosphatases. Acc. Chem. Res 2017, 50, 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He R; Zeng LF; He Y; Zhang S; Zhang ZY, Small Molecule Tools for Functional Interrogation of Protein Tyrosine Phosphatases. FEBS J. 2013, 280, 731–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose PW; Prlic A; Altunkaya A; Bi C; Bradley AR; Christie CH; Costanzo LD; Duarte JM; Dutta S; Feng Z; Green RK; Goodsell DS; Hudson B; Kalro T; Lowe R; Peisach E; Randle C; Rose AS; Shao C; Tao YP; Valasatava Y; Voigt M; Westbrook JD; Woo J; Yang H; Young JY; Zardecki C; Berman HM; Burley SK, The RCSB Protein Data Bank: Integrative View of Protein, Gene and 3D Structural Information. Nucleic Acids Res. 2017, 45, D271–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedersen AK; Peters GG; Moller KB; Iversen LF; Kastrup JS, Water-Molecule Network and Active-Site Flexibility of Apo Protein Tyrosine Phosphatase 1B. Acta Crystallogr. D Biol. Crystallogr 2004, 60, 1527–1534. [DOI] [PubMed] [Google Scholar]
- 19.Sheriff S; Beno BR; Zhai W; Kostich WA; McDonnell PA; Kish K; Goldfarb V; Gao M; Kiefer SE; Yanchunas J; Huang Y; Shi S; Zhu S; Dzierba C; Bronson J; Macor JE; Appiah KK; Westphal RS; O’Connell J; Gerritz SW, Small Molecule Receptor Protein Tyrosine Phosphatase Gamma (RPTPgamma) Ligands That Inhibit Phosphatase Activity via Perturbation of the Tryptophan-Proline-Aspartate (WPD) Loop. J. Med. Chem 2011, 54, 6548–6562. [DOI] [PubMed] [Google Scholar]
- 20.Barr AJ; Ugochukwu E; Lee WH; King ON; Filippakopoulos P; Alfano I; Savitsky P; Burgess-Brown NA; Muller S; Knapp S, Large-Scale Structural Analysis of the Classical Human Protein Tyrosine Phosphatome. Cell 2009, 136, 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu G; Xin Z; Liang H; Abad-Zapatero C; Hajduk PJ; Janowick DA; Szczepankiewicz BG; Pei Z; Hutchins CW; Ballaron SJ; Stashko MA; Lubben TH; Berg CE; Rondinone CM; Trevillyan JM; Jirousek MR, Selective Protein Tyrosine Phosphatase 1B Inhibitors: Targeting the Second Phosphotyrosine Binding Site with Non-Carboxylic Acid-Cntaining Ligands. J. Med. Chem 2003, 46, 3437–3440. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X; He Y; Liu S; Yu Z; Jiang ZX; Yang Z; Dong Y; Nabinger SC; Wu L; Gunawan AM; Wang L; Chan RJ; Zhang ZY, Salicylic Acid Based Small Molecule Inhibitor for the Oncogenic Src Homology-2 Domain Containing Protein Tyrosine Phosphatase-2 (SHP2). J. Med. Chem 2010, 53, 2482–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fox SJ; Li J; Sing Tan Y; Nguyen MN; Pal A; Ouaray Z; Yadahalli S; Kannan S, The Multifaceted Roles of Molecular Dynamics Simulations in Drug Discovery. Curr. Pharm. Des 2016, 22, 3585–3600. [DOI] [PubMed] [Google Scholar]
- 24.Durrant JD; McCammon JA, Molecular Dynamics Simulations and Drug Discovery. BMC Biol. 2011, 9, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortier J; Rakers C; Bermudez M; Murgueitio MS; Riniker S; Wolber G, The Impact of Molecular Dynamics on Drug Design: Applications for The Characterization of Ligand-Macromolecule Complexes. Drug Discov. Today 2015, 20, 686–702. [DOI] [PubMed] [Google Scholar]
- 26.Nichols SE; Baron R; Ivetac A; McCammon JA, Predictive Power of Molecular Dynamics Receptor Structures in Virtual Screening. J. Chem. Inf. Model 2011, 51, 1439–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uehara S; Tanaka S, Cosolvent-Based Molecular Dynamics for Ensemble Docking: Practical Method for Generating Druggable Protein Conformations. J. Chem. Inf. Model 2017, 57, 742–756. [DOI] [PubMed] [Google Scholar]
- 28.Yang TY; Wu JC; Yan CL; Wang YF; Luo R; Gonzales MB; Dalby KN; Ren PY, Virtual Screening Using Molecular Simulations. Proteins 2011, 79, 1940–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacob ST; Motiwala T, Epigenetic Regulation of Protein Tyrosine Phosphatases: Potential Molecular Targets for Cancer Therapy. Cancer Gene Ther. 2005, 12, 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motiwala T; Kutay H; Ghoshal K; Bai S; Seimiya H; Tsuruo T; Suster S; Morrison C; Jacob ST, Protein Tyrosine Phosphatase Receptor-Type O (PTPRO) Exhibits Characteristics of A Candidate Tumor Suppressor in Human Lung Cancer. Proc. Natl. Acad. Sci. U S A 2004, 101, 13844–13849. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Motiwala T; Ghoshal K; Das A; Majumder S; Weichenhan D; Wu YZ; Holman K; James SJ; Jacob ST; Plass C, Suppression of The Protein Tyrosine Phosphatase Receptor Type O Gene (PTPRO) by Methylation in Hepatocellular Carcinomas. Oncogene 2003, 22, 6319–6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W; Hou J; Wang X; Jiang R; Yin Y; Ji J; Deng L; Huang X; Wang K; Sun B, PTPRO-Mediated Autophagy Prevents Hepatosteatosis and Tumorigenesis. Oncotarget 2015, 6, 9420–9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stepanek L; Stoker AW; Stoeckli E; Bixby JL, Receptor Tyrosine Phosphatases Guide Vertebrate Motor Axons During Development. J. Neurosci 2005, 25, 3813–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gatto G; Dudanova I; Suetterlin P; Davies AM; Drescher U; Bixby JL; Klein R, Protein Tyrosine Phosphatase Receptor Type O Inhibits Trigeminal Axon Growth and Branching by Repressing Trkb and Ret Signaling. J. Neurosci 2013, 33, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoker AW, RPTPs in Axons, Synapses and Neurology. Semin. Cell Dev. Biol 2015, 37, 90–97. [DOI] [PubMed] [Google Scholar]
- 36.Gobert RP; van den Eijnden M; Szyndralewiez C; Jorand-Lebrun C; Swinnen D; Chen L; Gillieron C; Pixley F; Juillard P; Gerber P; Johnson-Leger C; Halazy S; Camps M; Bombrun A; Shipp M; Vitte PA; Ardissone V; Ferrandi C; Perrin D; Rommel C; Hooft van Huijsduijnen R, GLEPP1/Protein-Tyrosine Phosphatase Phi Inhibitors Block Chemotaxis in Vitro and in Vivo and Improve Murine Ulcerative Colitis. J. Biol. Chem 2009, 284, 11385–11395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Almo SC; Bonanno JB; Sauder JM; Emtage S; Dilorenzo TP; Malashkevich V; Wasserman SR; Swaminathan S; Eswaramoorthy S; Agarwal R; Kumaran D; Madegowda M; Ragumani S; Patskovsky Y; Alvarado J; Ramagopal UA; Faber-Barata J; Chance MR; Sali A; Fiser A; Zhang ZY; Lawrence DS; Burley SK, Structural Genomics of Protein Phosphatases. J. Struct. Funct. Genomics 2007, 8, 121–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rooklin D; Modell AE; Li H; Berdan V; Arora PS; Zhang Y, Targeting Unoccupied Surfaces on Protein-Protein Interfaces. J. Am. Chem. Soc 2017, 139, 15560–15563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rooklin D; Wang C; Katigbak J; Arora PS; Zhang Y, AlphaSpace: Fragment-Centric Topographical Mapping To Target Protein-Protein Interaction Interfaces. J. Chem. Inf. Model 2015, 55, 1585–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He R; Wang J; Yu ZH; Zhang RY; Liu S; Wu L; Zhang ZY, Inhibition of Low Molecular Weight Protein Tyrosine Phosphatase by an Induced-Fit Mechanism. J. Med. Chem 2016, 59, 9094–9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiederstein M; Sippl MJ, Prosa-Web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 2007, 35, W407–W410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baell JB; Holloway GA, New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem 2010, 53, 2719–2740. [DOI] [PubMed] [Google Scholar]
- 43.Pettersson I; Liljefors T, Structure-Activity Relationships for Apomorphine Congeners. Conformational Energies vs. Biological Activities. J. Comput. Aided Mol. Des 1987, 1, 143–152. [DOI] [PubMed] [Google Scholar]
- 44.Bostrom J; Norrby PO; Liljefors T, Conformational Energy Penalties of Protein-Bound Ligands. J. Comput. Aided Mol. Des 1998, 12, 383–396. [DOI] [PubMed] [Google Scholar]
- 45.Hou TJ; Wang JM; Li YY; Wang W, Assessing the Performance of the Molecular Mechanics/Poisson Boltzmann Surface Area and Molecular Mechanics/Generalized Born Surface Area Methods. II. The Accuracy of Ranking Poses Generated From Docking. J. Comput. Chem 2011, 32, 866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Genheden S; Kuhn O; Mikulskis P; Hoffmann D; Ryde U, The Normal-Mode Entropy in the MM/GBSA Method: Effect of System Truncation, Buffer Region, and Dielectric Constant. J. Chem. Inf. Model 2012, 52, 2079–2088. [DOI] [PubMed] [Google Scholar]
- 47.Hou TJ; Wang JM; Li YY; Wang W, Assessing the Performance of the MM/PBSA and MM/GBSA Methods. 1. The Accuracy of Binding Free Energy Calculations Based on Molecular Dynamics Simulations. J. Chem. Inf. Model 2011, 51, 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L; Li YY; Zhang LL; Hou TJ, Theoretical Studies on the Susceptibility of Oseltamivir against Variants of 2009 A/H1N1 Influenza Neuraminidase. J. Chem. Inf. Model 2012, 52, 2715–2729. [DOI] [PubMed] [Google Scholar]
- 49.Sun H; Li Y; Shen M; Tian S; Xu L; Pan P; Guan Y; Hou T, Assessing the Performance of MM/PBSA and MM/GBSA Methods. 5. Improved Docking Performance Using High Solute Dielectric Constant MM/GBSA and MM/PBSA Rescoring. Phys. Chem. Chem. Phys 2014, 16, 22035–22045. [DOI] [PubMed] [Google Scholar]
- 50.Sun H; Li Y; Tian S; Xu L; Hou T, Assessing the Performance of MM/PBSA And MM/GBSA Methods. 4. Accuracies of MM/PBSA And MM/GBSA Methodologies Evaluated by Various Simulation Protocols Using Pdbbind Data Set. Phys. Chem. Chem. Phys 2014, 16, 16719–16729. [DOI] [PubMed] [Google Scholar]
- 51.Fischer M; Coleman RG; Fraser JS; Shoichet BK, Incorporation of Protein Flexibility and Conformational Energy Penalties in Docking Screens to Improve Ligand Discovery. Nat. Chem 2014, 6, 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gupta A; Chaudhary N; Kakularam KR; Pallu R; Polamarasetty A, The Augmenting Effects of Desolvation and Conformational Energy Terms on the Predictions of Docking Programs against mPGES-1. Plos One 2015, 10, e0134472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perola E; Charifson PS, Conformational Analysis of Drug-Like Molecules Bound to Proteins: An Extensive Study of Ligand Reorganization upon Binding. J. Med. Chem 2004, 47, 2499–2510. [DOI] [PubMed] [Google Scholar]
- 54.Maier JA; Martinez C; Kasavajhala K; Wickstrom L; Hauser KE; Simmerling C, ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput 2015, 11, 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mark P; Nilsson L, Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. B 2001, 105, 9954–9960. [Google Scholar]
- 56.Wang JM; Wang W; Kollman PA; Case DA, Automatic Atom Type and Bond Type Perception in Molecular Mechanical Calculations. J. Mol. Graph. Model 2006, 25, 247–260. [DOI] [PubMed] [Google Scholar]
- 57.Wang JM; Wolf RM; Caldwell JW; Kollman PA; Case DA, Development and Testing of A General Amber Force Field. J. Comput. Chem 2004, 25, 1157–1174. [DOI] [PubMed] [Google Scholar]
- 58.Jakalian A; Bush BL; Jack DB; Bayly CI, Fast, Efficient Generation of High-Quality Atomic Charges. AM1-BCC Model: I. Method. J. Comput. Chem 2000, 21, 132–146. [DOI] [PubMed] [Google Scholar]
- 59.Jakalian A; Jack DB; Bayly CI, Fast, Efficient Generation of High-Quality Atomic Charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem 2002, 23, 1623–1641. [DOI] [PubMed] [Google Scholar]
- 60.Cerutti DS; Duke RE; Darden TA; Lybrand TP, Staggered Mesh Ewald: An Extension of the Smooth Particle-Mesh Ewald Method Adding Great Versatility. J. Chem. Theory Comput 2009, 5, 2322–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lippert RA; Bowers KJ; Dror RO; Eastwood MP; Gregersen BA; Klepeis JL; Kolossvary I; Shaw DE, A Common, Avoidable Source of Error in Molecular Dynamics Integrators. J. Chem. Phys 2007, 126, 046101. [DOI] [PubMed] [Google Scholar]
- 62.Salomon-Ferrer R; Gotz AW; Poole D; Le Grand S; Walker RC, Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput 2013, 9, 3878–3888. [DOI] [PubMed] [Google Scholar]
- 63.Berendsen HJC; Postma JPM; Vangunsteren WF; Dinola A; Haak JR, Molecular-Dynamics with Coupling to an External Bath. J. Chem. Phys 1984, 81, 3684–3690. [Google Scholar]
- 64.Dolinsky TJ; Nielsen JE; McCammon JA; Baker NA, PDB2PQR: An Automated Pipeline for The Setup of Poisson-Boltzmann Electrostatics Calculations. Nucleic Acids Res. 2004, 32, W665–W667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trott O; Olson AJ, AutoDock Vina: Improving the Speed and Accuracy of Docking with A New Scoring Function, Efficient Optimization,aAnd Multithreading. J. Comput. Chem 2010, 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verdonk ML; Cole JC; Hartshorn MJ; Murray CW; Taylor RD, Improved Protein-Ligand Docking Using GOLD. Proteins: Struct., Funct., Genet 2003, 52, 609–623. [DOI] [PubMed] [Google Scholar]
- 67.Yu X; Sun JP; He Y; Guo X; Liu S; Zhou B; Hudmon A; Zhang ZY, Structure, Inhibitor, and Regulatory Mechanism of Lyp, A Lymphoid-Specific Tyrosine Phosphatase Implicated in Autoimmune Diseases. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 19767–19772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu J; Chen M; Li R; Yang F; Shi X; Zhu L; Wang HM; Yao W; Liu Q; Meng FG; Sun JP; Pang Q; Yu X, Biochemical and Functional Studies of Lymphoid-Specific Tyrosine Phosphatase (Lyp) Variants S201F and R266W. PLoS One 2012, 7, e43631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huijbers IJ; Bin Ali R; Pritchard C; Cozijnsen M; Kwon MC; Proost N; Song JY; de Vries H; Badhai J; Sutherland K; Krimpenfort P; Michalak EM; Jonkers J; Berns A, Rapid Target Gene Validation in Complex Cancer Mouse Models using Re-Derived Embryonic Stem Cells. EMBO Mol. Med 2014, 6, 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gloria-Bottini F; Ammendola M; Saccucci P; Pietropolli A; Magrini A; Bottini E, The Association of PTPN22 Polymorphism with Endometriosis: Effect of Genetic and Clinical Factors. Eur. J. Obstet. Gynecol. Reprod. Biol 2013, 169, 60–63. [DOI] [PubMed] [Google Scholar]
- 71.Pan C; Liu HD; Gong Z; Yu X; Hou XB; Xie DD; Zhu XB; Li HW; Tang JY; Xu YF; Yu JQ; Zhang LY; Fang H; Xiao KH; Chen YG; Wang JY; Pang Q; Chen W; Sun JP, Cadmium is A Potent Inhibitor of PPM Phosphatases and Targets the M1 Binding Site. Sci. Rep 2013, 3, 2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun JP; Fedorov AA; Lee SY; Guo XL; Shen K; Lawrence DS; Almo SC; Zhang ZY, Crystal Structure of PTP1B Complexed with A Potent and Selective Bidentate Inhibitor. J. Biol. Chem 2003, 278, 12406–12414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.