Summary
Retinoic acid (RA), a vitamin A metabolite, regulates transcriptional programs that drive protective or pathogenic immune responses in the intestine, in a manner dependent on RA concentration. Vitamin A is obtained from diet and is metabolized by intestinal epithelial cells (IECs), which operate in intimate association with microbes and immune cells. Here we found that commensal bacteria belonging to class Clostridia modulate RA concentration in the gut by suppressing the expression of retinol dehydrogenase 7 (Rdh7) in IECs. Rdh7 expression and associated RA amounts were lower in the intestinal tissue of conventional mice, as compared to germ-free mice. Deletion of Rdh7 in IECs diminished RA signaling in immune cells, reduced the IL-22-dependent antimicrobial response, and enhanced resistance to colonization by Salmonella Typhimurium. Our findings define a regulatory circuit wherein bacterial regulation of IEC-intrinsic RA synthesis protects microbial communities in the gut from excessive immune activity, achieving a balance that prevents colonization by enteric pathogens.
eTOC blurb
Grizotte-Lake et al define a mechanism that regulates production of the vitamin A metabolite retinoic acid (RA), a key immunomodulator in the gut. Commensal bacteria suppress RA synthesis by the intestinal epithelium, which in turn results in the control of IL-22 activity and the prevention of microbial dysbiosis during pathogen colonization.
Introduction
Metabolites derived from dietary vitamin A, such as retinoic acid (RA), bind to receptors in the nucleus and regulate diverse transcriptional programs that direct varied physiological responses, ranging from eyesight and organogenesis to metabolism and immunological fitness. During embryonic stages, RA is instrumental in the development of primary and secondary lymphoid tissues (van de Pavert and Mebius, 2010). RA amounts in utero, dictated by the maternal intake of dietary vitamin A, are fundamental for controlling the size of the lymphocyte pool and the resistance to infection in the offspring (van de Pavert et al., 2014). In adults, RA is crucial for multiple adaptive and innate immune responses such as lymphocyte activation and proliferation, T-helper cell differentiation, tissue-specific lymphocyte homing, and the production of specific antibody isotypes (Larange and Cheroutre, 2016).
RA is necessary and sufficient to induce the gut homing receptors CCR9 and α4β7 on T cells and B cells, thereby regulating their numbers in the intestine (Iwata et al., 2004; Mora et al., 2006). Additionally, RA has a concentration dependent effect on T cell differentiation. At high concentrations, RA drives the differentiation of naïve T cells into regulatory T (Treg) cells in both mice and humans. At low concentrations, RA is essential for the production of the proinflammatory cytokines interferon (IFN)-γ and interleukin (IL)-17A by T helper (Th)1 and Th17 cells in response to infection, and for the coordination of the inflammatory immune response (Hall et al., 2011; Hill et al., 2008; Mucida et al., 2007; Nolting et al., 2009; Pino-Lagos et al., 2011). Studies also show that under infectious condition associated with induction of IL-6 and IL-15, RA acts as an adjuvant that promote rather than prevent inflammatory responses to fed antigen (DePaolo et al., 2011). The overall picture that emerges from the role of RA in regulating immune responses in the gut is that 1) RA-dependent immune responses can be protective or pathogenic in nature depending on the context, and 2) that concentration of RA in the intestinal tissue might play a critical role in tipping the balance from regulatory to inflammatory immune response (Erkelens and Mebius, 2017). Thus, to maintain immune homeostasis in the gut where immune cells and immunogenic cues from microbiota coexist, there must be mechanisms that modulate RA concentration within a narrow physiological range. Elucidating the cellular and molecular mechanisms that ensure optimal concentration of RA in the gut is crucial in understanding the context and tissue dependent role of RA in immunomodulation.
To maintain sufficient concentration of RA, the body relies on the uptake of vitamin A or retinol from the intestinal lumen by intestinal epithelial cells (IECs). After uptake by IECs, retinol can either be processed for storage or be further metabolized into RA (D’Ambrosio et al., 2011). This vitamin A uptake and metabolism in IECs operates in the context of intimate association with microbes on one side and immune cells on the other. Bacterial interactions with IECs are critical for inducing a broad spectrum of immune responses that include enhanced production of cytokines and chemokines, induction of B-cell class switching to IgA producing plasma cells and stimulating Th17 effector function (Peterson and Artis, 2014). Here we asked if commensal bacteria could regulate vitamin A metabolism and thereby influence the immunomodulatory functions of RA in the gut.
Results
Commensal bacteria regulate vitamin A metabolism and storage.
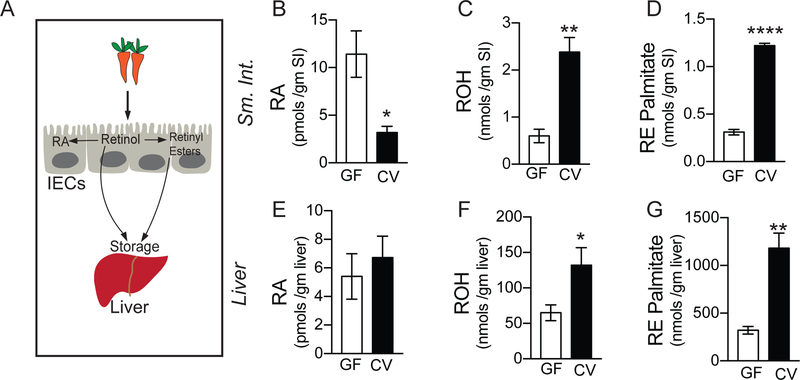
Animals cannot synthesize vitamin A de novo, but obtain the molecules as micronutrients from the diet. IECs are the main cell type responsible for the uptake of dietary vitamin A (Chelstowska et al., 2016). After uptake by IECs, diet-derived vitamin A or retinol can either be processed into retinyl esters or further metabolized to RA by two sequential oxidation steps (Figure 1A). To assess whether gut bacteria play a role in modulating vitamin A metabolism, we quantified the amount of various vitamin A metabolites (collectively called retinoids) in intestinal and extra-intestinal tissue of germ-free (GF) and conventional (CV) mice by liquid chromatography coupled to mass spectrometry (LC-MS). We found that GF mouse small intestine had significantly higher concentration of active metabolite (RA) compared to small intestine of conventionally raised mouse (Figure 1B). Concurrently, we observed lower concentration of precursor form (retinol) (Figure 1C) and storage form (retinyl esters) (Figure 1D) in the intestinal tissue of GF mouse compared to CV mouse. This indicates that gut bacteria suppressed the conversion of vitamin A into RA and promoted conversion into retinyl esters in the intestine. To assess if this accumulation of retinyl esters in the intestinal tissue of CV mice corresponded with higher retinoids in liver where 80% of vitamin A of the whole body is stored, we also quantified retinoids in the liver. We detected significantly higher quantities of retinol and retinyl esters (Figures 1F and1G), but not RA in the liver of CV mice (Figure 1E), suggesting that the presence of bacteria directed vitamin A metabolism away from RA synthesis in the intestinal tissue and towards vitamin A storage in liver. These results imply that bacteria regulate the amount of active metabolite of vitamin A locally in the intestine and impact storage of vitamin A in the liver.
Figure 1. Gut commensals modulate intestinal and extra-intestinal vitamin A metabolism.
(A) Diagram representing vitamin A absorption by the intestinal epithelial cells (IECs) and storage in the liver. Once taken up by IECs, Vitamin A (retinol) can be directly or after being processed into retinyl esters stored in the liver. Retinol can be also be metabolized into its active form, retinoic acid (RA) by IECs.
(B) Quantification of retinoic acid (RA) in the small intestine (SI) of germ-free (GF) and conventional mice (CV) performed using LC-MS.
(C) Quantification of retinol (ROH) in the small intestine (SI) of germ-free (GF) and conventional mice (CV) performed using LC-MS.
(D) Quantification of retinyl ester palmitate (RE) in the small intestine (SI) of germ-free (GF) and conventional mice (CV) performed using LC-MS.
(E) Quantification of retinoic acid (RA) in the liver of germ-free (GF) and conventional mice (CV) performed using LC-MS.
(F) Quantification of retinol (ROH) in the liver of germ-free (GF) and conventional mice (CV) performed using LC-MS.
(G) Quantification of retinyl ester palmitate (RE) in the liver of germ-free (GF) and conventional mice (CV) performed using LC-MS, (N=4 per group). Student’s t-test. Error bars represent SEM. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
Also see Supplemental Table 1
Commensal bacteria modulate vitamin A metabolic machinery in the intestinal epithelium.
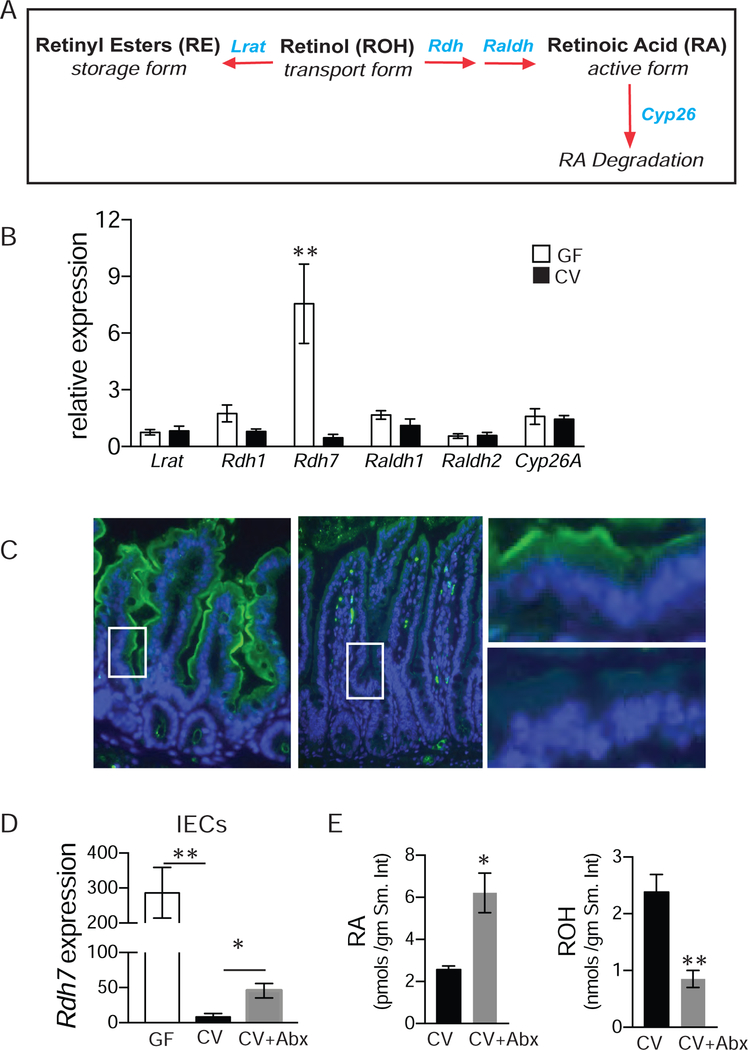
RA concentration in tissues is determined by strict temporal and spatial regulation of genes involved in vitamin A metabolism, retinoid transport, and RA catabolism (Figure 2A) (Lampen et al., 2000) . Next we wanted to determine if differing amounts of retinoids in GF and CV mice were due to bacterial modulation of vitamin A metabolic machinery in the gut. Panel of genes involved in vitamin A metabolism, storage and RA degradation were assessed in small intestine of germ-free and conventional mice by qPCR (Figure 2B). Our analysis revealed that expression of Rdh7, a gene that has previously been shown to catalyze the oxidation of retinol to retinaldehyde is significantly reduced in the presence of gut bacteria (Figure 2B) (Su et al., 1998). Expression of Rdh7 is limited to gastrointestinal tract during later embryonic development and adult stage and is conspicuously absent in early embryonic stage (Tomita et al., 2000). We did not detect differences in expression of genes encoding retinaldehyde dehydrogenases (Raldh) that mediate conversion of retinaldehyde to RA and lecithin retinyl acetate transferase (Lrat) (Figure 2B) that mediates conversion of retinol to retinyl ester for storage. We also did not observe any difference in expression of cytochrome P450 superfamily of genes (Cyp26a) that catabolize active RA to inactive metabolites. Therefore, we inferred that reduced concentration of RA in the intestinal tissue of conventional mice is due to lower expression of Rdh7. Similar analysis comparing expression of vitamin A metabolic genes in the liver tissue of GF and CV mice revealed that bacterial regulation Rdh7 expression was specific to the intestinal tissue, although we did observe that CV liver has higher expression of Cyp26a and Lrat compared to GF liver (Figures S1A and S1B).
Figure 2. Gut commensal bacteria modulate vitamin A machinery in the intestinal epithelium.
(A) Schematic representation of vitamin A metabolism. Vitamin A/retinol is absorbed from the diet can be processed into retinyl ester for storage, by the enzyme lrat. It can be metabolized into retinoic acid (RA) through a two-step reaction, with the help of retinol dehydrogenases (rdhs) as well as retinaldehyde dehydrogenases (raldhs). RA, when in excess, is degraded by cyp26.
(B) mRNA quantification by qPCR of genes involved in Vitamin A metabolism in small intestinal tissue.
(C) Immunofluorescence of Rdh7 (green) in the small intestinal tissue, DAPI was used to stain for the nuclei (blue).
(D) Rdh7 mRNA quantification by qPCR in the intestinal epithelial cells (IECs) isolated using laser capture microdissection (LCM) from small intestine of GF, CV, and CV mice treated with antibiotics for 7 days to deplete their microbiome and recapitulate germ-free conditions (CV +ABX).
(E) Using LC-MS, retinoic acid (RA) and retinol (ROH) were also quantified in CV and CV+ABX mice.
A–D : Representative figure of an experiment done 3 individual times. (N=4 per group). E: n=4. Student’s t-test; Error bars represent SEM. *P < 0.05, **P < 0.01.
Also see Figure S1
While dietary vitamin A is absorbed exclusively by the IECs, multiple cell types in the intestine such as dendritic cells and stromal cells in addition to IECs are capable of metabolizing vitamin A into RA (Hurst and Else, 2013; Jaensson et al., 2008; Vicente-Suarez et al., 2015). To determine cellular localization of Rdh7 we performed immuno-fluorescence assay using an anti-Rdh7 antibody. Immuno-staining of small intestine and colon tissue sections from GF and CV mice revealed that gut bacteria decreased Rdh7 protein specifically in the IECs in both small intestine and colon (Figure 2C and Figure S1C and S1D).
Commensal bacteria regulate several aspects of gut physiology including angiogenesis and fat uptake which could secondarily effect vitamin A metabolism in the gut (Nicholson et al., 2012).To assess if bacteria actively modulate RA concentration via Rdh7 expression in the IECs we partially depleted gut bacteria by orally treating CV mice with a cocktail of antibiotics for 1 week. Quantitative PCR analysis of laser captured IECs revealed that partial depletion of gut bacteria significantly increased Rdh7 expression in antibiotic treated mice (Figure 2D) compared to the untreated mice but not to the same extent as GF animals. The increased expression of Rdh7 in IECs coincided with increased RA concentration and decreased ROH concentration in the intestinal tissue of antibiotic treated mice compared to untreated mice (Figure 2E). These results establish that lower amounts of RA in the CV intestinal tissue are the result of gut bacteria actively suppressing vitamin A metabolic gene Rdh7 in the IECs.
Bacterial communities differentially regulate Rdh7 expression and RA synthesis in the gut.
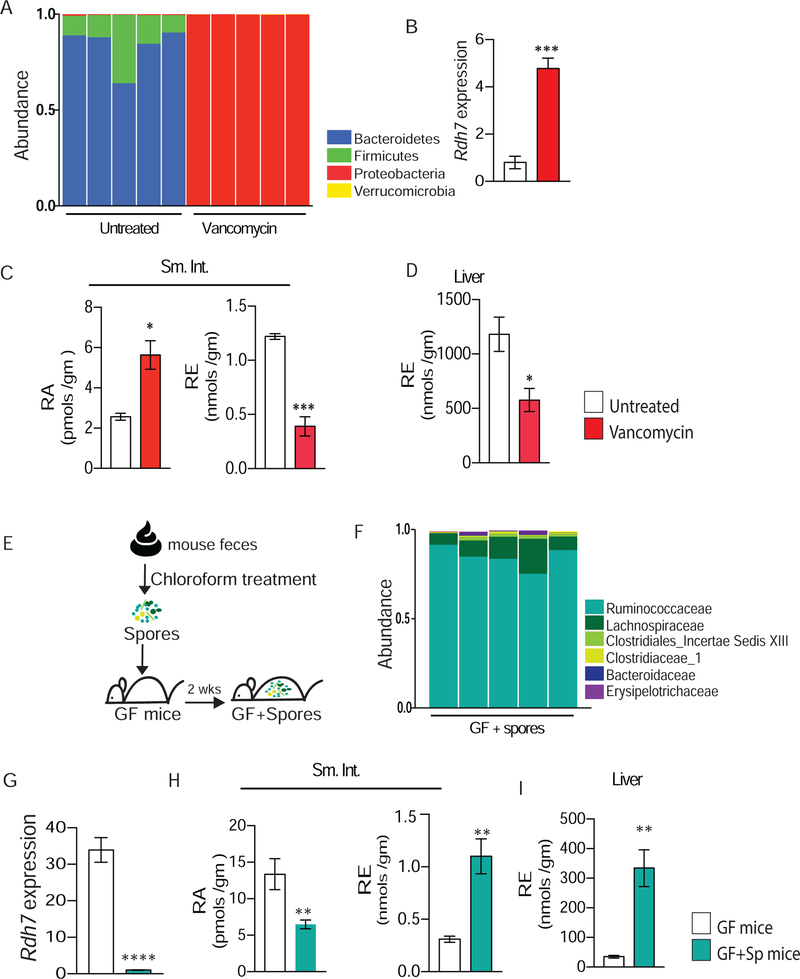
In a healthy state, more than 80% of the gut microbiome is constituted of obligate anaerobes belonging to phyla Firmicutes and Bacteroidetes and less than 5% belong to phylum Proteobacteria (Xiao et al., 2015). However, inflammation or treatment with specific antibiotics can result in dysbiotic microbial communities where obligate anaerobes diminish, and Proteobacteria blooms to constitute most of the microbiome (Winter et al., 2013). We wanted to evaluate if healthy and dysbiotic gut communities modulate Rdh7 gene expression and RA synthesis in the gut analogously. To do this we treated CV mice with vancomycin, an antibiotic that preferentially targets gut anaerobes (Vrieze et al., 2014)and polymyxin B, an antibiotic to which gut bacteria are highly resistant (Cullen et al., 2015). Gut microbiomes of mice treated with vancomycin for 4 weeks in drinking water had a drastic bloom in Proteobacteria and depletion of anaerobic bacteria while polymyxin B had minimal effect compared to untreated mice (Figure 3A and S2A). We observed that in comparison to untreated mice, vancomycin treated mice had a significantly higher expression of Rdh7 while polymyxin B treatment did not result in any change (Figure 3B and S2B). Bacterial load did not account for these differences, as the bacterial load was similar between both antibiotic treatments (Figure S2C). High Rdh7 expression in vancomycin treated group coincided with significantly higher amounts of RA in the intestinal tissue (Figure 3C). Conversely, lower amounts of retinyl esters (RE) were observed in both small intestine and liver of vancomycin treated group compared to untreated (Figures 3C and 3D). These data illustrate that while symbiotic microbial communities suppressed Rdh7 expression and RA synthesis in the gut, dysbiotic communities do not.
Figure 3. Microbial dysbiosis promotes Rdh7 expression and RA synthesis in the gut.
(A) Relative abundance of bacteria at phylum level determined by 16s rRNA analysis of fecal microbiome from untreated and vancomycin treated mice. Mice were treated with vancomycin (500mg/L) in drinking water for four weeks.
(B) Rdh7 mRNA quantification by qPCR in small intestine tissue mice treated with untreated versus vancomycin treated mice.
(C) Quantification of retinoic acid (RA) and retinyl ester (RE) in the small intestine (Sm. Int.) of untreated versus vancomycin treated mice using LC-MS.
(D) Quantification of retinoic acid (RA) and retinyl ester (RE) in the liver of untreated versus vancomycin treated mice using LC-MS.
(E) Diagram illustrating how spore-forming bacteria were given to germ-free mice. Fecal content isolated from female conventional mice went through chloroform treatment, and it was given to mice orally.
(F) Relative abundance of bacteria at family level determined by 16s rRNA analysis of fecal microbiome from mice that received spore-forming bacteria (GF+Sp).
(G) Rdh7 mRNA quantification by qPCR in small intestine tissue of germ-free (GF )and germ-free mice that received spore-forming bacteria (GF+Sp). (N= 5 per group) and Student’s t-test. Error bar represents SEM. ****P < 0.001 (H) Quantification of retinoic acid (RA) and retinyl ester (RE) in the small intestine (SI) of germ-free (GF) and germ-free +spores (GF+Sp) performed using LC-MS.
(I) Quantification of retinoic acid (RA) and retinyl ester (RE) in the liver of germ-free (GF) and germ-free +spores (GF+Sp) performed using LC-MS.
For figures A and B we are showing a representative figure of a single experiment that was done 3 individual times (N=4 per group). C and D : n=4 per group. Student’s t-test. For figures G-I Student’s t-test was used . Error bars represent SEM. *P < 0.05, **P < 0.01, ***P < 0.0001.
Also see Figure S2
To further understand the microbial trigger in a symbiotic community leading to Rdh7 regulation, we isolated spore- forming bacteria (Figure 3E) by treating mouse feces with chloroform as it has been previously described (Velazquez et al., 2017). GF mice were inoculated with the fecal spore preparation and after 2 weeks, 16S fecal microbiome analysis confirmed that these mice were colonized specifically with spore-forming bacteria belonging to class Clostridia (Figure 3F). qPCR analysis revealed that bacteria belonging to class Clostridia significantly diminished Rdh7 expression in the intestinal tissue (Figure 3G). The Rdh7 decrease was accompanied by lower RA concentration and higher RE concentration in the small intestine (Figure 3H), as well as higher RE concentration in the liver (Figure 3I) of compared to GF mice.
Epithelial cell intrinsic Rdh7 expression controls RA signaling in gut resident lymphocytes.
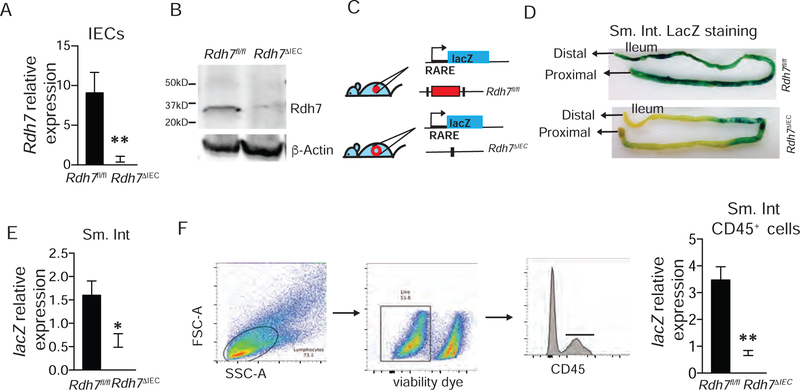
Given its unique tissue distribution and differential regulation by gut bacteria, we hypothesized that Rdh7 expression in IECs may serve as the control point for regulating RA-dependent immune responses in the gut. To evaluate the functional contributions of IEC intrinsic Rdh7 in terms of RA biosynthesis, and the potential downstream impact on intestinal immunity we generated a cell-specific loss of function mouse model of Rdh7 (Figures S3A–E). By breeding mice harboring Rdh7 “floxed” ( Rdh7fl/fl) allele with mice that express Cre recombinase specifically in intestinal epithelial cells (Villin-Cre) we generated mice that lack Rdh7 in IECs (Rdh7∆IEC) The loss of Rdh7 gene expression and protein in Rdh7∆IEC was validated by Qpcr and western blot (Figures 4A and4B). It is important to highlight that lack of Rdh7 in the epithelium did not affect epithelial cells differentiation as the numbers of specialized epithelial cells such as goblet and Paneth cells were similar (Figures S3F and S3G). To test if loss of Rdh7 in IECs had consequences for RA synthesis, we crossed reporter mice that harbor a RA response element (RARE) upstream of β-galactosidase (lacZ) transgene to Rdh7∆IEC and Rdh7fl/fl mice such that LacZ expression serves as a proxy for RA concentration in the tissue (Figure 4C). The LacZ expression and activity was measured by LacZ qPCR and β-galactosidase assay in small intestine. We found that LacZ expression as well β-galactosidase staining in Rdh7∆IEC mice small intestine was significantly reduced compared to Rdh7fl/fl mice indicating that Rdh7 is required for RA signaling in the small intestinal tissues (Figures 4D and 4E). Quantification of LacZ expression as well as intestine specific homeobox (ISX), an RA responsive gene expressed in the IECs established that loss of Rdh7 in IECs did not affect IEC specific RA signaling (Figure S3H and S3I). Furthermore, we observed lower expression of another RA responsive gene, stimulated by retinoic acid 6 (Stra6) that is not specific to IECs in Rdh7∆IEC (Figure S3J). Stra6 is expressed in multiple cell types including activated lymphocytes and is required for cellular vitamin A uptake (Kawaguchi et al., 2007). These data suggest that Rdh7 dependent RA production by IECs acts in a paracrine manner rather than autocrine. Next we assessed if loss of IEC intrinsic Rdh7 caused reduced RA signaling in underlying immune cells by quantifying LacZ mRNA in small intestinal lamina propria lymphocytes. Our results showed that RA signaling in small intestinal lymphocytes is indeed significantly diminished in Rdh7∆IEC mice compared to Rdh7fl/fl mice (Figure 4F).
Figure 4. Epithelial cell intrinsic RDH7 regulates RA signaling in the intestinal tissue including lamina propria lymphocytes.
(A) Rdh7 mRNA quantification in intestinal epithelial cells (IECs) from Rdh7fl/fl and Rdh7∆IEC . IECs were extracted using laser capture microdissection (LCM).
(B) Western blot analysis of Rdh7 protein in small intestinal tissue from Rdh7fl/fl and Rdh7∆IEC mice.
(C) RA signaling reporter mice to assess RA signaling in Rdh7fl/fl and Rdh7∆IEC mice. Mice that harbor a RA response element (RARE) upstream of β-galactosidase (lacZ) transgene were crossed to Rdh7∆IEC and Rdh7flox/flox (D) β-galactosidase staining of small intestine from RARE_lacZ reporter transgene carrying Rdh7fl/fl and Rdh7∆IEC mice.
(E) lacZ mRNA quantification in the small intestine of RARE_lacZ reporter transgene carrying Rdh7fl/fl and Rdh7∆IEC mice.
(F) Gating strategy to isolate lamina propria lymphocytes by sorting live CD45+ cells from small intestine of RARE_lacZ reporter transgene carrying Rdh7fl/fl and Rdh7∆IEC mice. lacZ mRNA quantification in lamina propria lymphocytes isolated by sorting live CD45+ cells from small intestine of RARE_lacZ reporter transgene carrying Rdh7fl/fl and Rdh7∆IEC mice.
Figures are representing a single experiment that was repeated 3 individual times ( n=4 per group) Student’s t-test; Error bars represent SEM. *P < 0.05, **P < 0.01.
Also see Figure S3
Epithelial cell RA dictates IL-22 amounts and activity in the gut.
Next, we wanted to assess if loss of RA synthesis in the IECs resulted in defect in specific RA-dependent intestinal immune responses in Rdh7∆IEC mice compared to Rdh7fl/fl mice. We observed no significant difference in numbers of Th17 and Treg cells in the small intestine and colon (Figures S4A and S4B), nor the number of IgA+ cells in the small intestine of Rdh7∆IEC and Rdh7fl/fl mice (Figure S4C), indicating that RA produced by IECs did not regulate T-cell differentiation and IgA class switching.
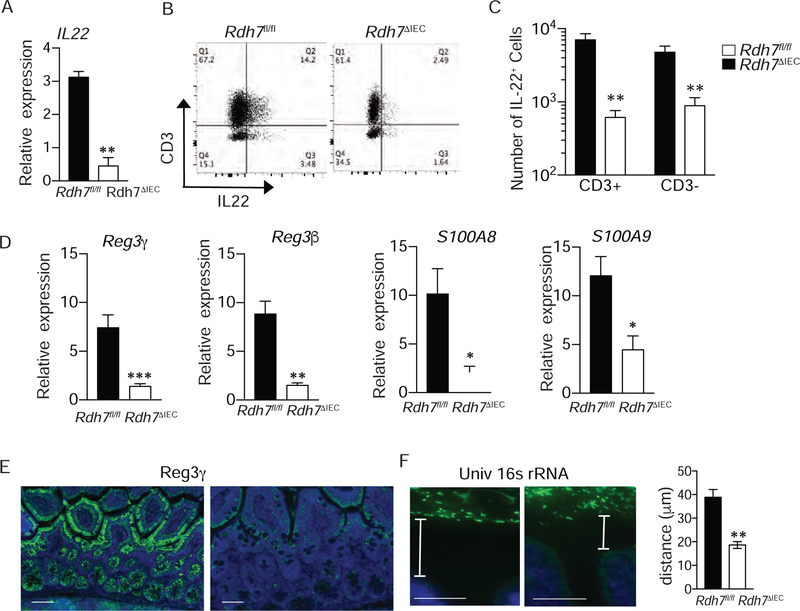
RA has recently been shown to play a key role in promoting innate immunity by enhancing the expression of Il-22 in the gut through multiple mechanisms, including promoting maturation and proliferation of IL-22 producing RORγt+ innate lymphoid cells (ILC3), direct binding to the il-22 locus, and upregulation of IL-23 from DCs that acts as a potent inducer of IL-22 (DePaolo et al., 2011; Goverse et al., 2016; Mielke et al., 2013). To assess if IEC intrinsic RA synthesis is required for maintaining ILC3 cells in the gut we performed flow cytometric analysis on small intestinal lamina propria lymphocytes from Rdh7ΔIEC and Rdh7fl/fl mice. We found that the numbers of NCR+ ILC3 in the small intestine of Rdh7ΔIEC was significantly less than Rdh7fl/fl mice, although there was no difference in NCR- ILC3 or lymphoid tissue inducer cells (LTi) (Figure S4D). Comparison of Rdh7ΔIEC and Rdh7fl/fl small intestinal tissue revealed that Il-22 expression was significantly diminished in mice lacking Rdh7 (Figure 5A). We also observed a decrease in the number of cells that secrete IL-22 in the small intestine (Figures 5B and 5C) as well as in the colon (Figure S4E). Together, our results establish that IEC intrinsic RA modulates the amount of IL-22 in the intestinal tissue by regulating Il-22 expression and/or numbers of IL-22 producing cells in the gut.
Figure 5. Epithelial Cell Intrinsic Rdh7 expression controls IL-22 levels and activity in the gut.
(A) Il-22 mRNA quantification in small intestine of Rdh7fl/fl and Rdh7∆IEC mice.
(B) Representative plots showing IL-22 producing CD3+ and CD3- lamina propria lymphocytes.
(C) Frequencies of CD3+ IL-22+ and CD3- IL-22+ lymphocyte populations in small intestine of Rdh7fl/fl and Rdh7∆IEC littermate mice.
(D) mRNA quantification of antimicrobials peptides in the intestine controlled by IL-22.
(E) mmunofluorescent of Reg3γ (green) on small intestine tissue, and DAPI was used to visualize nuclei (blue).
(F) Fluorescence In-situ hybridization (FISH) analysis using 16S rDNA universal probe (green), DAPI was used to visualize nuclei (blue)
Representative figures of single experiments that were repeated 3 individual times (N=4 per group). Student’s t-test; Error bars represent SEM. *P < 0.05, **P < 0belo.01 and ***P < 0.001.
Also see Figure S4
IL-22 produced by ILCs and T-cells acts on intestinal epithelial cells and upregulates mucosal antimicrobial response (Sonnenberg et al., 2011). Accordingly, Rdh7ΔIEC mice had significant reduction in IL-22 dependent antimicrobials such as Reg3γ, Reg3β, and calprotectin subunits, S100A8 and S100A9 in the small intestine compared to Rdh7fl/fl mice (Figure 5D), and the same was true in the colon ( Figure S4F) . In accordance with these findings we saw that reduced Reg3γ in the small intestinal tissue Rdh7ΔIEC mice coincided with diminished spatial segregation between intestinal microbiota and epithelial cells (Figures 5E and 5F). These data suggest that intestinal RA production regulatesIL-22 dependent antimicrobial response to modulate interactions with gut bacteria.
Rdh7 ΔIEC mice have enhanced resistance to colonization by gut pathogen.
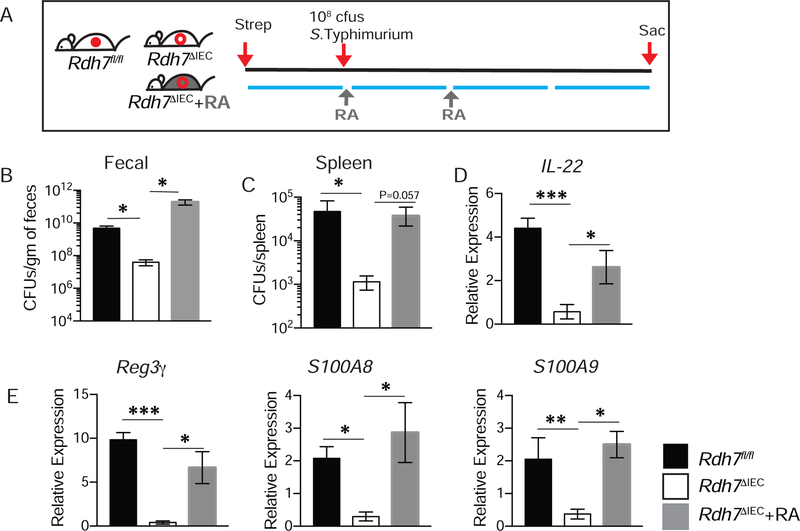
High amounts of IL-22 is produced by intestinal immune cells upon infection with enteric pathogens. Depending on the pathogen, IL-22 can either protect the host from colonization as in the case of attaching and effacing bacteria such as Citrobacter rodentium (Zheng et al., 2008), or promote pathogen colonization as in the case of Salmonella Typhimurium (Behnsen et al., 2014b). Rdh7ΔIEC mice did not show increased susceptibility to C. rodentium infection compared to Rdh7fl/fl mice even though they exhibited reduced Il-22 expression during infection (Figure S5A –S5C). This implies that reduced IL-22 due to defective IEC intrinsic RA synthesis did not render the host susceptible to C. rodentium infection. This is in line with studies showing that complete and not partial loss of IL-22 is necessary for susceptibility to C. rodentium infection (Rankin et al., 2016). On the other hand, Rdh7ΔIEC mice upon infection by S. Typhimurium showed significantly reduced pathogen load in the feces and pathogen dissemination to the spleen compared to Rdh7fl/fl mice (Figures 6A, 6B and 6C). Lower pathogen loads in the spleen were not because of difference in systemic immunity since both Rdh7fl/fl and Rdh7ΔIEC were equality susceptible to S. Typhimurium when challenged intraperitoneally (Figure S5 F). This resistance to S. Typhimurium colonization corresponded with reduced expression of Il-22 and IL-22 induced antimicrobials in Rdh7ΔIEC compared to Rdh7fl/fl mice (Figures 6D and 6E). More importantly, treatment of Rdh7ΔIEC mice with exogenous RA reversed the deficiency in Il-22 expression and IL-22 dependent antimicrobial response (Figures, 6D and 6E) making them equally susceptible to S. Typhimurium colonization (Figures 6B and 6C). This susceptibility was not genotype specific and was dependent on RA, since CV mice treated with exogenous RA showed enhanced susceptibility to S. Typhimurium colonization compared to untreated mice (Figure S5 G). We did not observe significant differences in neutrophil recruitment between the groups indicating that higher bacterial load in Rdh7fl/fl mice and Rdh7ΔIEC +RA mice were not due to differences in neutrophil recruitment (Figure S5 D and E). To confirm that increase of IL-22 was specifically required to render susceptibility to pathogen colonization in our mouse model we performed IL-22 supplementation assay (Figure S5 H). We observed that exogenous IL-22 supplementation also reversed the colonization resistance phenotype in Rdh7ΔIEC mice comparable to Rdh7fl/fl mice (Figure S5I and J). These results demonstrate that RA deficiency due to Rdh7 deletion in IECs afforded protection against pathogen colonization specifically due to IL-22 deficiency. Taken together these results underscore the significance of suppression of IEC intrinsic RA synthesis by commensals in preventing expansion of pathogenic bacteria in the gut.
Figure 6. RA deficiency in Rdh7∆IEC mice is protective against pathogen colonization.
(A)Diagram illustrating S.Typhimurium infection and RA treatment timeline. 24 hours prior to infection, mice were treated with 20mg of streptomycin via oral gavage, followed by 108cfu/mL 24 post streptomycin. 250µg RA was administered twice intraperitoneally to Rdh7∆IEC. Mice were sacrificed 72 hours post infection.
(B) Quantification of S. Typhimurium bacterial burden in feces from streptomycin treated Rdh7fl/fl, Rdh7∆IEC and Rdh7∆IEC+RA mice at 72 hours post infection.
(C) S. Typhimurium burden in spleen after 72 hours post infection Rdh7fl/fl, Rdh7∆IEC and Rdh7∆IEC+RA streptomycin treated mice.
(D) mRNA quantification of Il-22 in the colon of Rdh7fl/fl, Rdh7∆IEC and Rdh7∆IEC+RA streptomycin treated mice 72 hours post infection with S. Typhimurium.
(E) mRNA quantification of IL-22 dependent antimicrobials, Reg3γ and calprotectin subunits S100A8 and S100A9 in the colon of Rdh7fl/fl, Rdh7∆IEC and Rdh7∆IEC+RA mice 72 hours post infection with S. Typhimurium.
All the mice used for this experiment were littermate controls that were co-housed before infection and housed separately after infection (N=6). Figures represent an individual experiment that was repeated four times. Error bar represent SEM. One-way ANOVA. *P < 0.05, **P < 0.01 and ***P < 0.001.
Also see Figure S5
Reduced antimicrobial response in Rdh7 ΔIEC mice protects against microbial dysbiosis.
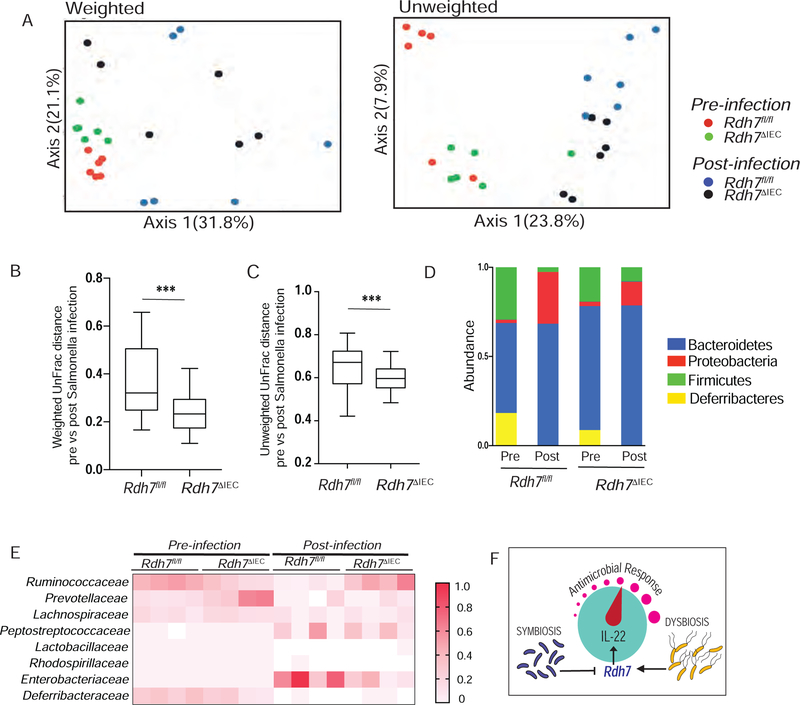
One of the drivers of Salmonella-induced dysbiosis is the depletion of Clostridia, SCFA producing anaerobic commensals within the phylum Firmicutes. Decrease in SCFA due to Clostridia depletion leads to elevated oxygenation and increased aerobic proliferation of S. Typhimurium (Rivera-Chavez et al., 2016). Therefore, next we wanted find out if differences in gut microbiota composition might explain lower Salmonella burden in Rdh7ΔIEC compared to Rdh7fl/fl mice. 16s rRNA analysis of fecal microbial communities revealed that pre-infection fecal microbial communities in both Rdh7ΔIEC and Rdh7fl/fl were constituted of mainly of phylum Bacteroidetes and Firmicutes (Figures 7A–7D). However, we found that post Salmonella infection, fecal communities of Rdh7ΔIEC mice suffered less shift relative to their pre-infection state than Rdh7fl/fl mice (Figures 7A–7D). Moreover, we observed that compared to Rdh7fl/fl, Rdh7ΔIEC mice preserved higher proportion of obligate anaerobes belonging to phylum Firmicutes (Figure 7D). Specifically, bacteria belonging Ruminococcaceae family were significantly higher in Rdh7ΔIEC mice post-infection compared to Rdh7fl/fl mice. Concurrently, post-infection Rdh7ΔIEC mice had significantly less proportion of bacteria belonging to class Enterobacteriaceae which corresponded with our data that Rdh7ΔIEC mice showed enhanced colonization resistance to Salmonella (Figure 7E). Our data suggested that preservation of commensal bacteria specifically of those belonging to phylum Firmicutes prevented efficient pathogen colonization of Rdh7ΔIEC mice compared to Rdh7fl/fl mice. Based on our current findings we propose a model where IEC intrinsic Rdh7 expression and RA synthesis act as a “control knob” to regulate IL-22 activity in the gut (Figure 7F). Decrease of Rdh7 gene expression and RA synthesis by obligate anaerobes promotes symbiosis by “dialing down” IL-22 dependent antimicrobial response, whereas RA synthesis and IL-22 activity is “dialed up” by pathogens promoting microbial dysbiosis and pathogen colonization.
Figure 7. IEC intrinsic Rdh7 ablation protects the host against microbial dysbiosis during infection.
(A) PCoA plot of the fecal microbiota composition (weighted and unweighted UniFrac distances) (B) Box-and-whisker plot (boxes show median, first and third quartiles, whisker denotes minimum to maximum range) of intercommunity β-diversity within the fecal microbiomes of Rdh7fl/fl and Rdh7∆IEC pre and post Salmonella infection determined by weighted 16S UniFrac distances.
(C) Box-and-whisker plot (boxes show median, first and third quartiles, whisker denotes minimum to maximum range) of intercommunity β-diversity within the fecal microbiomes of Rdh7fl/fl and Rdh7∆IEC pre and post Salmonella infection determined by unweighted 16S UniFrac distances.
(D) Phylum-level microbiota composition of Rdh7fl/fl and Rdh7∆IEC pre and post Salmonella infection.
(E) Heat map of the fecal microbiome composition at the family level of Rdh7fl/fl and Rdh7∆IEC pre and post Salmonella infection.
(F) Diagram illustrating how microbial regulation of Rdh7 in the intestinal epithelium controls the IL-22 levels and antimicrobial response.
All the mice used for this experiment were littermate controls that were co-housed before infection and housed separately after infection (N=6). Error bar represent SEM. Mann-Whitney test. *P < 0.05, **P < 0.01 and ***P < 0.001.
Discussion
In this study we demonstrated a direct role of bacteria in modulating the concentration of the vitamin A metabolite RA in the gut. Specifically, we showed that gut bacteria differentially regulate expression of Rdh7, a key gene involved in conversion of retinol into RA in the intestinal epithelium. We found that bacteria belonging to class Clostridia suppressed Rdh7 expression and RA synthesis in the gut whereas bacteria belonging to phylum Proteobacteria did not. By employing genetic mouse models harboring a deletion in Rdh7 in IECs (Rdh7ΔIEC) coupled to RA-signaling reporter mouse, we showed that Rdh7 expression is required for regulating RA signaling and IL-22 production by gut residing lymphocytes. Furthermore, Rdh7ΔIEC mice compared to Rdh7fl/fl mice exhibited enhanced resistance to Salmonella Typhimurium, an enteric pathogen that exploits heightened IL-22 dependent antimicrobial response to outcompete commensal bacteria to colonize the gut.
Vitamin A metabolite, RA is crucial in inducing gut tropism in lymphocytes and in modulating T helper cell differentiation (Hill et al., 2008; Iwata et al., 2004; Mora et al., 2006; Mucida et al., 2007). In addition to its widely recognized role in adaptive immunity, increasing evidence identifies RA as an important modulator of innate immune cells, such as tolerogenic dendritic cells (DCs) and innate lymphoid cells (ILCs) (Goverse et al., 2016; Jaensson-Gyllenback et al., 2011; McDonald et al., 2012; Mielke et al., 2013). Studies examining the role of vitamin A in mucosal immunity thus far have largely depended on acute or chronic deprivation of vitamin A through dietary or pharmacologic means. Since RA regulates the functional fate of almost every cell type involved in innate and adaptive immunity, such global strategies of modulating RA-signaling have produced inconsistent roles for RA in promoting immune homeostasis in the gut (Jason et al., 2002; Mullin, 2011; Ross, 2012; Sirisinha, 2015). In contrast to previous studies, our work focused on bacterially modulated vitamin A metabolic gene in the intestinal epithelium. In doing so our work provides key insights into the tissue specific and context dependent role of RA in regulating immune homeostasis with gut bacteria. We have uncovered that spore forming gut commensals known to be dominated by Clostridial species (Atarashi et al., 2011) provide the microbial stimuli that inhibited RA synthesis by IECs. On the other hand microbial communities that suffer from dysbiosis, characterized by lack of Clostridia species and bloom of Proteobacteria due to antibiotic treatment, did not suppress RA synthesis.
Complex consortia of bacteria colonize the gut and constantly interact with the intestinal epithelium as well as the underlying immune cells. Intestinal epithelium intrinsic antimicrobials are upregulated in response to bacterial presence to protect host tissue from bacterial encroachment (Duerkop et al., 2009). A tight regulation of intestinal antimicrobial response in turn is needed for maintaining homeostatic host-microbe interactions in the gut. Insufficient or impaired antimicrobial response results in inability to maintain spatial segregation between commensal bacteria and mucosal surfaces (Vaishnava et al., 2011), whereas excessive antimicrobial response during an infection favors microbial dysbiosis and pathogen colonization (Behnsen et al., 2014b; Miki et al., 2017). Multiple feedback loops involving direct and indirect microbial sensing by the intestinal epithelium have been shown to upregulate antimicrobial response in the gut. Direct feedback loop operates by sensing bacterial ligands via epithelial cell intrinsic innate immune receptors such as Toll-like and Nod-like receptors to upregulate antimicrobial response (Vaishnava et al., 2008; Voss et al., 2006). Indirect feedback loop involves sensing of bacterial ligands by gut residing immune cells leading to secretion of cytokines such as IL-22 and IL-17 that then further reinforces intestinal epithelium intrinsic antimicrobial response (Kinnebrew et al., 2010; Zheng et al., 2007).
Although IL-22 has been shown to be key in regulating barrier function against intestinal bacteria and other insults by upregulating antimicrobial response, cell proliferation and tissue repair(Costa et al., 2013; Eidenschenk et al., 2014; Monteleone et al., 2011; Rubino et al., 2012), IL-22 induction is not always beneficial for the host. Several studies have shown that IL-22 can be a potent inducer of pathological inflammation. Indeed, IL-22 can promote tissue inflammation and self-destruction and is involved in the pathophysiology of several immune-mediated inflammatory diseases, such as psoriasis, celiac and rheumatoid arthritis(DePaolo et al., 2011; Geboes et al., 2009; Sonnenberg et al., 2010; Zheng et al., 2007). Moreover, IL-22 induction can be exploited by pathogens such as Salmonella to suppress the growth of commensal bacteria, thereby enhancing pathogen colonization of mucosal surfaces (Behnsen et al., 2014b). Although for some other pathogens such as Citrobacter rodentium IL-22 activity is essential for host protection (Qiu et al., 2012; Zheng et al., 2008).
A tight regulation of the IL-22 is therefore critical in maintaining the beneficial effects of IL-22 and avoiding deleterious inflammatory effects. Previous studies have demonstrated that IL-22 can be negatively regulated by IL-22 Binding Protein (IL-22BP, also known as IL-22RA2), a soluble receptor secreted by gut DCs that is able to capture IL-22 from IL-22 receptor (IL-22RA1) complex (Huber et al., 2012). Additionally, epithelial expression of IL-25 is able to suppress IL-22 production by RORyt+ ILCs (Sawa et al., 2011). Since RA has been shown to enhance IL-22 responses in mice through multiple mechanisms (Goverse et al., 2016; Mielke et al., 2013), regulating RA levels would be an effective strategy in normalizing IL-22 activity in the gut. Our studies now show that RA sourced from IECs is required for maintaining IL-22 production and activity in the gut. It is interesting to note that both IL-25 secretion and RA production by IECs that can regulate IL-22 are modulated by commensal bacteria. Taken together these data suggest that a direct dialogue between IL-22 responsive cells and IL-22 producing cells mediated by commensal bacteria is essential for titrating antimicrobial response in the gut.
The ability of RA to induce differentiation of T-reg cells at the expense of Th17 cells has made RA an attractive therapeutic candidate for IBD. Studies show that treatment of DSS-treated mice with RA reduces the severity of the disease (Hong et al., 2014). Studies further show that treatment with RA results in increased expression of T-reg cells and inhibited IL-17 production during 2,4,6-trinitrobenzenesulfonic acid (TNBS)-induced colitis, another chemically induced colitis mouse model (Bai et al., 2009). These observations imply that RA can restore the balance between Th17 and T-reg cells in IBD-like diseases in mice. In support of the notion that RA-mediated signaling may beneficially affect the course of IBD, vitamin A-deficient mice developed more severe colitis and recovered more slowly in both DSS and TNBS models. However the role of RA in IBD is not as straightforward as it previously seemed. In humans, a reduced activity of RA synthetic enzymes in DCs and macrophages was noted in patients with UC (Magnusson et al., 2016). Moreover, polymorphism in RA degrading enzyme CYP26B1 that results in higher levels of RA was recently shown to be associated with an increased risk of Crohn’s disease (Bhattacharya et al., 2016; Fransen et al., 2013) making the role of RA in promoting anti-inflammatory immune response contentious.
In IBD patients pathophysiology of colitis often coincides with aberrant interactions between the gut microbiota and the host mucosa and an heightened antimicrobial response in the intestinal tissue (Fahlgren et al., 2003; Ho et al., 2013). As a result many of the IL-22 induced antimicrobial proteins such those belonging to Reg family, Calprotectin and Hepcidins are routinely used as disease markers of colitis severity or response to therapy in humans (Fengming and Jianbing, 2014). It is plausible that heightened antimicrobial response feeds into microbial dysbiosis that is associated with IBD further promoting inflammatory immune response. Our work provokes the hypothesis that limiting IL-22 amounts via suppression of intestinal RA synthesis could be a mechanism by which the microbial self is protected from host antimicrobial response. Accordingly, one can envision lowering heightened antimicrobial response during disease states pharmacologically or microbially via modulation of IEC intrinsic RA synthesis to re-establish host-microbial symbiosis at the intestinal mucosa.
Although our current work establishes a clear link between type of microbial stimuli and its effect on RA synthesis in the gut, exact signaling mechanism that mediates Rdh7 gene expression in IECs remains unclear. Future work will determine how Clostridia species suppress Rdh7 and RA production in the gut. Whether it can be attributed to Clostridia’s ability to produce short chain fatty acids (SFCA) that affect global gene expression and the epigenome through inhibiting histone deacetylases (HDACs) remains to be seen(Koh et al., 2016; Krautkramer et al., 2016). Nevertheless, current work has direct implications for developing approaches that exploit gut microbiota to control tissue specific RA levels and modulate RA dependent mucosal immune response such as IL-22 production in the gut to provide clinical benefits.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and reasonable requests for reagents may be directed to, and will be fulfilled by the corresponding author, Shipra Vaishnava (shipra_vaishnava@brown.edu)
Experimental Model and Subject Details
Mice
Wild-type C57BL/6J mice were bred in the SPF barrier facility at the Brown University. RARE-Hspa1b/lacZ mice were purchased from Jackson Laboratory and bred to Rdh7fl/fl mice in the SPF barrier facility at Brown University. All mice used throughout this study were 6–8 weeks old. Germ-free C57BL/6J mice were raised and bred in flexible film isolators gnotobiotic isolators as described previously. For every experiment, mice were either littermates or were co-housed to make sure that they shared the same microbiota. Experiments were performed according to protocols approved by the Institutional Animal Care and Use Committees of the Brown University.
Generation of Rdh7fl/fl mice
To generate Rdh7-floxed mice, Rdh7-tm1a (KOMP) Wtsi ES cells (JM8A agouti) were purchased from and analyzed by KOMP. Then ES cells were microinjected into C57BL/6 blastocysts at UT Southwestern Medical Center transgenic core. Chimeric offsprings were backcrossed to C57BL/6 mice, and germline transmission was confirmed by southern blot and PCR of tail genomic DNA using primers P1, P2 and P3 as shown in (Fig S3). Founder mice were then crossed to FlpO mice to deplete the FRT-Neo-FRT cassette to generate Rdh7-floxed pups. Genotyping of Rdh7-floxed mice was performed using primers P1 and P2 with amplicons of a 1.1-kb product from the wild-type allele, and a 1.3-kb product from the targeted allele.
Quantitation of atRA, retinol, and retinyl esters in liver and small intestine
Methanol, acetonitrile, and water used for quantitative analysis were LC-MS grade and from EMD Millipore (Billerica, MA). Formic acid (LC-MS grade) and hexanes (HLPC grade) were from Fisher Scientific (Hampton, NH). All retinoid standards were purchased from Toronto Research Chemicals, except retinol-d8 and retinyl palmitate-d4, which were from Cambridge isotopes (Tewksbury, MA).
Retinoids were extracted from liver and intestine under yellow light to prevent retinoid isomerization and degradation using a two-step liquid-liquid extraction method adapted from Kane et al (Kane and Napoli, 2010). 30–120 mg tissue was homogenized with 5x volume of 0.9% saline on ice and 10 µL of internal standards in ethanol were added. For atRA quantitation, 4 µM atRA-d5 was added, for RE and ROH quantitation in intestinal tissue 1.5 µM of RE palmitate-d4 and ROH-d8 were added and for RE and ROH quantitation in liver 1.6 mM of RE acetate was added. To extract retinoids 2 mL of ethanol containing 0.025 M KOH was added, the sample was briefly vortexed and 10 mL hexanes was added. After centrifugation at 480 x g for 2 minutes the hexanes layer containing RE and ROH was collected and evaporated to dryness. To the aqueous layer, 120 µL of 4 M HCl was added, the sample vortexed, and 10 mL hexanes was added to the acidified sample to extract atRA. After centrifugation at 480 x g for 2 minutes, the top hexanes layer containing atRA was collected and evaporated to dryness at 40° C under a gentle stream of nitrogen. The RE/ ROH containing residue was resuspended in 80–200 µL ACN. The atRA containing residue was resuspended in 60 µL ACN for intestinal samples and 70 µL for liver samples. The resuspended extracts were transferred to amber glass vials with glass inserts. On the day of LC-MS analysis, 40 µL water was added to the small intestine atRA extracts to improve chromatographic resolution.
All retinoids from intestinal samples and atRA in livers were analyzed by LC-MS using an Agilent 1290 UHPLC (Agilent, Santa Clara, CA) coupled to a AB SCIEX 5500 QTRAP Q-Lit mass spectrometer (AB Sciex, Foster City, CA). For atRA analysis, 20 µL of sample was injected whereas for ROH and RE analysis 3 µL of sample was injected. The retinoids were separated using a 150 x 2.1 mm Supelco Ascentis Express reverse phase amide column (Sigma, St. Louis, MO) with 2.7 µm particle size and an Ascentis Express reverse phase amide 2.7 µm guard cartridge. A gradient elution with solvent A containing H2O + 0.1% formic acid and solvent B containing ACN + 0.1% formic acid was used. For atRA analysis mobile phase flow rate was 0.4 mL/min and the gradient was from initial conditions of 70% B held to 3 min to 78% B at 12 min then increasing to 100% B at 12.5 min and held at 100% B until 15 min before returning to initial conditions. For RE and ROH analysis the mobile phase flow rate was 0.5 mL/min and the gradient was from initial conditions of 60% B held until 2 min then increasing to 66% B at 9.22 min then increasing to 100% B at 13 min and held at 100% B until 25 min before returning to initial conditions.
The MS was operated either in MS/MS/MS (MS3) mode (for atRA liver analysis) or selected reaction monitoring (SRM) mode (for atRA, retinol, retinyl ester analysis in small intestine), in positive ion polarity using atmospheric pressure chemical ionization (APCI). The MS source temperature was 350° C and capillary curr ent was 5 µA. Ion source gas 1 was 80 and curtain gas was set to 35. Nitrogen was used for both the collision and curtain gases. The optimized SRM conditions are provided in Table S1. In MS3 mode, Q1 resolution was set to low, Q0 trapping was turned on, and scan rate was 10,000 Da/s. The m/z 205.2+ fragment generated from m/z 301.2 was further fragmented with the linear ion trap, with a fill time of 200 ms, and three ions, m/z 119.1, 159.1, 161.1 were summed for atRA quantitation. For the internal standard, the mass transition m/z 306.2>208.2>162.1 was monitored with a trap fill time of 5 ms. Q3 entry barrier of 8.0 volts, auxiliary frequency 27 mV, and excitation time 50 ms were used for MS3. The declustering potential was 62 V. All other parameters were the same as for MRM and listed in Table S1.
REs and ROHs were measured in liver by LC-UV using an Agilent 1200 series HPLC with a multi-wavelength UV detector and identical chromatographic conditions as described above. REs and ROHs were detected at 325 nm with a bandwidth of 10 nm. The reference wavelength was set to 425 nm with a bandwidth of 100 nm. The slit was set to 4 nm. Injection volume was 1 µL. The column was held at 45° C and the autosample r at 4° C.
Antibiotic treatment
Mice were given ampicillin (0.5g/ml), vancomycin (0.0.25g/ml), neomycin sulfate (0.5g/ml), gentamycin (0.5g/ml) and metronidazole (0.5g /ml) in drinking water for 7 days, as described previously (Abt et al., 2012). Sucralose-based artificial sweetener (Splenda) was added at 4 g/l to both antibiotic-treated and control mice drinking water. Microbiota depletion was verified by aerobic and anaerobic culture of intestinal contents. For vancomycin and polymyxin B treatment alone, mice received 500mg/L of vancomycin or 100mg/L pf polymyxin B in drinking water for 4 weeks. The solution was renewed every 3 days.
Preparation of lamina propria lymphocytes and isolation of intestinal epithelial cells
Lamina propria lymphocytes were isolated as described in the literature (Ivanov et al., 2006). In summary, mice were euthanized using isoflurane followed by cervical dislocation. Small intestine was removed and Peyer’s patches were removed. Intestine was cut longitudinally and cut into 16 pieces and thoroughly washed with ice-cold PBS. Intestinal epithelium was removed from the underlying tissue by incubation for 30 min at 37°C in 1 mM EDTA with calcium- and magnesium-free PBS, followed by vigorous shaking. Remaining tissues were digested by Collagenase I (Sigma- Aldrich), DNase I (Sigma- Aldrich) and Dispase (Sigma- Aldrich) for 25 min at 37°C twice. Cells were filtered through 70 µm cell strainers, re-suspended in RPMI complete media ( 3% FBS) and applied onto a 40%: 80% Percoll gradient (GE Healthcare, Pittsburgh, Pennsylvania), in which lamina propria lymphocytes were found at the interface of 40% and 80% fractions and collected for staining.
Staining, Antibodies and Flow Cytometry Analysis
For intracellular cytokine staining, cells obtained from in vitro cultures or isolated LPLs were incubated for 4 hours with 1X Cell Stimulation cocktail and 1X Protein Transport Inhibitor (eBioscience) in a tissue culture incubator at 37°C. Surface staining was performed for 30 min with the corresponding cocktail of fluorescently labeled antibodies. After surface staining, cells were re-suspended in Fixation/Permeabilization solution (eBioscience Foxp3 Staining Buffer Set), and intracellular cytokine staining was performed according to the manufacturer’s protocol. For sorting CD45+ and CD45- cells, we followed the same protocol for extracting the LPLs, and we then stained for CD45+ and CD45-. Flow cytometry data, as well as sorting, were collected using the Aria IIIu and analyzed using FlowJo V10. All the antibodies used are listed on the Key Resources Table.
Laser Capture Microdissection and Quantitative real-time- PCR
Small intestine tissues were flushed with ice-cold PBS and embedded in Fisher Healthcare™ Tissue-Plus™ O.C.T Compound, frozen using dry-ice and stored at −80°C. Sections were then cut at 8 µm thick. Methyl Green and Eosin staining were used to visualize sections. Intestinal epithelial cells were captured using Arcturus LCM System from Thermo Fisher. RNA from IECs was immediately isolated using RNAqueous™-Micro Total RNA Isolation Kit, and cDNA was synthesized using iScript cDNA Synthesis Kit from BioRad. Whole tissue RNA was extracted using Qiagen RNeasy Mini Kit and cDNA was synthesized with and M-MLV Reverse Transcriptase (ThermoFisher). Gene expression was normalized to GAPDH or 18S and fold changes in gene expression were relative to uninfected controls and calculated using the ∆∆ Ct method (All primers are listed on Key Resource Table).
Immunofluorescence
For detection of Reg3γ, and Rdh7, and lysozyme, tissues were fixed in Bouin’s or formalin, and embedded in paraffin. Using the microtome, paraffin blocks were cut at 8 µm, and tissues were de-paraffinized using xylenes, ethanol and water. Citrate buffer was used for antigen retrieval and slides were blocked with 1% bovine serum albumin (BSA). They were then stained with anti-Reg3γ antibody (1:250) raised against purified recombinant Reg3γ in rabbit (Cash et al., 2006), anti- Rdh7 antibody(1:200) or anti-lysozyme antibody (1:500 from abcam ) . Anti-Rdh7 antibody was generated by injecting rabbit with Rdh7 peptide (KGAEQLRNKTSDRLETV) by Pacific Immunology, Ramona, CA. Coverslips were mounted in Fluoro-Gel with Tris Biuffer) (Electron Microscopy Sciences) and viewed on a Zeiss fluorescence microscope.
Fluorescence In Situ hybridization (FISH)
FISH was done as described by Vaishanava et.al (Vaishnava et al., 2011b). In summary, small intestine tissues were removed and fixed in Methacarn Fixative (Fisher Scientific) and embedded in paraffin. Tissues were sectioned at a 7µm thickness and hybridized to a universal bacterial probe directed against the 16S rRNA gene. Probes used are listed on the Key Resources Table.
Western Blot
Whole tissue protein was extracted using RIPA lysis buffer (RIPA; Millipore Sigma). Protein samples were separated by 12.5% SDS-PAGE and transferred to trans-blot turbo transfer packs (BioRad) and blocked for 1 h with 5% dry milk in 0.1% Tween-20 in TBS (TTBS). The membranes were then washed and incubated with primary antibody overnight. The goat α-rabbit secondary antibody (1:5000 dilution) was added and incubated at room temperature for 2 hours. After washing 4 times with TTBS, immunoreactive bands were visualized by chemiluminescent detection (ECL; Roche Diagnostics, Penzberg, Germany) and exposure to ChemiDoc (BioRad).
Pathogen Challenge
Salmonella Typhimurium (S. enterica serovar Typhimurium SL1344) challenge was done as described by Barthel et. al(Barthel et al., 2003a). In short, mice were not allowed to eat or drink 4 hours prior to receiving 20 mg of streptomycin. 24 hours post streptomycin, food and water were withdrawn from mice for a period of 4 hours, followed by an oral gavage of 108 CFU of Salmonella. Fecal contents were collected daily, weighted, serially diluted and plated on ampicillin laced Brain Heart Infusion Agar plates to determine the number of S.typhimurium. The colonization in spleen was determined by removing the organ, homogenizing (using Omni Tissue Homogenizer) in ice-cold PBS and then plating the homogenate ampicillin laced Brain Heart Infusion Agar plates. Retinoic acid was given to mice via i.p with 250 µg RA or DMSO on the day of infection and 48 hours after, mice were euthanized 72 hours post treatment (Jie et al., 2017). To supplement IL-22 to Rdh7∆IEC mice, 30 µg of recombinant IL-22 (IL22-Fc, Genentech PRO312045) was administered every other day starting the day of streptomycin treatment (Behnsen et al., 2014a). In order to create a systemic infection, Salmonella Typhimurium (S. enterica serovar Typhimurium SL1344) was given to mice intraperitoneally (5000 CFU), and mice were sacrificed 24 hours post infection.
Lynn Bry at Brigham and Women’s Hospital (Boston, MA, USA) kindly provided the Citrobacter rodentium expressing GFP (Belzer et al., 2011). GFP-C. rodentium was grown in LB medium (containing 100 µg/ml piperacillin for GFP–C. rodentium). Mice were challenged with 2x109 cfu/ml of C.rodentium by oral gavage. The survival of infected mice ( 2x109 cfu/ml) and changes in their body weight were measured daily over the course of the 10 days of infection . Survival and body weight data are from a representative experiment out of three experiments showing similar results. Body weight data are presented as the mean of the percent start weight of 8 mice at each time point.
Spore-forming bacteria Isolation
To enrich for spore-forming bacteria, we followed Velazquez et.al`s protocol (Velazquez et al., 2017), where cecum content of female mice were extracted and treated with 3% chloroform in order to kill all vegetative bacteria, except for spores. Once the aqueous layer containing the spores was removed, germ-free mice were gavaged with 200µl of the same. Upon completion of the two weeks, mice were sacrificed, and 16S sequencing was done on fecal content to confirm that mice are colonized with spore forming bacteria.
β—Galactosidase staining
Small intestines were dissected out and flushed three times with 10ml cold PBS to remove gut bacteria. Tissues were fixed for 30 min in 0.2% glutaraldehyde, containing 0.1 M NaH2PO4 (pH 7.3), 5 mM EGTA, and 3 mMMgCl2, washed three times in wash buffer [0.1 M NaH2PO4 (pH 7.3), 2 mMMgCl2, 0.02% NP-40], and incubated at 37°C overnight in an Xgal solution containing 1 mg/ml Xgal (Boehringer Mannheim, Indianapolis, IN), 5 mMK3Fe(CN)6, and 5mM K4Fe(CN)6·3 H2O dissolved in wash buffer.
Histopathology and Alcian Blue Staining
Tissues samples were fixed in formalin and embedded in paraffin following standard procedures. Paraffin blocks were sectioned at 8µm. For histopathology, H&E staining (hematoxylin and eosin) was performed. Tissues were also stained with hematoxylin and eosin to assess morphology and with Alcian blue to identify goblet cells.
16s rRNA sequencing and microbiome analysis
DNA extraction and amplification
Genomic DNA extracted from fecal material using the QIAamp DNA Stool Mini Kit. PCR amplification performed using 518F/926R Illumina primers targeting the V4/V5 region of the 16S rRNA gene. PCR reactions consist of 1X Phusion HF Buffer, 200µM dNTPs, 1µM forward and reverse primers, and 1 unit Phusion DNA Polymerase for a 50µL reaction, split into three for triplicate reactions. Triplicates pooled and submitted to the Genomics and Sequencing Center at the University of Rhode Island for PrepX NGS library preparation. Amplicons sequenced using Illumina MiSeq platform, yielding paired-end, 250-base-pair reads. Data uploaded to Illumina’s genomics cloud computing environment, BaseSpace, providing demultiplexed Fastq files.
Processing of Sequenced Data
DADA2 pipeline (Callahan et al. 2016) used in R (version 3.3.4) to truncate reads where average Phred scores <30, and to infer ribosomal sequence variants (RSVs). The RDP classifier algorithm with RDP training set 14 used to perform taxonomic assignment.
16S rRNA Gene Sequencing and Microbial Community Analysis
RSV table is imported into R using phyloseq package (McMurdie and Holmes, 2013). Bar plots made using phyloseq after converting sample counts into percentage of total sample to account for variations in sampling depth. Principal Coordinates Analysis (PCoA) plots also generated using phyloseq and also normalized by converting counts into relative abundance. Distance matrices generated using both weighted and unweighted UniFrac distance metrics (Lozupone and Knight, 2005). Analysis of variance using distance matrices calculated using adonis from the vegan package (Oksanen, Blanchet et al. 2017).
Statistical analysis
The difference between the two different groups was determined by using Student t test. One-way ANOVA was used for multiple group comparisons (GraphPad Software v7). The p values <0.05 were considered significant, <0.01 as very significant, and <0.001 as highly significant.
Data Availability
Raw reads were deposited into the NCBI Sequence Read Archive (SRA) database under the BioProject ID number PRJNA400781.
Supplementary Material
| Reagents or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Anti-Mouse CD45 BV605 | eBioscience | Cat#: 86–0451-41 |
| Anti-Mouse CD4 BV785 | BioLegend | Cat#: 100551 |
| Anti-Mouse CD3 eF450 | ThermoFisher Scientific | Cat#: 48–0032-82 |
| Anti-Mouse CD19 Evolve605 | ThermoFisher Scientific | Cat#: 83–0193-41 |
| Anti-Mouse CD335 BV510 | BioLegend | Cat#: 137621 |
| Anti-Mouse CD8 Evolve605 | BioLegend | Cat#: 100743 |
| Anti-Mouse GATA-3 PerCPeF710 | eBioscience | Cat#: 46–9966-42 |
| Anti-Mouse IL-17 Alexa Fluor 488 | eBioscience | Cat#: 53–7177-81 |
| Anti-Mouse IL-22 PE | eBioscience | Cat#: 12–7221 |
| Anti-Mouse Rorγt APC | eBioscience | Cat#: 176981–82 |
| Anti-Mouse Foxp3 PeCy5 | eBioscience | Cat#: 15–5773-82 |
| Anti-Mouse Viability Dye APCeF780 | ThermoFisher Scientific | Cat#: 65–0865-14 |
| Anti-Mouse Streptavidin APCeF780 | ThermoFisher Scientific | Cat#: 47–4317-82 |
| Anti- Mouse T-Bet | BioLegend | Cat#: 644819 |
| Anti-Mouse Biotin Lineage Panel | BioLegend | Cat#: 133307 |
| Anti-Reg3γ | Hooper Lab | NA |
| Anti- Mouse IgA FITC | eBioscience | Cat #: 11–4204-82 |
| Anti-lysozyme antibody | abcam | Cat #: ab108508 |
| Universal bacterial Probe (16S rRNA) Alexa 488 Green GCTGCCTCCCGTAGGAGT | ThermoFisher Scientific | NA |
| Universal bacterial Probe (16S rRNA) Alexa 488 Green GCAGCCACCCGTAGGTGT Alexa 488 Green | ThermoFisher Scientific | NA |
| Universal bacterial Probe (16S rRNA) GCCTTCCCACATCGTTT | ThermoFisher Scientific | NA |
| Cell Stimulation cocktail | eBioscience | Cat#: 00–4970-93 |
| DAPI | Life tech | Cat#: D1306 |
| Primers: Genotyping | ||
| Vil-Cre forward 5’ GTGTGGGACAGAGAACAAACC 3’ | Life tech | n/a |
| Vil-Cre reverse 5’ ACATCTTCAGGTTCTGCGGG 3’ | Life tech | n/a |
| LacZ forward 5’ ATCCTCTGCATGGTCAGGTC 3’ | Life tech | n/a |
| LacZ reverse 5’ CGTGGCCTGATTCATTCC 3’ | Life tech | n/a |
| Rdh7 forward 5’ CAGCCCCTACCTTCTGTCTCCC 3’ | Life tech | n/a |
| Rdh7 reverse 5’ CACACCAGGCGGTGGGCTGAG 3’ | Life tech | n/a |
| Chemicals, Peptides and Recombinant Proteins | ||
| Ampicillin | Fisher Scientific | Cat#: BP1760–25 |
| Gentamycin | Fisher Scientific | Cat#: 50–247-622 |
| Metronidazole | Fisher Scientific | Cat#: 50–213-513 |
| Neomycin | Fisher Scientific | Cat#: BP26695 |
| Vancomycin | Fisher Scientific | Cat#: AAJ6279006 |
| Sucralose | Fisher Scientific | Cat#: AAJ6673618 |
| Polymyxin B sulfate salt | Sigma Aldrich | Cat#: 1405–20-5 |
| HBSS | Life tech | Cat#: 14170161 |
| EDTA (0.5M), ph 8 | Fisher Scientific | Cat#: BP120–500 |
| RPMI 1640 Medium | ThermoFisher | Cat#: 61870127 |
| Collagenase VIII | Sigma-Aldrich | Cat#: C2139–100MG |
| DNase I | Sigma-Aldrich | Cat#: 4536282001 |
| Dispase | Sigma- Aldrich | Cat#: D4848–2MG |
| Isoflurane | Piramal Healthcare | NDC 66794–013-25 |
| Fetal Bovine Serum (FBS) | Genesee Scientific | Cat#: 25–514H |
| Trans- Retinoic Acid | Sigma-Aldrich | Cat#: 302–79-4 |
| Percoll | Sigma-Aldrich | Cat#: GE17–0891-01 |
| Eosin-Y | Fisher HealthCare | Cat#: 22220104 |
| Hematoxylin | Fisher | Cat#: 22220102 |
| Methyl Green | Fisher Scientific | Cat#: 7114–03-6 |
| Dimethyl Sulfoxide | Fisher Scientific | Cat#: 67–68-5 |
| Methacarn | Fisher Scientific | Cat#: NC0547175 |
| Bovine Serum Albumin (BSA) | Fisher Scientific | Cat#: BP9706100 |
| RIPA Lysis Buffer | Millipore Sigma | Cat#: 20–188 |
| Tween-20 | Fisher Scientific | Cat#: 900–64-5 |
| M-MLV Reverse Transcriptase | ThermoFisher Scientific | Cat#: 28025013 |
| Fluoro Gel ( With Tris Buffer) | Electron Microscopy Sciences | Cat#: 17985–10 |
| Trans- Retinoic Acid | Sigma Aldrich | Cat#: 554720 |
| IL-22 recombinant Protein | Genentech | IL22-Fc, Genentech PRO312045 |
| Retinoid Quantification Reagents | ||
| All-trans-retinoic acid | Sigma-Aldrich | Cat#: R2625 |
| 13-cis-retinoic acid | Sigma-Aldrich | Cat#: R3255 |
| All-trans-retinyl palmitate | Toronto Research Chemicals | Cat#: R275450 |
| All-trans-retinol | Toronto Research Chemicals | Cat#: R252000 |
| All-trans-retinoic acid-d5 | Toronto Research Chemicals | Cat#: R250202 |
| 13-cis-retinoic acid-d5 | Toronto Research Chemicals | Cat#: R250004 |
| All-trans-retinyl palmitate-d4 | Cambridge Isotope Laboratory | Cat#: DLM-4902-PK |
| All-trans-retinol-d8 | Cambridge Isotope Laboratory | Cat#: DLM-9306-PK |
| Formic Acid | Fisher Scientific | Cat#: A117–50 |
| Acetonitrile (LC-MS grade) | Fisher Scientific | Cat#: A996–4 |
| Water (LC-MS grade) | Fisher Scientific | Cat#: W6–4 |
| DC MASS SPECT GOLD serum | Golden West Diagnostics | Cat#: MSG4000 |
| Hexanes | Fisher Scientific | Cat#: H292–4 |
| Hydrochloric acid | Fisher Scientific | Cat#: A144SI-212 |
| Potassium hydroxide | Fisher Scientific | Cat#: P250–500 |
| Sodium chloride | Fisher Scientific | Cat#: S271–1 |
| Ethanol 200 proof | Decon Labs | Cat#: 2701 |
| Ascentis® RP-amide column (2.7µm, 15cm x 2.1 mm) | Sigma-Aldrich | Cat#: 53914-U |
| Ascentis® RP-amide Guard Cartridge (2.7 µm, 5 mm x 2.1 mm) | Sigma-Aldrich | Cat#: 53514-U |
| Critical Commercial Assay | ||
| Foxp3 Staining Buffer Set Kit | eBioscience | Cat#: 00–5523-00 |
| RNeasy Mini Kit | QIagen | Cat#: 74106 |
| RNAqueous ™- Micro Total RNA Isolation Kit | Thermo Fisher Scientific | Cat#: AM1931 |
| iScript RT Mix | Bio-Rad | Cat#: 1708841 |
| Experimental Model: Organisms/ Strains | ||
| C57Bl/6 | Jackson Laboratories | Cat#: 000664 |
| Rdh7 | This study | |
| Salmonella Typhimurium (S. enterica serovar Typhimurium) | UT. South Western Dr. Vanessa Sperandio | Strain: SL1344 |
| C. rodentium | Lynn Bry, MD, PhD | Strain: DBS100 GFP |
| Softwares and Algorithms | ||
| FlowJo (v10) | FlowJo LLC | https://www.flowjo.com/solutions/flowjo |
| GraphPad Prism 7 | GraphPad Software Inc | |
| DADA2 | The DADA2 R package | https://benjjneb.github.io/dada2/index.html |
| Phyloseq | https://joey711.github.io/phyloseq/ | |
| FastQC | FastQC- Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Plotly | Plotly | https://plot.ly/ |
| Deposited Data | ||
| Sequencing data | NCBI Sequence Read Archive (SRA) | SUB3841090 (https://www.ncbi.nlm.nih.gov/bioproject/445734) |
| Other | ||
| Brain Heart Infusion | Thermo Scientific | Code: CM1135 |
| MacConkey Agar | Sigma-Aldrich | Cat#: M7408 |
| O.C.T. Compound (Tissue – Plus) | Fisher Healthcare | Cat# 23–730-571 |
Highlights.
Gut commensals curb retinoic acid production by suppressing expression of Rdh7 in IECs.
IEC specific Rdh7 expression is required for IL-22 production by gut lymphocytes.
Rdh7-/- mice have diminished IL-22 induced antimicrobial response.
Rdh7-/- mice are protected from pathogen colonization and microbial dysbiosis.
Acknowledgement:
We thank Sohini Mukherjee and Felix Yarovinsky for their intellectual input. We thank Lora Hooper for providing anti-Reg3γ antibody and UT Southwestern mouse transgenic core for help in developing Rdh7fl/fl mouse models. We thank Kevin Carlson for flow cytometry analysis. This work was supported by NIH (P20GM10903 and 1R01DK113265 to S.V. and GM111772 to N.I.) and CCFA career development award to S.V.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest: The authors declare no competing interests
References:
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai A, Lu N, Guo Y, Liu Z, Chen J, and Peng Z (2009). All-trans retinoic acid down-regulates inflammatory responses by shifting the Treg/Th17 profile in human ulcerative and murine colitis. J Leukoc Biol 86, 959–969. [DOI] [PubMed] [Google Scholar]
- Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, and Raffatellu M (2014a). The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 40, 262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, and Raffatellu M (2014b). The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 40, 262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzer C, Liu Q, Carroll MC, and Bry L (2011). THE ROLE OF SPECIFIC IgG AND COMPLEMENT IN COMBATING A PRIMARY MUCOSAL INFECTION OF THE GUT EPITHELIUM. Eur J Microbiol Immunol (Bp) 1, 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya N, Yuan R, Prestwood TR, Penny HL, DiMaio MA, Reticker-Flynn NE, Krois CR, Kenkel JA, Pham TD, Carmi Y, et al. (2016). Normalizing Microbiota-Induced Retinoic Acid Deficiency Stimulates Protective CD8(+) T Cell-Mediated Immunity in Colorectal Cancer. Immunity 45, 641–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelstowska S, Widjaja-Adhi MA, Silvaroli JA, and Golczak M (2016). Molecular Basis for Vitamin A Uptake and Storage in Vertebrates. Nutrients 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MM, Saraceni PR, Forn-Cuni G, Dios S, Romero A, Figueras A, and Novoa B (2013). IL-22 is a key player in the regulation of inflammation in fish and involves innate immune cells and PI3K signaling. Developmental and comparative immunology 41, 746–755. [DOI] [PubMed] [Google Scholar]
- Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, Degnan PH, Booth CJ, Yu H, and Goodman AL (2015). Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 347, 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio DN, Clugston RD, and Blaner WS (2011). Vitamin A metabolism: an update. Nutrients 3, 63–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePaolo RW, Abadie V, Tang F, Fehlner-Peach H, Hall JA, Wang W, Marietta EV, Kasarda DD, Waldmann TA, Murray JA, et al. (2011). Co-adjuvant effects of retinoic acid and IL-15 induce inflammatory immunity to dietary antigens. Nature 471, 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop BA, Vaishnava S, and Hooper LV (2009). Immune responses to the microbiota at the intestinal mucosal surface. Immunity 31, 368–376. [DOI] [PubMed] [Google Scholar]
- Eidenschenk C, Rutz S, Liesenfeld O, and Ouyang W (2014). Role of IL-22 in microbial host defense. Current topics in microbiology and immunology 380, 213–236. [DOI] [PubMed] [Google Scholar]
- Erkelens MN, and Mebius RE (2017). Retinoic Acid and Immune Homeostasis: A Balancing Act. Trends Immunol 38, 168–180. [DOI] [PubMed] [Google Scholar]
- Fahlgren A, Hammarstrom S, Danielsson A, and Hammarstrom ML (2003). Increased expression of antimicrobial peptides and lysozyme in colonic epithelial cells of patients with ulcerative colitis. Clin Exp Immunol 131, 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fengming Y, and Jianbing W (2014). Biomarkers of inflammatory bowel disease. Dis Markers 2014, 710915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen K, Franzen P, Magnuson A, Elmabsout AA, Nyhlin N, Wickbom A, Curman B, Torkvist L, D’Amato M, Bohr J, et al. (2013). Polymorphism in the retinoic acid metabolizing enzyme CYP26B1 and the development of Crohn’s Disease. PLoS One 8, e72739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geboes L, Dumoutier L, Kelchtermans H, Schurgers E, Mitera T, Renauld JC, and Matthys P (2009). Proinflammatory role of the Th17 cytokine interleukin-22 in collagen-induced arthritis in C57BL/6 mice. Arthritis and rheumatism 60, 390–395. [DOI] [PubMed] [Google Scholar]
- Goverse G, Labao-Almeida C, Ferreira M, Molenaar R, Wahlen S, Konijn T, Koning J, Veiga-Fernandes H, and Mebius RE (2016). Vitamin A Controls the Presence of RORgamma+ Innate Lymphoid Cells and Lymphoid Tissue in the Small Intestine. Journal of immunology 196, 5148–5155. [DOI] [PubMed] [Google Scholar]
- Hall JA, Cannons JL, Grainger JR, Dos Santos LM, Hand TW, Naik S, Wohlfert EA, Chou DB, Oldenhove G, Robinson M, et al. (2011). Essential role for retinoic acid in the promotion of CD4(+) T cell effector responses via retinoic acid receptor alpha. Immunity 34, 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JA, Hall JA, Sun CM, Cai Q, Ghyselinck N, Chambon P, Belkaid Y, Mathis D, and Benoist C (2008). Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi Cells. Immunity 29, 758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S, Pothoulakis C, and Koon HW (2013). Antimicrobial peptides and colitis. Curr Pharm Des 19, 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K, Zhang Y, Guo Y, Xie J, Wang J, He X, Lu N, and Bai A (2014). All-trans retinoic acid attenuates experimental colitis through inhibition of NF-kappaB signaling. Immunol Lett 162, 34–40. [DOI] [PubMed] [Google Scholar]
- Huber S, Gagliani N, Zenewicz LA, Huber FJ, Bosurgi L, Hu B, Hedl M, Zhang W, O’Connor W Jr., Murphy AJ, et al. (2012). IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 491, 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst RJ, and Else KJ (2013). The retinoic acid-producing capacity of gut dendritic cells and macrophages is reduced during persistent T. muris infection. Parasite immunology 35, 229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, and Song SY (2004). Retinoic acid imprints gut-homing specificity on T cells. Immunity 21, 527–538. [DOI] [PubMed] [Google Scholar]
- Jaensson E, Uronen-Hansson H, Pabst O, Eksteen B, Tian J, Coombes JL, Berg PL, Davidsson T, Powrie F, Johansson-Lindbom B, et al. (2008). Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. The Journal of experimental medicine 205, 2139–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaensson-Gyllenback E, Kotarsky K, Zapata F, Persson EK, Gundersen TE, Blomhoff R, and Agace WW (2011). Bile retinoids imprint intestinal CD103+ dendritic cells with the ability to generate gut-tropic T cells. Mucosal immunology 4, 438–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jason J, Archibald LK, Nwanyanwu OC, Sowell AL, Buchanan I, Larned J, Bell M, Kazembe PN, Dobbie H, and Jarvis WR (2002). Vitamin A levels and immunity in humans. Clinical and diagnostic laboratory immunology 9, 616–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, Wiita P, Bok D, and Sun H (2007). A Membrane Receptor for Retinol Binding Protein Mediates Cellular Uptake of Vitamin A. Science 315, 820–825. [DOI] [PubMed] [Google Scholar]
- Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, and Pamer EG (2010). Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. The Journal of infectious diseases 201, 534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A, De Vadder F, Kovatcheva-Datchary P, and Backhed F (2016). From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 165, 1332–1345. [DOI] [PubMed] [Google Scholar]
- Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, Keller MP, Attie AD, Rey FE, and Denu JM (2016). Diet-Microbiota Interactions Mediate Global Epigenetic Programming in Multiple Host Tissues. Mol Cell 64, 982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampen A, Meyer S, Arnhold T, and Nau H (2000). Metabolism of vitamin A and its active metabolite all-trans-retinoic acid in small intestinal enterocytes. The Journal of pharmacology and experimental therapeutics 295, 979–985. [PubMed] [Google Scholar]
- Larange A, and Cheroutre H (2016). Retinoic Acid and Retinoic Acid Receptors as Pleiotropic Modulators of the Immune System. Annu Rev Immunol 34, 369–394. [DOI] [PubMed] [Google Scholar]
- Magnusson MK, Brynjolfsson SF, Dige A, Uronen-Hansson H, Borjesson LG, Bengtsson JL, Gudjonsson S, Ohman L, Agnholt J, Sjovall H, et al. (2016). Macrophage and dendritic cell subsets in IBD: ALDH+ cells are reduced in colon tissue of patients with ulcerative colitis regardless of inflammation. Mucosal immunology 9, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KG, Leach MR, Brooke KW, Wang C, Wheeler LW, Hanly EK, Rowley CW, Levin MS, Wagner M, Li E, et al. (2012). Epithelial expression of the cytosolic retinoid chaperone cellular retinol binding protein II is essential for in vivo imprinting of local gut dendritic cells by lumenal retinoids. Am J Pathol 180, 984–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke LA, Jones SA, Raverdeau M, Higgs R, Stefanska A, Groom JR, Misiak A, Dungan LS, Sutton CE, Streubel G, et al. (2013). Retinoic acid expression associates with enhanced IL-22 production by gammadelta T cells and innate lymphoid cells and attenuation of intestinal inflammation. The Journal of experimental medicine 210, 1117–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Goto R, Fujimoto M, Okada N, and Hardt WD (2017). The Bactericidal Lectin RegIIIbeta Prolongs Gut Colonization and Enteropathy in the Streptomycin Mouse Model for Salmonella Diarrhea. Cell host & microbe 21, 195–207. [DOI] [PubMed] [Google Scholar]
- Monteleone I, Rizzo A, Sarra M, Sica G, Sileri P, Biancone L, MacDonald TT, Pallone F, and Monteleone G (2011). Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology 141, 237–248, 248 e231. [DOI] [PubMed] [Google Scholar]
- Mora JR, Iwata M, Eksteen B, Song SY, Junt T, Senman B, Otipoby KL, Yokota A, Takeuchi H, Ricciardi-Castagnoli P, et al. (2006). Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science 314, 1157–1160. [DOI] [PubMed] [Google Scholar]
- Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, and Cheroutre H (2007). Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science 317, 256–260. [DOI] [PubMed] [Google Scholar]
- Mullin GE (2011). Vitamin A and immunity. Nutrition in clinical practice : official publication of the American Society for Parenteral and Enteral Nutrition 26, 495–496. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, and Pettersson S (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. [DOI] [PubMed] [Google Scholar]
- Nolting J, Daniel C, Reuter S, Stuelten C, Li P, Sucov H, Kim BG, Letterio JJ, Kretschmer K, Kim HJ, et al. (2009). Retinoic acid can enhance conversion of naive into regulatory T cells independently of secreted cytokines. The Journal of experimental medicine 206, 2131–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson LW, and Artis D (2014). Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 14, 141–153. [DOI] [PubMed] [Google Scholar]
- Pino-Lagos K, Guo Y, Brown C, Alexander MP, Elgueta R, Bennett KA, De Vries V, Nowak E, Blomhoff R, Sockanathan S, et al. (2011). A retinoic acid-dependent checkpoint in the development of CD4+ T cell-mediated immunity. The Journal of experimental medicine 208, 1767–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX, and Zhou L (2012). The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 36, 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin LC, Girard-Madoux MJ, Seillet C, Mielke LA, Kerdiles Y, Fenis A, Wieduwild E, Putoczki T, Mondot S, Lantz O, et al. (2016). Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nature immunology 17, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chavez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, et al. (2016). Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell host & microbe 19, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AC (2012). Vitamin A and retinoic acid in T cell-related immunity. The American journal of clinical nutrition 96, 1166S–1172S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino SJ, Geddes K, and Girardin SE (2012). Innate IL-17 and IL-22 responses to enteric bacterial pathogens. Trends in immunology 33, 112–118. [DOI] [PubMed] [Google Scholar]
- Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Berard M, Kleinschek M, Cua D, Di Santo JP, and Eberl G (2011). RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nature immunology 12, 320–326. [DOI] [PubMed] [Google Scholar]
- Sirisinha S (2015). The pleiotropic role of vitamin A in regulating mucosal immunity. Asian Pacific journal of allergy and immunology / launched by the Allergy and Immunology Society of Thailand 33, 71–89. [PubMed] [Google Scholar]
- Sonnenberg GF, Fouser LA, and Artis D (2011). Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nature immunology 12, 383–390. [DOI] [PubMed] [Google Scholar]
- Sonnenberg GF, Nair MG, Kirn TJ, Zaph C, Fouser LA, and Artis D (2010). Pathological versus protective functions of IL-22 in airway inflammation are regulated by IL-17A. The Journal of experimental medicine 207, 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Chai X, Kahn B, and Napoli JL (1998). cDNA cloning, tissue distribution, and substrate characteristics of a cis-Retinol/3alpha-hydroxysterol short-chain dehydrogenase isozyme. J Biol Chem 273, 17910–17916. [DOI] [PubMed] [Google Scholar]
- Tomita K, Sato M, Kajiwara K, Tanaka M, Tamiya G, Makino S, Tomizawa M, Mizutani A, Kuwano Y, Shiina T, et al. (2000). Gene structure and promoter for Crad2 encoding mouse cis-retinol/3alpha-hydroxysterol short-chain dehydrogenase isozyme. Gene 251, 175–186. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, and Hooper LV (2008). Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proceedings of the National Academy of Sciences of the United States of America 105, 20858–20863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, and Hooper LV (2011). The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pavert SA, Ferreira M, Domingues RG, Ribeiro H, Molenaar R, Moreira-Santos L, Almeida FF, Ibiza S, Barbosa I, Goverse G, et al. (2014). Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature 508, 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pavert SA, and Mebius RE (2010). New insights into the development of lymphoid tissues. Nat Rev Immunol 10, 664–674. [DOI] [PubMed] [Google Scholar]
- Velazquez EM, Rivera-Chávez F, and Bäumler AJ (2017). Spore Preparation Protocol for Enrichment of Clostridia from Murine Intestine. Bio-protocol 7, e2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Suarez I, Larange A, Reardon C, Matho M, Feau S, Chodaczek G, Park Y, Obata Y, Gold R, Wang-Zhu Y, et al. (2015). Unique lamina propria stromal cells imprint the functional phenotype of mucosal dendritic cells. Mucosal immunology 8, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss E, Wehkamp J, Wehkamp K, Stange EF, Schroder JM, and Harder J (2006). NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J Biol Chem 281, 2005–2011. [DOI] [PubMed] [Google Scholar]
- Vrieze A, Out C, Fuentes S, Jonker L, Reuling I, Kootte RS, van Nood E, Holleman F, Knaapen M, Romijn JA, et al. (2014). Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol 60, 824–831. [DOI] [PubMed] [Google Scholar]
- Winter SE, Lopez CA, and Baumler AJ (2013). The dynamics of gut-associated microbial communities during inflammation. EMBO Rep 14, 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Feng Q, Liang S, Sonne SB, Xia Z, Qiu X, Li X, Long H, Zhang J, Zhang D, et al. (2015). A catalog of the mouse gut metagenome. Nature biotechnology 33, 1103–1108. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Danilenko DM, Valdez P, Kasman I, Eastham-Anderson J, Wu J, and Ouyang W (2007). Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature 445, 648–651. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, et al. (2008). Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nature medicine 14, 282–289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads were deposited into the NCBI Sequence Read Archive (SRA) database under the BioProject ID number PRJNA400781.