Abstract
Rationale:
Graft-derived-cell-free DNA (Gcf-DNA) in plasma was a promising biomarker to monitor graft-rejection after liver transplantation. However, little is known about the application of Gcf-DNA in living-donor-liver-transplantation (LDLT).
Patients concern:
In this study, 2 patients diagnosed with Ornithine Transcarbamylase Deficiency (OTCD) were enrolled and indicated for LDLT.
Diagnoses:
Two patients were genetically diagnosed with OTCD, and they suffered from recurrent and uncontrollable hyper-ammonemia and failed in accepting the normalized OTCD treatments, such as decreasing dietary nitrogen intake and increasing waste-nitrogen excretion.
Interventions:
LDLT was performed in the 2 patients uneventfully, and we collected circulating cell-free DNA from plasma in specific postoperative time points (day 1, day 7, day 14, day 30, day 60). Since both of the recipients were sex-mismatch with the donors, we measured Gcf-DNA through the Y-chromosome method and compared it with the routine liver function.
Outcomes:
The result showed that Gcf-DNA had the similar discrimination of graft injury trend while compared to routine liver function. The follow-up showed these 2 patients’ status is stable.
Lessons:
Applying Gcf-DNA to monitor graft injury in LDLT is promising, but still long term follow-up and more samples are needed for validation.
Keywords: graft injury, graft-derived cell-free DNA, living donor liver transplantation, Y-chromosome method
1. Introduction
Metabolic disorders have become the indication for liver transplantation because of poor prognosis, while living donor liver transplantation (LDLT) was reported to treat inherited metabolic disorders can provide an acceptable survival rate over 15 years.[1] From this, post-transplant management have increasingly become the research emphasis, such as monitoring the graft-function and the adjustment of immunosuppressant. Routine liver function tests (including ALT, AST, ALP, GGT, and BIL) were monitored after LDLT; however, some research reported that graft injury had initiated earlier than a liver function tests showed,[2] which indicated that liver function tests were not sensitive enough to detect early graft-injury or rejection. More specific, the gold standard to diagnosis graft injury and rejection requires graft-biopsy for histopathological examination, which is an invasive procedure.[3] Accordingly, an accurate biomarker was needed to monitor graft function so that early graft injury can be detected, and timely interventions can be given.
Graft derived cell-free DNA (Gcf-DNA) was recently reported as a promising noninvasive biomarker to detect graft damage or even rejection after LTx.[4–8] The presence of Gcf-DNA in the blood-stream was firstly detected by Polymerase Chain Reaction (PCR) amplification of Y-chromosome specific genes in female recipients from male donors.[9] However, no study showed that the change of Gcf-DNA in LDLT, here we report 2 cases of the change of Gcf-DNA after LDLT.
2. Methods
2.1. Ethical approval and consent for publication
All of the procedures and informed consent were approved by the Department of Ethics committee at the Beijing Friendship Hospital of the Capital Medical University (Beijing, China) (approval document number: 2017-P2-080-02). The patient of their legal guardian was provided written informed consent before each undergoing examination and surgery. We explained the study purpose and all the laboratory test of this study to the patient and their parents in detail, which we aimed to detect the change of Gcf-DNA in liver transplantation, and they decided to participate in or not. The patient of their legal guardian has provided informed consent for publication of the case.
2.2. Measurement of Gcf-DNA
Around 5 mL blood specimens were collected by cell-free DNA collection tube (Roche, Germany) from 2 recipient at day 0, day 1, day 7, day 14, day 30 and day 60. Cell-free plasma was obtained from blood samples using a 2-step centrifugation process (4°C at 2500 × g for 10 minutes and 4°C at 2500 × g for 10 minutes) within 6 hours after sampling.[10] The resultant plasma was stored at −80°C until analysis. DNA fragments from 600 μL of cell-free plasma were extracted using Circulating Nucleic Acid Kit (Qiagen, Germany). Y-chromosome DNA concentration was determined by using the formula reported by Chiu et al,[11]:
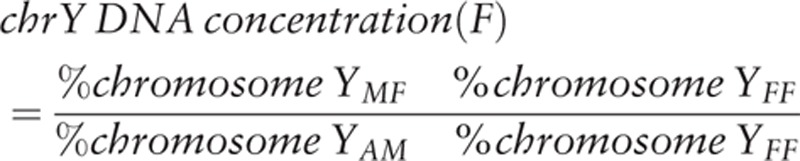 |
where %chromosome YMF is the Y-chromosome percentage of samples with sex-mismatch, %chromosome YFF is the background average Y chromosome percentage of female samples with sex match (containing 100% female DNA), and %chromosome-YAM is the average Y chromosomal percentage among cell-free DNA in the plasma of 3 adult men (containing 100% male DNA, 0.170%).
2.3. Case reports
2.3.1. Case 1
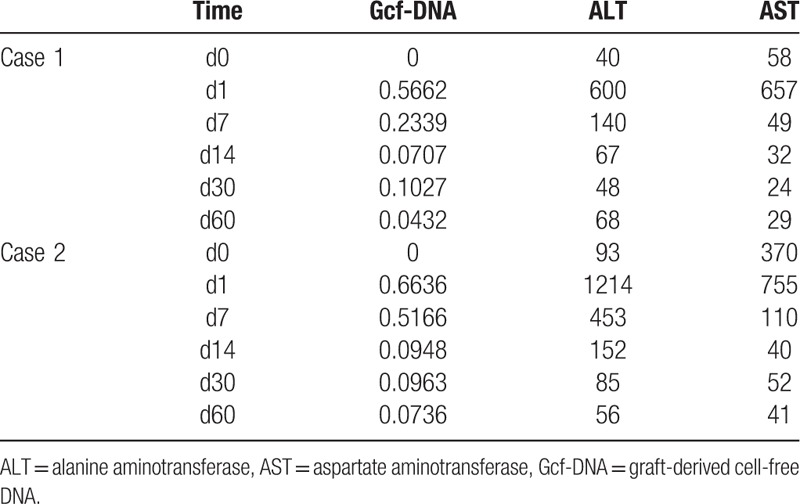
In October, 2016, a 2 months male came to the hospital because of convulsion. Blood test results showed normal except hyper-ammonemia (237 μmol/L), the patient was treated by decrease-ammonia treatment and became stable. The patient showed weak, slow to react and vomiting. Family history showed 2 older brothers of the patients died in 7 days and 1 year after birth with uncertain cause. Then, the patient went through the genetic test, which showed Ornithine Transcarbamylase Deficiency (OTCD), mutated gene as OTC, c.587A>T (p.D196V). The patient came to our hospital because of recurrent and uncontrollable hyper-ammonemia (over 200 μmol/L each time). The legal guardian of the patient chose LDLT as the surgery style and mother of the patient as the donor. The operation went through successfully and we collected the patient's specimens based on the protocol (Table 1). As Figure 1A showed Gcf-DNA climbed up the highest on day 1 and gradually decreased to about 0.1 from day 14 to day 30. At the same time, the curve of ALT and AST also had the similar trend with the Gcf-DNA (Table 2). The follow-up showed the patient status was stable with immunosuppressant treatment.
Table 1.
Characteristics of 2 patients.
Figure 1.
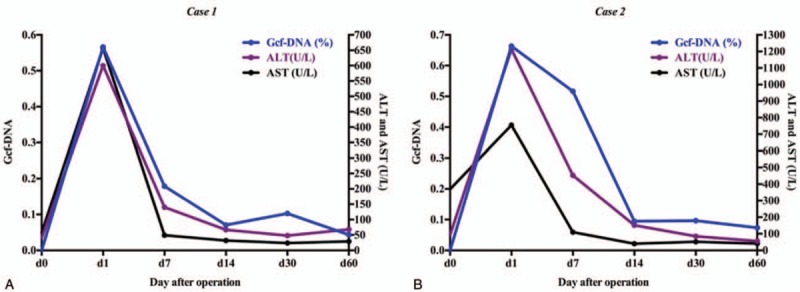
Gcf-DNA were elevated most at day 1 after operation, and gradually dropped down to a stable baseline (<0.1). Thus, we compared the change of the normal liver-function and the change of Gcf-DNA, which showed the same declined-trend. Gcf-DNA = graft-derived cell-free DNA.
Table 2.
Gcf-DNA quantification results and liver function information of Case 1 and Case 2.
2.3.2. Case 2
In February, 2017, a 2-year-old female showed weak and sluggishness with unknown reason and her parents sent her to the hospital. Blood test results showed ALT was 1412 U/L and AST 748.8 U/L and hyper-ammonemia (226 μmol/L). The patient was treated by glutathione, arginine and lactulose. Because of the hyper-ammonemia occurred frequently, the patient went through genetic test, which showed OTCD (mutated gene as OTC, c.540+265G>A het∗). The patient came to our hospital because of OTCD and recurrent hyper-ammonemia. Then, after completely health-evaluation, the legal guardians of the patient chose LDLT as the surgery style and father of the patient as the donor. LDLT went through uneventfully and the specimens were collected following by the protocol (Table 1). As Figure 1B showed, Gcf-DNA climbed up the highest on day 1 and gradually decreased to lower than 0.1 from day 14 to day 60. The curve of ALT and AST also showed the same trend with the curve of Gcf-DNA (Table 2). The follow-up of this patient also showed stable status with immunosuppressant treatment.
3. Discussion
Liver transplantation is an effective therapeutic option for a variety of inborn errors of metabolism, of which urea cycle disorders are the most common indication for transplantation. It accounts for 25.6% of cases.[12] Ornithine transcarbamylase deficiency (OTCD), the most common urea cycle disorder in human, represents 20% of the patients with metabolic diseases indicated for living donor LTx.[13] In this case report, we measured Gcf-DNA using chromosome Y sequence changes in 2 donor-recipient-sex-mismatch pairs, and confirmed that Gcf-DNA was highly elevated in day 1 because of ischemia and reperfusion injury, then it gradually decreased to a relative stable level of 0.1, which was consistent with Schütz E's team reported.[4] More, we compared time-matched Gcf-DNA and liver function tests samples showed that Gcf-DNA can also have the same sensitivity to reflect graft damage, and we did not show others liver function index (BIL, ALP, and GGT) because all of them were among the normal range before and after the operation (Fig. 1). Although the result of 2 cases were similar, we observed little difference between case 1 and case 2, which the curve-change of Gcf-DNA almost had the same variance with ALT and AST in Figure 1A; however, we observed Gcf-DNA still remained high but ALT and AST dropped down obviously in day 7. Interestingly, although this fine distinction only appeared in case 2, it prompted that Gcf-DNA might be more sensitive than routine liver function test because Gcf-DNA were only derived from the graft but ALT and AST were not only derived from the liver. According to the same trend of Gcf-DNA and ALT and AST, we considered that Gcf-DNA could also be a potential and sensitive method to monitor the graft function in LDLT. However, more cases are needed to testify the reproducibility of Y-chromosome method in monitoring graft injury and more samples should be further collected to identify the stable baseline of Gcf-DNA in LDLT patients.
However, an obvious limitation of the Y-chromosome method is that it is only apply for donor-recipient-sex-mismatch pairs. In order to solve this problem, different techniques have been adopted to detect and quantify Gcf-DNA, including quantitative-PCR,[14–16] digital droplet PCR (ddPCR)[16] and massively parallel sequencing.[17–18] Although with extensive development of methods, but the related technological limitations and high-cost still impeded the application of Gcf-DNA as a clinical routine test. Even though the Next-Generation-Sequencing (NGS) assay has been developed to quantify Gcf-DNA without recipient or donor genotype, but it was only reported in heart transplant recipients,[18–19] but it is too expensive to apply in the routine clinical practice. Nevertheless, the price will decrease in the future time as the NGS detecting method become more universal.
Author contributions
Conceptualization: Zhi-Jun Zhu, Li-Ying Sun
Recourses: Hoi-Ioi Ng
Visualization: Hoi-Ioi Ng
Writing-original draft: Hoi-Ioi Ng
Writing-review and editing: Hoi-Ioi Ng, Zhi-Jun Zhu, Li-Ying Sun
Conceptualization: Zhi-Jun Zhu.
Data curation: Hoi-Ioi Ng.
Investigation: Hoi-Ioi Ng.
Validation: Hoi-Ioi Ng.
Visualization: Hoi-Ioi Ng.
Writing – original draft: Hoi-Ioi Ng.
Writing – review & editing: Hoi-Ioi Ng, Li-Ying Sun, Zhi-Jun Zhu.
Footnotes
Abbreviations: ALT = alanine aminotransferase, AST = aspartate aminotransferase, ALP = alkaline phosphatase, GGT = γ-Glutamyl transpeptidase, BIL = bilirubin, cfDNA = cell-free DNA, Gcf-DNA = graft-derived cell-free DNA, LTx = liver transplantation, LDLT = living donor liver transplantation, OTCD = ornithine transcarbamylase deficiency.
The work was supported by Capital Special Program for Health Research and Development (No. 2016-1-2021) and Beijing Municipal Administration of Hospitals Ascent Plan (Code: DFL20150101).
The authors have no conflicts of interest to disclose.
References
- [1].Kasahara M, Sakamoto S, Horikawa R, et al. Living donor liver transplantation for pediatric patients with metabolic disorders: the Japanese multicenter registry. Pediatr Transplant 2014;18:6–15. [DOI] [PubMed] [Google Scholar]
- [2].Kanzow P, Kollmar O, Schutz E, et al. Graft-derived cell-free DNA as an early organ integrity biomarker after transplantation of a marginal HELLP syndrome donor liver. Transplantation 2014;98:E43–5. [DOI] [PubMed] [Google Scholar]
- [3].Boyum JH, Atwell TD, Schmit GD, et al. Incidence and risk factors for adverse events related to image-guided liver biopsy. Mayo Clin Proc 2016;91:329–35. [DOI] [PubMed] [Google Scholar]
- [4].Schutz E, Fischer A, Beck J, et al. Graft-derived cell-free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: A prospective, observational, multicenter cohort study. PLoS Med 2017;14:e1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].De Vlaminck I, Valantine HA, Snyder TM, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med 2014;6:241ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Beck J, Oellerich M, Schulz U, et al. Donor-derived cell-free DNA is a novel universal biomarker for allograft rejection in solid organ transplantation. Transpl P 2015;47:2400–3. [DOI] [PubMed] [Google Scholar]
- [7].Snyder TM, Khush KK, Valantine HA, et al. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci U S A 2011;108:622962–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Burnham P, Khush K, De Vlaminck I. Myriad applications of circulating cell-free DNA in precision organ transplant monitoring. Ann Am Thorac Soc 2017;14:S237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lo YM, Tein MS, Pang CC, et al. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. Lancet 1998;351:1329–30. [DOI] [PubMed] [Google Scholar]
- [10].Wong D, Moturi S, Angkachatchai V, et al. Optimizing blood collection, transport and storage conditions for cell free DNA increases access to prenatal testing. Clin Biochem 2013;46:1099–104. [DOI] [PubMed] [Google Scholar]
- [11].Chiu RW, Akolekar R, Zheng YW, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ 2011;342:c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Arnon R, Kerkar N, Davis MK, et al. Liver transplantation in children with metabolic diseases: the studies of pediatric liver transplantation experience. Pediatr Transplant 2010;14:796–805. [DOI] [PubMed] [Google Scholar]
- [13].Oishi K, Arnon R, Wasserstein MP, et al. Liver transplantation for pediatric inherited metabolic disorders: considerations for indications, complications, and perioperative management. Pediatr Transplant 2016;20:756–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Macher HC, Suarez-Artacho G, Guerrero JM, et al. Monitoring of transplanted liver health by quantification of organ-specific genomic marker in circulating DNA from receptor. PLoS One 2014;9:e113987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sigdel TK, Vitalone MJ, Tran TQ, et al. A rapid noninvasive assay for the detection of renal transplant injury. Transplantation 2013;96:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Beck J, Bierau S, Balzer S, et al. Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft Injury. Clin Chem 2013;59:1732–41. [DOI] [PubMed] [Google Scholar]
- [17].Gordon PM, Khan A, Sajid U, et al. An algorithm measuring donor cell-free DNA in plasma of cellular and solid organ transplant recipients that does not require donor or recipient genotyping. Front Cardiovasc Med 2016;3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Grskovic M, Hiller DJ, Eubank LA, et al. Validation of a clinical-grade assay to measure donor-derived cell-free DNA in solid organ transplant recipients. J Mol Diagn 2016;18:890–902. [DOI] [PubMed] [Google Scholar]
- [19].Hidestrand M, Tomita-Mitchell A, Hidestrand PM, et al. Highly sensitive noninvasive cardiac transplant rejection monitoring using targeted quantification of donor-specific cell-free deoxyribonucleic acid. J Am Coll Cardiol 2014;63:1224–6. [DOI] [PMC free article] [PubMed] [Google Scholar]


