Abstract
Purpose:
Despite a growing arsenal of approved drugs, therapeutic resistance remains a formidable and, often, insurmountable challenge in cancer treatment. The mechanisms underlying therapeutic resistance remain largely unresolved and, thus, examples of effective combinatorial or sequential strategies to combat resistance are rare. Here, we present Differential Sensitivity Analysis for Resistant Malignancies (DISARM), a novel, integrated drug screen analysis tool designed to address this dilemma.
Experimental Design:
DISARM, a software package and web-based application, analyzes drug response data to prioritize candidate therapies for models with resistance to a reference drug and to assess whether response to a reference drug can be utilized to predict future response to other agents. Using cisplatin as our reference drug, we applied DISARM to models from nine cancers commonly treated with frontline platinum chemotherapy including recalcitrant malignancies such as small cell lung cancer (SCLC) and pancreatic adenocarcinoma (PAAD).
Results:
In cisplatin-resistant models, DISARM identified novel candidates including multiple inhibitors of PI3K, MEK and BCL-2, among other classes, across unrelated malignancies. Additionally, DISARM facilitated the selection of predictive biomarkers of response and identification of unique molecular subtypes, such as contrasting ASCL1-low/cMYC-high SCLC targetable by AURKA inhibitors and ASCL1-high/cMYC-low SCLC targetable by BCL-2 inhibitors. Utilizing these predictions, we assessed several of DISARM’s top candidates including inhibitors of AURKA, BCL-2, and HSP90 to confirm their activity in cisplatin-resistant SCLC models.
Conclusions:
DISARM represents the first validated tool to analyze large-scale in vitro drug response data to statistically optimize candidate drug and biomarker selection aimed at overcoming candidate drug resistance.
Introduction
Cancer is the second leading cause of death in the United States (1), despite extensive efforts to improve prevention, detection and treatment of malignancies. Survival for many patients with inoperable cancer remains poor, largely owing to resistance to standard of care (SOC) therapies and the lack of biomarkers to guide treatment with novel agents. Medical oncologists have an ever-expanding repertoire of approved and experimental treatment options from which to choose. Therefore, in many cases, the challenge has become selecting the optimal treatment, or treatment sequence, for each patient--rather than a lack of available therapies from which to choose.
Clinical and translational researchers have access to unprecedented volumes of drug response data (2–5). Unfortunately, there is no standard approach for integrating response data for multiple drugs. Preclinical data may show that in an unselected population of lung adenocarcinoma (LUAD) models, 10% respond to drug X, while 5% respond to drug Y. If, for example, drugs X and Y are found to both target the same population (e.g. EGFR-mutant LUAD), then drug Y may have little utility. If, however, drug X targets EGFR-mutant LUAD, while drug Y targets LUAD with ALK rearrangements, then the responding populations are largely mutually exclusive (6) and drug Y may have a role for many tumors that would not have responded to drug X. The distinction between overlapping and mutually exclusive sensitive models would be elusive amidst the standard presentation of large-scale drug screen data, especially in cases where the predictive biomarkers were not yet established.
The lack of a systematic, unbiased approach to large-scale drug response data that allows one to compare the activity of two or more drugs is a challenge that appears straightforward. If we consider two drugs, one the SOC and another an experimental agent, a researcher can quickly ascertain if the experimental agent has efficacy among instances where there is resistance to the SOC. However, without well-characterized biomarkers or molecular targets to narrow the researcher’s focus down to this single drug pair, the abundance of potential pairs necessitates a formal approach to answer the unexpectedly challenging question, “If a tumor model is resistant to a drug X, to what other drug(s) may it respond?”. To address this problem computationally, we have developed DISARM (Differential Sensitivity Analysis for Resistant Malignancies), a bioinformatics tool engineered to identify drugs with efficacy in models where there is resistance to a drug of interest by iteratively comparing in vitro IC50 (half-maximal inhibitory concentration) measurements between a designated reference drug and all other available candidate agents assessed in equivalent models. In doing so, DISARM is intended to not only streamline and standardize the identification and prioritization of candidate drugs among scenarios of therapeutic resistance, but also to assess the hypothesis that prior drug response data may be used to predict future responses to novel agents across tumor types. DISARM has been designed as both a software package and as an interactive web-based application intended to make DISARM accessible for the scientific community at large.
Materials and Methods
DISARM formulation and workflow
DISARM generates a 2×2 table for each drug pair to identify those instances in which a significant number of cell lines are sensitive to a candidate drug, despite resistance to the reference drug. Here, Drug X is defined as the drug of interest to which all other Drug(s) Y are compared. Let X denote the response to Drug X where X=0 and X=1 indicate sensitivity and resistance, respectively, with the response to Drug(s) Y similarly represented with the variable Y. Each sample is categorized into one of the four quadrants, where A, B, C and D are the observed numbers of cell lines in each quadrant with total sample size N = A+B+C+D. Thus, D represents the cell lines that are resistant to Drug X, but sensitive to Drug Y, while C is the cell lines that are sensitive to both drugs, etc.
| Drug X (Reference) | |||
| X=0 (Sensitive) | X=1 (Resistant) | ||
|
Drug Y (Candidate) |
Y=1 (Resistant) | A | B |
| Y=0 (Sensitive) | C | D | |
DISARM aims to identify those drug pairs in which D represents a significant portion of the cell lines, as this suggests a niche in which samples are sensitive to Drug Y but not Drug X.
Specifically, let D represent the random variable associated with the number of samples resistant to Drug X but sensitive to Drug Y (X=1 and Y=0) and the value of D=μ10. If μ10 is sufficiently large (for example, significantly larger than zero), this suggests Drug Y is a potential alternative (or, complementary) treatment to Drug X.
To identify such candidates, we formulate the following one-sided hypothesis testing:
Here γ is a threshold value, specified by the user, for the minimum number of samples being both resistant to Drug X and sensitive to Drug Y.
The Wald test statistic which follows the standard Gaussian distribution is defined below:
| (1) |
When the response is randomly distributed, we have μ10 = Npx(1 − py) where px is the probability of a sample being resistant to Drug X and py is the probability of a sample being resistant to Drug Y. We can estimate , where and . can be estimated by the Delta-method.
| (2) |
Thus, the test statistic becomes:
| (3) |
The test statistics described here differs from Fisher’s exact test in the purposes and hypotheses. Fisher’s exact test is a test of independence between rows and columns in a two-way contingency table, while our test concerns one of the 4 quadrants only (i.e., quadrant D), and tests the number in that quadrant against a threshold value γ.
We defined the value computed from equation (3) as DISARM score. This score follows a standard normal distribution. It is just as a standard score or Z-score and can be placed on a normal distribution curve. A higher DISARM score corresponds to higher significance level.
IC50 values have limitations as a measure of drug sensitivity, such as the fact that this value is undefined for some drug-cell line pairs in which IC50 exceeded highest tested concentration. However, the widespread availability of this value, particularly for tumors of interest in this analysis, outweighs these shortcomings. Usually, drug sensitivity data such as IC50 and GI50 (50% growth inhibition) are continuous. However, DISARM requires the specification of a boundary between sensitivity and resistance. To permit flexibility in defining such a cut-off, DISARM users can discretize IC50 values based on quantiles or other approaches (e.g. known maximum serum concentration, or Cmax). This process is intentionally arbitrary to accommodate a variety of applications of DISARM – applications that may range from users casting a broad net in initial exploratory analyses with expected high false discovery rates to efforts to identify only the most extreme outliers for expensive and time intensive animal based studies. We employ a filter on the mean IC50 value of both Drug X and Drug Y for samples in quadrant D to ensure that these values accurately reflect resistance to Drug X and sensitivity to Drug Y, when this data is available. For example, in the subsequent analyses using cisplatin as a reference drug, we ensure that IC50 cut-off for cisplatin resistance exceeds the Cmax of cisplatin (7.04 μM) in all cases. DISARM was designed to allow the user to easily test multiple thresholds in rapid succession to identify the most appropriate criteria for their application. Candidates can then be prioritized based upon a number of variables that may be of particular importance to the user including the DISARM score and its associated p-value, the proportion of samples in quadrant D or mean IC50 values for selected candidate drugs. Each analysis that follows employs a unique set of selection criteria, filters and cut-offs that are outlined below.
Sensitivity groups were based on quantiles of IC50 values and results represented here were generated using the highest 25% of IC50 values for each drug as threshold for resistance. For instances where IC50 was not reached experimentally, the highest tested concentration is considered the IC50 and the cell line is, therefore, considered resistant. After initial results were generated, an additional filter was applied to minimize false positive selections. To further prioritize drug candidates, we applied several filters based on mean IC50 values of cell lines in quadrant D for drug A (μ1) and drug B (μ2), DISARM score and proportion of samples in quadrant D (proportion score, or prop D). For the analysis of SCLC alone, we required μ1 fall in the upper quantile among all candidate drugs (> 60%) and μ2 in the lower quantile among all candidate drugs (< 40% in our case). For the analysis of multiple cancer types, we required μ1 fall in the upper quantile among all candidate drugs (> 45%) and μ2 in the lower quantile among all candidate drugs (< 40% in our case). For all analyses, we required a DISARM score larger than 2.0 and prop D larger than 0.2. As with discretization process, we encourage users to exploit the ease of applying various filtering cut-offs to yield the scale and stringency of results they desire. The DISARM web application is not currently designed to incorporate cut-offs for DISARM score, proportion of samples in quadrant D, or mean IC50 values. This is intentional, since different scenarios will call for more or less stringent application of such cut-offs to yield manageable candidate lists. However, the web application is designed to make the use of such filters and cut-offs straightforward (see Supplementary Figs. S1 and S2).
Gene and protein expression.
Gene expression data includes publicly available data (7,8) as assessed by exon array, as well as data that we generated via Affymetrix Human Genome U133 Plus 2 arrays as previously described (9). Protein expression was assessed for 171 total and phospho-proteins in small cell lung cancer cell lines by reverse phase protein array (RPPA) as previously described (9).
Statistical analyses.
In addition to the statistical analysis inherent to DISARM (described above), biomarker analysis employed student’s T-test when performing binary comparisons between sensitive and resistant groups. Elsewhere, we used Spearman rank correlation to assess the correlations between IC50 values and individual RPPA protein markers. To account for multiple hypotheses testing, we applied a beta-uniform mixture model (BUM) to the resulting p-values computed from test statistics in order to estimate false discovery rates (FDRs). Appropriate FDR cut-offs were applied to identify significant biomarkers(10). We used R packages to perform all statistical analyses (https://cran.r-project.org/web/packages/ and https://www.bioconductor.org/).
Cell culture
Cell lines were grown in RPMI unless otherwise mentioned by the provider (9) with 10% fetal bovine serum and antibiotics, cultured at 37⁰C in a humidified chamber with 5% CO2. All cell lines included in the study were profiled at passage 4–8 to abrogate the heterogeneity introduced by long-term culture. All cell lines were tested for Mycoplasma at the time of thawing as previously described (11) and the characteristic phenotype (floating aggregates and colony formation) of SCLC cell lines.
DNA fingerprinting to confirm cell line identity
DNA from 5–6 × 106 cells was isolated using a QIAamp DNA mini kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol. DNA was eluted in 100μl of elution buffer (Buffer AE, Qiagen, Hilden, Germany). The concentration of the eluted DNA was measured by the absorbance at 260 nm, and the purity of the eluted DNA was determined by the ratio of the absorbance at 260 nm to the absorbance at 280 nm. About 50 ng of DNA was used for DNA fingerprint analysis of short tandem repeat profiling (PowerPlex 16 hs, Promega; Madison, WI, USA) to authenticate each cell line. The analysis system used covers at least eight short tandem repeat loci. Fingerprinting results for each cell line were compared to reference fingerprints from the Cancer Cell Line Encyclopedia (CCLE) (12).
Cell viability assay treatments
Cell lines were plated at 2000 cells per well in 96 well plates 24 hours prior to treatment with drug using cell culture conditions described above. Each line was treated, in triplicate, with respective agents (cisplatin, obatoclax, 17-AAG, and alisertib) starting at 33 μM followed by 1:10 serial dilutions (3.3 μM, 0.33 μM, 0.033 μM) and DMSO-only control. Following 120 hour incubations, cell viability was assessed using CellTiter-Glo Luminescent Cell Viability Assay (Promega; Wisconsin, USA).
Calculation of drug parameters
For single-agent analysis, we estimated IC50 values by using the software program drexplorer, which fitted multiple dose-response models and selected the best model using the residual standard error (13).
Results
DISARM validation
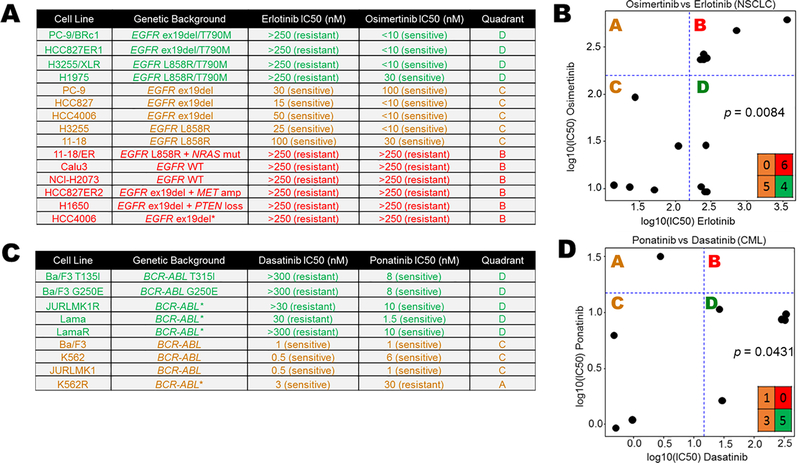
To assess DISARM’s design, we tested drug pairs in which the reference drug is a SOC therapy and the candidate drug is an approved option following resistance to the SOC agent. In the case of metastatic non-small cell lung cancer (NSCLC) with exon 19 deletions or L858R mutation in EGFR, SOC therapy includes tyrosine-kinase inhibitors (TKIs) such as erlotinib, gefitinib or afatinib (14), which provide high initial response rates (RR) but to which resistance almost inevitably develops. In approximately 50% of patient tumors (15), this resistance is due to the acquisition of a second EGFR mutation, T790M. Response data (Fig. 1A) were adapted from a prior study (15) wherein osimertinib, initially approved by the United States Food & Drug Administration (FDA) only for patients who have developed resistance via T790M, was identified by DISARM as a viable candidate in this setting (Fig. 1B). Specifically, 40% of erlotinib-resistant NSCLC cell lines (IC50 values > ~160 nanomolar (nM)) were sensitive to osimertinib (IC50 values < ~160 nM) and these were the cell lines with T790M EGFR mutations.
Figure 1. DISARM design and validation.
(A) Adapted IC50 data (15) for NSCLC cell lines treated with the EGFR inhibitors erlotinib and osimertinib, the latter of which is effective in instances of EGFR T790M erlotinib resistance mutations. (C) DISARM-generated 2×2 plot for erlotinib (reference) and osimertinib (candidate) pair, in which DISARM accurately selects osimertinib as an effective agent in erlotinib-resistant NSCLC. Each black dot in 2×2 plot indicates a specific cell line. (D) Similarly, adapted IC50 data for CML cell lines (16) treated with the BCR-ABL inhibitors dasatinib (reference) and ponatinib (candidate), the latter of which is an effective therapy in instances of BCR-ABL T315I resistance mutations. (E) DISARM correctly selects ponatinib as an effective therapy for dasatinib-resistant CML. (B-E) IC50 and log10(IC50) values are expressed in nanomolar (nM) concentration.
A second example is demonstrated for chronic myelogenous leukemia (CML), wherein treatment typically begins with a tyrosine-kinase inhibitor targeting the BCR-ABL fusion protein, including dasatinib, nilotinib and imatinib. For patients whose leukemia develops resistance to these agents, often via a T315I mutation in BRC-ABL, ponatinib was identified by DISARM as an attractive candidate (Fig. 1C and D) with data adapted from a prior study (16), and ultimately FDA approved for this clinical setting. In this case, all five CML lines that were resistant to dasatinib (IC50 > ~15 nM) were sensitive to ponatinib (IC50 < ~15 nM). In each case, our analysis accurately reflects the clinically established relationships between these drug pairs.
DISARM identifies common vulnerabilities in platinum-resistant SCLC
The need for active drugs for platinum-resistant cancers is especially urgent in the case of small cell lung cancer, the most aggressive form of lung cancer. Platinum chemotherapy represents the backbone of all SOC first-line therapies in SCLC. While initial response rates exceed 50%, platinum-resistance develops rapidly and nearly universally, often within months (17). Furthermore, platinum-resistance is associated with cross-resistance to second-line chemotherapy agents with only one FDA approved second-line treatment (topotecan) to which less than 5% of platinum-resistant and refractory patients respond (17).
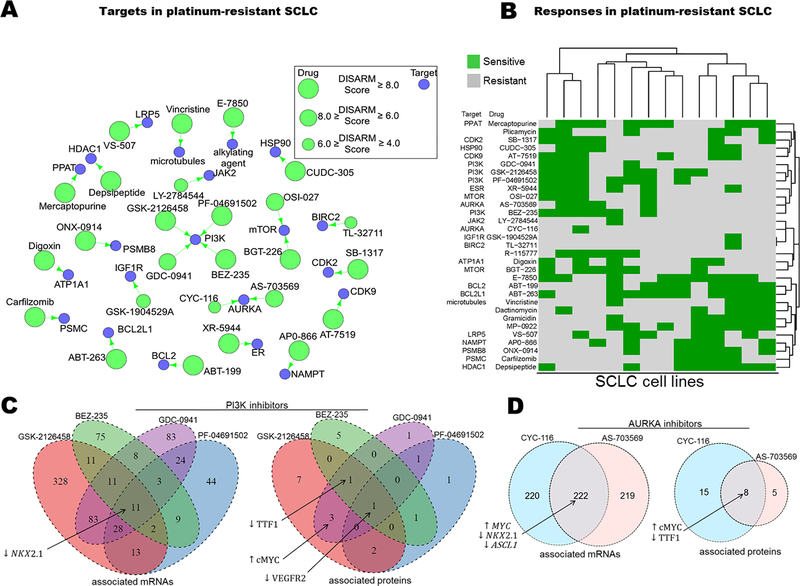
Using cisplatin, the most widely prescribed platinum agent, as our reference drug, we applied DISARM to the problem of platinum-resistance in small cell lung cancer (SCLC). We employed IC50 data for cisplatin generated by our lab for SCLC cell lines (18), along with publicly available IC50 data (7) for 526 FDA-approved and investigational anti-cancer agents in the same cell lines. After filtering (as described in DISARM formulation and workflow), DISARM selected 31 candidate drugs, including 26 with defined molecular targets (Fig. 2A). The top 31 drugs share numerous targets, including six drugs which target PI3K or mTOR, and multiple drugs targeting AURKA, BCL-2 family proteins, proteasomes and cyclin-dependent kinases. Notably, the 526 analyzed candidate drugs included 23 drugs which target PI3K or mTOR and, of these 23 drugs, 19 met initial selection criteria with DISARM scores >2.0 and p-values <0.05. The top six candidates of this class, determined by subsequent filtering selection, are illustrated here. The reoccurrence of similar drug classes from a highly diverse pool of candidates suggest that cisplatin-refractory models of SCLC share common vulnerabilities that warrant further pursuit.
Figure 2. DISARM identifies candidate drugs for cisplatin-resistant SCLC including inhibitors of PI3K, mTOR and AURKA.
(A) Drug-target constellation (DTECT) map highlighting top candidates selected by DISARM for cisplatin-resistant SCLC using previously reported IC50 values (7) (B) Heatmap clustering cell lines on the basis of similarity in sensitivity to candidate agents reveals two distinct subsets of cisplatin-resistant SCLC, a subset that is cisplatin-resistant but sensitive to inhibitors of PI3K, AURKA and others (upper left) and another subset that is cisplatin-resistant but sensitive to BCL-2 and proteasome inhibitors, among others (lower right). (C) Venn diagrams depicting shared mRNA (left) and protein (right) predictive biomarkers of sensitivity of SCLC cell lines to four PI3K inhibitors selected as candidates by DISARM (see also Tables S1-S2). (D) Similar Venn diagrams for sensitivity to two AURKA inhibitors selected as candidates by DISARM (see also Tables S3–4).
Among cisplatin-resistant cell lines, we observed distinct subgroups that respond to multiple drugs from similar classes. For example, cell lines H1930 and DMS-273 are sensitive to all six selected inhibitors of PI3K and mTOR, while H196 is sensitive to five of them. Hierarchical clustering of cell lines on the basis of similarity of categorical sensitivity (or resistance) to DISARM-selected candidates revealed two predominant clusters (Fig. 2B). The first cluster is comprised of lines sensitive to inhibitors of PI3K/mTOR, AURKA and CDKs, among other targets. The second cluster is comprised of lines sensitive to inhibitors of the BCL-2 pathway, proteasomes and HDACs, among others. Of the cell lines that are sensitive to none of the PI3K/mTOR inhibitors, all five (H2029, H1417, H128, H2330, COR-L88) are sensitive to both agents targeting the BCL-2 pathway. Similarly, of the lines that are sensitive to none of the agents targeting the BCL-2 pathway, all four (H378, SHP-77, DMS-273 and H196) are sensitive to at least two inhibitors of PI3K/mTOR. These observations suggest that subsets of SCLC cisplatin-resistant models may be defined by distinct molecular vulnerabilities.
Candidate drugs identified by DISARM share common predictive biomarkers in SCLC models
We then tested whether the distinct groups of cell lines identified on the basis of sensitivity to candidate drugs that target the same molecule also shared common molecular profiles that could be leveraged to identify predictive biomarkers. For the four drugs targeting PI3K, we compared publically available mRNA (7) and protein expression generated by our lab (18,19) between sensitive and resistant cell lines, as determined by DISARM, to identify predictive biomarkers of sensitivity (Fig. 2C; Supplementary Tables S1 and S2). These analyses suggest that low expression of the gene NKX2–1and its associated protein, TTF-1, is predictive of response to PI3K inhibitors. Notably, most tumors of suspected lung origin, including SCLC, are assessed for TTF-1 status by immunohistochemical (IHC) staining performed as part of the pathologic diagnosis with approximately 80–85% considered TTF-1 positive based on IHC (20). A similar analysis of AURKA sensitive and resistant cell lines again identifies low NKX2–1/TTF-1 expression as a predictive biomarker of AURKA inhibitor sensitivity (Fig. 2D; Supplementary Tables S3 and S4). AURKA sensitive cell lines were also characterized by low expression of ASCL1, a transcription factor that regulates neuroendocrine differentiation and is required for tumor formation in murine SCLC models (21). These findings suggest the presence of a TTF-1 low, platinum-resistant niche within SCLC, consistent with the observation that TTF-1 negative extensive-stage SCLC patients have poorer response to platinum-based therapy (20). High levels of MYC and c-MYC mRNA and protein expression were also identified as markers of AURKA inhibitor sensitivity (Fig. 2D). This is compatible with recent data suggesting that in SCLC, MYC is induced by NEUROD1, a transcription factor that defines a cellular population that is largely mutually exclusive with ASCL1 expressing cells in SCLC (21). The resulting MYC-high, neuroendocrine-low (ASCL1-low/NEUROD1-high) SCLC subset is characterized by susceptibility to AURKA inhibition (22–24).
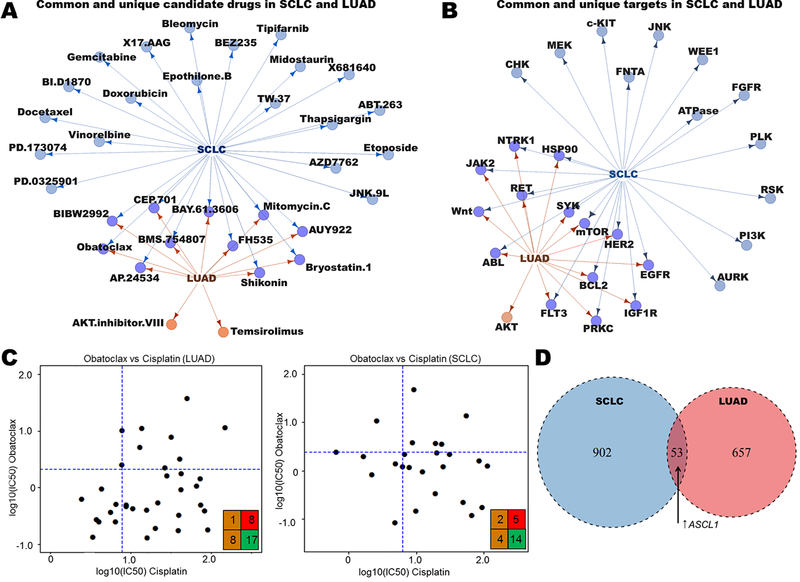
DISARM identifies common candidates and biomarkers in SCLC and LUAD
Platinum-resistance is a challenge that extends beyond SCLC. Thus, we sought to broaden the scope of DISARM’s application to additional tumor types commonly treated with first-line platinum therapy. We began by applying DISARM to drug treatment data from models of LUAD, the most common form of lung cancer. Although SCLC and LUAD are commonly treated with cisplatin-containing regimens, they are otherwise clinically and biologically distinct. Employing drug response and expression data for 138 drugs in 44 SCLC and 39 LUAD cell lines from the Genomics of Drug Sensitivity in Cancer (GDSC) database (25), we identified candidate drugs (Fig. 3A) and drug targets (Fig. 3B) for platinum-resistant models, including shared targets such as mTOR, BCL-2 and HSP90. These results demonstrate that there are both shared and cancer type-specific targets among platinum-resistant LUAD and SCLC. While the unique targets may reflect fundamental differences in tumor biology, the common drugs and classes highlight examples of common vulnerabilities in platinum-resistant lung cancer models. We present one such example in the BCL-2 inhibitor obatoclax, selected by DISARM as a candidate therapy for both platinum-resistant LUAD and SCLC (Fig. 3C). Using a common drug or target, we can repeat a similar analysis of mRNA expression, this time between tumor types, to identify predictive biomarkers of obatoclax sensitivity common to both LUAD and SCLC (Fig. 3D; Supplementary Table S5). Here we find that many of the predictive biomarkers of sensitivity are histology-specific, while others, including increased gene expression of the transcription factor ASCL1 (21), are common between the two (SCLC: FC = 6.4, p = 0.04; LUAD: FC = 1.4, p = 0.05). We also observed a trend toward increased expression of BCL-2 protein predicting sensitivity in both histologies (SCLC: FC = 4.6, p = 0.051; LUAD: FC = 1.67, p = 0.08) consistent with others’ observations (7,24). Examining tumor-specific biomarkers in SCLC, we observed decreased MYC expression as a significant predictor of sensitivity to both obatoclax (FC = −5.3, p = 0.03) and another BCL-2 inhibitor selected by DISARM in this analysis, TW-37 (FC −5.2, p = 0.02). In opposition to the prior biomarker analysis for AURKA inhibitors in SCLC wherein low ASCL1 and high MYC predicted sensitivity (Fig. 2D), these data suggest that BCL-2 inhibitors target a unique subset of SCLC with contrasting (i.e. ASCL1-high, MYC-low) molecular determinants. This is consistent with prior observations regarding ASCL1 and BCL-2 inhibitors(7) and tracks with the observation from Fig. 2B that largely non-overlapping subsets of platinum-resistant SCLC respond to AURKA inhibitors and BCL-2 inhibitors.
Figure 3. DISARM identifies common candidates for cisplatin-resistant SCLC and LUAD.
(A-B) Maps illustrating common and unique drugs (A) and drug targets (B) selected as top candidates by DISARM for SCLC and LUAD using IC50 data from GDSC. (C) Examples of DISARM-generated 2×2 plots for obatoclax, selected by DISARM for both cisplatin-resistant SCLC and LUAD. IC50 values are reported as μM. (D) Venn diagram illustrating numbers of unique and common mRNA-based predictive biomarkers of obatoclax sensitivity between SCLC and LUAD, including increased expression of the transcription factor ASCL1 (See also Table S5).
DISARM reveals shared vulnerabilities across platinum-resistant solid tumor models
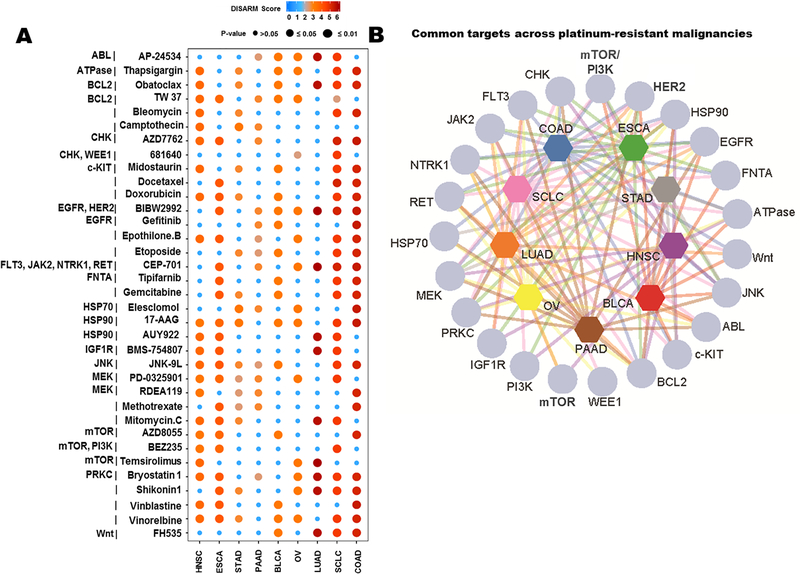
In an effort to extend this analysis beyond lung cancers, we selected nine tumor types for which a platinum-based therapy is an established first-line therapy according to NCCN guidelines (14,26–33), and for which adequate drug response data were available in GDSC. These included SCLC (n=44 cell lines), LUAD (n=39), stomach adeno-(STAD; n=17), pancreatic adeno-(PAAD; n = 15), ovarian (OV; n = 19), head and neck squamous cell (HNSC; n=19), esophageal (ESCA; n = 21), colon adeno-(COAD; n = 34) and bladder carcinoma (BLCA; n = 17). There were 138 total drugs with adequate IC50 data for each of these nine tumor types available in GDSC. DISARM’s analyses revealed unique (Supplementary Tables S6-S14) and common (Fig. 4A and B) drugs and targets across cisplatin-resistant models of different tumor types. Multiple DISARM-selected candidates were agents commonly paired with platinum as SOC therapy. For example, in SCLC, etoposide, which is partnered with cisplatin in SOC frontline therapy for SCLC, is selected by DISARM as a candidate (27,34). Similarly, gemcitabine, selected by DISARM as a candidate for BLCA, is paired with cisplatin as a standard frontline regimen for locally advanced or metastatic BLCA (26,35), while the alternative standard frontline regimen pairs cisplatin with three drugs - methotrexate, vinblastine and doxorubicin (as MVAC), the latter two also having been selected by DISARM in this analysis. Meanwhile, numerous other DISARM-selected candidates are acceptable therapies for the tumor type in question, often following prior platinum treatment, including vinorelbine and docetaxel for SCLC (27) and docetaxel for ESCA (32). DISARM’s demonstration that these agents are effective despite cisplatin resistance is consistent with clinical observations for cisplatin combination and post-cisplatin treatment.
Figure 4. DISARM identifies common candidates across cisplatin-resistant solid tumors.
(A) Plot depicting drugs selected by DISARM as candidates in cisplatin-resistant models from at least two of the nine malignancies analyzed via IC50 data from GDSC. Dot colors and sizes indicate DISARM scores and statistical significance. (B) Map illustrating interrelatedness of targets identified in multiple tumor types in this same analysis.
Selection as a candidate therapy by DISARM implies that a drug is effective in cisplatin-resistant models, but DISARM’s analyses also permit a determination as to whether any of these candidates are more effective in cisplatin-resistant models. By comparing mean IC50 values for cell lines in quadrants A and C (cisplatin-sensitive) to quadrants B and D (cisplatin-resistant) for each DISARM candidate from Fig. 4A across each malignancy, we were able to identify a number of agents with lower mean IC50 values in cisplatin-resistant models (Supplementary Fig. S3). While superiority in cisplatin-resistant versus cisplatin-sensitive models varies by malignancy for many agents, some patterns do emerge. For example, several conventional chemotherapeutic agents seem to consistently perform better in cisplatin-sensitive models, including vinblastine and etoposide for every malignancy in which DISARM selected these agents as candidates. Meanwhile, bryostatin-1, a protein kinase-C (PKC) inhibitor, has a superior mean IC50 value in six different malignancies, consistent with previous data suggesting that depletion of PKC can restore cisplatin sensitivity(36).
DISARM’s analyses also revealed several targeted therapies with potentially novel utility across platinum-resistant malignancies including multiple inhibitors of PI3K, mTOR, BCL-2, HSP90, and MEK (Fig. 4A and B). Agents targeting MEK were common candidates throughout this analysis, as seven of the nine tumor types analyzed had at least one MEK inhibitor selected by DISARM. In several cases, numerous MEK inhibitors were selected by DISARM for the same tumor type, including two each for HNSC and STAD, and four (of only 18 total post-filtering candidates selected) for PAAD. Independent analyses further supported a role for the MAPK (Raf-MEK-ERK) pathway in mediating cisplatin resistance. We correlated the IC50 of cisplatin for many of our SCLC cell lines with total and phosphorylated proteins quantified by our lab using reverse-phase protein array (RPPA) (Supplementary Fig. S4A). Using Spearman correlation, we found that MAPK pathway effectors were significantly higher in cisplatin resistant cell lines, as compared to platinum sensitive lines (Supplementary Fig. S4A and B). Specifically, high expression of pS217 MEK1/2 (p=0.01), pT202_Y204 MAPK (p=0.008), pT180 p38 (p=0.003), and p90RSK (p=0.03) (Supplementary Fig. S4B) were all correlated with cisplatin resistance. While these cisplatin-resistant cell line models are derived from a range of treatment naïve and treatment refractory patients, we can directly investigate whether MAPK signaling is upregulated during the acquisition of cisplatin resistance. To do this, we used human SCLC H69 cells (cisplatin IC50 < 1 μM), which were originally derived from a treatment naïve SCLC patient, and treated with increasing doses of cisplatin until cells developed resistance to cisplatin (H69/CR (cisplatin resistant); cisplatin IC50 >10μM) (37). Similar to the previous analysis, comparisons of RPPA-based protein expression profiles between the H69 (parental) and H69/CR lines revealed a nearly two-fold increase in expression of pT202_Y204 ERK (p=0.008; FC = 1.95) in the platinum-resistant lines as compared to the platinum-sensitive parental cell line (Supplementary Fig. S4C). These data suggest increased expression and activation of MAPK components including MEK are characteristic of cisplatin resistance in SCLC and that inhibition of MEK may be a viable treatment strategy for overcoming resistance to cisplatin.
Experimental validation of DISARM candidates
All nine tumor types from GDSC analyzed by DISARM had at least one BCL-2 inhibitor selected, while eight of nine had at least one HSP90 inhibitor selected. The most commonly selected candidates from each class were obatoclax and 17-AAG (tanespimycin), which were identified in six and seven tumor types, respectively, including SCLC. To assess obatoclax we identified two cell lines not included in the previous GDSC obatoclax analysis (H1672 and H841) that were expected to be cisplatin resistant with the expectation that obatoclax may be effective in these models. As predicted, we found that both lines were sensitive to obatoclax and resistant to cisplatin according to DISARM’s classification (Supplementary Fig. S5A-C). For 17-AAG, we again selected two cell lines predicted to have cisplatin resistance (H1436 and H2227) that did not appear in our prior GDSC 17-AAG analysis and similarly found that these cell lines were sensitive to 17-AAG but resistant to cisplatin per DISARM (Supplementary Fig. S5D-F).
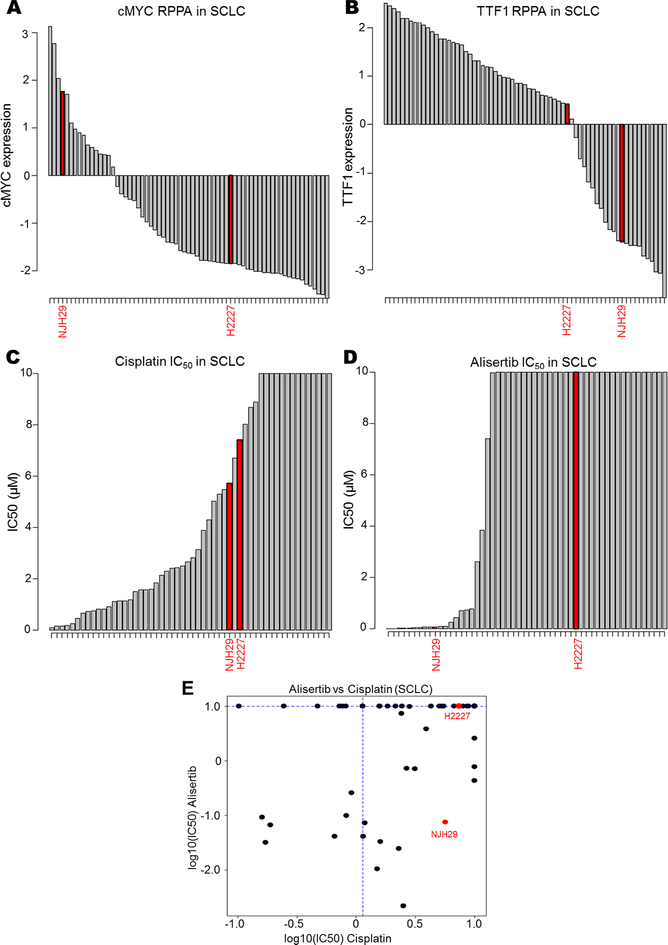
DISARM’s analysis of SCLC (Fig. 2A-B, D) identified AURKA inhibitors as a candidate drug class in cisplatin resistant disease. To validate this prediction, we utilized previously published IC50 data for alisertib (24), an AURKA inhibitor not included in our previous analysis to generate a DISARM 2×2 plot for alisertib. In light of the prediction that AURKA inhibitors would be effective in cisplatin-resistant models with high cMYC and low TTF1 expression, we selected two additional models beyond the available data set for experimental validation – NJH29, which is cisplatin-resistant, cMYC-high, and TTF1-low (Fig. 5A-B), and H2227, which is cisplatin-resistant, cMYC-low, and TTF1-high (Fig. 5A-B). Cell proliferation assays determined IC50 values for alisertib in each model. While we confirmed that both models are cisplatin-resistant, we find that NJH29 is alisertib sensitive, while H2227 is alisertib resistant, as predicted by our biomarker analysis (Fig. 5C-E). Together, these experimental observations support DISARM’s capacity for predicting effective therapies using reference-drug sensitivity as a selection criteria, as well as more conventional biomarker-driven approaches.
Figure 5. DISARM accurately predicts biomarkers for AURKA sensitivity.
(A,B) Bar graphs depicting RPPA-based expression of cMYC and TTF1 for NJH29 and H2227 relative to other SCLC cell lines. Bar graphs illustrating experimentally observed IC50 values for NJH29 and H2227 for cisplatin (C) and alisertib (D) relative to other SCLC cell lines along with classification according to DISARM 2×2 plot (E).
DISARM Web Tool
In order to make DISARM available to the research community to explore other reference drugs, cancer types and datasets, we have designed a JavaScript-based web tool that allows users to utilize DISARM interactively according to their specific needs (Supplementary Figs. S1 and S2). This tool allows the users to select an applicable database for query, assign a reference drug (i.e. Drug X) of their choice and select either quantile-based or specific IC50 value cut-offs for sensitivity and resistance of their reference drugs and all candidate drugs. For example, an investigator interested in PAAD could query the DISARM application for a list of active drugs in the setting of resistance to gemcitabine, one of the SOC frontline chemotherapy agents for PAAD. The investigator is then provided with a searchable and sortable list of candidate drugs from the queried database, with multiple sortable options for prioritization including p-values and DISARM scores. Bonferroni correction is applied on the resulting p-values in order to correct for multiple hypotheses testing. Selection of a candidate drug of interest yields an interactive scatter plot and 2×2 table for each reference-candidate drug pair. The tool is packaged into a Docker container to facilitate sharing and deployment across multiple operating systems. DISARM web-based tool is available at http://ibl.mdanderson.org/DISARM/index.html and requires Mozilla Firefox or Google Chrome browser for access. Note that any discoveries made using DISARM software package or web-based tool will belong solely to the users, although we do request that users reference the application
DISCUSSION
In this study, we introduce a new computational tool that can identify candidate drugs with activity in resistant cancer models. As a proof of concept, we first show that DISARM accurately identifies targeted treatments (X and Y) with clinically established activity in tumors with resistance to erlotinib and dasatinib. Osimertinib and ponatinib are rarities in current clinical practice – oncology drugs with clear clinical indications following resistance to SOC therapy and with known predictive biomarkers of response. In contrast, there is a critical, unmet need for therapeutic strategies that overcome or delay resistance to platinum chemotherapy, the most widely prescribed chemotherapeutic drug class (38). In its analyses of multiple databases and tumor histologies vis-à-vis cisplatin resistance, DISARM identified multiple agents and classes, including several targets previously implicated in mediating platinum resistance. For example, PI3K pathway activation and BCL-2 overexpression are observed in platinum-resistant ovarian cancer (39–41), while cisplatin resistance in ovarian and bladder cancer cells can be overcome via the addition of HSP90 inhibitors (42,43). MEK inhibitors are an especially intriguing result from this analysis as targeting MEK has a well-characterized role in overcoming treatment resistance in the case of BRAF inhibitors in BRAF mutant melanoma and lung cancer (44–46), ultimately leading to FDA approval of trametinib (melanoma; NSCLC) and cobimetinib (melanoma) for these indications. Previous data have also supported MAPK pathway activation as a mediator of cisplatin resistance in HNSC – resistance that was successfully reversed with MEK inhibition (47).
While these cases offer some validation to DISARM’s selections, further expanding the repertoire of active agents is critical for clinical progress, as eradication, or even long-term control, of tumors using systemic treatment typically requires multiple drugs, with multiple mechanisms of action and resistance. Unfortunately, in most cancers, countless trials combining SOC chemotherapy with targeted therapy have failed to yield significant improvements over SOC. Should effective treatment combinations already exist among agents previously assessed, there is currently no efficient way of identifying them. Although current drug databases largely contain single drug response data, DISARM may help to identify rational combinations for further exploration and biomarker-selected populations where they may have the greatest activity. To determine which of the 526 drugs in the existing dataset (7) would best complement cisplatin in SCLC requires 526 combination treatment experiments, while DISARM, as applied here, offers an alternative 31 experiment shortcut by selecting the most probable candidates. Our analyses demonstrate that DISARM can serve to initiate and facilitate rapid progress from therapeutic impasse to rational, biomarker-driven combinatorial or sequential treatment paradigms for all cancer types. For example, DISARM’s analysis suggests the use of PI3K and AURKA inhibitors specifically in TTF-1 negative SCLC. As TTF-1 testing is already ubiquitous for lung tumors, subsequent preclinical validation of TTF-1 negative status as a predictor of PI3K and AURKA inhibitor sensitivity could immediately be translated into clinical use without repeat biopsies or the optimization of new biomarker assays. Furthermore, DISARM’s approach with respect to cisplatin resistance and PI3K and AURKA inhibitors demonstrated its utility for elucidate novel molecular subtypes. In these cases, we were able to uncover several uncommon SCLC subtypes that were until recently, largely obscure, including overlapping ASCL1-low, TTF1-low, and cMYC-high subtypes (21,23,24), all of which had been previously shown to possess platinum resistance and possess unique targets relative to the dominant ASCL1-high, cMYC-low subtypes. The latter subtype, in our analysis as in others’ (7), appears targetable by BCL-2 inhibitors such as obatoclax and TW-37. The success of our approach to resolve even rare subtypes suggests that a DISARM-initiated approach, wherein a researcher begins with a subset of cells with defined drug response and then works in reverse through molecular analysis may be an efficient way to dissect molecular subtypes.
Our results for cisplatin resistance across multiple solid tumors reveal many alternative drug candidates with common predictive biomarkers. While there exists considerable disagreement in the literature regarding the reproducibility of in vitro drug data between multiple data sets (48,49), raising concerns about the reproducibility of DISARM’s results when applied to the same reference drug across multiple data sets, the observation that similar agents and drug classes emerge as candidates from DISARM’s analysis of multiple data sets is reassuring. These common results imply that platinum resistance may be a biological state (or, more accurately, series of states) shared across multiple histologies. In other words, resistance to platinum may predict alternative, active therapeutic options independent of tumor histology. The DISARM approach does possess all of the potential limitations of analyses that focus exclusively on in vitro data, as well as those associated with excessive reliance on IC50 data as a measure of drug efficacy However, several DISARM-selected candidates present opportunities for clinical validation of our in vitro-derived results. For example, in BLCA, doxorubicin and vinblastine, which constitute two of the three drugs added to cisplatin in the frontline SOC regimen MVAC, were identified as top candidates in cisplatin-resistant disease by DISARM. Previous prospective trial data found that RR in advanced urothelial cancer patients was more than three-fold higher (39% and 12%) with MVAC than with cisplatin alone, along with significant improvements in progression-free survival (PFS) and overall survival (OS) (50). Another DISARM-selected candidate in BLCA, gemcitabine, when paired with cisplatin, represents the other SOC in advanced urothelial cancer, in which gemcitabine/cisplatin offers comparable RR, PFS, and OS to MVAC(35). While at this time DISARM is intended as an efficient method to narrow a researcher’s initially broad net and to streamline subsequent, more sophisticated analyses in vitro and in vivo,ultimately, DISARM could be adapted to in vivo or even human clinical data, substituting tumor measurements or PFS for multiple lines of therapy in place of IC50. With appropriate validation, it is realistic to imagine a clinical trial designed not around a specific histology but instead targeting patients with multiple tumor types who share resistance to a common reference therapy.
Supplementary Material
Statement of Translational Relevance.
Resistance to standard-of-care therapy is a fundamental challenge of cancer care. Following resistance to many of the most commonly prescribed chemotherapies and targeted therapies, selection of subsequent lines of therapy is rarely optimized to overcome this resistance. Differential Sensitivity Analysis for Resistant Malignancies (DISARM) is a computational tool specifically designed to address the issue of optimizing the selection of therapy following resistance to a drug of interest. Using DISARM, we identified several agents that retain efficacy, or even have improved efficacy, following the development of cisplatin resistance, including inhibitors of AURKA, PI3K, BCL-2, and HSP90. DISARM’s analyses allow us to identify both previously validated and novel biomarkers that predict improved response to DSIARM-identified candidates relative to cisplatin, predictions that we independently validate. DISARM offers a high-throughput approach to optimizing preclinical and clinical experimental design to approach resistance to any drug of interest.
Acknowledgments and Grant Support
This work was supported by: NIH/NCI T32 CA-009666 (CMG) and Conquer Cancer Foundation of ASCO/Lung Cancer Alliance Young Investigator Award (CMG); NIH/NCI 1-R01-CA207295 (LAB); the MDACC CCSG (P30CA016672); the LUNGevity Foundation (LAB); the MD Anderson Physician Scientist Award (LAB); Rexanna’s Foundation for Fighting Lung Cancer (LAB; JVH); the David Bruton Jr. Chairmanship in Cancer Research (JVH); The Sheikh Khalifa Bin Zayed Al Nahyan Institute for the Personalized Cancer Therapy’s (IPCT’s) Center for Professional Education and Training (LAB), and by generous philanthropic contributions to The University of Texas MD Anderson Moon Shots Program (LAB; JVH).
Footnotes
Disclosure of Potential Conflicts of Interest
Dr. Heymach reports consulting/advisory board relationships with AstraZeneca, Boehringer Ingelheim, Bristol-Myers Squibb, Medivation, ARIAD, Synta, Oncomed, Novartis, Genentech, Calithera Biosciences, in addition to research funding from AstraZeneca. Dr. Byers reports consulting/advisory board relationships with AstraZeneca and Abbvie, in addition to research funding from AbbVie, AstraZeneca, and Tolero Pharmaceuticals. The remaining authors have no relevant conflicts of interest to report.
REFERENCES
- 1.Xu J, Murphy SL, Kochanek KD, Arias E. Mortality in the United States, 2015. NCHS Data Brief 2016(267):1–8. [PubMed] [Google Scholar]
- 2.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a Cancer Dependency Map. Cell 2017;170(3):564–76 e16 doi 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonald ER 3rd, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR, et al. Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell 2017;170(3):577–92 e10 doi 10.1016/j.cell.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Bester AC, Lee JD, Chavez A, Lee YR, Nachmani D, Vora S, et al. An Integrated Genome-wide CRISPRa Approach to Functionalize lncRNAs in Drug Resistance. Cell 2018;173(3):649–64 e20 doi 10.1016/j.cell.2018.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keenan AB, Jenkins SL, Jagodnik KM, Koplev S, He E, Torre D, et al. The Library of Integrated Network-Based Cellular Signatures NIH Program: System-Level Cataloging of Human Cells Response to Perturbations. Cell Syst 2018;6(1):13–24 doi 10.1016/j.cels.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gainor JF, Varghese AM, Ou SH, Kabraji S, Awad MM, Katayama R, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res 2013;19(15):4273–81 doi 10.1158/1078-0432.CCR-13-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polley E, Kunkel M, Evans D, Silvers T, Delosh R, Laudeman J, et al. Small Cell Lung Cancer Screen of Oncology Drugs, Investigational Agents, and Gene and microRNA Expression. J Natl Cancer Inst 2016;108(10) doi 10.1093/jnci/djw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016;166(3):740–54 doi 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byers LA, Wang J, Nilsson MB, Fujimoto J, Saintigny P, Yordy J, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov 2012;2(9):798–811 doi 10.1158/2159-8290.CD-12-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics 2003;19(10):1236–42. [DOI] [PubMed] [Google Scholar]
- 11.Johnson FM, Saigal B, Talpaz M, Donato NJ. Dasatinib (BMS-354825) tyrosine kinase inhibitor suppresses invasion and induces cell cycle arrest and apoptosis of head and neck squamous cell carcinoma and non-small cell lung cancer cells. Clin Cancer Res 2005;11(19 Pt 1):6924–32 doi 10.1158/1078-0432.CCR-05-0757. [DOI] [PubMed] [Google Scholar]
- 12.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483(7391):603–7 doi 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong P, Coombes KR, Johnson FM, Byers LA, Diao L, Liu DD, et al. drexplorer: A tool to explore dose-response relationships and drug-drug interactions. Bioinformatics 2015;31(10):1692–4 doi 10.1093/bioinformatics/btv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman J, Chirieac LR, et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2017;15(4):504–35. [DOI] [PubMed] [Google Scholar]
- 15.Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov 2014;4(9):1046–61 doi 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cassuto O, Dufies M, Jacquel A, Robert G, Ginet C, Dubois A, et al. All tyrosine kinase inhibitor-resistant chronic myelogenous cells are highly sensitive to ponatinib. Oncotarget 2012;3(12):1557–65 doi 10.18632/oncotarget.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byers LA, Rudin CM. Small cell lung cancer: where do we go from here? Cancer 2015;121(5):664–72 doi 10.1002/cncr.29098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allison Stewart C, Tong P, Cardnell RJ, Sen T, Li L, Gay CM, et al. Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget 2017;8(17):28575–87 doi 10.18632/oncotarget.15338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardnell RJ, Feng Y, Mukherjee S, Diao L, Tong P, Stewart CA, et al. Activation of the PI3K/mTOR Pathway following PARP Inhibition in Small Cell Lung Cancer. PLoS One 2016;11(4):e0152584 doi 10.1371/journal.pone.0152584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misch D, Blum T, Boch C, Weiss T, Crolow C, Griff S, et al. Value of thyroid transcription factor (TTF)-1 for diagnosis and prognosis of patients with locally advanced or metastatic small cell lung cancer. Diagn Pathol 2015;10:21 doi 10.1186/s13000-015-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, et al. ASCL1 and NEUROD1 Reveal Heterogeneity in Pulmonary Neuroendocrine Tumors and Regulate Distinct Genetic Programs. Cell Rep 2016;16(5):1259–72 doi 10.1016/j.celrep.2016.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sos ML, Dietlein F, Peifer M, Schottle J, Balke-Want H, Muller C, et al. A framework for identification of actionable cancer genome dependencies in small cell lung cancer. Proc Natl Acad Sci U S A 2012;109(42):17034–9 doi 10.1073/pnas.1207310109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mollaoglu G, Guthrie MR, Bohm S, Bragelmann J, Can I, Ballieu PM, et al. MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell 2017;31(2):270–85 doi 10.1016/j.ccell.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardnell RJ, Li L, Sen T, Bara R, Tong P, Fujimoto J, et al. Protein expression of TTF1 and cMYC define distinct molecular subgroups of small cell lung cancer with unique vulnerabilities to aurora kinase inhibition, DLL3 targeting, and other targeted therapies. Oncotarget 2017;8(43):73419–32 doi 10.18632/oncotarget.20621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2013;41(Database issue):D955–61 doi 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark PE, Spiess PE, Agarwal N, Bangs R, Boorjian SA, Buyyounouski MK, et al. NCCN Guidelines Insights: Bladder Cancer, Version 2.2016. J Natl Compr Canc Netw 2016;14(10):1213–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalemkerian GP, Akerley W, Bogner P, Borghaei H, Chow LQ, Downey RJ, et al. Small cell lung cancer. J Natl Compr Canc Netw 2013;11(1):78–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ajani JA, D’Amico TA, Almhanna K, Bentrem DJ, Chao J, Das P, et al. Gastric Cancer, Version 3.2016, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2016;14(10):1286–312. [DOI] [PubMed] [Google Scholar]
- 29.Tempero MA, Malafa MP, Behrman SW, Benson AB, 3rd, Casper ES, Chiorean EG, et al. Pancreatic adenocarcinoma, version 2.2014: featured updates to the NCCN guidelines. J Natl Compr Canc Netw 2014;12(8):1083–93. [DOI] [PubMed] [Google Scholar]
- 30.Morgan RJ Jr., Armstrong DK, Alvarez RD, Bakkum-Gamez JN, Behbakht K, Chen LM, et al. Ovarian Cancer, Version 1.2016, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2016;14(9):1134–63. [DOI] [PubMed] [Google Scholar]
- 31.Pfister DG, Spencer S, Brizel DM, Burtness B, Busse PM, Caudell JJ, et al. Head and Neck Cancers, Version 1.2015. J Natl Compr Canc Netw 2015;13(7):847–55; quiz 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ajani JA, D’Amico TA, Almhanna K, Bentrem DJ, Besh S, Chao J, et al. Esophageal and esophagogastric junction cancers, version 1.2015. J Natl Compr Canc Netw 2015;13(2):194–227. [DOI] [PubMed] [Google Scholar]
- 33.Benson AB 3rd, Venook AP, Cederquist L, Chan E, Chen YJ, Cooper HS, et al. Colon Cancer, Version 1.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2017;15(3):370–98. [DOI] [PubMed] [Google Scholar]
- 34.Evans WK, Shepherd FA, Feld R, Osoba D, Dang P, Deboer G. VP-16 and cisplatin as first-line therapy for small-cell lung cancer. J Clin Oncol 1985;3(11):1471–7 doi 10.1200/JCO.1985.3.11.1471. [DOI] [PubMed] [Google Scholar]
- 35.von der Maase H, Hansen SW, Roberts JT, Dogliotti L, Oliver T, Moore MJ, et al. Gemcitabine and cisplatin versus methotrexate, vinblastine, doxorubicin, and cisplatin in advanced or metastatic bladder cancer: results of a large, randomized, multinational, multicenter, phase III study. J Clin Oncol 2000;18(17):3068–77 doi 10.1200/JCO.2000.18.17.3068. [DOI] [PubMed] [Google Scholar]
- 36.Isonishi S, Ohkawa K, Tanaka T, Howell SB. Depletion of protein kinase C (PKC) by 12-O-tetradecanoylphorbol-13-acetate (TPA) enhances platinum drug sensitivity in human ovarian carcinoma cells. Br J Cancer 2000;82(1):34–8 doi 10.1054/bjoc.1999.0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sen T, Tong P, Stewart CA, Cristea S, Valliani A, Shames DS, et al. CHK1 Inhibition in Small-Cell Lung Cancer Produces Single-Agent Activity in Biomarker-Defined Disease Subsets and Combination Activity with Cisplatin or Olaparib. Cancer Res 2017;77(14):3870–84 doi 10.1158/0008-5472.CAN-16-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McWhinney SR, Goldberg RM, McLeod HL. Platinum neurotoxicity pharmacogenetics. Mol Cancer Ther 2009;8(1):10–6 doi 10.1158/1535-7163.MCT-08-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee S, Choi EJ, Jin C, Kim DH. Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA amplification contributes to cisplatin resistance in an ovarian cancer cell line. Gynecol Oncol 2005;97(1):26–34 doi 10.1016/j.ygyno.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 40.Cai Y, Tan X, Liu J, Shen Y, Wu D, Ren M, et al. Inhibition of PI3K/Akt/mTOR signaling pathway enhances the sensitivity of the SKOV3/DDP ovarian cancer cell line to cisplatin in vitro. Chin J Cancer Res 2014;26(5):564–72 doi 10.3978/j.issn.1000-9604.2014.08.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai Y, Jin S, Li X, Wang D. The involvement of Bcl-2 family proteins in AKT-regulated cell survival in cisplatin resistant epithelial ovarian cancer. Oncotarget 2017;8(1):1354–68 doi 10.18632/oncotarget.13817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Xie Z, Sun G, Yang P, Li J, Yang H, et al. Reversing drug resistance of cisplatin by hsp90 inhibitors in human ovarian cancer cells. Int J Clin Exp Med 2015;8(5):6687–701. [PMC free article] [PubMed] [Google Scholar]
- 43.Tatokoro M, Koga F, Yoshida S, Kawakami S, Fujii Y, Neckers L, et al. Potential role of Hsp90 inhibitors in overcoming cisplatin resistance of bladder cancer-initiating cells. Int J Cancer 2012;131(4):987–96 doi 10.1002/ijc.26475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012;367(18):1694–703 doi 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med 2014;371(20):1867–76 doi 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 46.Planchard D, Besse B, Groen HJM, Souquet PJ, Quoix E, Baik CS, et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol 2016;17(7):984–93 doi 10.1016/S1470-2045(16)30146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong LR, Chua KN, Sim WJ, Ng HC, Bi C, Ho J, et al. MEK Inhibition Overcomes Cisplatin Resistance Conferred by SOS/MAPK Pathway Activation in Squamous Cell Carcinoma. Mol Cancer Ther 2015;14(7):1750–60 doi 10.1158/1535-7163.MCT-15-0062. [DOI] [PubMed] [Google Scholar]
- 48.Haverty PM, Lin E, Tan J, Yu Y, Lam B, Lianoglou S, et al. Reproducible pharmacogenomic profiling of cancer cell line panels. Nature 2016;533(7603):333–7 doi 10.1038/nature17987. [DOI] [PubMed] [Google Scholar]
- 49.Rao JS, Liu H. Discordancy Partitioning for Validating Potentially Inconsistent Pharmacogenomic Studies. Sci Rep 2017;7(1):15169 doi 10.1038/s41598-017-15590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loehrer PJ Sr., Einhorn LH, Elson PJ, Crawford ED, Kuebler P, Tannock I, et al. A randomized comparison of cisplatin alone or in combination with methotrexate, vinblastine, and doxorubicin in patients with metastatic urothelial carcinoma: a cooperative group study. J Clin Oncol 1992;10(7):1066–73 doi 10.1200/JCO.1992.10.7.1066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.