Abstract
We report the detection of endometrial and ovarian cancers based on genetic analyses of DNA recovered from the fluids obtained during a routine Papanicolaou (Pap) test. The new test, called PapSEEK, incorporates assays for mutations in 18 genes as well as an assay for aneuploidy. In Pap brush samples from 382 endometrial cancer patients, 81% [95% confidence interval (CI), 77 to 85%] were positive, including 78% of patients with early-stage disease. The sensitivity in 245 ovarian cancer patients was 33% (95% CI, 27 to 39%), including 34% of patients with early-stage disease. In contrast, only 1.4% of 714 women without cancer had positive Pap brush samples (specificity, ~99%). Next, we showed that intrauterine sampling with a Tao brush increased the detection of malignancy over endocervical sampling with a Pap brush: 93% of 123 (95% CI, 87 to 97%) patients with endometrial cancer and 45% of 51 (95% CI, 31 to 60%) patients with ovarian cancer were positive, whereas none of the samples from 125 women without cancer were positive (specificity, 100%). Finally, in 83 ovarian cancer patients in whom plasma was available, circulating tumor DNA was found in 43% of patients (95% CI, 33 to 55%). When plasma and Pap brush samples were both tested, the sensitivity for ovarian cancer increased to 63% (95% CI, 51 to 73%). These results demonstrate the potential of mutation-based diagnostics to detect gynecologic cancers at a stage when they are more likely to be curable.
INTRODUCTION
The Papanicolaou (Pap) test has dramatically decreased the mortality of cervical cancer in the screened population. Unfortunately, the Pap test is generally unable to detect endometrial or ovarian cancers (1–4). In light of the success of the Pap test in detecting early-stage, curable cervical cancers, ovarian and endometrial cancers are currently the most lethal and most common gynecologic malignancies, respectively, in countries where Pap tests are routinely performed (5). Together, endometrial and ovarian cancers account for about 25,000 deaths each year and are the third leading cause of cancer-related mortality in women in the United States (5). Most of these deaths are caused by high-grade tumor subtypes, which tend to metastasize before the onset of symptoms (6, 7).
Endometrial cancer is the most common gynecologic malignancy, with 61,380 estimated new cases in 2017 in the United States (5). The incidence of endometrial cancer has been rising with higher prevalence of obesity and increased life expectancy (8). At the same time, relative survival has not improved over the past decades (5, 9). Much effort has been directed toward developing a screening test for this cancer type. The most common diagnostic test is transvaginal ultrasound (TVUS), which measures the thickness of the endometrium. The potential of TVUS as a screening test is undermined by its inability to reliably distinguish between benign and malignant lesions, subjecting women without cancer to unnecessary invasive procedures and their associated complications. Its high false-positive rate is demonstrated by the fact that as few as 1 in 50 women who tested positive by TVUS was proven to have endometrial cancer after undergoing additional diagnostic procedures (10).
Ovarian cancer is the second most common gynecologic malignancy in the United States and Europe. It is often diagnosed at a late stage, when the 5-year survival rate is less than 30% (5). The high mortality has made the development of an effective screening test a high priority. Large randomized trials have assessed the use of CA-125 and TVUS as potential screening tests for ovarian cancer (11–14). However, screening with current diagnostic approaches is not recommended for the general population because it leads to “important harms, including major surgical interventions in women who do not have cancer” (15). Thus, the development of new diagnostic approaches is important.
Among ovarian cancers, high-grade serous carcinomas (HGSCs) account for 90% of all ovarian cancer deaths. Increasing evidence suggests that most HGSCs arise in the fallopian tube and subsequently implant on the ovarian surface (16–21). A recent prospective study of symptomatic women reported that most early-diagnosed HGSCs have extraovarian origins (22). This might explain the low sensitivity of TVUS for early disease, when no ovarian abnormalities are detectable. Multimodal screening with serum CA-125 improves sensitivity; however, with the way it is currently used, CA-125 lacks specificity and is elevated in a variety of common benign conditions (23).
Unlike markers associated with neoplasia, cancer driver gene mutations are causative agents. It has been shown that tumor DNA could be detected in the vaginal tract of women with ovarian cancer (24). Furthermore, a recent proof-of-principle study showed that endometrial and ovarian cancers shed cells that collect at the cervix, allowing detectable amount of tumor DNA to be found in the fluids obtained during routine Pap tests (25). These cells are sampled with a brush (a “Pap brush”) that is inserted into the endocervical canal. The brush is then dipped into preservative fluid. For the detection of cervical cancers, cells from the fluid are applied to a slide for cytologic examination (the classic Pap smear). In addition, DNA is often purified from the fluid to search for human papillomavirus sequences. Here, we used the DNA from this fluid in a polymerase chain reaction (PCR)–based, multiplex test to simultaneously assess genetic alterations that commonly occur in endometrial or ovarian cancers. In addition, we explored two ways to increase sensitivity. First, we tested intrauterine sampling (with a “Tao brush”), a method that allows sample collection closer to the anatomical sites of the tumors. Second, in a recent study, we showed that testing for mutations in both saliva and plasma from the same individual increased the sensitivity of detecting head and neck tumors (26). On the basis of this precedent, we assessed whether testing for mutations in both the plasma and Pap test fluid would increase sensitivity for ovarian cancers.
RESULTS
Evaluation of somatic mutations in Pap brush samples from patients with endometrial or ovarian cancer
Overall, 1915 samples from 1658 individuals were included in this study, including 656 patients with endometrial or ovarian cancers and 1002 healthy controls. The age, race, histopathologic diagnosis, stage, and other clinical information for the cancer patients are provided in table S1. The samples tested from these patients are listed in table S2.
The amount of DNA shed from neoplastic cells was expected to be a minor fraction of the total DNA in the Pap brush samples, with most DNA emanating from normal cells. We therefore used a sensitive, PCR-based error-reduction technology, called Safe-Sequencing System (Safe-SeqS), to identify mutations in these samples (27). In brief, primers were designed to amplify 139 regions, covering 9392 distinct nucleotide positions within the 18 genes of interest (table S3). Three multiplex PCRs, each containing nonoverlapping amplicons, were then performed on each sample.
We applied this assay to Pap brush samples of 382 women with endometrial cancer, 245 women with ovarian cancer, and 714 women without cancer. We found that 81% [95% confidence interval (CI), 76 to 84%] of the patients with endometrial cancers had detectable mutations, including 78% of patients with early-stage disease (stages I and II) and 89% of the patients with late-stage disease (stages III and IV; table S2). The most commonly mutated genes were PTEN (64%), TP53 (41%), PIK3CA (31%), PIK3R1 (29%), CTNNB1 (21%), KRAS (18%), FGFR2 (11%), POLE (9%), APC (9%), FBXW7 (8%), RNF43 (7%), and PPP2R1A (5%), consistent with previous genome-wide studies of endometrial cancers (25, 28, 29). The median mutant allele fraction (MAF) was 4.0% [interquartile range (IQR), 1.3 to 12%] (table S4).
Twenty-nine percent of 245 ovarian cancer patients harbored detectable mutations in their Pap brush samples (95% CI, 24 to 36%). These included 28% of patients with early-stage disease and 30% of patients with late-stage disease (table S2). The most commonly mutated gene was TP53 (74%), consistent with previous genome-wide studies of this tumor type (25, 30). The median MAF was 0.54% (IQR, 0.22 to 2.6%) (table S4). We also applied this assay to 714 women without cancer and found that 1.3% had a detectable mutation, yielding a specificity of ~99% (Fig. 1).
Fig. 1. Detection of aneuploidy and somatic mutations (PapSEEK) in Pap or Tao brush samples from healthy controls and patients with endometrial or ovarian cancers.
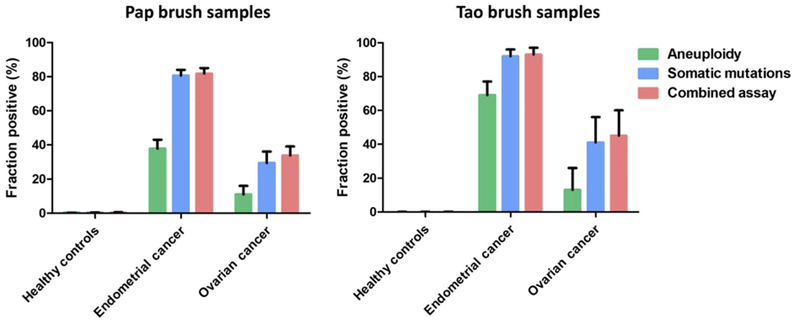
Data shown as means ± 95% confidence intervals.
Tumor tissue was available from 83 and 84% of endometrial and ovarian cancer patients who donated Pap brush samples, respectively. Using the same multiplex assay applied to the Pap brush samples, a driver gene mutation could be identified in 98 and 82% of the endometrial and ovarian cancer tissues, respectively (table S5). Of the endometrial and ovarian cancer patients with a driver mutation identified in their primary tumor, 85 and 29%, respectively, had mutations in their Pap brush samples. Conversely, of the positive Pap brush samples from patients with endometrial or ovarian cancers, 93% contained at least one driver gene mutation that was identical to that observed in their primary tumor. The fraction of Pap brush samples with mutations that were also found in the primary tumors was higher in endometrial cancer patients (97%) than in ovarian cancer patients (73%).
Evaluation of aneuploidy in Pap brush samples
In addition to somatic mutations, aneuploidy is found in the great majority of endometrial and ovarian cancers (28, 30, 31). To assess aneuploidy, we used a PCR-based method to amplify ~38,000 loci of long interspersed nucleotide elements (LINEs) with a single primer pair (32). LINEs have spread throughout the genome via retrotransposition and are found on all 39 nonacrocentric autosomal arms. After sequencing, the data are processed to identify gains or losses on single chromosome arms (see Materials and Methods).
Aneuploidy was detected in the Pap brush samples of 38% (95% CI, 33 to 43%) of the 382 patients with endometrial cancer, including 34 and 51% of those with early- and late-stage disease, respectively (table S2). Aneuploidy was also detected in the Pap brush samples of 11% (95% CI, 7 to 16%) of the 245 ovarian cancer patients, including 15 and 9.3% of those with early- and late-stage disease, respectively (table S2). In endometrial and ovarian cancers, the most commonly altered arms were 4p, 7q, 8q, and 9q, consistent with previous reports (28, 30). In contrast, when we applied the aneuploidy assay to the Pap brush samples of 714 women without cancer, only one woman was positive (specificity, ~100%; Fig. 1).
Even if a sample does not contain a genetic alteration in 1 of the 18 genes assessed, it might still be aneuploid and detectable by our test. This conjecture was supported by our identification of six patients (three with endometrial and three with ovarian cancers) who had no mutations in their Pap brush samples or primary tumors (when available) but whose Pap brush samples displayed aneuploidy. The combined test, incorporating the above-described assays for mutations plus aneuploidy, was dubbed “PapSEEK.” PapSEEK scores a sample as positive if it harbors either a mutation or an abnormal chromosome arm number. Eighty-one percent (95% CI, 77 to 85%) of the Pap brush samples from women with endometrial cancers were PapSEEK-positive, including 78% of patients with early-stage disease and 92% of patients with late-stage disease (Figs. 2 and 3). Thirty-three percent (95% CI, 27 to 39%) of the Pap brush samples from women with ovarian cancers were PapSEEK-positive, including 34% of patients with early-stage disease and 33% of patients with late-stage disease (Figs. 2 and 3). Only 1.4% of the Pap brush samples from 714 women without cancer were PapSEEK-positive, yielding a specificity of ~99% (table S6 and Fig. 1).
Fig. 2. Venn diagrams showing increased detection with combined testing for somatic mutations and aneuploidy, as well as combined testing of Pap brush and plasma samples.
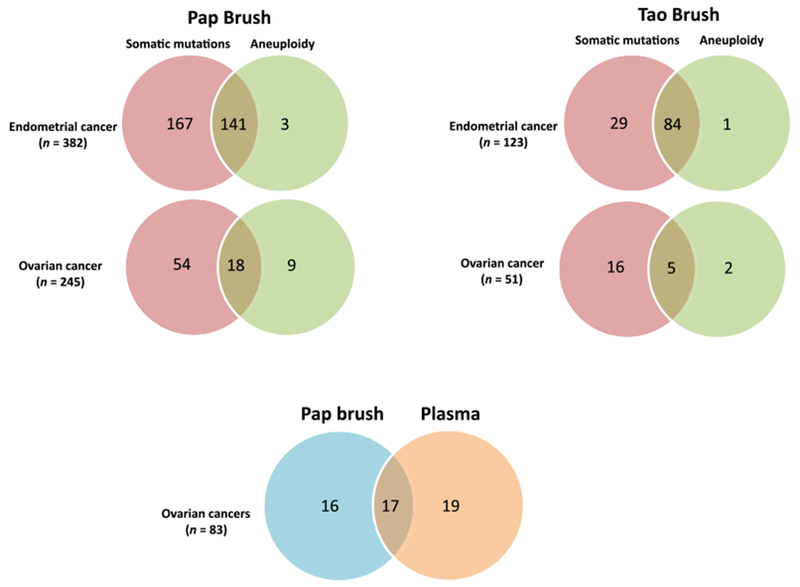
For both endometrial and ovarian cancers, combined testing for somatic mutations and aneuploidy increased sensitivity in the Pap and Tao brush samples. For ovarian cancer, combined testing of Pap brush and plasma samples also increased sensitivity compared to testing either sample type alone.
Fig. 3. Detection of endometrial or ovarian cancers In Pap or Tao brush samples with PapSEEK by stage.
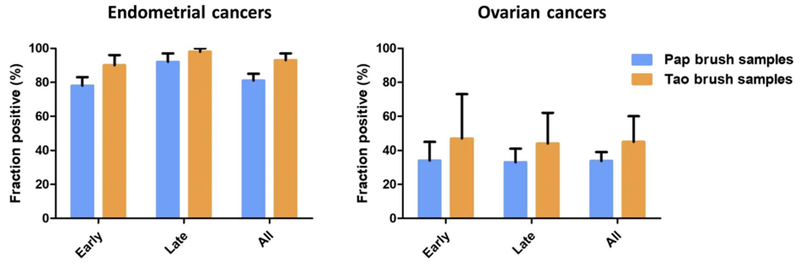
Data shown as means ± 95% confidence intervals.
Evaluation of Tao brush samples from patients with ovarian or endometrial cancers
We wondered whether more direct, minimally invasive sampling of the intrauterine cavity (rather than the endocervical canal) could increase the sensitivity of this approach for detecting gynecologic cancers. To explore this possibility, we collected intrauterine samples using a Tao brush, which is a flexible, narrow brush covered by a retractable outer sheath that allows direct sampling of the entire endometrial cavity without injury to the myometrium or contamination from the cervical canal (33). It has been approved by the Food and Drug Administration for endometrial sampling and can be used in an outpatient setting without the need for anesthesia. Importantly for a potential screening test, it is well tolerated by patients (33, 34).
We applied PapSEEK to Tao brush samples collected from 123 patients with endometrial cancers, 51 patients with ovarian cancers, and 125 women without cancer. Ninety-three percent (95% CI, 87 to 97%) of the Tao brush samples from endometrial cancer patients contained genetic alterations detected by PapSEEK, including 90 and 98% of patients with early- and late-stage disease, respectively (Fig. 3). The most commonly mutated genes in the Tao brush samples were PTEN (63%), TP53 (42%), PIK3CA (36%), PIK3R1 (20%), KRAS (17%), CTNNB1 (15%), FGFR2 (15%), RNF43 (11%), PPP2R1A (7%), POLE (7%), and FBXW7 (6%), similar to those observed in the Pap brush samples. The median MAF was 24.7% (IQR, 10.4 to 35.4%), considerably higher than that observed in the Pap brush samples, in which the median MAF was 4.0% (IQR, 1.3 to 12%; table S4).
Genetic alterations detectable by PapSEEK were found in 45% (95% CI, 31 to 60%) of the Tao brush samples from 51 women with ovarian cancers, including 47 and 44% of patients with early- and late-stage cancers, respectively (Fig. 3). The most commonly mutated gene was TP53 (86%), consistent with that observed in the Pap brush samples. The median MAF was 0.88% (IQR, 0.35 to 3.8%), which was higher than that in the Pap brush samples (median, 0.54%; IQR, 0.22 to 2.6%; table S4).
PapSEEK was applied to the Tao brush samples from 125 women without cancer. None (0%) of these women tested positive for mutations, yielding a specificity of 100% (table S6 and Fig. 1).
Tao brush and Pap brush samples from the same women were available in 145 patients (103 with endometrial and 42 with ovarian cancers). In endometrial cancers, PapSEEK was positive in 91% of the Tao brush samples and in 82% of the Pap brush samples (P = 0.02, mid-P McNemar test). Similarly, the fraction of ovarian cancer patients with a positive PapSEEK test was higher for Tao brush (45%) than for Pap brush [17%; P = 0.002, mid-P McNemar test (table S1)].
Tumor tissue was available from 90 and 88% of patients with endometrial and ovarian cancers who donated Tao brush samples, respectively. PapSEEK identified driver gene mutations in 97 and 80% of the endometrial and ovarian cancer tissues, respectively (table S5). Of the endometrial and ovarian cancer patients with a driver mutation identified in their primary tumor, 93 and 42%, respectively, had mutations detectable in their Tao brush samples. Conversely, of the positive Tao brush samples from patients with endometrial or ovarian cancers, 91% contained at least one driver gene mutation that was identical to that observed in their primary tumor. The fraction of Tao brush samples with mutations that were also found in the primary tumors was higher in endometrial cancer patients (97%) than in ovarian cancer patients (53%).
Evaluation of ctDNA in patients with ovarian cancers
We hypothesized that ovarian cancers that were inaccessible by Pap or Tao brush sampling due to anatomical or other factors might be detectable by the presence of circulating tumor DNA (ctDNA) in plasma (35). We were able to test this hypothesis in 83 ovarian cancer patients who had donated both Pap brush and plasma samples. Because of the smaller size of degraded ctDNA, primers were designed to amplify short 67– to 81–base pair (bp) DNA fragments, covering 1933 distinct nucleotide positions within 16 genes of interest, as described previously (36). When this assay was applied to plasma samples from 192 healthy individuals, none (0%) tested positive, yielding a specificity of 100%.
We found that 43% (95% CI, 33 to 55%) of the plasma from the 83 patients with ovarian cancers had detectable ctDNA. The mutations detected are listed in table S7. As expected, the sensitivity for ctDNA in plasma was higher in patients with late-stage tumors than in patients with early-stage tumors (56 versus 35%; Fig. 4). For early-stage disease, the median MAF in the plasma was 0.85% (IQR, 0.40 to 3.4%), which was less than the median MAF (5.7%; IQR, 0.83 to 12%) in the Pap brush samples. At least one of the mutations identified in the plasma could be identified in 88% of the corresponding primary tumors.
Fig. 4. Detection of ovarian cancer in Pap and plasma samples.
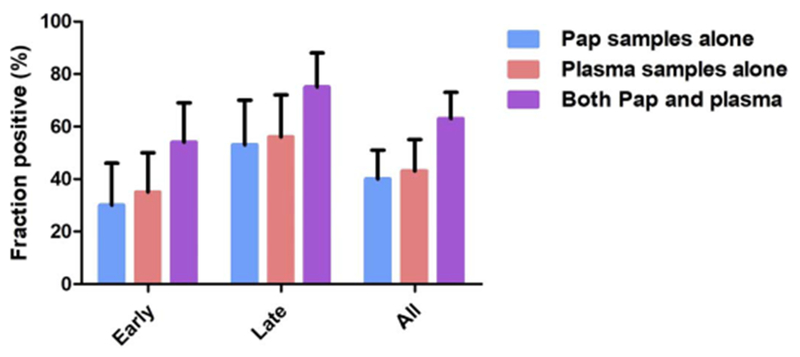
Data shown as means ± 95% confidence intervals.
In the Pap brush samples from this same cohort of 83 patients, 40% were positive by the PapSEEK test. The individuals scoring positive in their Pap brush and plasma samples only partially overlapped (Fig. 2). As a result, 63% (95% CI, 51 to 73%) of patients were positive with at least one of the two tests. Those who tested positive included 54% of patients with early-stage disease and 75% with late-stage disease, respectively (table S1 and Fig. 4).
DISCUSSION
Here, we designed and applied a multiplex PCR-based test (PapSEEK) to detect genetic alterations in Pap brush or Tao brush samples (Fig. 5). These samples are minimally invasively and conveniently obtained during routine office visits. Most endometrial cancers could be detected with PapSEEK: 93% with Tao Brush and 81% with Pap brush. A substantial fraction of ovarian cancers could also be detected with PapSEEK: 45% with Tao Brush and 33% with Pap brush. The specificity of PapSEEK was high, with only 0 and 1.4% of women without cancer testing positive with Tao and Pap brush samples, respectively. We also showed that assays for ctDNA in plasma could be used in conjunction with PapSEEK on Pap brush samples, increasing the sensitivity of detecting ovarian cancer to 63%. We have not yet tested whether combining ctDNA analysis with PapSEEK analysis of Tao brush would further increase sensitivity.
Fig. 5. PapSEEK test for the detection of tumor DNA in the Pap brush, Tao brush, and plasma samples of patients with endometrial or ovarian cancers.
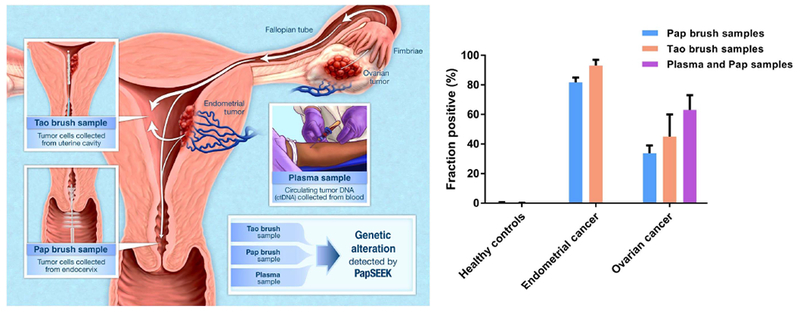
Tumor cells shed from ovarian or endometrial cancers are carried into the uterine cavity, where they can be collected by the Tao brush. The tumor cells that pass down into the endocervical canal can be captured by the Pap brush used in the routine Pap test. These brushes are dipped into a liquid fixative, from which DNA is isolated and sequenced. The sequences are analyzed for somatic mutations and aneuploidy. In addition, tumor DNA shed into the bloodstream can be detected by circulating tumor DNA (ctDNA) analysis. Detection of endometrial and ovarian cancers with PapSEEK in the Pap brush, Tao brush, and plasma samples is shown as means ± 95% confidence intervals.
It was particularly notable that the sensitivity for detecting early-stage ovarian cancers was as high as that for late-stage disease (47 versus 44% for Tao; 34 versus 33% for Pap). There are two possible explanations for this unexpected but enticing finding. First, it has been shown that some ovarian cancers originate in the fallopian tubes, which could facilitate their early detection with PapSEEK when tumor cells are shed into the uterine cavity. Second, in late-stage tumors, the fallopian tubes are often matted and obliterated by the disease and, thus, less likely to serve as a conduit for tumor cells to pass into the uterus or endocervical canal. In this setting, the addition of ctDNA analysis in plasma to Pap or Tao brush sampling may be particularly beneficial.
An important subset of our samples was composed of high-grade, early-stage cancers. Currently available diagnostic modalities have low sensitivities for these lesions (37–39). Although the high-grade subtypes comprise only about 10% of incident endometrial cancers, they account for more than 40% of deaths from the disease (7). Because these high-grade cancers often arise from a background of atrophic endometrium and can metastasize before visible abnormalities on imaging, TVUS has a limited role in screening and early diagnosis. Thus, it was encouraging that PapSEEK detected 85% (n = 34) and 89% (n = 9) of high-grade endometrial cancers confined to the endometrium in the Pap and Tao brush samples, respectively. In the case of ovarian cancers, our cohort included only a small number of early-stage, high-grade cases, consistent with the unfortunate fact that these cancers are often diagnosed only at advanced stages. Nevertheless, our finding that 36% (n = 11) were positive with combined Pap and plasma sample testing and that 80% (n = 5) were positive in Tao brush samples is notable.
Although promising, our study has several limitations that are important to acknowledge. First, it was retrospective rather than prospective. The samples we examined were derived from patients with known cancers, even though a substantial fraction was from patients with early-stage lesions. In a screening setting, the cancers would hopefully be at an earlier stage, and the sensitivities for detection would be expected to be closer to the sensitivity for early-stage cancers observed in our study. Furthermore, it is conceivable that the combined testing of PapSEEK in conjunction with conventional methods, such as CA-125 testing or TVUS, would provide an additional increase in sensitivity. A considerable proportion of patients in our retrospective cohort were initially diagnosed using these conventional methods and, therefore, preselected for having abnormal results based on these tests. This precluded an accurate assessment of combined testing of PapSEEK with conventional detection methods in our study. A prospective, unbiased cohort would be more appropriate for such an assessment. In addition, in a prospective study, the age ranges of the controls and cases would be better matched than in our retrospective study, and would include patients with benign as well as malignant tumors.
Second, some of the ovarian cancer patients who had mutations detectable in their Pap brush or Tao brush samples did not have the identical mutations in their primary tumors. This was not an issue with endometrial cancers, wherein at least one mutation in the brush samples was nearly always (97%) found in the corresponding primary tumors. However, it was an issue for the ovarian cancer patients, particularly with the Tao brush. At least one mutation identifiable in the Pap brush could be identified in 73% of the corresponding primary ovarian tumors, whereas the same was true for only 53% of the Tao brush samples.
One possible explanation for the discordance between the mutations in brush samples and ovarian cancers from the same patients is that the assay detects mutations that do not exist in vivo, representing technical artifacts. We do not believe that this is likely, given that the specificity of our assays was 100 and 99% in Tao brush and Pap brush samples, respectively. Another possible explanation is tumor heterogeneity (40). Only a small portion of the primary tumors that we analyzed was sampled and sequenced; the additional mutations found in the Pap or Tao brush samples could represent mutations from other parts of the tumor. It is also possible that some mutations were from small synchronous endometrial cancers or premalignant endometrial lesions that were unnoted by the pathologist. A nontrivial proportion of women with ovarian cancer have synchronous endometrial cancer (41–43), with risk factors including Lynch syndrome, polycystic ovarian syndrome, perimenopause, obesity, nulliparity, and unopposed estrogen replacement therapy (41, 44).
Although tumor heterogeneity or multiple synchronous tumors are feasible explanations that are often used to explain discordances in liquid biopsy studies, we are skeptical that this is the major cause. We believe that clonal expansions of nonmalignant cells may be more important. Clonal proliferations that are not considered neoplastic have been described in the bone marrow, skin, and other tissues (45–48). Of particular interest are the clonal proliferations of endometrial cells that cause endometriosis, a potentially debilitating condition that affects millions of women. It has recently been shown that these lesions, which can occur throughout the pelvis and are derived from the endometrium, are clonal proliferations that can be driven by the same mutations that we detect in endometrial cancers (49). The possibility that these mutations might reflect benign or noncancerous endometrial lesions is also consistent with the recent report of cancer-associated mutations found in uterine lavages of women without cancer (50). Finally, it is possible that the hormonal and physiologic changes contributing to or resulting from ovarian cancers stimulate or select for such clonal proliferations in the endometrial lining. On the one hand, this explanation is worrisome, because it argues against the exquisite specificity that is the conceptual basis for all liquid biopsies. On the other hand, it could actually enhance the sensitivity of detection of ovarian cancers, without diminishing specificity, if large clonal proliferations are almost exclusively found in women with gynecologic malignancies. Only clonal proliferations that account for >0.03% of the total cells in the endometrial lining are detectable by our assay.
Our study lays the foundation for evaluating PapSEEK in a large prospective study. The most natural cohort for such a study would include patients who are at high risk for gynecologic cancers because of hereditary factors, obesity, or symptoms such as postmenopausal or dysfunctional uterine bleeding. The cost of a PapSEEK test would be more than the cost of a Pap test but comparable to colonoscopy, mammography, and computed tomography imaging. There are many issues to be investigated in such large-scale trials. For example, is Tao brush sampling superior to Pap brush sampling, both with respect to sensitivity and patient compliance? Is it feasible to combine plasma ctDNA analysis with PapSEEK? Although plasma ctDNA analysis can improve sensitivity in combination with PapSEEK, positive ctDNA results can come from a variety of cancer types, thus raising issues about the appropriate follow-up. Would combining serial serum CA-125 measurements (14) or other protein markers (51) with PapSEEK, or PapSEEK with ctDNA analysis, offer advantages over either alone? Will repeat testing increase the sensitivity of PapSEEK, as it does for CA-125 (14), and what is the appropriate time interval for such repeats? Finally, what is the best way to manage patients with a positive PapSEEK test? Should such women undergo hysteroscopy as well as TVUS, or other imaging procedures? Moreover, if negative, how often should they be retested, either with PapSEEK or with imaging modalities? Although the answers to all these questions must await future trials, PapSEEK adds another dimension to screening for gynecologic cancers.
MATERIALS AND METHODS
Study design
This was a retrospective study with samples collected from 1658 individuals, including 656 patients with endometrial or ovarian cancers and 1002 healthy controls. Data analysis was performed in a blinded fashion, and all patient samples were de-identified.
Patient samples
All samples for this study were obtained according to protocols approved by the Institutional Review Boards of the Johns Hopkins Medical Institutions (Baltimore, MD), McGill University (Montreal, Quebec, Canada), Sahlgrenska University Hospital (Gothenburg, Sweden), BioreclamationIVT (Chestertown, MD), Memorial Sloan Kettering Cancer Center (New York City, NY), and Danish Scientific Ethical Committee (Copenhagen, Denmark). Demographic, clinical, and pathologic staging data were collected for each patient with cancer and are listed in table S1. The average age of 714 women without cancer who underwent Pap brush analysis was 34 (range, 17 to 67 years). The average age of 125 women without cancer who underwent Tao brush analysis was 29 (range, 18 to 74 years). All histopathology was rereviewed by board-certified pathologists. DNA was extracted from tumors, Pap brush, and plasma samples as previously described (27, 52). The patients evaluated in this study were completely different than those evaluated in (36). For intrauterine sampling, Tao Brush IUMC Endometrial Sampler (Cook Medical Inc.) was gently inserted to the level of the uterine fundus. The outer sheath was then pulled back, and the brush was rotated 360° clockwise and counterclockwise. Then, the outer sheath was pushed in again, and the device was removed. The sample was placed into thin-prep buffer, from which DNA was purified using DNA purification kits (Qiagen) according to the manufacturer’s instructions. Purified DNA from all samples was quantified as previously described (53).
Healthy controls included patients with normal cytology findings on Pap smears and no history of gynecologic tumors. Ovarian cancer patients with history of tubal ligation were excluded from the study.
Aneuploidy detection and analysis
For each sample, a single primer pair was used to amplify ~38,000 loci of LINEs throughout the genome (32). One of the primers included a unique identifier sequence (UID) as a molecular barcode to reduce error rates associated with PCR and sequencing. Massively parallel sequencing was performed on Illumina instruments. The sequencing data were then processed to identify single chromosomal arm gains or losses, as well as allelic imbalance on 39 chromosome arms, using the Within-Sample AneupLoidy DetectiOn (WALDO) software (54). WALDO incorporates a support vector machine (SVM) to discriminate between aneuploid and euploid samples. The SVM was trained using 3150 synthetic aneuploid samples with low neoplastic content and 677 euploid peripheral white blood cell samples. A sample was scored as positive (aneuploid) if the SVM discriminant score exceeded a given threshold or if gains of chromosome arms 7q and 8q were observed. These chromosome arms are frequently gained in both endometrial and ovarian cancers (28, 30).
Somatic mutation detection and analysis
DNA from Pap brush samples, Tao brush samples, or primary tumors was amplified in three multiplex PCRs with 139 primer pairs that were designed to amplify 110- to 142-bp segments. These segments contain regions of interest from the following 18 genes: AKT1, APC, BRAF, CDKN2A, CTNNB1, EGFR, FBXW7, FGFR2, KRAS, MAPK1, NRAS, PIK3CA, PIK3R1, POLE, PPP2R1A, PTEN, RNF43, and TP53. For each sample, three multiplex reactions, each containing nonoverlapping amplicons, were performed, as previously described (55). Each sample was assessed in two duplicate wells. DNA from plasma was amplified in two multiplex PCRs consisting of 61 primer pairs that were designed to amplify 67- to 81-bp segments. Each sample was assessed in six duplicate wells. These segments contained regions of interest from the following genes: AKT1, APC, BRAF, CDKN2A, CTNNB1, EGFR, FBXW7, FGFR2, GNAS, HRAS, KRAS, NRAS, PIK3CA, PPP2R1A, PTEN, and TP53 (36).
Safe-SeqS, an error-reduction technology for detection of low-frequency mutations (27), was used for all sequencing analyses. One primer in each pair included a UID, consisting of 14 degenerate bases with an equal chance of being an A, C, T, or G. High-quality sequence reads were selected on the basis of quality scores, which were generated by the sequencing instrument to indicate the probability that a base was called in error. Redundant reads arising from optical duplication were eliminated by requiring reads with the same UID and sample index to be at least 5000 pixels apart when located on the same tile. Reads from a common template molecule were then grouped on the basis of the UIDs that were incorporated as molecular barcodes. Artefactual mutations introduced during the sample preparation or sequencing steps were reduced by requiring a mutation to be present in >90% of reads in each UID family (to be scored as a “supermutant”) (27).
Statistical analysis of sequencing data
All Pap brush and Tao brush samples were analyzed using a MAF-based approach. Mutations that met one of the two following criteria were considered: (i) present in the COSMIC (Catalogue of Somatic Mutations in Cancer) database (56) or (ii) predicted to be inactivating in tumor suppressor genes (nonsense mutations, out-of-frame insertions or deletions, and canonical splice site mutations). Synonymous mutations [except those at exon ends (57)] and intronic mutations (except for those at splice sites) were excluded. Finally, mutations that could not be uniquely mapped to hg39 were excluded from the analysis. The MAF of each mutation in the sample of interest was first normalized on the basis of how the distribution of MAFs of the same mutation in the control group compared to the distribution of MAFs of all mutations in the control group. After this mutation-specific normalization, a P value was obtained by comparing the normalized MAF of each mutation in each well with a reference distribution of normalized MAFs built from normal controls where all mutations were included. The Stouffer’s Z score was then calculated from the P values of two independent wells, weighted by their number of UIDs. Stouffer’s method was used because the sample aliquot in each well was assessed independently of the other wells. For this assessment, 10 ng of DNA was aliquoted into each of two wells, which were then amplified independently, sequenced independently (through the use of well barcodes), and analyzed independently. The assumption (null hypothesis) on which the P value was calculated for each well was the following: We assumed that the wells did not contain driver mutations and that any background mutations identified were actually PCR or sequencing artifacts, which followed the reference distribution built from the normal controls. The null hypothesis was therefore that the background mutations in the two wells came from the same reference distribution, but the two wells were independent (independent and identically distributed random variables).
A sample was scored as positive when any of its mutations had a value above the corresponding thresholds for any of the following three criteria: (i) the difference between its MAF and the corresponding maximum MAF observed for that mutation in the controls, (ii) the ratio of its Stouffer’s Z score to the average of the highest six nonzero Stouffer’s Z scores for the same mutation in the controls, or (iii) its Stouffer’s Z score alone when the mutation was not seen in the controls. The thresholds were determined to ensure a desired overall specificity.
Sensitivity and specificity were obtained from a 10-fold crossvalidation. In each of the 10 rounds, Pap brush samples from 90% of the 714 women without cancer served as controls in the training set, with the remaining 10% of the Pap brush samples from women without cancer scored to obtain specificity. The controls in each round were randomly selected to ensure that each of the 714 normal Pap brush samples is scored exactly once after 10 rounds of cross-validation to obtain an overall specificity. All other samples were scored once in each of the 10 rounds for a total of 10 times and were considered to be positive overall if they scored positive more than half of the time (five or more rounds). The mutations in the Pap and Tao brush samples that scored positive are listed in table S4.
For plasma samples, sensitivity and specificity were also obtained from a 10-fold cross-validation. In each of the 10 rounds, plasma samples from 90% of the 192 healthy individuals served as controls in the training set, with the remaining 10% scored to obtain specificity, as for the Pap and Tao brush samples described above. The controls in the training set in each round were randomly selected in a way to ensure that each of the 192 normal plasma samples was scored exactly once in 10 rounds of cross-validation to obtain an overall specificity. In addition, the analysis of the plasma samples was performed using an empirical Bayes approach. In each round of cross-validation, a β distribution was fitted on the basis of the MAFs in the normal controls (90% of the 192 plasma samples from healthy individuals used in the particular round) using maximum likelihood estimation (MLE). Next, the MAFs of all mutations in the controls, as well as the samples to be scored, were adjusted as follows
where α and β are parameters obtained from MLE.
A P value was then calculated for each mutation in each independent well by comparison to the distribution of adjusted MAFs among the controls. An overall P value for every mutation was obtained as the product of the P values from all six independent wells. A sample was considered to be positive if it was positive in five or more rounds of the 10-fold cross-validation. The mutations in the samples that scored positive are listed in table S7.
Confidence intervals for sensitivities and specificities were calculated assuming binomial distributions, with the actual sensitivities and specificities set as the corresponding success probabilities.
Supplementary Material
Table S1. Patient and tumor characteristics.
Table S2. Samples tested with PapSEEK for somatic mutations and aneuploidy.
Table S3. Genomic regions covered in multiplex assays for Pap and Tao brush samples.
Table S4. Mutations identified in Pap and Tao brush samples from patients with ovarian or endometrial cancer.
Table S5. Mutations identified in the primary tumors.
Table S6. Contingency tables for PapSEEK in patients with ovarian and endometrial cancers, as well as healthy controls.
Table S7. Mutations identified in the plasma of ovarian cancer patients.
Acknowledgments:
We thank our patients for their courage and generosity. We thank C. Blair, K Judge, and S. Lio for technical and clinical assistance. We thank E. Cook for artistic contribution. We also thank X. V. Le, B. Kohl, D. Nevriouev, M. Clare, R. Thorp, N. V. Tien, N. V. Bang, B. D. Phu, P. H. Nguyen, L. Catrinici, S. Stepa, V. Cernat, L. Gutu, V. Bucinschi, S. Doruc, M. Ciobanu, S. Mura, M. Cernat, A. Clipca, G. Gorincioi, D. Tcaciuc, N. Botnariuc, I. Chemencedji, I. Stancu, I. Caraman, and M. Pirtac for help with sample procurement.
Funding: This work was supported by the Virginia and D.K. Ludwig Fund for Cancer Research, the John Templeton Foundation, Swim Across America, the Sol Goldman Sequencing Facility at Johns Hopkins, the Commonwealth Foundation, the Conrad R. Hilton Foundation, the U.S. Department of Defense (W81XWH-11-2-0230), the NIH, the National Cancer Institute (CA06973, CA200469, CA215483, and U24CA204817), the Basser Center for BRCA Gray Foundation, the Swedish Cancer Foundation, the Göteborg Medical Society, the Royal Victoria Hospital Foundation, the Carole Epstein Foundation, the Doggone Foundation, the MERMAID Project, the Novo Nordisk Foundation (NNF14OC0012483), the Honorable Tina Brozman Foundation, Stand Up to Cancer Colorectal Cancer Dream Team Translational Research Grant (SU2C-AACR-DT22-17), and the Early Detection Research Network (UO1 CA200469).
Author contributions: B.V., N.P., L.G., A.N.F., K.W.K., L.A.D., and Y.W. designed the study; B.V., J.P., J.S., N.S., L. Dobbyn, and M.P. performed the sample preparation and massively parallel sequencing; Y.W., C.D., J.D.C., I.K., S.S., and R.K. performed the analysis of the sequencing data; L.L., B.A., L. Danilova, and C.T. developed the algorithm for statistical analysis; R.H.H., I.-M.S., T.-L.W., and R.J.K. performed the pathology assessment of the tumor tissue; K.S., S.K.K., T.-T.Y., E.J.T., A.A., M.L., K. Jochumsen, D.A.L., K. Jardon, X.Z., J.A., L.F., L.A.D., B.V., A.N.F., and L.G. contributed to patient recruitment and sample acquisition; Y.W., C.T., K.W.K., B.V., A.N.F., L.G., and N.P. interpreted the data; and all the authors contributed to the writing and reviewing of the manuscript.
Footnotes
Competing interests: Under agreements between the Johns Hopkins University (JHU), Genzyme, Sysmex Inostics, Qiagen, Invitrogen, and Personal Genome Diagnostics, B.V., K.W.K., N.P., and L.A.D. are entitled to a share of the royalties received by the university on the sales of products related to genes and technologies described in this manuscript. B.V., K.W.K., N.P., and L.A.D. are cofounders of Personal Genome Diagnostics and PapGene Inc., are members of the Scientific Advisory Boards of Sysmex Inostics, Personal Genome Diagnostics, and PapGene Inc., and own Personal Genome Diagnostics and PapGene Inc. stock, which is subject to certain restrictions under JHU policy. I.K. is a cofounder and chief scientific officer of PapGene Inc. The company has licensed previously described technologies related to the work described in this paper. The terms of these arrangements are managed by the JHU in accordance with its conflict of interest policies. Part of the technology described in U.S. Patent 20150292027 (Papanicolaou Test for Ovarian and Endometrial Cancers) was applied in this study. Y.W., K.W.K., N.P., C.T., and B.V. are inventors on a patent application on the use of biomarker combinations for the detection of gynecologic cancers. This application will be submitted by the JHU and managed in accordance with its conflict of interest policies. Y.W., K.W.K., N.P., B.V., R.K., I.K., L.D., and C D. are inventors of technologies that are related to those described in this paper and that are associated with equity or royalty payments to the inventors. The terms of these arrangements are being managed by JHU in accordance with its conflict of interest policies. L.G. is listed as a co-inventor on U.S. Provisional Patent application no. 62/656,525 (Uterine Brush and Sample Collection Kit, and Method of Collecting Endometrial Cells from the Uterus) that partially describes the intrauterine sampling method outlined in this paper.
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/10/433/eaap8793/DC1
REFERENCES AND NOTES
- 1.Geldenhuys L, Murray ML, Sensitivity and specificity of the Pap smear for glandular lesions of the cervix and endometrium. Acta Cytol 51, 47–50 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Ng AB, Reagan JW, Hawliczek S, Wentz BW, Significance of endometrial cells in the detection of endometrial carcinoma and its precursors. Acta Cytol 18, 356–361 (1974). [PubMed] [Google Scholar]
- 3.Schnatz PF, Guile M, O’Sullivan DM, Sorosky JI, Clinical significance of atypical glandular cells on cervical cytology. Obstet. Gynecol. 107, 701–708 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Zhao C, Florea A, Onisko A, Austin RM, Histologic follow-up results in 662 patients with Pap test findings of atypical glandular cells: Results from a large academic womens hospital laboratory employing sensitive screening methods. Gynecol. Oncol. 114, 383–389 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Howlader N, Noone AM, Krapcho M, Miller D, Bishop K, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA, SEER Cancer Statistics Review, 1975–2014 (National Cancer Institute, 2017). [Google Scholar]
- 6.Kurman RJ, Shih I-M, The origin and pathogenesis of epithelial ovarian cancer: A proposed unifying theory. Am. J. Surg. Pathol. 34, 433–443 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore KN, Fader AN, Uterine papillary serous carcinoma. Clin. Obstet. Gynecol. 54, 278–291 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Arnold M, Pandeya N, Byrnes G, Renehan AG, Stevens GA, Ezzati M, Ferlay J, Miranda JJ, Romieu I, Dikshit R, Forman D, Soerjomataram I, Global burden of cancer attributable to high body-mass index in 2012: A population-based study. Lancet Oncol 16, 36–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM, Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 74, 2913–2921 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Jacobs I, Gentry-Maharaj A, Burnell M, Manchanda R, Singh N, Sharma A, Ryan A, Seif MW, Amso NN, Turner G, Brunell C, Fletcher G, Rangar R, Ford K, Godfrey K, Lopes A, Oram D, Herod J, Williamson K, Scott I, Jenkins H, Mould T, Woolas R, Murdoch J, Dobbs S, Leeson S, Cruickshank D, Skates SJ, Fallowfield L, Parmar M, Campbell S, Menon U, Sensitivity of transvaginal ultrasound screening for endometrial cancer in postmenopausal women: A case-control study within the UKCTOCS cohort. Lancet Oncol. 12, 38–48 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Buys SS, Partridge E, Black A, Johnson CC, Lamerato L, Isaacs C, Reding DJ, Greenlee RT, Yokochi LA, Kessel B, Crawford ED, Church TR, Andriole GL, Weissfeld JL, Fouad MN, Chia D, O’Brien B, Ragard LR, Clapp JD, Rathmell JM, Riley TL, Hartge P, Pinsky PF, Zhu CS, Izmirlian G, Kramer BS, Miller AB, Xu JL, Prorok PC, Gohagan JK, Berg CD; PLCO Project Team, Effect of screening on ovarian cancer mortality: The Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening randomized controlled trial. JAMA 305, 2295–2303 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi H, Yamada Y, Sado T, Sakata M, Yoshida S, Kawaguchi R, Kanayama S, Shigetomi H, Haruta S, Tsuji Y, Ueda S, Kitanaka T, A randomized study of screening for ovarian cancer: A multicenter study in Japan. Int. J. Gynecol. Cancer 18, 414–420 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Jacobs IJ, Menon U, Ryan A, Gentry-Maharaj A, Burnell M, Kalsi JK, Amso NN, Apostolidou S, Benjamin E, Cruickshank D, Crump DN, Davies SK, Dawnay A, Dobbs S, Fletcher G, Ford J, Godfrey K, Gunu R, Habib M, Hallett R, Herod J, Jenkins H, Karpinskyj C, Leeson S, Lewis SJ, Liston WR, Lopes A, Mould T, Murdoch J, Oram D, Rabideau DJ, Reynolds K, Scott I, Seif MW, Sharma A, Singh N, Taylor J, Warburton F, Widschwendter M, Williamson K, Woolas R, Fallowfield L, McGuire AJ, Campbell S, Parmar M, Skates SJ, Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): A randomised controlled trial. Lancet 387, 945–956 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menon U, Ryan A, Kalsi J, Gentry-Maharaj A, Dawnay A, Habib M, Apostolidou S, Singh N, Benjamin E, Burnell M, Davies S, Sharma A, Gunu R, Godfrey K, Lopes A, Oram D, Herod J, Williamson K, Seif MW, Jenkins H, Mould T, Woolas R, Murdoch JB, Dobbs S, Amso NN, Leeson S, Cruickshank D, Scott I, Fallowfield L, Widschwendter M, Reynolds K, McGuire A, Campbell S, Parmar M, Skates SJ, Jacobs I, Risk algorithm using serial biomarker measurements doubles the number of screen-detected cancers compared with a single-threshold rule in the united kingdom collaborative trial of ovarian cancer screening. J. Clin. Oncol. 33, 2062–2071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moyer VA; Preventive US Services Task Force., Screening for ovarian cancer: U.S. Preventive Services Task Force reaffirmation recommendation statement. Ann. Intern. Med. 157, 900–904 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Kurman RJ, Shih IM, Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer—Shifting the paradigm. Hum. Pathol. 42, 918–931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y, Miron A, Drapkin R, Nucci MR, Medeiros F, Saleemuddin A, Garber J, Birch C, Mou H, Gordon RW, Cramer DW, McKeon FD, Crum CP, A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J. Pathol. 211, 26–35 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Eckert MA, Pan S, Hernandez KM, Loth RM, Andrade J, Volchenboum SL, Faber P, Montag A, Lastra R, Peter ME, Yamada SD, Lengyel E, Genomics of ovarian cancer progression reveals diverse metastatic trajectories including intraepithelial metastasis to the fallopian tube. Cancer Discov. 6, 1342–1351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karst AM, Levanon K, Drapkin R, Modeling high-grade serous ovarian carcinogenesis from the fallopian tube. Proc. Natl. Acad. Sci. U.S.A. 108, 7547–7552 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhai Y, Wu R, Kuick R, Sessine MS, Schulman S, Green M, Fearon ER, Cho KR, High-grade serous carcinomas arise in the mouse oviduct via defects linked to the human disease. J. Pathol. 243, 16–25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurman RJ, Shih I-M, The dualistic model of ovarian carcinogenesis: Revisited, revised, and expanded. Am. J. Pathol 186, 733–747 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert L, Basso O, Sampalis J, Karp I, Martins C, Feng J, Piedimonte S, Quintal L, Ramanakumar AV, Takefman J, Grigorie MS, Artho G, Krishnamurthy S; DOvE Study Group, Assessment of symptomatic women for early diagnosis of ovarian cancer: Results from the prospective DOvE pilot project. Lancet Oncol. 13, 285–291 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Meden H, Fattahi-Meibodi A, CA 125 in benign gynecological conditions. Int. J. Biol. Markers 13, 231–237 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Erickson BK, Kinde I, Dobbin ZC, Wang Y, Martin JY, Alvarez RD, Conner MG, Huh WK, Roden RBS, Kinzler KW, Papadopoulos N, Vogelstein B, Diaz LA Jr., Landen CN Jr., Detection of somatic TP53 mutations in tampons of patients with highgrade serous ovarian cancer. Obstet. Gynecol. 124, 881–885 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinde I, Bettegowda C, Wang Y, Wu J, Agrawal N, Shih IM, Kurman R, Dao F, Levine DA, Giuntoli R, Roden R, Eshleman JR, Carvalho JP, Marie SK, Papadopoulos N, Kinzler KW, Vogelstein B, Diaz LA Jr., Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci. Transl. Med. 5, 167ra4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Springer S, Mulvey CL, Silliman N, Schaefer J, Sausen M, James N, Rettig EM, Guo T, Pickering CR, Bishop JA, Chung CH, Califano JA, Eisele DW, Fakhry C, Gourin CG, Ha PK, Kang H, Kiess A, Koch WM, Myers JN, Quon H, Richmon JD, Sidransky D, Tufano RP, Westra WH, Bettegowda C, Diaz LA Jr., Papadopoulos N, Kinzler KW, Vogelstein B, Agrawal N, Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci. Transl. Med. 7, 293ra104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B, Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 108, 9530–9535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research Network, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, Yau C, Laird PW, Ding L, Zhang W, Mills GB, Kucherlapati R, Mardis ER, Levine DA, Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheung LWT, Hennessy BT, Li J, Yu S, Myers AP, Djordjevic B, Lu Y, Stemke-Hale K, Dyer MD, Zhang F, Ju Z, Cantley LC, Scherer SE, Liang H, Lu KH, Broaddus RR, Mills GB, High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov. 1, 170–185 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research Network., Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW, Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kinde I, Papadopoulos N, Kinzler KW, Vogelstein B, FAST-SeqS: A simple and efficient method for the detection of aneuploidy by massively parallel sequencing. PLOS ONE 7, e41162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao LC, Direct intrauterine sampling: The IUMC endometrial sampler. Diagn. Cytopathol. 17, 153–159 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Wu HHJ, Harshbarger KE, Berner HW, Elsheikh TM, Endometrial brush biopsy (Tao brush): Histologic diagnosis of 200 cases with complementary cytology: An accurate sampling technique for the detection of endometrial abnormalities. Am. J. Clin. Pathol 114, 412–418 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Siravegna G, Marsoni S, Siena S, Bardelli A, Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 14, 531–548 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, Hruban RH, Wolfgang CL, Goggins MG, Dal Molin M, Wang T-L, Roden R, Klein AP, Ptak J, Dobbyn L, Schaefer J, Silliman N, Popoli M, Vogelstein JT, Browne JD, Schoen RE, Brand RE, Tie J, Gibbs P, Wong H-L, Mansfield AS, Jen J, Hanash SM, Falconi M, Allen PJ, Zhou S, Bettegowda C, Diaz LA Jr., Tomasetti C, Kinzler KW, Vogelstein B, Lennon AM, Papadopoulos N, Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fishman DA, Cohen L, Blank SV, Shulman L, Singh D, Bozorgi K, Tamura R, Timor-Tritsch I, Schwartz PE, The role of ultrasound evaluation in the detection of earlystage epithelial ovarian cancer. Am. J. Obstet. Gynecol. 192, 1214–1221 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Sharma A, Apostolidou S, Burnell M, Campbell S, Habib M, Gentry-Maharaj A, Amso N, Seif MW, Fletcher G, Singh N, Benjamin E, Brunell C, Turner G, Rangar R, Godfrey K, Oram D, Herod J, Williamson K, Jenkins H, Mould T, Woolas R, Murdoch J, Dobbs S, Leeson S, Cruickshank D, Fourkala E-O, Ryan A, Parmar M, Jacobs I, Menon U, Risk of epithelial ovarian cancer in asymptomatic women with ultrasound-detected ovarian masses: A prospective cohort study within the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS). Ultrasound Obstet. Gynecol 40, 338–344 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Hamilton CA, Cheung MK, Osann K, Chen L, Teng NN, Longacre TA, Powell MA, Hendrickson MR, Kapp DS, Chan JK, Uterine papillary serous and clear cell carcinomas predict for poorer survival compared to grade 3 endometrioid corpus cancers. Br. J. Cancer 94, 642–646 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C, Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.AlHilli MM, Dowdy SC, Weaver AL, St Sauver JL, Keeney GL, Mariani A, Podratz KC, Bakkum-Gamez JN, Incidence and factors associated with synchronous ovarian and endometrial cancer: A population-based case-control study. Gynecol. Oncol. 125, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walsh C, Holschneider C, Hoang Y, Tieu K, Karlan B, Cass I, Coexisting ovarian malignancy in young women with endometrial cancer. Obstet. Gynecol. 106, 693–699 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Zaino R, Whitney C, Brady MF, DeGeest K, Burger RA, Buller RE, Simultaneously detected endometrial and ovarian carcinomas—A prospective clinicopathologic study of 74 cases: A gynecologic oncology group study. Gynecol. Oncol. 83, 355–362 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Song T, Seong SJ, Bae D-S, Kim J-H, Suh DH, Lee K-H, Park S-Y, Lee TS, Prognostic factors in women with synchronous endometrial and ovarian cancers. Int. J. Gynecol. Cancer 24, 520–527 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, Ebert BL, Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 126, 9–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, Hyman DM, Solit DB, Robson ME, Baselga J, Arcila ME, Ladanyi M, Tallman MS, Levine RL, Berger MF, Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell 21, 374–382.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young AL, Challen GA, Birmann BM, Druley TE, Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat. Commun. 7, 12484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krimmel JD, Schmitt MW, Harrell MI, Agnew KJ, Kennedy SR, Emond MJ, Loeb LA, Swisher EM, Risques RA, Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in noncancerous tissues. Proc. Natl. Acad. Sci. U.S.A. 113, 6005–6010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anglesio MS, Papadopoulos N, Ayhan A, Nazeran TM, Noe M, Horlings HM, Lum A, Jones S, Senz J, Seckin T, Ho J, Wu R-C, Lac V, Ogawa H, Tessier-Cloutier B, Alhassan R, Wang A, Wang Y, Cohen JD, Wong F, Hasanovic A, Orr N, Zhang M, Popoli M, McMahon W, Wood LD, Mattox A, Allaire C, Segars J, Williams C, Tomasetti C, Boyd N, Kinzler KW, Gilks CB, Diaz L, Wang T-L, Vogelstein B, Yong PJ, Huntsman DG, Shih I-M, Cancer-associated mutations in endometriosis without cancer. N. Engl. J. Med. 376, 1835–1848 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nair N, Camacho-Vanegas O, Rykunov D, Dashkoff M, Camacho SC, Schumacher CA, Irish JC, Harkins TT, Freeman E, Garcia I, Pereira E, Kendall S, Belfer R, Kalir T, Sebra R, Reva B, Dottino P, Martignetti JA, Genomic analysis of uterine lavage fluid detects early endometrial cancers and reveals a prevalent landscape of driver mutations in women without histopathologic evidence of cancer: A prospective cross-sectional study. PLOS Med. 13, e1002206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore RG, Brown AK, Miller MC, Skates S, Allard WJ, Verch T, Steinhoff M, Messerlian G, DiSilvestro P, Granai CO, Bast RC Jr., The use of multiple novel tumor biomarkers for the detection of ovarian carcinoma in patients with a pelvic mass. Gynecol. Oncol. 108, 402–408 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong S-M, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SKN, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih I-M, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang T-L, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz LA Jr., Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rago C, Huso DL, Diehl F, Karim B, Liu G, Papadopoulos N, Samuels Y, Velculescu VE, Vogelstein B, Kinzler KW, Diaz LA Jr., Serial assessment of human tumor burdens in mice by the analysis of circulating DNA. Cancer Res. 67, 9364–9370 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Douville C, Springer S, Kinde I, Cohen JD, Hruban RH, Lennon AM, Papadopolous N, Kinzler KW, Vogelstein B, Karchin R, Detection of aneuploidy in patients with cancer through amplification of long interspersed nucleotide elements (LINEs). Proc. Natl. Acad. Sci. U.S.A. 115, 1871–1876 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y, Sundfeldt K, Mateoiu C, Shih I-M, Kurman RJ, Schaefer J, Silliman N, Kinde I, Springer S, Foote M, Kristjansdottir B, James N, Kinzler KW, Papadopoulos N, Diaz LA, Vogelstein B, Diagnostic potential of tumor DNA from ovarian cyst fluid. eLife 5, e15175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, Stefancsik R, Harsha B, Kok CY, Jia M, Jubb H, Sondka Z, Thompson S, De T, Campbell PJ, COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 45, D777–D783 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jung H, Lee D, Lee J, Park D, Kim YJ, Park W-Y, Hong D, Park PJ, Lee E, Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat. Genet. 47, 1242–1248 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Patient and tumor characteristics.
Table S2. Samples tested with PapSEEK for somatic mutations and aneuploidy.
Table S3. Genomic regions covered in multiplex assays for Pap and Tao brush samples.
Table S4. Mutations identified in Pap and Tao brush samples from patients with ovarian or endometrial cancer.
Table S5. Mutations identified in the primary tumors.
Table S6. Contingency tables for PapSEEK in patients with ovarian and endometrial cancers, as well as healthy controls.
Table S7. Mutations identified in the plasma of ovarian cancer patients.


