Abstract
Rationale:
Although lung cancer is the leading cause of cancer-related death in the world, targeted therapy plays an essential role in improving the survival of lung cancer. Next-generation sequencing (NGS) technology can dynamically monitor the genomic profiles of tumors and assist cancer diagnosis and treatment.
Patient concerns:
We reported on a 55-year-old man who presented with chest tightness and wheezing for 1 month.
Diagnoses:
The patient was diagnosed with stage cT4N2M1a non-small cell lung cancer (NSCLC) and was found to have wild-type EGFR by pleural effusion cytology.
Interventions:
The patient received systemic treatments, including chemotherapy, targeted therapy, and radiotherapy. During the cancer development, sequential DNA sequencing data that used circulating cell-free tumor DNA, and NGS revealed EGFR L858R and T790M mutations, MYC amplification, and other gene variations.
Outcomes
: The patient died of brain and lung metastases, and had an overall survival as long as 37 months.
Lessons:
The dynamic monitoring of tumor genomic profiles has important implications for NSCLC diagnosis, treatment, and prognosis.
Keywords: circulating cell-free tumor DNA, drug resistance, epidermal growth factor receptor tyrosine kinase inhibitors, next-generation sequencing, non-small cell lung cancer, targeted therapy
1. Introduction
Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR TKIs) play an important role in the improvement in survival of patients with lung adenocarcinoma. Currently, the most common mechanism of resistance to 1st-generation EGFR TKIs is the EGFR T790M mutation, as this accounts for approximately 60% of cases of resistance.[1,2] The 3rd-generation EGFR TKI AZD9291 has been shown to be effective against both EGFR TKI-sensitizing and T790M resistance mutations, with a response rate of 61% and a median progression-free survival of approximately 9.6 months.[3] However, drug resistance is also an inevitable problem for 3rd-generation EGFR TKIs. Recently, next-generation sequencing (NGS) technology has been widely used in cancer diagnosis and treatment, as it can help with the dynamic monitoring of genomic profiles, the early detection of tumor resistance, improvement in patient survival, and cancer precision medicine.[4–6] In this report, we present a 55-year-old man with stage cT4N2M1a non-small cell lung cancer (NSCLC) who had an overall survival as long as 37 months by virtue of dynamic monitoring of genomic profiles during the cancer process.
2. Case presentation
A 55-year-old man with chest tightness and wheezing for 1 month was admitted to our hospital in November 2013; he was diagnosed with stage cT4N2M1a left-sided lung cancer and was found to have wild-type EGFR by pleural effusion cytology and a total-body positron-emission tomography/computed tomography (PET/CT) scan (Fig. 1). He initially received 4 cycles of bevacizumab-based chemotherapy with gemcitabine, nedaplatin, and bevacizumab, and a partial regression treatment effect was obtained. Palliative radiation therapy to the left lung lesion was sequentially administered. On August 23, 2014, the pleural effusion recurred, and pleural effusion cytology revealed EGFR T790M and L858R mutations with no fusion of EML4-ALK (Table 1). He received 4 cycles of pemetrexed, nedaplatin and bevacizumab, but the lung lesion progressed on March 17, 2015. The patient was further treated with 4 cycles of paclitaxel liposome (Lipusu), nedaplatin, and bevacizumab. On September 10, 2015, the patient experienced dizziness, nausea, and vomiting for 1 week and was found to have meningeal metastasis by brain magnetic resonance imaging (MRI). The symptoms decreased shortly after AZD9291 was administered in October 2015. In April 2016, a new brain metastasis was found (Fig. 2). Subsequent treatment included cyberknife radiotherapy for brain metastases and 2 cycles of pemetrexed, carboplatin, and bevacizumab. To evaluate the molecular characteristics, DNA extracted from the plasma was used for DNA sequencing by NGS on April 5, 2016. Interestingly, the patient had EGFR T790M and L858R mutations, an ALK Y1278N mutation, a TP53 LPPGS299 frame-shift mutation and MYC amplification (Table 1). He continued to receive AZD9291 and was also treated with PF299804 (Dacomitinib) for only 3 weeks beginning in September 2016 due to the occurrence of side effects (in this case, severe dermatitis). Evidence of the progression of brain metastases was found again in November 2016. Cell-free plasma DNA was collected again for DNA sequencing by NGS on November 18, 2016, while EGFR T790M and L858R mutations, a TP53 LPPGS299 frame-shift mutation, and EGFR, MET, and MYC amplifications were found (Table 1). The patient was treated with AZD9291 and INC280 (Capmatinib) for 2 weeks based on the finding of MET amplification; however, his performance status continuously deteriorated. On December 4, 2016, he died of brain and lung metastases. His overall survival was 37 months (Fig. 3).
Figure 1.

Total-body positron-emission tomography/computed tomography scan before treatment.
Table 1.
Summary of gene-sequencing data.
Figure 2.

Brain metastasis in the left temporal pole as seen on magnetic resonance imaging.
Figure 3.
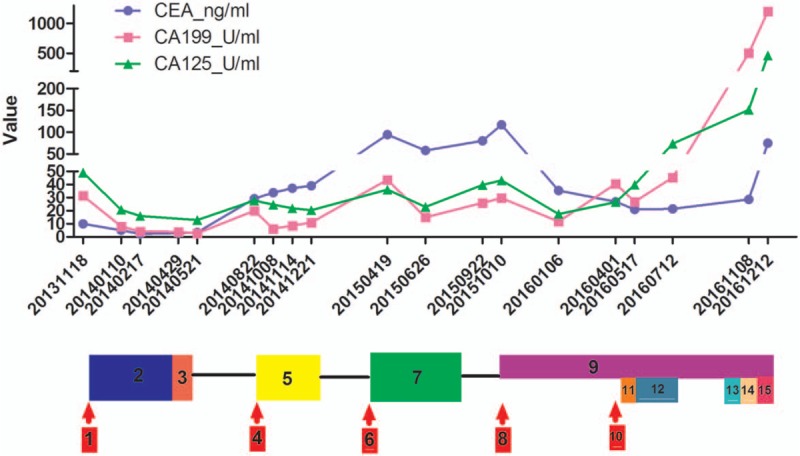
The fluctuation of tumor marker levels during the cancer process. 1: Stage cT4N2M1a lung cancer with wild-type epidermal growth factor receptor (EGFR). 2: Four cycles of gemcitabine, nedaplatin, and bevacizumab. 3: Palliative radiation therapy to the left lung lesion. 4: Pleural effusion reoccurred; pleural effusion cytology: EGFR T790M and L858R mutations. 5: Four cycles of pemetrexed, nedaplatin, and bevacizumab. 6: Progression of lung lesions. 7: Four cycles of paclitaxel liposome, nedaplatin, and bevacizumab. 8: Meningeal metastasis. 9: AZD9291. 10: Brain metastasis in the left temporal pole. 11: Cyberknife radiotherapy for brain metastases. 12: Two cycles of pemetrexed, nedaplatin, and bevacizumab. 13: AZD9291 + PF299804. 14: AZD9291 + Tarceva. 15: AZD9291 + INC280.
3. Discussion
With the rapid development of gene sequencing technology, many studies have illustrated that dynamic monitoring of tumor-driven genes has extensive applications in cancer diagnosis and treatment, including in the identification of tumor genomic profiles, the monitoring of treatment responses, the early detection of drug resistance, the identification of minimal residual disease, and the prediction of prognosis.[7]
Recent studies have revealed the mechanisms of resistance to third-generation EGFR TKIs. They are categorized as follows: (1) EGFR dominant: EGFR mutations (L718Q, L844V, C797S) and the EGFR T790M mutation; (2) activation of by-pass signaling tracts: RAS amplification, RAS mutations (Q61K, G12S, E63K), MET amplification, and HER2 amplification or mutations, among others; (3) phenotypic alterations: small-cell lung cancer transition and epithelial-mesenchymal transition.[8] Of these, the EGFR C797S mutation is the main cause of AZD9291 resistance, as it accounts for approximately 40% of cases.[9]
As the pleural effusion cytology of this patient showed the coexistence of EGFR L858R and T790M mutations, which indicated primary drug resistance to 1st-generation EGFR TKIs, the patient initially received chemotherapy. Later, after the patient was treated with AZD9291 for 6 months, his condition worsened due to a brain metastasis. The results of 2 sequential rounds of NGS revealed a TP53 Leu299 frame-shift mutation and MYC amplification; amplifications in EGFR, MET, and MYC as well as a TP53 LPPGS299 frame-shift mutation were found in that order. This implies that the resistance of this patient to AZD9291 was primarily caused by activation of by-pass signaling tracts and EGFR amplification. At a later stage, the patient received AZD9291 and INC280 but did not exhibit good effects, which indicated that MET amplification was not the primary mechanism of drug resistance in this case, but rather, other mechanisms such as activation of by-pass signaling tracts and EGFR amplification may have been responsible.
At present, the molecular landscapes of solid tumors are established using surgical or biopsy tissue samples. This process can frequently put the patient at risk and remains complicated by sample availability and tumor heterogeneity. Genomic profiles of circulating cell-free tumor DNA (ctDNA) have been shown to closely match those of the corresponding tumors, which has important implications for both molecular pathology and clinical oncology.[7] In a recent retrospective analysis, Oxnard et al found that the sensitivity of plasma genotyping for the detection of the T790M mutation in advanced NSCLC was 70%. The objective response rate and the median progression-free survival were similar in patients with T790M-positive plasma or T790M-positive tumors.[10] The patient in this case refused an invasive tumor biopsy, and therefore, we based the cancer diagnosis and treatment decision on pleural effusion cytology and plasma ctDNA results. Through the dynamic monitoring of his tumor genomic profile, this patient with stage IV lung cancer had an overall survival of more than 3 years after he received chemotherapy, radiotherapy, and targeted therapy, which indicates the significance of the dynamic monitoring of genomic profiles in cancer treatment.
In summary, with the development of gene sequencing technology and the continuous reduction in its cost, the dynamic monitoring of tumor genomic profiles can detect driver genes, which can guide cancer treatment. Dynamic monitoring may also reveal potential target genes that can be used to reverse drug resistance for the purpose of cancer precision medicine.
Author contributions
Conceptualization: Bo-Hua Kuang, Bi-Cheng Wang, Ru-Bo Cao, Li Liu.
Data curation: Bo-Hua Kuang, Bi-Cheng Wang, Ru-Bo Cao.
Formal analysis: Bo-Hua Kuang, Fan Tong.
Funding acquisition: Bo-Hua Kuang, Li Liu.
Investigation: Bo-Hua Kuang.
Methodology: Bo-Hua Kuang, Yu-Lan Zeng.
Project administration: Fan Tong.
Resources: Fan Tong, Jin-Song Yang, Yu-Lan Zeng.
Supervision: Ru-Bo Cao, Li Liu.
Validation: Bo-Hua Kuang, Fan Tong, Jin-Song Yang.
Writing – original draft: Bo-Hua Kuang.
Writing – review & editing: Bo-Hua Kuang, Ru-Bo Cao, Li Liu.
Footnotes
Abbreviations: ctDNA = circulating cell-free tumor DNA, EGFR TKIs = epidermal growth factor receptor tyrosine kinase inhibitors, MRI = magnetic resonance imaging, NGS = next-generation sequencing, NSCLC = non-small cell lung cancer, PET/CT = positron-emission tomography/computed tomography.
B-HK and B-CW contributed equally to this work.
The patient has provided informed consent for publication of the case.
This work was supported by grants from Open Funds of State Key Laboratory of Oncology in South China (HN2017-02) and the National Scientific Foundation of China (no: 81372260).
The authors have no conflicts of interest to disclose.
References
- [1].Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:26r–75r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Janne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689–99. [DOI] [PubMed] [Google Scholar]
- [4].Hagemann IS, Devarakonda S, Lockwood CM, et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer 2015;121:631–9. [DOI] [PubMed] [Google Scholar]
- [5].Takeda M, Sakai K, Terashima M, et al. Clinical application of amplicon-based next-generation sequencing to therapeutic decision making in lung cancer. Ann Oncol 2015;26:2477–82. [DOI] [PubMed] [Google Scholar]
- [6].DiBardino DM, Saqi A, Elvin JA, et al. Yield and clinical utility of next-generation sequencing in selected patients with lung adenocarcinoma. Clin Lung Cancer 2016;17:517–22. [DOI] [PubMed] [Google Scholar]
- [7].Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531–48. [DOI] [PubMed] [Google Scholar]
- [8].Kim TM, Song A, Kim DW, et al. Mechanisms of acquired resistance to AZD9291: a mutation-selective, irreversible EGFR inhibitor. J Thorac Oncol 2015;10:1736–44. [DOI] [PubMed] [Google Scholar]
- [9].Wang S, Song Y, Yan F, et al. Mechanisms of resistance to third-generation EGFR tyrosine kinase inhibitors. Front Med 2016;10:383–8. [DOI] [PubMed] [Google Scholar]
- [10].Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol 2016;34:3375–82. [DOI] [PMC free article] [PubMed] [Google Scholar]


