Supplemental Digital Content is available in the text
Keywords: ABCA1, lipid, polymorphism, rs2066714, rs2230806, rs2230808
Abstract
Background:
Studies on the associations of the adenosine triphosphate-binding cassette transporter A1 gene (ABCA1) rs2230806, rs2230808, and rs2066714 polymorphisms with plasma lipid levels have reported apparently conflicting findings. This meta-analysis aimed to clarify the relationships between the 3 polymorphisms and fasting lipid levels.
Methods:
A comprehensive search of the literature was carried out by using the databases including Medline, Google Scholar, Web of Science, Embase, Cochrane Library, CNKI, Wanfang, and VIP. The studies that presented mean lipids and standard deviations or standard errors according to the rs2230806, rs2230808, and/or rs2066714 genotypes were examined and included. The random effects model was used. Standardized mean difference and 95% confidence interval were used to assess the differences in lipid levels between the genotypes. Heterogeneity among studies was tested by Cochran's χ2-based Q-statistic, and Galbraith plots were used to detect the potential sources of heterogeneity. Publication bias was assessed by Begg's rank correlation test as well as funnel plots.
Results:
Sixty-two studies (48,452 subjects), 12 studies (9853 subjects) and 14 studies (10,727 subjects) were identified for the rs2230806, rs2230808, and rs2066714 polymorphisms, respectively. A dominant model was used for all the polymorphisms in this meta-analysis. The A allele carriers of the rs2230806 polymorphism had higher levels of high-density lipoprotein cholesterol (HDL-C) (P <.001), and lower levels of low-density lipoprotein cholesterol (LDL-C) (P =.03) and triglycerides (TG) (P <.01) than the non-carriers. The A allele carriers of the rs2230808 polymorphism had higher levels of total cholesterol (TC) (P <.001) than the non-carriers. The G allele carriers of the rs2066714 polymorphism had higher levels of TC (P <.01) and HDL-C (P = .02) than the non-carriers.
Conclusion:
The ABCA1 rs2230806, rs2230808, and rs2066714 polymorphisms are significantly associated with plasma lipid levels in the present meta-analysis.
1. Introduction
Coronary heart disease (CHD) is a common, multifactorial disease that causes significant morbidity and mortality worldwide.[1] A number of cardiovascular risk factors have been identified. Dyslipidemia is one of the most important risk factors for CHD and accounts for at least 50% of the population-attributable risk.[2] Dyslipidemia is characterized by elevated levels of triglycerides (TG), total cholesterol (TC) and low-density lipoprotein cholesterol (LDL-C), and/or reduced levels of high-density lipoprotein cholesterol (HDL-C) in circulation. Over the past several decades, intensive efforts have been made in the scientific community to investigate the genetic polymorphisms or mutations that affect plasma lipid levels. However, the research results were inconclusive and inconsistent due to various reasons such as small sample sizes and differences in ethnicities and health conditions.
Adenosine triphosphate-binding cassette transporter A1 (ABCA1) plays a critical role in reverse cholesterol transport (RCT) and has anti-atherosclerosis effects.[3] RCT is a process in which cholesterol is effluxed from peripheral tissues, arterial wall macrophages and atherosclerotic plaques to high-density lipoproteins and subsequently delivered to the liver for clearance. ABCA1 is one of the most important lipid transporters that significantly affect plasma HDL-C levels.[4] Low HDL-C and isolated low HDL-C constitute an important risk factor for CHD. Singaraja et al.[5] demonstrated that a 50% increase in ABCA1-mediated cholesterol efflux can result in a 30% increase in HDL-C concentrations and consequently a 35% to 50% reduction in the risk of CHD. The ABCA1 gene is located on the long arm of human chromosome 9 (9q22–31) and contains 50 exons and 49 introns with a total length of approximately 50 kB. The ABCA1 gene is highly polymorphic. There are over 5000 polymorphisms in or around the ABCA1 gene according to the NCBI database of genetic variation (https://www.ncbi.nlm.nih.gov/SNP). Most of the polymorphisms are located in the introns of this gene, and some polymorphisms located in the promoter region and exons usually have great effects on the expression and function of ABCA1 protein. Three nonsynonymous polymorphisms (rs2230806, rs2230808, and rs2066714) within the exons of ABCA1 gene have been extensively studied in terms of their associations with CHD risk and plasma lipid levels over the past 2 decades. The rs2230806 polymorphism (also known as R219K or G1051A) is located in exon 7 and formed by a transition from G to A. The 219th amino acid residue is changed accordingly from arginine (R) to lysine (K). The frequency range of the variant A allele is 0.21 to 0.27 in Caucasians, 0.34 to 0.50 in Asians, and 0.16 to 0.23 in Arabs (https://www.ncbi.nlm.nih.gov/SNP). The rs2230808 polymorphism (also known as R1587K or G5155A) is located in exon 35 of ABCA1 gene and formed by a transition from G to A, and the 1587th amino acid residue of ABCA1 polypeptide is changed accordingly from arginine to lysine. The frequency range of A allele is 0.23 to 0.27 in Caucasians and 0.28 to 0.46 in Asians (https://www.ncbi.nlm.nih.gov/SNP). The rs2066714 polymorphism (also known as I883 M, A3044G, rs4149313, rs2853570, or rs58387182) is located in exon 18 of ABCA1 gene, and its 2 alleles, A and G, encode an isoleucine (I) and a methionine (M) in ABCA1 polypeptide, respectively. There is a big difference in allele frequencies between Caucasians and Asians. The G allele is the minor allele with a frequency range of 0.11 to 0.15 in Caucasians, whereas it is the major allele with a frequency range of 0.49 to 0.73 in Asians (https://www.ncbi.nlm.nih.gov/SNP). Some studies have demonstrated that the rs2230806,[6–11] rs2230808[7] and rs2066714[11–13] polymorphisms are significantly associated with CHD, but whether these polymorphisms are also associated with dyslipidemia remains to be examined. A large body of literature has investigated the associations of these polymorphisms with plasma lipid levels, but the results were inconsistent and inconclusive. In some of these studies, A allele of the rs2230806 polymorphism was reported to be significantly associated with lower levels of TG,[14–28] TC[29–34] and LDL-C,[24–27] and higher levels of HDL-C;[35–49] A allele of the rs2230808 polymorphism was associated with higher levels of TG,[50] TC[51,52] and LDL-C,[52–54] and lower levels of HDL-C;[6] G allele of the rs2066714 polymorphism was associated with higher levels of TC,[29] LDL-C,[29,50,55] and HDL-C.[56,57] However, the results from other studies did not support these findings.[58–77] Hence, a meta-analysis is required to clarify the relationships of the 3 polymorphisms with plasma lipid levels.
In the present study, a meta-analysis was performed based on previous publications to investigate the associations of the rs2230806, rs2230808, and rs2066714 polymorphisms with fasting lipid levels. Our analysis results can provide an opportunity to unveil the interrelationships among the rs2230806, rs2230808, and rs2066714 polymorphisms, dyslipidemia, and CHD.
2. Methods
2.1. Search strategy
A comprehensive search of the literature was carried out by using the databases including Medline, Google Scholar, Web of Science, Embase, Cochrane Library, China National Knowledge Infrastructure (CNKI), Wanfang and VIP. The keywords used for the search are “adenosine triphosphate-binding cassette transporter A1 or ATP-binding cassette transporter A1 or ABCA1” and “polymorphism or variant or mutation or SNP” and “lipid or triglyceride or cholesterol”. The variables of this meta-analysis were limited to TG, TC, LDL-C, and HDL-C. All articles published before July 2018 on the associations of the rs2230806, rs2230808, and rs2066714 polymorphisms with plasma lipid levels were identified. The languages of the articles were limited to English and Chinese. All references cited in the included articles were reviewed to check the published work which was not indexed by Medline, Google Scholar, Web of Science, Embase, Cochrane Library, CNKI, Wanfang, or VIP database. Email, phone, and fax were used to contact authors to identify studies and to request missing data. Ethical approval is not necessary since this study is a meta-analysis.
2.2. Inclusion criteria
The studies that fulfilled the following criteria were included:
-
(1)
studies in which mean lipids and standard deviations (SD) or standard errors (SE) according to the rs2230806, rs2230808, and rs2066714 genotypes were available;
-
(2)
data which reported at least one of the 4 variables (TG, TC, LDL-C, and HDL-C);
-
(3)
data which reported fasting lipid variables.
Baseline data were used for interventional studies. Reports with incomplete data, studies based on pedigree data, case reports, review articles, abstracts, and animal studies were excluded from the meta-analysis.
2.3. Data extraction
Data were extracted by using a structured collection form. The irrelevant studies or studies that did not meet the inclusion criteria were excluded after being reviewed independently by 2 reviewers. The data were double-checked and compared after extraction. Data uncertainty was discussed and solved by the whole group members. For the overlapping articles, only those publications that presented the most detailed information were included. In the present meta-analysis, the data extracted from each of the studies are as follows: first author, year of publication, age, ethnicity, gender, health condition, genotype and lipid assay methods, sample size, mean lipid variables and SD or SE by genotypes.
2.4. Meta-analysis
The STATA software package (Version 10, Stata Corporation, College Station, TX) was used for the meta-analysis. All data were presented as mean ± SD in this meta-analysis. For those articles in which mean ± SE was given, the value of SD was calculated. The unit “mmol/L” was used for all the lipid variables in the meta-analysis, and unit conversion was conducted for the articles in which other units were used. Hardy–Weinberg equilibrium (HWE) of the populations was tested by χ2 test, and the significance level was defined as α <.05. Since most of the included studies reported results in a dominant way [i.e., GG vs (GA + AA) for the rs2230806 polymorphism; GG vs (GA + AA) for the rs2230808 polymorphism; AA vs (AG + GG) for the rs2066714 polymorphism], a dominant model was employed to ensure adequate statistical power. When data were presented for more than one subpopulation (e.g., male or female subjects, the subjects from different ethnicities, or the subjects with different health statuses) in 1 article, each subpopulation was treated as a separate comparison in this meta-analysis. Subgroup analyses were conducted according to gender, ethnicity, and health condition. Ethnic subgroups were defined as Caucasians, Chinese, and other ethnicities. Health condition subgroups were defined as healthy/control subjects, CHD patients, diabetic patients, and so on. The subgroup analyses were performed with at least 3 comparisons to ensure adequate statistical power.
The random effects model was used in the meta-analysis in that:
-
(1)
both between-study and within-study heterogeneity is considered in random effects model;
-
(2)
the random effects model provides a more conservative evaluation of the significance of the associations than the fixed effects model.[78]
Standardized mean difference (SMD) and 95% confidence interval (CI) were used to assess the differences in lipid levels between the genotypes. Heterogeneity among studies was tested by Cochran's χ2-based Q-statistic at a significance level of P <.05, and Galbraith plots were used to detect the potential sources of heterogeneity. Publication bias was assessed by Begg rank correlation test and funnel plots, and a significance level of .05 was used to indicate the presence of potential publication bias.[79]
3. Results
3.1. Literature search and characteristics of included studies
The literature selection process is presented in Figure 1. Initial search of the databases yielded 1249 articles. One thousand and seventy-eight studies were excluded according to the titles and abstracts. Then full-text articles were retrieved and assessed on the basis of the inclusion criteria. One hundred and 5 articles were ineligible for the following reasons: 64 articles did not provide lipid data; 21 articles provided invalid data; 16 articles presented data for other polymorphisms, 1 article had subjects overlapping with other publication; and 3 articles were based on pedigree analysis. In the end, 66 studies[6,7,14–77] were selected for this meta-analysis. Among the included studies, 48 studies[6,7,14,16,23,25–30,32,34–39,44,45,49–61,63–77] were published in English, and 18 studies[15,17–22,24,31,33,40–43,46–48,62] were published in Chinese.
Figure 1.

Flow diagram of study selection process.
The characteristics of the included 66 studies are summarized in Table in Supplementary S1 Table. Sixty-two studies[6,7,14–49,53–75,77] presented lipid data for the rs2230806 polymorphism according to genotypes. Among them, 48 studies,[6,14–33,35–43,46–49,55,58–62,64,66,69,72–75,77] 49 studies,[6,14–24,26–37,39–43,46–49,53–55,58–62,64,69,71–75,77] 48 studies[6,14–33,35–37,39–43,46–49,53–55,58–62,64,69,71–75] and 59 studies[6,7,14–33,35–49,55–75,77] presented the data for TG, TC, LDL-C and HDL-C, respectively. Twelve studies[6,7,38,50–54,56,61,67,68] presented lipid data for the rs2230808 polymorphism according to genotypes, and 5 studies,[6,38,50,51,61] 7 studies,[6,50–54,61] 7 studies[6,50–54,61] and 10 studies[6,7,38,50–52,56,61,67,68] of which presented the data for TG, TC, LDL-C and HDL-C, respectively. Fourteen studies[6,7,14,29,38,50,53–57,61,67,76] presented lipid data for the rs2066714 polymorphism according to genotypes, and 9 studies,[6,14,29,38,50,53,55,61,76] 7 studies,[14,29,50,54,55,61,76] 8 studies[14,29,50,53–55,61,76] and 12 studies[6,7,14,29,38,50,55–57,61,67,76] of them presented the data for TG, TC, LDL-C and HDL-C, respectively. Twenty-two studies,[6,7,25,30,34,44,45,50,52–56,58,60,61,63–66,71,72] 36 studies[14,15,17–25,28,31–33,35–43,46–49,51,59,62,68–70,73,74] and 11 studies[16,25–27,29,57,66,67,75–77] involved Caucasians, Chinese and the subjects of other ethnic origins, respectively. Four studies[6,19,55,60] and 2 studies[50,52] involved males and females, respectively, and the rest 60 studies[7,14–18,20–49,51,53,54,56–59,61–77] involved both genders. Twenty-two studies,[6,14,17–19,21–23,26,27,30–32,43,45–48,56,64,72,77] 5 studies,[15,33,35,39,40] 2 studies,[61,76] 4 studies,[29,44,49,57] 5 studies,[7,20,42,68,69] 5 studies,[16,24,34,58,65] 2 studies[37,75] and 1 study[36] involved CHD, stroke, dyslipidemia, overweight/obesity, type 2 diabetes mellitus (T2DM), hypertension, Alzheimer's disease and abdominal aortic aneurysm, respectively. Twenty-two studies[16,18,22,24,26,28,32,35,40,44,45,47,49,57,59,63,66,67,69,70,72,75] separately provided data for more than one subpopulation, and each subpopulation was treated as a comparison.
3.2. Summary statistics
Eighty-seven comparisons, 13 comparisons, and 16 comparisons were distinguished for the rs2230806, rs2230808, and rs2066714 polymorphisms, respectively, according to the categories such as gender, ethnicity and health condition. Sixty-seven, 67, 66 and 84 comparisons were included to compare the differences in TG, TC, LDL-C, and HDL-C, respectively, for the rs2230806 polymorphism (Table in Supplementary S2 Table). Five, 6, 6 and 10 comparisons were included to compare the differences in TG, TC, LDL-C, and HDL-C, respectively, for the rs2230808 polymorphism (Table in Supplementary S3 Table). Nine, 7, 8, and 14 comparisons were included to compare the differences in TG, TC, LDL-C, and HDL-C, respectively, for the rs2066714 polymorphism (Table in Supplementary S4 Table).
Forty-eight thousand four hundred fifty-two subjects, 9853 subjects and 10,727 subjects were enrolled in the analyses for the rs2230806, rs2230808, and rs2066714 polymorphisms, respectively. For the rs2230806 polymorphism, 42.3% of the subjects (20,511 subjects) had GG genotype, and 57.7% of them (27,941 subjects) had GA or AA genotype. For the rs2230808 polymorphism, 49.8% of the subjects (4906 subjects) had GG genotype, and 50.2% of them (4947 subjects) had GA or AA genotype. For the rs2066714 polymorphism, 61.1% of the subjects (6553 subjects) had AA genotype, and 38.9% of them (4174 subjects) had AG or GG genotype.
3.3. Associations of the ABCA1 rs2230806 polymorphism with plasma lipid levels
The details of the associations of the ABCA1 rs2230806 polymorphism with plasma lipid levels are presented in Table 1. The analyses on all comparisons showed that the A carriers have higher levels of HDL-C [SMD = 0.18, 95% CI = (0.13, 0.24), P <.001] and lower levels of TG [SMD = −0.10, 95% CI = (−0.16, −0.03), P <.01] and LDL-C [SMD = −0.05, 95% CI = (−0.09, −0.00), P =.03] than the non-carriers (Table 1, Figs. 2–4). There was no significant difference in TC levels between the genotypes (Table 1, Fig. 5). When the analyses were limited to the studies in HWE, the significant associations between the rs2230806 polymorphism and plasma levels of HDL-C [SMD = 0.13, 95% CI = (0.08, 0.18), P <.001], TG [SMD = −0.10, 95% CI = (−0.18, −0.02), P = .01] and LDL-C [SMD = −0.06, 95% CI = (−0.11, −0.02), P = .01] were also detected.
Table 1.
Meta-analysis of the rs2230806 polymorphism in ABCA1 with plasma lipid levels.

Figure 2.
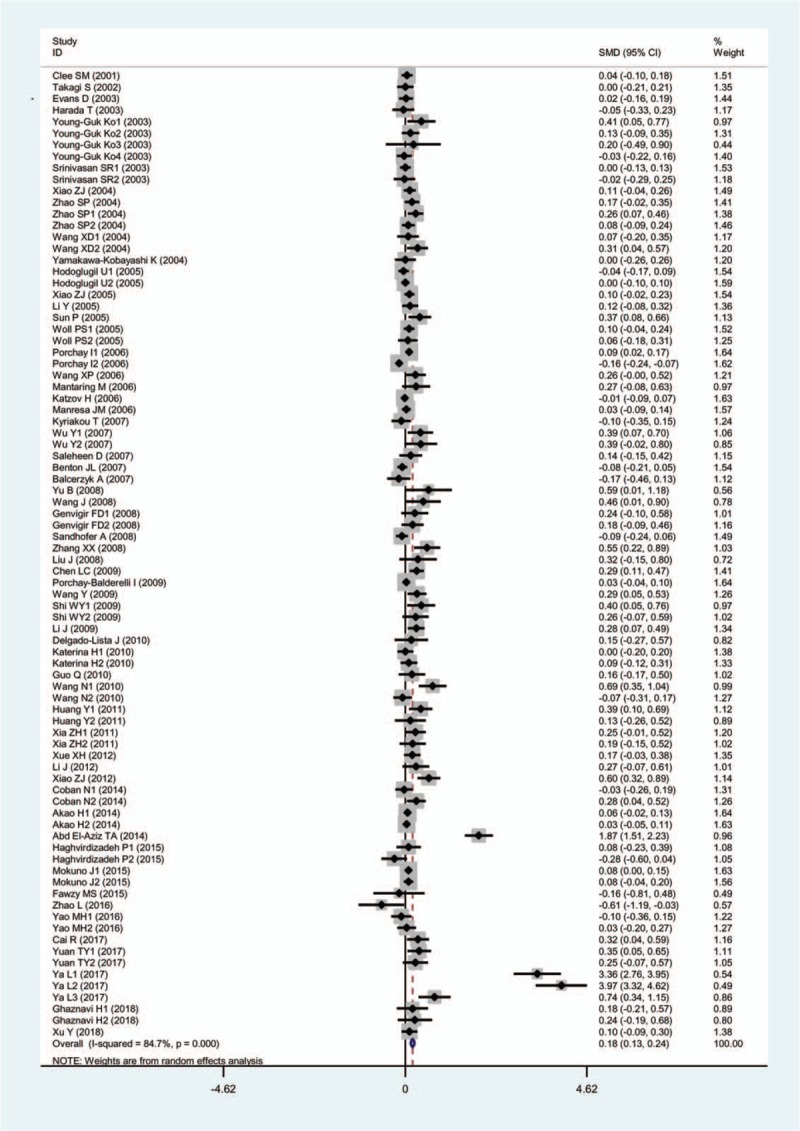
Forest plot of the meta-analysis between ABCA1 rs2230806 polymorphism and plasma HDL-C levels.
Figure 4.

Forest plot of the meta-analysis between ABCA1 rs2230806 polymorphism and plasma LDL-C levels.
Figure 5.

Forest plot of the meta-analysis between ABCA1 rs2230806 polymorphism and plasma TC levels.
Figure 3.

Forest plot of the meta-analysis between ABCA1 rs2230806 polymorphism and plasma TG levels.
Then the subgroup analyses stratified by the characteristics of the subjects were performed. The significant associations of the rs2230806 polymorphism with lower levels of TG and higher levels of HDL-C were detected in males, but not in females. The significant associations of the rs2230806 polymorphism with lower levels of TG and LDL-C, and higher levels of HDL-C were detected in Chinese. In Caucasians, however, it was found that the rs2230806 polymorphism is only significantly associated with TG levels, but not with LDL-C and HDL-C levels. The association between the rs2230806 polymorphism and lower levels of TG was significant in hyperlipidemic patients and marginally insignificant in T2DM. In healthy/control subjects, the rs2230806 polymorphism was significantly associated with lower levels of TG and higher levels of HDL-C. The rs2230806 polymorphism was also significantly associated with higher levels of HDL-C, and lower levels of TG and LDL-C in CHD patients.
3.4. Associations of the ABCA1 rs2230808 polymorphism with plasma lipid levels
The details of the associations of the ABCA1 rs2230808 polymorphism with plasma lipid levels are listed in Table 2. The analyses on all comparisons showed that the variant A allele carriers had higher levels of TC [SMD = 0.15, 95% CI = (0.08, 0.22), P <.001] than the non-carriers (Table 2, Supplementary S1 Fig). The association between the rs2230808 polymorphism and higher levels of LDL-C was marginally insignificant [SMD = 0.13, 95% CI = (−0.01, 0.28), P = .08] (Table 2, Supplementary S2 Fig). There were no significant differences detected in TG and HDL-C levels between the genotypes (Table 2, Supplementary S3 and S4 Figs). The significant associations of the rs2230808 polymorphism with TC [SMD = 0.15, 95% CI = (0.08, 0.23), P <.001] and LDL-C [SMD = 0.18, 95% CI = (0.05, 0.31), P = .01] were detected when the analyses were limited in the studies in HWE.
Table 2.
Meta-analysis between the rs2230808 polymorphism in ABCA1 and plasma lipid levels.

The subgroup analyses stratified by the characteristics of the subjects were performed, and the significant association between the rs2230808 polymorphism and higher levels of TC was detected in Caucasians and healthy/control subjects. The significant association between the rs2230808 polymorphism and higher levels of LDL-C was detected in healthy/control subjects. No other significant associations were detected in the subgroup analyses.
3.5. Associations of the ABCA1 rs2066714 polymorphism with plasma lipid levels
The details of the associations of the ABCA1 rs2066714 polymorphism with lipid levels are listed in Table 3. The analyses on all comparisons showed that the carriers of the variant G allele had higher levels of TC [SMD = 0.13, 95% CI = (0.04, 0.21), P <.01] and HDL-C [SMD = 0.10, 95% CI = (0.02, 0.18), P = .02] than the non-carriers (Table 3, Supplementary S5 and S6 Fig). There were no significant differences detected in TG and LDL-C levels between the genotypes (Table 3, Supplementary S7 and S8 Fig). When the analyses were limited to the studies in HWE, the associations of the rs2066714 polymorphism with high levels of TC [SMD = 0.13, 95% CI = (0.04, 0.22), P <.01] and HDL-C [SMD = 0.09, 95% CI = (0.00, 0.18), P = .03] were also detected.
Table 3.
Meta-analysis between the rs2066714 polymorphism in ABCA1 and plasma lipid levels.

The subgroup analyses stratified by the characteristics of the subjects were performed, and the significant associations between the rs2066714 polymorphism and higher levels of TC and HDL-C were detected in Caucasians. No other significant associations were detected in the subgroup analyses.
3.6. Heterogeneity analysis
In the analyses for the rs2230806 polymorphism, there was significant heterogeneity in the total comparisons for TG, TC, LDL-C, and HDL-C (Table 1). Nine comparisons (Harada T 2003, Li J 2009, Coban N1 2014, Coban N2 2014, Fawzy MS 2015, Yuan TY1 2017, Ya L1 2017, Ya L2 2017 and Ya L3 2017), 6 comparisons (Kolovou G 2013, Abd El-Aziz TA 2014, Yuan TY1 2017, Ya L1 2017, Ya L2 2017 and Ya L3 2017), 6 comparisons (Sun P 2005, Coban N2 2014, Abd El-Aziz TA 2014, Fawzy MS 2015, Ya L2 2017 and Ya L3 2017) and 15 comparisons (Sun P 2005, Porchay I2 2006, Benton JL 2007, Sandhofer A 2008, Zhang XX 2008, Chen LC 2009, Wang N1 2010, Huang Y1 2011, Xiao ZJ 2012, Abd El-Aziz TA 2014, Haghvirdizadeh P2 2015, Zhao L 2016, Ya L1 2017, Ya L2 2017 and Ya L3 2017) were identified as the main contributors to the heterogeneity for TG, TC, LDL-C and HDL-C, respectively, by using Galbraith plots (Supplementary S9-S12 Figs). The heterogeneity was effectively removed or decreased after exclusion of the outlier studies, but the SMD values and their 95% CIs did not change substantially [TG: SMD = −0.07, 95% CI = (−0.10, −0.04), PSMD <.001, PHeterogeneity = .11; TC: SMD = 0.00, 95% CI = (−0.03, 0.03), PSMD = .91, PHeterogeneity = .54; LDL-C: SMD = −0.03, 95% CI = (−0.06, −0.00), PSMD = .04, PHeterogeneity = .27; HDL-C: SMD = 0.08, 95% CI = (0.06, 0.11), PSMD <.001, PHeterogeneity = .07] after exclusion of the outlier studies.
In the analyses for the rs2230808 polymorphism, there was significant heterogeneity in the total comparison for LDL-C (Table 2), and 2 comparisons (Mantaring M 2006 and Jensen MK 2007) were identified as the main contributors to the heterogeneity by using Galbraith plot (Supplementary S13 Fig). The heterogeneity was removed after exclusion of the outlier studies, and the SMD value and its 95% CI changed significantly [SMD = 0.23, 95% CI = (0.14, 0.32), PSMD <.001, PHeterogeneity = .38].
In the analyses for the rs2066714 polymorphism, there was significant heterogeneity in the total comparison for HDL-C (Table 3), and 1 comparison (Kyriakou T 2007) was identified as the main contributor to the heterogeneity by using Galbraith plot (Supplementary S14 Fig). The heterogeneity was removed after exclusion of the outlier study, and the SMD value and its 95% CI did not change significantly [SMD = 0.07, 95% CI = (0.01, 0.13), PSMD = .04, PHeterogeneity = .11].
3.7. Publication bias test
In the present study, Begg test did not find any publication bias in the analyses for the rs2230808 and rs2066714 polymorphisms (Supplementary S15-S22 Figs). Regarding the rs2230806 polymorphism, no publication bias was detected except HDL-C (Supplementary S23-S26 Figs). A trim-and-fill method was employed to adjust the result for HDL-C, and the SMD value and its 95% CI did not change substantially [SMD = 0.18, 95% CI = (0.13, 0.24), PSMD <.001] after adjustment.
4. Discussion
The present meta-analysis suggests that A allele of the rs2230806 polymorphism is associated with higher levels of HDL-C and lower levels of LDL-C and TG in the total population. A number of case-control studies[6–11] demonstrated that A allele of the rs2230806 polymorphism has a protective role in CHD risk. In combination with our findings, it is possible that the association between the rs2230806 polymorphism and a lower risk of CHD is mediated by the increased levels of HDL-C and decreased levels of LDL-C and TG caused by A allele of the rs2230806 polymorphism. It is well-known that HDL has atheroprotective properties such as RCT, anti-oxidation, and anti-inflammation.[80] In addition, A allele the rs2230806 polymorphism was also associated with lower levels of apolipoprotein B (APOB) (data not shown). It indicates that the carriers of A allele have less atherogenic potentials than the non-carriers because each particle of the atherogenic lipoproteins [i.e., low-density lipoprotein, very low-density lipoprotein, intermediary density lipoprotein and lipoprotein (a)] carries one APOB molecule. This meta-analysis also demonstrates that A allele of the rs2230808 polymorphism and G allele of the rs2066714 polymorphism is associated with higher levels of TC, which partly explains why the 2 polymorphisms were associated with a higher risk of CHD in several case-control studies.[7,12,13] We also found that the G allele carriers of the rs2066714 polymorphism have higher levels of HDL-C, which is contradictive with the finding that G allele is a risk allele for CHD reported by previous studies.[11–13] The relationships among the rs2066714 polymorphism, dyslipidemia, and CHD risk need to be further investigated.
Subgroup analyses by gender, ethnicity and health condition were performed since they might be important variables in determining associative risk with lipid levels. For example, the present meta-analysis indicates that ethnicity might modulate the associations of the rs2230806 polymorphism with LDL-C and HDL-C levels since the significant associations only exist in Chinese, but not in Caucasians (Table 1). Our finding on the association between the rs2230806 polymorphism and HDL-C is consistent with the meta-analysis by Ma et al[81] in which the investigators demonstrated that A allele is associated with higher HDL-C levels in Asians, but not in Caucasians. A recent meta-analysis[8] revealed that A allele carriers of the rs2230806 polymorphism have a lower risk of CHD than the non-carriers in various ethnicities. However, another study[9] demonstrated that A allele of the rs2230806 polymorphism is associated with a lower risk of CHD only in Asian populations, but not in Caucasian populations. A meta-analysis[10] which focused only on Chinese population reported that A allele of the rs2230806 polymorphism is associated with a lower risk of CHD in Chinese. In combination with our findings, it is possible that the association between A allele of the rs2230806 polymorphism and a lower risk of CHD in Asians is mediated by atheroprotective lipid profiles (i.e., increased HDL-C and decreased LDL-C and TG levels) caused by A allele of the rs2230806 polymorphism.
Significant heterogeneity was detected in the analyses for the rs2230806 (TG, TC, LDL-C, and HDL-C), rs2230808 (LDL-C), and rs2066714 (HDL-C) polymorphisms. Subgroup analyses stratified by the characteristics of the subjects were performed to explore the potential sources of the observed heterogeneity, and the results showed that the sources of heterogeneity were mainly from the ethnic origins of the subjects and health conditions. The ethnic origins in the present study were very diverse, including Caucasians, Asians, Africans, Turkish, Egyptians, and so on. The health statuses of the included populations were also diverse, including CHD, stroke, dyslipidemia, overweight/obese, diabetes, hypertension, Alzheimer's disease, abdominal aortic aneurysm, and so forth. Galbraith plots were employed to figure out the specific comparisons which produced heterogeneity. Outlier comparisons were identified by using Galbraith plots, and the heterogeneity was effectively removed or decreased after exclusion of the outlier comparisons. No significant changes in the SMD values and 95% CIs were found for most of the analyses after excluding the outlier studies. However, the difference in LDL-C levels became significant in the analysis for the rs2230808 polymorphism after excluding the outlier studies. By using the Galbraith plot, the heterogeneity was produced from the studies by Jensen et al[50] and Mantaring et al[61] In the study by Jensen et al,[50] only women were included and the characteristics of the subjects included diabetes, hypertension, hypercholesterolemia, CHD, overweight, and smoking. In the study by Mantaring et al,[61] 70% of the subjects had abnormal HDL-C levels. Therefore, the result after excluding the outlier studies was reasonable since there was a selection bias in the study populations in the 2 studies. Begg test showed that there might be a publication bias in the pooling analysis for HDL-C for the rs2230806 polymorphism. The possible reason for the observed publication bias might be the strong association between the rs2230806 polymorphism and HDL-C levels, and high heterogeneity such as different ethnicities and health conditions. To clarify this problem, a trim-and-fill method was employed to adjust the result, and the result did not change substantially after including the “19 missing comparisons”.
The possible mechanisms in which the rs2230806, rs2230808, and rs2066714 polymorphisms modulate the plasma lipid levels have not been clarified yet. One possible explanation is that the rare alleles of these polymorphisms influence the stability and abundance of ABCA1 mRNA, or the molecular structure and activities of ABCA1 protein. Some rare alleles in exons have been reported to affect mRNA structure and stability. Lee et al[82] reported that TT genotype of the rs1800137 polymorphism in low-density lipoprotein receptor-related protein 1 (LRP1) gene is associated with high plasma levels of factor VIII, and further demonstrated that T allele of the rs1800137 polymorphism decreased the LRP1 mRNA stability via translation-dependent mRNA degradation associated with codon optimality. Akdeli et al.[83] found that the rs27770 polymorphism in the 3′untranslated region (3′UTR) of Polo-like Kinase 1 (PLK1) gene influenced the expression of PLK1, and that the alleles of the rs27770 polymorphism displayed different secondary mRNA structures and mRNA stabilities. The rs2230806, rs2230808, and rs2066714 polymorphisms are nonsynonymous genetic variations in which the amino acid residues are replaced with other amino acid residues. Nonsynonymous polymorphisms in exons might influence the structure and function of proteins due to the replacement of amino acid residues with different molecular properties. Lee et al[84] evaluated the relationships between genetic variations in serotonin transporter gene and degenerative mitral valve disease, and identified 3 nonsynonymous genetic polymorphisms in exons as probable causes of damage to protein function, and which was confirmed by protein structure model verification.
The associations of the rs2230806, rs2230808, and rs2066714 polymorphisms with plasma lipid levels are not likely to be type I errors (false-positive results). Firstly, the results from this meta-analysis were based on the random effects model. Comparing with fixed effects model, the random effects model is a more conservative method and less likely to produce false-positive results. Secondly, 48,452 subjects, 9853 subjects and 10,727 subjects were identified for rs2230806, rs2230808 and rs2066714 polymorphisms, respectively. Among the subjects, 57.7% (rs2230806), 50.2% (rs2230808), and 38.9% (rs2066714) of them are the carriers of the variant allele. Since the incidence of the variant allele carriers is very high, type I error could have been prevented for all of the polymorphisms.
Major human diseases, such as cardiovascular disease, cancer and diabetes, are closely related to genetic variations. It is estimated that there are about 3 to 4 million polymorphic loci in human genome, with an average of 1 variant per 1000 to 2000 nucleotides.[85,86] It is these polymorphic loci that lead to differences in susceptibility to disease, sensitivity to drugs and tolerance to poisons. Therefore, the studies on genetic polymorphisms will fundamentally elucidate the pathogenesis of major human diseases, and lay the foundation for the thorough conquest of these diseases. In this study, the 3 polymorphisms, rs2230806, rs2230808, and rs2066714, were found to be significantly associated with blood lipid levels by meta-analysis. They can be used for genetic diagnosis in patients with CHD to determine the etiology, and for screening the high-risk individuals of CHD. For those who are carriers of risk alleles, they can be intervened early to postpone or prevent the occurrence of CHD. Our proposals for future studies are as follows:
-
1.
identify all the genes which are closely related to hyperlipidemia and CHD in human genome;
-
2.
identify all the polymorphisms in the hyperlipidemia- and CHD-related genes and construct polymorphism databases specifically for hyperlipidemia and CHD;
-
3.
screen the hyperlipidemia- and CAD-related polymorphisms early in life to accurately assess the risk of CHD, and give lifestyle suggestions and medical interventions for the high-risk individuals as early as possible.
The present meta-analysis has several limitations. At first, dyslipidemia involves in a large number of genes as well as some environmental factors. However, the interactions of the rs2230806, rs2230808, and rs2066714 polymorphisms with other polymorphisms or environmental factors on plasma lipid levels have not been investigated in this meta-analysis due to the lack of original data from the included studies. In other words, more precise results could have been gained if more detailed individual data were available, or the stratification analyses based on the environmental factors such as diet, exercise, smoking, and so on, were performed. Secondly, a relatively small number of subjects were included in the association analyses for the rs2230808 and rs2066714 polymorphisms due to the limited studies that met the inclusion criteria, which might reduce the statistic power and even cause type I error (false-positive results). Thirdly, this meta-analysis only included the studies published in English and Chinese as it is very difficult to get the full papers published in various languages.
5. Conclusions
A allele of rs2230806 polymorphism is significantly associated with higher levels of HDL-C, and lower levels of LDL-C and TG in the present meta-analysis. In addition, the significant association between A allele of the rs2230808 polymorphism and higher levels of TC, and the significant associations between G allele of the rs2066714 polymorphism and higher levels of TC and HDL-C were also detected.
Author contributions
Song YY, Lu Z, and Luo Z conceived of the study, participated in the design, and drafted the manuscript. Lu Z, Luo Z, Jia AM, Yu LQ, Muhammad I, and Zeng W carried out the study searches and collected the data. Song YY, Lu Z, and Luo Z performed the statistical analyses. All authors reviewed and approved the final manuscript.
Conceptualization: Zhan Lu, Zhi Luo, Yongyan Song.
Data curation: Zhan Lu, Zhi Luo, Aimei Jia, Liuqin Yu, Irfan Muhammad, Wei Zeng, Yongyan Song.
Formal analysis: Zhan Lu, Zhi Luo, Yongyan Song.
Methodology: Zhan Lu, Zhi Luo, Yongyan Song.
Resources: Liuqin Yu, Wei Zeng.
Writing – original draft: Zhan Lu, Zhi Luo, Yongyan Song.
Writing – review & editing: Yongyan Song.
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: 95 CI% = 95% confidence interval, ABCA1 = adenosine triphosphate-binding cassette transporter A1 gene, CHD = coronary heart disease, HDL-C = high-density lipoprotein cholesterol, HWE = Hardy–Weinberg equilibrium, LDL-C = low-density lipoprotein cholesterol, RCT = reverse cholesterol transport, SMD = standardized mean difference, T2DM = type 2 diabetes mellitus, TC = total cholesterol, TG = triglycerides.
ZL and ZL contributed equally to this work and should be considered as co-first authors.
All relevant data are within the paper and its supporting information files.
This research was supported by the grants from the Key Project of Education Department of Sichuan Province, People's Republic of China (17ZA0172) and the Cooperative Project on Scientific Research between Nanchong city and North Sichuan Medical College, People's Republic of China (NSMC20170403). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that there is no conflict of interest regarding the publication of this article.
Supplemental Digital Content is available for this article.
References
- [1].Mozaffarian D, Benjamin EJ, Go AS, et al. Executive summary: heart disease and stroke statistics—2016 update: a report from the American Heart Association. Circulation 2016;133:447–54. [DOI] [PubMed] [Google Scholar]
- [2].Yusuf S, Hawken S, Ounpuu S, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet 2004;364:937–52. [DOI] [PubMed] [Google Scholar]
- [3].He XW, Yu D, Li WL, et al. Anti-atherosclerotic potential of baicalin mediated by promoting cholesterol efflux from macrophages via the PPAR(-LXR(-ABCA1/ABCG1 pathway. Biomed Pharmacother 2016;83:257–64. [DOI] [PubMed] [Google Scholar]
- [4].Lake NJ, Taylor RL, Trahair H, et al. TRAK2, a novel regulator of ABCA1 expression, cholesterol efflux and HDL biogenesis. Eur Heart J 2017;38:3579–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Singaraja RR, Brunham LR, Visscher H, et al. Efflux and atherosclerosis: the clinical and biochemical impact of variations in the ABCA1 gene. Arterioscler Thromb Vasc Biol 2003;23:1322–32. [DOI] [PubMed] [Google Scholar]
- [6].Clee SM, Zwinderman AH, Engert JC, et al. Common genetic variation in ABCA1 is associated with altered lipoprotein levels and a modified risk for coronary artery disease. Circulation 2001;103:1198–205. [DOI] [PubMed] [Google Scholar]
- [7].Porchay-Baldérelli I, Péan F, Emery N, et al. DIABHYCAR Study Group. Relationships between common polymorphisms of adenosine triphosphate-binding cassette transporter A1 and high-density lipoprotein cholesterol and coronary heart disease in a population with type 2 diabetes mellitus. Metabolism 2009;58:74–9. [DOI] [PubMed] [Google Scholar]
- [8].Li JF, Peng DY, Ling M, et al. Evaluation of adenosine triphosphate-binding cassette transporter A1 (ABCA1) R219K and C-reactive protein gene (CRP) +1059G/C gene polymorphisms in susceptibility to coronary heart disease. Med Sci Monit 2016;22:2999–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Liu N, Hou M, Ren W, et al. The R219K polymorphism on ATP-binding cassette transporter A1 gene is associated with coronary heart disease risk in Asia population: evidence from a meta-analysis. Cell Biochem Biophys 2015;71:49–55. [DOI] [PubMed] [Google Scholar]
- [10].Li YY, Zhang H, Qin XY, et al. ATP-binding cassette transporter A1 R219K polymorphism and coronary artery disease in Chinese population: a meta-analysis of 5,388 participants. Mol Biol Rep 2012;39:11031–9. [DOI] [PubMed] [Google Scholar]
- [11].Yin YW, Li JC, Gao D, et al. Influence of ATP-binding cassette transporter 1 R219K and M883I polymorphisms on development of atherosclerosis: a meta-analysis of 58 studies. PLoS One 2014;9:e86480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Song C, Pedersen NL, Reynolds CA, et al. CARDIoGRAMplusC4D Consortium Genetic variants from lipid-related pathways and risk for incident myocardial infarction. PLoS One 2013;8:e60454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nebel A, Croucher PJ, El Mokhtari NE, et al. Common coding polymorphisms in the ABCA1 gene and risk of early-onset coronary heart disease in northern Germany. Atherosclerosis 2007;193:458–60. [DOI] [PubMed] [Google Scholar]
- [14].Harada T, Imai Y, Nojiri T, et al. A common Ile 823 Met variant of ATP-binding cassette transporter A1 gene (ABCA1) alters high density lipoprotein cholesterol level in Japanese population. Atherosclerosis 2003;169:105–12. [DOI] [PubMed] [Google Scholar]
- [15].Xue XH, Huang SE, Hong JC, et al. Association of the R219K polymorphism in ABCAl gene with carotid atherosclerosis and atherosccrotic cerebral infarction. Zhong Xi Yi Jie He Xin Guan Bing Za Zhi 2012;10:574–6. [Google Scholar]
- [16].Genvigir FD, Soares SA, Hirata MH, et al. Effects of ABCA1 SNPs, including the C-105T novel variant, on serum lipids of Brazilian individuals. Clin Chim Acta 2008;389:79–86. [DOI] [PubMed] [Google Scholar]
- [17].Li Y, Zhang SZ, Ma YX, et al. Relationship between the R219K polymorphism of ATP-binding cassette transporter 1 gene and coronary heart disease. Yi Chuan 2005;27:549–52. [PubMed] [Google Scholar]
- [18].Wang XD, Fu Y, Jiang HJ. Polymorphism of R219K of ABCAl gene in patients with coronary artery disease. Lin Chuang Xin Xue Guan Bing Za Zhi 2004;20:215–8. [Google Scholar]
- [19].Yu B, Deng B. Correlation of polymorphism of R219K in ABCA1 with lipid metabolism and the risk of AMI. Tong Ji Da Xue Xue Bao (Yi Xue Ban) 2008;29:64–7. [Google Scholar]
- [20].Guo Q, Gao QG, Liu J. The relationship of R219K polymorphism in ABCA1 gene with characteristics of plasma lipids in patients with type 2 diabetes mellitus and coronary heart disease. Zhongguo Tang Niao Bing Za Zhi 2010;18:827–30. [Google Scholar]
- [21].Li J, Wang LF, Huang YP, et al. Effect of R219K polymorphism of ABCA1 gene on lipid-lowering response to statin in patients with acute myocardial infarction. Xin Zang Za Zhi 2012;24:185–8. [Google Scholar]
- [22].Xia ZH, Huang J, Dai L. Study on relationship between coronary heart disease and the polymorphism of ABCA1 gene R219K. Zhongguo Quan Ke Yi Xue 2011;14:252–4. [Google Scholar]
- [23].Li J, Wang LF, Li ZQ, et al. Effect of R219K polymorphism of the ABCA1 gene on the lipid-lowering effect of pravastatin in Chinese patients with coronary heart disease. Clin Exp Pharmacol Physiol 2009;36:567–70. [DOI] [PubMed] [Google Scholar]
- [24].Wu Y, Bai H, Liu R, et al. Analysis of ATP binding cassette A1 gene R219K polymorphism in patients with endogenous hypertriglyceridemia in Chinese population. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2007;24:177–81. [PubMed] [Google Scholar]
- [25].Benton JL, Ding J, Tsai MY, et al. Associations between two common polymorphisms in the ABCA1 gene and subclinical atherosclerosis: Multi-Ethnic Study of Atherosclerosis (MESA). Atherosclerosis 2007;193:352–60. [DOI] [PubMed] [Google Scholar]
- [26].Çoban N, Onat A, Kömürcü Bayrak E, et al. Gender specific association of ABCA1 gene R219K variant in coronary disease risk through interactions with serum triglyceride elevation in Turkish adults. Anadolu Kardiyol Derg 2014;14:18–25. [DOI] [PubMed] [Google Scholar]
- [27].Abd El-Aziz TA, Mohamed RH, Hagrass HA. Increased risk of premature coronary artery disease in Egyptians with ABCA1 (R219K), CETP (TaqIB), and LCAT (4886C/T) genes polymorphism. J Clin Lipidol 2014;8:381–9. [DOI] [PubMed] [Google Scholar]
- [28].Yuan T, Wang Y, Liu X. Relationship between single nucleotide polymorphism of ABCA1 gene and susceptibility of coronary heart disease in mongolian/han population. Int J Clin Exp 2017;10:3478–85. [Google Scholar]
- [29].Fawzy MS, Alhadramy O, Hussein MH, et al. Functional and structural impact of ATP-binding cassette transporter A1 R219K and I883 M gene polymorphisms in obese children and adolescents. Mol Diagn Ther 2015;19:221–34. [DOI] [PubMed] [Google Scholar]
- [30].Katzov H, Bennet AM, Höglund K, et al. Quantitative trait loci in ABCA1 modify cerebrospinal fluid amyloid-beta 1-42 and plasma apolipoprotein levels. J Hum Genet 2006;51:171–9. [DOI] [PubMed] [Google Scholar]
- [31].Wang Y, Zhang XY, Xu XJ, et al. Association between R219K polymorphism of ATP-binding cassette transporter 1 gene in Xinjiang Uygur population and coronary heart disease. Xin Xue Guan Kang Fu Yi Xue Za Zhi 2009;18:35–9. [Google Scholar]
- [32].Ko YG, Cho EY, Park HY, et al. Association of R219K polymorphism in the ABCAl gene with plasma lipid levels and coronary artery disease in koreans. Korean Circulation J 2003;33:44–51. [Google Scholar]
- [33].Xiao ZJ, Zhao SP, Nie S, et al. The study on the ATP-binding cassette transporter 1 gene polymorphism in patients with cerebral infarction. Zhonghua Shen Jing Ke Za Zhi 2004;37:516–20. [Google Scholar]
- [34].Kolovou G, Kolovou V, Mihas C, et al. Cholesteryl ester transfer protein and ATP-binding cassette transporter A1 genotype alter the atorvastatin and simvastatin efficacy: time for genotype-guided therapy? Angiology 2013;64:266–72. [DOI] [PubMed] [Google Scholar]
- [35].Wang N, Xue XH, Lin Y, et al. The R219K polymorphism in the ATP-binding cassette transporter 1 gene has a protective on atherothrombotic cerebral infarction in Chinese Han ethnic population. Neurobiol Aging 2010;31:647–53. [DOI] [PubMed] [Google Scholar]
- [36].Zhao L, Jin H, Yang B, et al. Correlation between ABCA1 gene polymorphism and aopA-I and HDL-C in abdominal aortic aneurysm. Med Sci Monit 2016;22:172–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Xiao Z, Wang J, Chen W, et al. Association studies of several cholesterol-related genes (ABCA1, CETP and LIPC) with serum lipids and risk of Alzheimer's disease. Lipids Health Dis 2012;11:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yamakawa-Kobayashi K, Yanagi H, Yu Y, et al. Associations between serum high-density lipoprotein cholesterol or apolipoprotein AI levels and common genetic variants of the ABCA1 gene in Japanese school-aged children. Metabolism 2004;53:182–6. [DOI] [PubMed] [Google Scholar]
- [39].Xiao ZJ, Zhao SP, Nie S, et al. Effect of the interaction between paranoxonase 1 and ATP-binding cassette transporter 1 gene polymorphism on serum lipid level. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2005;22:272–6. [PubMed] [Google Scholar]
- [40].Zhao SP, Xiao ZJ, Li QZ, et al. Relationship between ATP-binding cassette transporter 1 R219K genetic variation and blood lipids. Zhonghua Yi Xue Za Zhi 2004;84:1421–5. [PubMed] [Google Scholar]
- [41].Sun P, Bo XP, Guo DP, et al. Study on the association of ABCA1 gene common variants with the risk of coronary atherosclerotic heart disease. Zhonghua Xin Xue Guan Bing Za Zhi 2005;33:627–30. [PubMed] [Google Scholar]
- [42].Liu J, Gao QG, Gao JY, et al. Study of correlations of ABCA1 R219K gene polymorphism with coronary heart disease in subjects with type 2 diabetes. Zhongguo Tang Niao Bing Za Zhi 2008;16:104–6. [Google Scholar]
- [43].Chen LC, Peng J, Lai WY, et al. Association of ATP-binding cassette transporter A1 R219K polymorphism with atrial fibrillation. Nan Fang Yi Ke Da Xue Xue Bao 2009;29:494–6. [PubMed] [Google Scholar]
- [44].Porchay I, Péan F, Bellili N, et al. ABCA1 single nucleotide polymorphisms on high-density lipoprotein-cholesterol and overweight: the D.E.S I R study. Obesity 2006;14:1874–9. [DOI] [PubMed] [Google Scholar]
- [45].Woll PS, Hanson NQ, Arends VL, et al. Effect of two common polymorphisms in the ATP binding cassette transporter A1 gene on HDL-cholesterol concentration. Clin Chem 2005;51:907–9. [DOI] [PubMed] [Google Scholar]
- [46].Wang XP, Qi XY, Li RY, et al. Association between ATP binding cassette transporter A1 R219K polymorphism and coronary heart disease. Lin Chuang Xin Xue Guan Bing Za Zhi 2006;22:516–8. [Google Scholar]
- [47].Shi WY, Zhao ZZ, Xiao DM, et al. Study on single -nucleotide polymorphisms of ABCA1 R219K in Han population. Shi Yong Yu Fang Yi Xue 2009;16:1057–60. [Google Scholar]
- [48].Zhao SP, Xiao ZJ, Nie S, et al. The study on ABCA1 R219K genetic variation in patients with coronary heart disease. Zhonghua Xin Xue Guan Bing Za Zhi 2004;32:712–6. [Google Scholar]
- [49].Huang Y, Wu Y, Liu R, et al. Differential effect of ATP binding cassette transporter A1 R219K and cholesteryl ester transfer protein TaqIB genotypes on HDL-C levels in overweight/obese and non-obese Chinese subjects. Acta Cardiol 2011;66:231–7. [DOI] [PubMed] [Google Scholar]
- [50].Jensen MK, Pai JK, Mukamal KJ, et al. Common genetic variation in the ATP-binding cassette transporter A1, plasma lipids, and risk of coronary heart disease. Atherosclerosis 2007;195:172–80. [DOI] [PubMed] [Google Scholar]
- [51].Lu Y, Liu Y, Li Y, et al. Association of ATP-binding cassette transporter A1 gene polymorphisms with plasma lipid variability and coronary heart disease risk. Int J Clin Exp Pathol 2015;8:13441–9. [PMC free article] [PubMed] [Google Scholar]
- [52].Kolovou V, Kolovou G, Marvaki A, et al. ATP-binding cassette transporter A1 gene polymorphisms and serum lipid levels in young Greek nurses. Lipids Health Dis 2011;10:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kolovou V, Marvaki A, Boutsikou M, et al. Effect of ATP binding Cassette Transporter A1 (ABCA1) gene polymorphisms on plasma lipid variables and common demographic parameters in Greek Nurses. Open Cardiovasc Med J 2016;10:233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Marvaki A, Kolovou V, Katsiki N, et al. Impact of 3 Common ABCA1 Gene Polymorphisms on Optimal vs Non-Optimal Lipid Profile in Greek Young Nurses. Open Cardiovasc Med J 2014;8:83–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sandhofer A, Iglseder B, Kaser S, et al. The influence of two variants in the adenosine triphosphate-binding cassette transporter 1 gene on plasma lipids and carotid atherosclerosis. Metabolism 2008;57:1398–404. [DOI] [PubMed] [Google Scholar]
- [56].Kyriakou T, Pontefract DE, Viturro E, et al. Functional polymorphism in ABCA1 influences age of symptom onset in coronary artery disease patients. Hum Mol Genet 2007;16:1412–22. [DOI] [PubMed] [Google Scholar]
- [57].Yao MH, He J, Ma RL, et al. Association between polymorphisms and haplotype in the ABCA1 Gene and overweight/obesity patients in the Uyghur population of China. Int J Environ Res Public Health 2016;13:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Evans D, Beil FU. The association of the R219K polymorphism in the ATP-binding cassette transporter 1 (ABCA1) gene with coronary heart disease and hyperlipidaemia. J Mol Med (Berl) 2003;81:264–70. [DOI] [PubMed] [Google Scholar]
- [59].Akao H, Polisecki E, Schaefer EJ, et al. PROspective Study of Pravastatin in the Elderly at Risk Investigator. ABCA1 gene variation and heart disease risk reduction in the elderly during pravastatin treatment. Atherosclerosis 2014;235:176–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Delgado-Lista J, Perez-Martinez P, Perez-Jimenez F, et al. ABCA1 gene variants regulate postprandial lipid metabolism in healthy men. Arterioscler Thromb Vasc Biol 2010;30:1051–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Mantaring M, Rhyne J, Ho Hong S, et al. Genotypic variation in ATP-binding cassette transporter-1 (ABCA1) as contributors to the high and low high-density lipoprotein-cholesterol (HDL-C) phenotype. Transl Res 2007;149:205–10. [DOI] [PubMed] [Google Scholar]
- [62].Zhang XX, Xu LX, Zhang HQ, et al. The association between ABCA1 gene R219K polymorphism and coronary heart disease. Xin Nao Xue Guan Bing Fang Zhi 2008;8:304–7. [Google Scholar]
- [63].Katerina H, Michaela S, Michal V, et al. Interaction of common sequence variants and selected risk factors in determination of HDL cholesterol levels. Clin Biochem 2010;43:754–8. [DOI] [PubMed] [Google Scholar]
- [64].Balcerzyk A, Zak I, Krauze J. Synergistic effect between polymorphisms of PPARA and ABCA1 genes on the premature coronary artery disease. Acta Cardiol 2007;62:233–8. [DOI] [PubMed] [Google Scholar]
- [65].Saleheen D, Khanum S, Haider SR, et al. A novel haplotype in ABCA1 gene effects plasma HDL-C concentration. Int J Cardiol 2007;115:7–13. [DOI] [PubMed] [Google Scholar]
- [66].Srinivasan SR, Li S, Chen W, et al. R219K polymorphism of the ABCA1 gene and its modulation of the variations in serum high-density lipoprotein cholesterol and triglycerides related to age and adiposity in white versus black young adults. The Bogalusa heart study. Metabolism 2003;52:930–4. [DOI] [PubMed] [Google Scholar]
- [67].Hodoğlugil U, Williamson DW, Huang Y, et al. Common polymorphisms of ATP binding cassette transporter A1, including a functional promoter polymorphism, associated with plasma high density lipoprotein cholesterol levels in Turks. Atherosclerosis 2005;183:199–212. [DOI] [PubMed] [Google Scholar]
- [68].Wang J, Bao YQ, Hu C, et al. Effects of ABCA1 variants on rosiglitazone monotherapy in newly diagnosed type 2 diabetes patients. Acta Pharmacol Sin 2008;29:252–8. [DOI] [PubMed] [Google Scholar]
- [69].Haghvirdizadeh P, Ramachandran V, Etemad A, et al. Association of ATP-binding cassette transporter A1 gene polymorphisms in type 2 diabetes mellitus among Malaysians. J Diabetes Res 2015;2015:289846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Mokuno J, Hishida A, Morita E, et al. ATP-binding cassette transporter A1 (ABCA1) R219K (G1051A, rs2230806) polymorphism and serum high-density lipoprotein cholesterol levels in a large Japanese population: cross-sectional data from the Daiko Study. Endocrine J 2015;62:543–9. [DOI] [PubMed] [Google Scholar]
- [71].Manresa JM, Zamora A, Tomás M, et al. Relationship of classical and non-classical risk factors with genetic variants relevant to coronary heart disease. Eur J Cardiovasc Prev Rehabil 2006;13:738–44. [DOI] [PubMed] [Google Scholar]
- [72].Ghaznavi H, Aali E, Soltanpour MS. Association study of the ATP binding cassette transporter A1 (ABCA1) Rs2230806 genetic variation with lipid profile and coronary artery disease risk in an Iranian population. Open Access Maced J Med Sci 2018;6:274–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Xu Y, Li Z. Relationship between ABCA1 gene polymorphism and lacunar infarction combined with arteriosclerosis in patients. Exp Ther Med 2018;16:1323–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Cai R, Han J, Sun J, et al. Effects of ABCA1 R219K Polymorphism and Serum Lipid Profiles on Mild Cognitive Impairment in Type 2 Diabetes Mellitus. Front Aging Neurosci 2017;9:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ya L, Lu Z. Differences in ABCA1 R219K polymorphisms and serum indexes in Alzheimer and Parkinson Diseases in Northern China. Med Sci Monit 2017;23:4591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Tao F, Weinstock J, Venners SA, et al. Associations of the ABCA1 and LPL gene polymorphisms with lipid levels in a hyperlipidemic population. Clin Appl Thromb Hemost 2018;24:771–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Takagi S, Iwai N, Miyazaki S, et al. Relationship between ABCA1 genetic variation and HDL cholesterol level in subjects withischemic heart diseases in Japanese. Thromb Haemost 2002;88:369–70. [PubMed] [Google Scholar]
- [78].Fleiss JL. The statistical basis of meta-analysis. Stat Methods Med Res 1993;2:121–45. [DOI] [PubMed] [Google Scholar]
- [79].Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994;50:1088–101. [PubMed] [Google Scholar]
- [80].Nosratola D Vaziri, Hamid Moradi, Madeleine V Pahl, et al. In vitro stimulation of HDL anti-inflammatory activity and inhibition of LDL pro-inflammatory activity in the plasma of patients with end-stage renal disease by an apoA-1 mimetic peptide. Kidney Int 2009;76:437–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ma XY, Liu JP, Song ZY. Associations of the ATP-binding cassette transporter A1 R219K polymorphism with HDL-C level and coronary artery disease risk: a meta-analysis. Atherosclerosis 2011;215:428–34. [DOI] [PubMed] [Google Scholar]
- [82].Lee JD, Hsiao KM, Chang PJ, et al. A common polymorphism decreases LRP1 mRNA stability and is associated with increased plasma factor VIII levels. Biochim Biophys Acta 2017;1863:1690–8. [DOI] [PubMed] [Google Scholar]
- [83].Akdeli N, Riemann K, Westphal J, et al. A 3′UTR polymorphism modulates mRNA stability of the oncogene and drug target Polo-like Kinase 1. Mol Cancer 2014;13:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lee CM, Han JI, Kang MH, et al. Polymorphism in the serotonin transporter protein gene in Maltese dogs with degenerative mitral valve disease. J Vet Sci 2018;19:129–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Sachidanandam R, Weissman D, Schmidt SC, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 2001;409:928–33. [DOI] [PubMed] [Google Scholar]
- [86].Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature 2008;452:872–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


