Summary
In addition to phosphodiester bond formation, RNA polymerase II has an RNA endonuclease activity, stimulated by TFIIS, which rescues complexes that have arrested and backtracked. How TFIIS affects transcription under normal conditions is poorly understood. We identified backtracking sites in human cells using a dominant-negative TFIIS (TFIISDN) that inhibits RNA cleavage and stabilizes backtracked complexes. Backtracking is most frequent within 2 kb of start sites, consistent with slow elongation early in transcription, and in 3’ flanking regions where termination is enhanced by TFIISDN, suggesting that backtracked pol II is a favorable substrate for termination. Rescue from backtracking by RNA cleavage also promotes escape from 5’ pause sites, prevents premature termination of long transcripts, and enhances activation of stress-inducible genes. TFIISDN slowed elongation rates genome-wide by half suggesting that rescue of backtracked pol II by TFIIS is a major stimulus of elongation under normal conditions.
Graphical Abstract
RNA polymerase II can backtrack and resume transcription after cleavage of the nascent RNA transcript. Sheridan et al show that in vivo human pol II backtracks frequently and that RNA cleavage enhances gene activation, 5’ pause release, normal termination and rapid transcription elongation.
Introduction
Both initiation of pol II transcription and post-initiation steps in the transcription cycle: 5’ pausing and release, elongation and termination can influence the control of gene expression. The best understood post-initiation control mechanism is exerted at the promoter-proximal pause site to regulate the flux of pol II into productive elongation within gene bodies (Jonkers and Lis, 2015; Mayer et al., 2017; Nechaev and Adelman, 2011). Following transit past the 5’ pause, transcription elongation within genes proceeds at rates that differ by at least an order of magnitude between genes (Fuchs et al., 2014; Jonkers et al., 2014). For reasons that are not understood, elongation starts out slowly and then accelerates within genes (Danko et al., 2013; Jonkers et al., 2014). This variation in elongation rate is of physiological significance because co-transcriptional processes including pre-mRNA processing and chromatin modification are kinetically coupled to ongoing transcription (Bentley, 2014; Naftelberg et al., 2015). Furthermore sites of termination at the ends of genes are determined by elongation rate as a result of kinetic competition between pol II and the termination factor Xrn2 (Fong et al., 2015; Hazelbaker et al., 2013; Proudfoot, 2016). The mechanisms that determine elongation rates within genes are poorly understood, but one potential rate-limiting event is pol II arrest and backtracking that can occur at positions where the enzyme mis-incorporates a base, or runs into a roadblock like a DNA binding factor or a nucleosome (Bondarenko et al., 2006; Churchman and Weissman, 2011; James et al., 2016; Thomas et al., 1998).
In vitro, the transcription factor TFIIS rescues pol II elongation complexes that have backtracked by stimulating the RNA endonuclease activity of the polymerase, which cleaves the nascent RNA to produce a new 3’ end situated in the active site (Izban and Luse, 1992; Reines, 1992; Thomas et al., 1998). TFIIS-stimulated endonucleolytic cleavage releases RNA fragments 2 – 14 bases long (Izban and Luse, 1993). In vitro, TFIIS does not affect the pause-free velocity of yeast or human pol II and stimulates elongation rate entirely by reducing the length of pauses (Ishibashi et al., 2014; Zhang and Burton, 2004). In yeast, TFIIS (Dst1) is normally dispensable but is essential when GMP synthesis is inhibited (Exinger and Lacroute, 1992; Gomez-Herreros et al., 2012; Malik et al., 2017; Shaw and Reines, 2000). In Δdst1 cells, over-expression of a dominant-negative TFIIS mutant (TFIISDN) that inhibits RNA cleavage is lethal, implying an important function of this activity even when NTP levels are unperturbed (Sigurdsson et al., 2010). TFIISDN has mutations in two conserved residues at the tip of domain III (D190A, E191A), that projects through the side channel of the polymerase and interacts with the active site (Cheung and Cramer, 2011; Jeon et al., 1994; Kettenberger et al., 2003; Kettenberger et al., 2004). These two acidic residues form part of a conserved motif, QTRSADEP that interacts with the pol II trigger loop and bridge helix and is identical in yeast and mammalian TFIIS (Fig. S1A)(Kettenberger et al., 2003). The DE residues in this motif coordinate a Mg2+ ion, (metal ion B) and a nucleophilic water molecule, that participate in the RNA cleavage reaction (Cheung and Cramer, 2011; Kettenberger et al., 2003). Orthologous acidic residues in bacterial GreA/GreB serve the same function in stimulating transcript cleavage by RNA polymerase (Sosunova et al., 2003). In the nucleotide addition reaction, metal ion B is coordinated in an alternate position by the incoming NTP (Cramer et al., 2001; Sosunov et al., 2003). Yeast TFIISDN binding affinity for pol II is similar to wild type TFIIS and like WT TFIIS (Ishibashi et al., 2014; Zhang and Burton, 2004), it does not affect the pause-free rate of nucleotide addition (Sigurdsson et al., 2010), suggesting that its dominant-negative effect is exerted specifically through inhibition of RNA endonuclease activity.
Pol II backtracking sites have been mapped genome-wide in budding yeast by comparing nascent RNA 3’ ends in WT and Δdst1 cells. This study showed that backtracking is ubiquitous and quite uniformly distributed throughout genes (Churchman and Weissman, 2011). TFIIS deletion has little effect on yeast under normal growth conditions and it remains unclear whether resolution of backtracked complexes has a widespread influence on elongation rates or gene activation. In vitro TFIIS facilitates elongation of pol II stalled at the Drosophila Hsp70 promoter, and in vivo it stimulates Hsp70 activation and pol II recruitment to the promoter (Adelman et al., 2005). Backtracking has been directly detected in Drosophila at the +1 nucleosome (Weber et al., 2014) and near promoter proximal pause sites (Nechaev et al., 2010). It has also been inferred within 3’ flanking regions in S. pombe where TFIIS delays transcription termination (Lemay et al., 2014), and in human cells at termination sites that occur near sequence elements predicted to destabilize the DNA-RNA hybrid (Schwalb et al., 2016).
The importance of resolving backtracked pol II complexes for the transition to productive elongation at 5’ pause sites in vivo, and for subsequent elongation and termination remains unclear, in part because sites of backtracking have not been mapped in a multicellular organism. It remains to be determined if backtracking occurs uniformly throughout metazoan genes or is more common at particular positions like 5’ pause sites or termination sites. Here we used an inducible TFIIS dominant negative mutant in human cells to identify sites of backtracking genome-wide, and to investigate the role of RNA cleavage in pol II transcription under normal and stressed conditions. The results demonstrate that backtracking is ubiquitous and has widespread effects on release of paused pol II, elongation rate within genes, completion of long transcripts, termination, and gene activation.
Results
Pol II RNA cleavage activity promotes escape from 5’ pause sites
To investigate the importance of the RNA cleavage activity that rescues backtracked pol II, we generated a HEK293 Flp-in cell line that inducibly over-expresses the mouse TFIISDN mutant with substitutions (D282A, E283A) homologous to those that inhibit cleavage activity in yeast (Fig. S1A, B) (Sigurdsson et al., 2010). The mouse and human homologues are 96.7% identical (Fig. S1A). Under the conditions of our experiments (24 hr induction) expression of TFIISDN did not detectably affect viability or proliferation as shown by flow cytometry (Fig. S1C). This result is consistent with the viability of yeast co-expressing wild type and mutant TFIIS (Sigurdsson et al., 2010). Because endogenous WT TFIIS will compete with TFIISDN for binding to pol II in our cells, the effects of the mutant will reflect a partial inhibition of RNA cleavage activity.
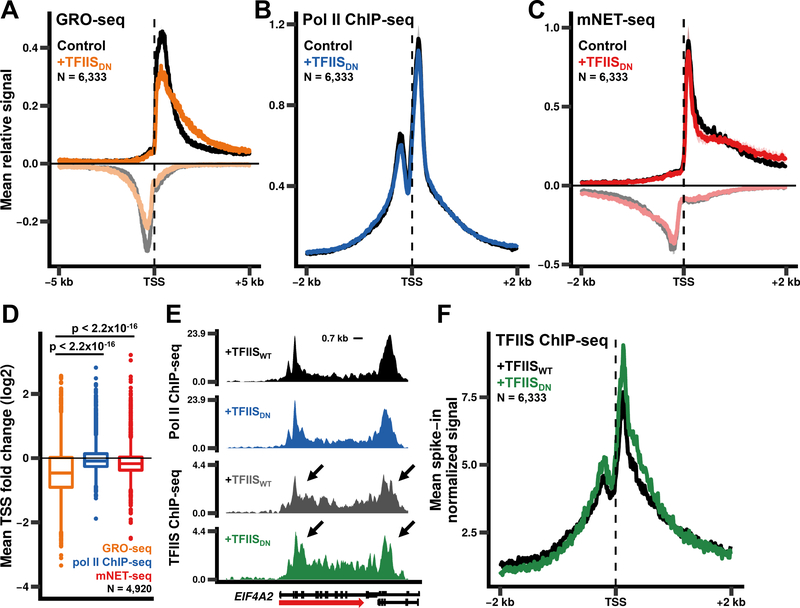
To establish whether rescue of backtracked complexes is indeed inhibited by TFIISDN in vivo, we performed global run-on sequencing (GRO-seq) (Core et al., 2008), anti-pol II ChIPseq, and nascent RNA 3’ end mapping (mNET-seq) (Nojima et al., 2016) before and after induction of the mutant protein. Only polymerases that can actively elongate in the run-on reaction will incorporate Br-UTP and produce GRO-seq signal, therefore if TFIISDN stabilizes backtracked pol II, it is predicted to reduce the GRO-seq signal relative to the total amount of pol II engaged on the gene. These experiments showed that expression of TFIISDN specifically reduced the GRO-seq signal at promoter proximal regions (Fig. 1A, D) with relatively little effect on total pol II occupancy and transcriptionally engaged pol II as measured by ChIP-seq and mNET-seq, respectively (Fig. 1B-D). These results were observed in each biological replicate (Fig. S2, S3A-E) and together show that TFIISDN prevents promoter proximal pol II from resuming elongation in the run-on assay, strongly suggesting that in vivo the mutant stabilizes backtracked complexes by preventing RNA cleavage as it does in vitro (Sigurdsson et al., 2010). Most importantly, the results in Figure 1 show that pol II commonly backtracks within 300 bp of the TSS and that cleavage activity helps maintain 5’ paused pol II in an elongationcompetent state.
Figure 1. Pol II RNA cleavage activity promotes escape from 5’ pause sites.
(A)Expression of TFIISDN results in genome-wide loss of GRO-seq signal from promoter proximal regions. Metaplots of relative GRO-seq signals (10 bp bins) for genes >5 kb long and separated by >2 kb. The relative signal for each gene was calculated by dividing the signal in each bin by the total signal within the region plotted. The mean relative signal was calculated for each individual replicate. The mean relative signal was then averaged for the replicates with the shaded region representing the standard error of the means (SEM) for two biological replicates. Negative values correspond to anti-sense signal.
(B)Metaplots of relative anti-pol II ChIP-seq signals as in A. The mean and SEM for three biological replicates is shown.
(C)Metaplots of relative anti-pol II mNET-seq signals as in A. The mean and SEM for two biological replicates is shown.
(D)Fold change in GRO-seq, pol II ChIP-seq, and mNET-seq signals for the region from the TSS to +300 bp. The average fold change for two (GRO-seq, mNET-seq) or three (pol II ChIPseq) biological replicates is shown for genes >1 kb long and separated by >2 kb. p-values were calculated using the Welch two sample t-test with Bonferroni-Holm correction.
(E)TFIIS localizes to 5’ and 3’ ends of genes. Avi-TFIISWT and Avi-TFIISDN ChIP-seq signals for EIF4A2 normalized to a yeast spike-in (see Methods).
(F)WT TFIIS and TFIISDN associate with pol II at similar levels. Metaplots of Avi-TFIIS ChIPseq signals (10 bp bins) as in E.
The reduced GRO-seq signal at 5’ pause sites caused by TFIISDN suggests a direct effect on paused pol II complexes. We tested this idea by ChIP mapping of wild type and dominant negative mutant TFIIS expressed in human breast cancer cells (Fig. S4A). This experiment showed that TFIIS strongly localizes to 5’ pause sites on human genes (Fig. 1E, F) consistent with previous work in mouse ES cells (Carrière et al., 2012). Quantitation of ChIP-seq signals relative to a spike-in control showed that WT and mutant TFIIS were recruited approximately equally well to 5’ pause sites when measured relative to pol II at these regions (Fig. 1E, S4B-E).
Unexpectedly the stabilization of backtracked pol II at promoter proximal pause sites did not detectably increase pol II occupancy in this region as measured by pol II ChIP-seq and mNET-seq (Fig. 1B-D, S3B-D) possibly because of turnover of backtracked complexes by premature termination that has been reported at 5’ pause sites (Erickson et al., 2018; Steurer et al., 2018). We also noted that TFIISDN elevated relative GRO-seq signals in the region between +1 and +3 kb possibly reflecting a problem in the maturation of pol II complexes into a fully elongation competent state (Fig. 1A, S3A).
Pol II cleavage activity antagonizes termination at gene 3’ ends
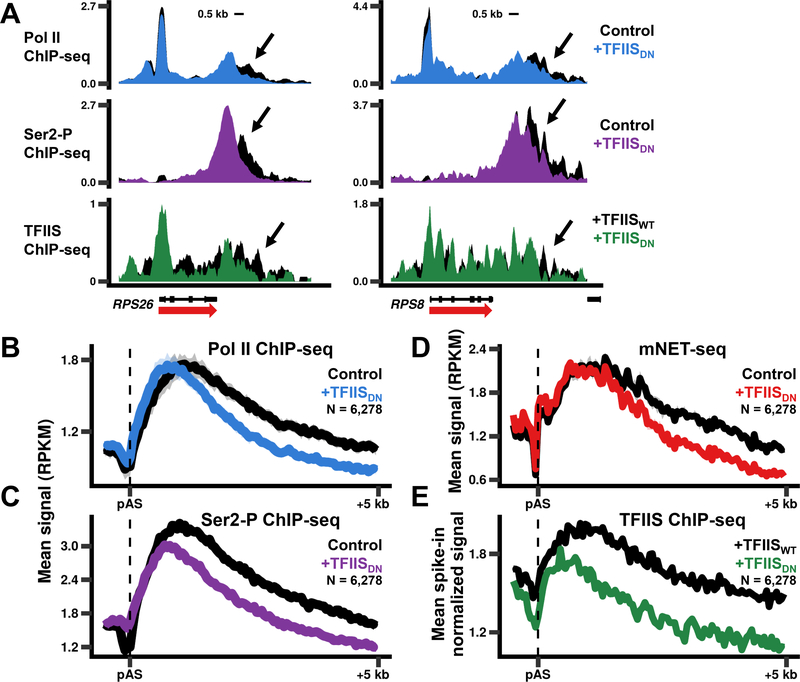
In addition to promoter proximal pausing which occurs within a short window downstream of the TSS, pol II undergoes a second major pause within a much larger window downstream of the poly(A) site preceding transcription termination (Anamika et al., 2012; Glover-Cutter et al., 2008). Poly(A) site cleavage exposes a 5’-PO4 that serves as a substrate for the 5’−3’ exonuclease Xrn2, which “torpedoes” the polymerase and promotes termination (Connelly and Manley, 1988; Kim et al., 2004; Proudfoot, 2016; West et al., 2004). Inhibition of cleavage activity by TFIISDN caused a reproducible upstream shift in both total pol II and the CTD Ser2 phosphorylated isoform associated with the 3’ pause (Fig. 2A-C, S3G). A shift of roughly equal magnitude was also observed for mNET-seq signal downstream of the poly(A) site indicating that transcription terminated earlier (Fig. 2D, S3F). Relief of backtracking at 3’ pause sites by TFIIS-activated RNA cleavage is consistent with the observation that TFIIS accumulates with pol II downstream of poly(A) sites (Fig. 1E, 2A, E, S4B-D, F). We conclude that inhibition of pol II cleavage activity impairs elongation in the 3’ flanking region and permits Xrn2 to catch and evict pol II closer to the poly(A) site.
Figure 2. Pol II cleavage activity antagonizes termination at 3’ ends of genes.
(A)Expression of TFIISDN causes early transcription termination. Note the early drop in Pol II, Ser2-P, and TFIIS ChIP signals at gene 3’ ends when TFIISDN is expressed.
(B)Metaplots of anti-pol II ChIP-seq signals near poly(A) sites (pAS) (50 bp bins) for genes >1 kb long and separated by >5 kb. The mean signal was calculated for each individual replicate. The replicates were then averaged with the shaded region representing the SEM for three replicates. RPKM, reads per kb per million mapped reads.
(C) Metaplots of anti-CTD Ser2-P pol II signals as in B for one replicate.
(D)Metaplots of mNET-seq signals as in B for two replicates.
(E) Metaplots of WT TFIIS and TFIISDN ChIP-seq signals (50 bp bins) normalized to a yeast spike-in as in Fig. 1F for genes shown in B.
Pol II cleavage activity is required to maintain rapid elongation within genes
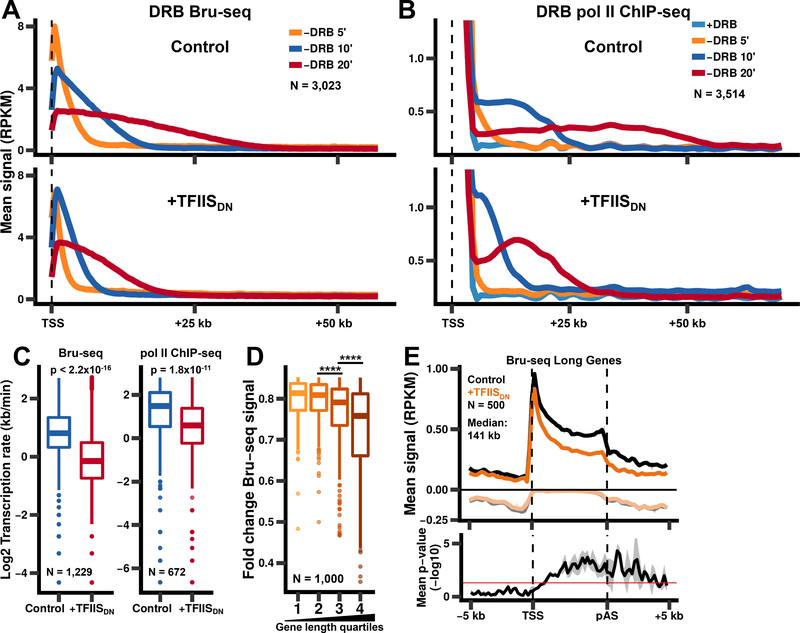
The results above suggest that resolution of backtracked pol II complexes by the pol II endonuclease is important for pol II release from 5’ pause sites and proper termination at 3’ ends. We next investigated whether RNA cleavage also influences productive elongation within gene bodies. Elongation rates were measured using the reversible Cdk9 inhibitor, 5,6dichlorobenzimidazone-1-β-D-ribofurano-side (DRB), which arrests pol II at the 5’ ends of genes while permitting it to clear from gene bodies. Removal of DRB permits the 5’ paused polymerases to be released into the gene body in a synchronous “wave” (Singh and Padgett, 2009). We monitored the pol II wave after DRB washout by sequencing nascent RNA pulselabeled with bromouridine (Bru-seq) (Veloso et al., 2014) and by anti-pol II ChIP-seq (Fig. 3A, B). By these two methods elongation rates were estimated for ~600–1,200 genes over 75 kb long. In control cells we observed a median elongation rate of ~2–3 kb/min (Fig. 3C), in agreement with previous reports (Jonkers and Lis, 2015). Remarkably, expression of TFIISDN reduced the median elongation rate to only ~1–1.5 kb/min (Fig. 3C). This reduction was observed for both proximal and distal regions of genes (Fig. S5A). We conclude that the RNA cleavage activity of pol II is a major positive effector of elongation rate through gene bodies.
Figure 3. Pol II cleavage activity is required for rapid elongation through the gene body.
(A) Inhibition of pol II cleavage activity reduces the rate of transcription elongation. Metaplots of Bru-seq signals (500 bp bins) after washout of DRB for non-overlapping genes >75 kb long.
(B) Metaplots of pol II ChIP-seq signals after washout of DRB as in A.
(C)Transcription rates were calculated using Bru-seq data shown in A (left) or pol II ChIP-seq data shown in B (right) by comparing the 10 min and 20 min timepoints for genes where waves were detected for both timepoints. p-values were calculated using the Welch two sample t-test.
(D)Transcription of long genes is more sensitive to inhibition of pol II RNA cleavage activity. The fold change in Bru-seq signal was plotted for the top 1,000 genes that show reduced Bru-seq signal after expression of TFIISDN, divided into quartiles based on gene length. Fold changes were calculated from two biological replicates for genes >1 kb long and separated by >2 kb. pvalues were calculated using the Welch two sample t-test with Bonferroni-Holm correction. **** p < 0.0001.
(E)Metaplots of Bru-seq signals for genes from the 3rd and 4th quartiles from E (N = 500, median length: 141 kb). The mean signal and SEM was plotted as in Fig. 2A for two biological replicates. Negative values correspond to anti-sense signal. p-values (bottom panel) were calculated by comparing the sense Bru-seq signals for uninduced and TFIISDN-expressing cells for each replicate using the Welch two sample t-test. p-values were then averaged across the two biological replicates with the shaded region representing the SEM.
If RNA cleavage is crucial for normal elongation within genes, one might expect that long genes would be most affected by TFIISDN since they have more potential backtrack sites. To study this question, we measured nascent RNA synthesis by Bru-seq and identified 1,000 genes with the greatest reduction in signal after induction of TFIISDN. Notably within this group, the longest genes showed the greatest decrease in nascent RNA levels after inhibition of pol II cleavage activity (Fig. 3D). Furthermore, the most significant reductions in nascent RNA abundance caused by TFIISDN occurred at the 3’ ends of long genes for both biological replicates, with little effect at 5’ ends (Fig. 3E, S5B, C) or in genes of average length (Fig. S5D). These observations suggest that cleavage activity is important not only to maintain rapid elongation rates but also to permit completion of long transcripts, presumably by preventing premature termination.
Pol II pauses and backtracks most frequently during early elongation and in the termination zone
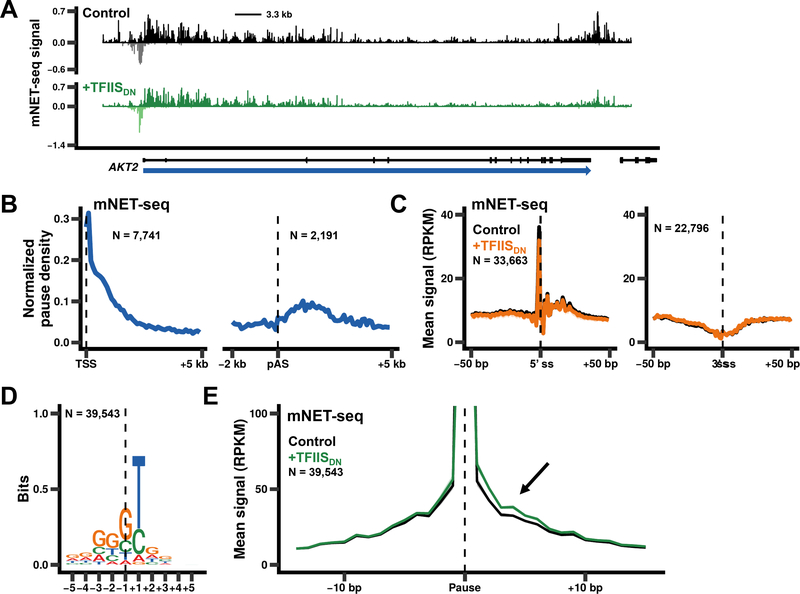
The widespread transcriptional defects caused by TFIISDN at early and late stages of the transcription cycle suggest that backtracking is a relatively ubiquitous event even in unstressed human cells. To determine exactly where backtracking occurs, we analyzed 3’ ends of nascent transcripts in immunopurified pol II complexes by mNET-seq (Churchman and Weissman, 2011; Nojima et al., 2015). Cross-correlation analysis, demonstrated a good correlation between replicate mNET-seq data sets at the single basepair level (Fig. S6A). A two-step procedure was employed to map positions of backtracking: 1) locate pause sites defined as positions with a mNET-seq signal at least three standard deviations above the mean for a 200 bp window and 2) identify those pause sites where inhibition of RNA cleavage by TFIISDN caused an accumulation of extended nascent 3’ ends immediately downstream. Using step 1 of this scheme, we identified 179,605 pausing events occurring between the TSS and +5 kb downstream of the poly(A) site in control cells that were common between two biological replicates. Notably there is specific enrichment of pauses within 2 kb of the TSS and in the termination zone downstream of poly(A) sites after normalizing for total mNET-seq signal (Fig. 4A, B).
Figure 4. Pol II pausing is most frequent near the 5’ ends and termination zones.
(A) mNET-seq signals are shown for AKT2. Note the high frequency of pauses that occur close to the 5’ end and downstream of the poly(A) site. Negative values correspond to anti-sense signal.
(B) Metaplots (100 bp bins) of pause density (pauses / bp) normalized to total mNET-seq signal. Pauses were identified as positions where there were >5 mapped reads and NET-seq signal rose three standard deviations above the mean for the surrounding 200 bp. Pauses shared between two control datasets (179,605 pauses) were used to calculate the pause density for the region downstream of the TSS for non-overlapping genes >5 kb long (left panel, N = 7,741 genes) and for the region around the poly(A) site (pAS) for genes >2 kb long and separated by >5 kb (right panel, N = 2,191 genes).
(C) Pausing is not enriched at splice sites. Metaplots of mNET-seq signals around 5’ and 3’ splice sites (ss, 1 bp bins) for sites contained within non-overlapping genes >1 kb long. The peak at the 5’ss corresponds to step 1 intermediates associated with pol II. The mean signal and SEM was plotted for two biological replicates as in Fig. 2A.
(D) The consensus sequence observed for pause sites present in both control datasets, separated by >30 bp, and containing signal within 15 bp of the pause (N = 39,543/179,605 pauses). Dotted line marks the 3’ end of nascent transcripts.
(E) Expression of TFIISDN causes an extension of nascent RNA 3’ ends associated with pause sites. Metaplots of mNET-seq signals (1 bp bins) around pause sites shown in D. The mean signal and SEM was plotted for two biological replicates as in Fig. 2A. Note the downstream shift in mNET-seq signal after inhibition of pol II cleavage activity.
We did not detect elevated pol II pausing near 5’ or 3’ splice sites, but consistent with cotranscriptional splicing, abundant RNA 3’ ends were reproducibly detected at splice donor sites corresponding to step 1 intermediates associated with pol II (Fig. 4C, S6B, left panels). In contrast, excised lariats which would appear as mNET-seq reads at splice acceptor sites were not present in any of our datasets implying they are not stably associated with pol II (Fig. 4C, S6B, right panels).
Analysis of the sequences flanking each pause site revealed a strong preference for a T at the +1 position on the coding strand immediately downstream of the nascent RNA 3’ end and a weaker preference for G’s at the −1, −2 and −3 positions (Fig. 4D) in both WT and TFIISDN datasets (Fig. S6C, D). This bias was not observed in GRO-seq libraries made using the same method of library preparation (Fig. S6E). A similar preference for T at +1 relative to pol II pause sites was also observed in yeast when TFIIS (Dst1) was deleted (Churchman and Weissman, 2011).
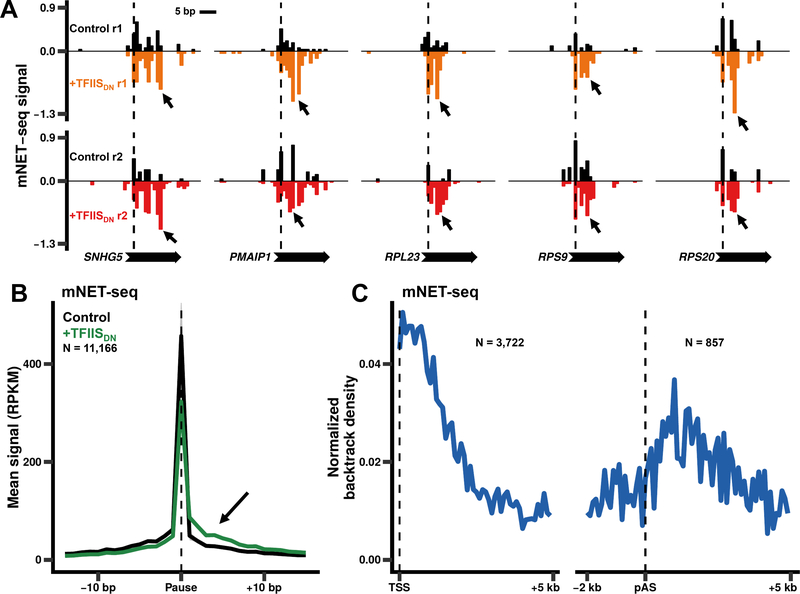
We next examined how inhibiting pol II cleavage activity affects the 3’ ends of nascent transcripts at pause sites. Notably, there is a small but reproducible increase in mNET-seq signal within 15 bp downstream of these pauses when TFIISDN was expressed (Fig. 4E, S7A) indicating that impaired RNA cleavage causes a lengthening of nascent RNA 3’ ends as expected for backtrack sites. To quantify this effect we calculated a “backtracking index”, which is the difference in NET-seq signal between the regions 15 bp downstream and upstream of the pause, for each pause separated by at least 30 bp where there is a signal in at least one of the regions (N = 39,543/179,605 pauses). At approximately 30% of these well-separated pauses (11,166/39,543), TFIISDN increased the backtracking index in both replicate datasets, and we identified these as high confidence backtracking sites (Fig. 5A, B, S7B). This is a minimum estimate of the fraction of pauses associated with backtracking and it therefore shows that backtracking occurs at a substantial number of pause sites throughout the length of genes and downstream of poly(A) sites. Like pause sites, backtracking sites are also enriched for a T at the +1 position just downstream of the RNA 3’ end (Fig. S7C). Backtracking sites are widely distributed, but after normalizing for total mNET-seq signal they are most highly enriched within 2 kb of the TSS and in the termination zone downstream of poly(A) sites (Fig. 5C). These data provide the first global picture of pol II backtracking in a metazoan and suggest that RNA cleavage by pol II is required to counteract backtracking at all post-initiation stages of the transcription cycle, but especially during early elongation and in the 3’ flanking region prior to termination.
Figure 5. Pol II backtracking occurs throughout the transcription cycle.
(A) Examples of pauses identified as high confidence backtracking sites in two biological replicates (r1, r2). The dotted line indicates the position of the pause site identified in the control datasets. Note the increase in RNA 3’ ends downstream of the pauses after expression of TFIISDN. Values represent reads per million mapped reads.
(B) Metaplots of mNET-seq signals (1 bp bins) around pauses identified as high confidence backtracking sites (N = 11,166/39,543 pauses). The mean signal and SEM was plotted for two biological replicates as in Fig. 2A.
(C) Backtracking is most frequent near 5’ ends and downstream of poly(A) sites. Metaplots (100 bp bins) of backtrack density (sites / bp) normalized to total mNET-seq signal as in Fig. 4B. High confidence backtrack sites shown in B (11,166/39,543 pauses) were used to calculate the density for the regions downstream of the TSS (left panel, N = 3,722 genes) and around the poly(A) site (right panel, N = 857 genes).
Pol II cleavage activity is required for rapid activation of stress-inducible genes
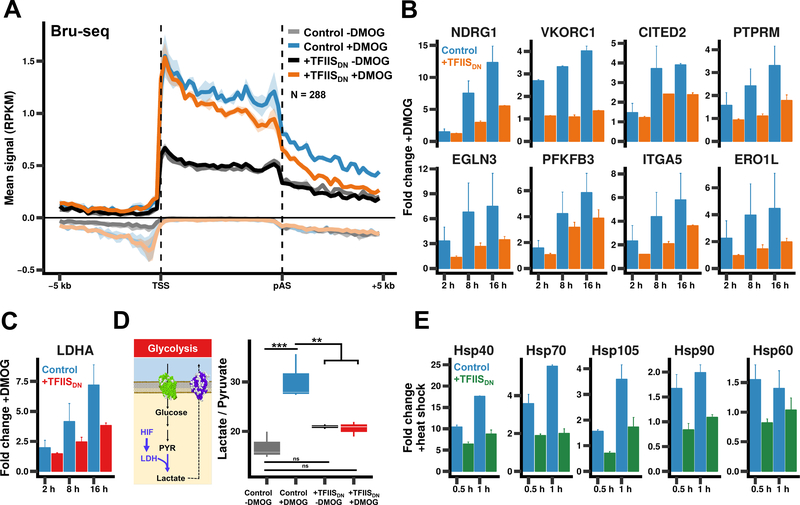
In light of how pol II cleavage activity affects pause release and elongation rate, we asked whether it influences gene activation. To investigate this question, we tested how TFIISDN impacts the upregulation of hypoxia inducible genes in response to dimethyloxalylglycine (DMOG), which stabilizes hypoxia inducible factor (HIF) (Semenza, 2011). TFIISDN expression prior to DMOG treatment substantially reduced nascent RNA synthesis at hypoxia inducible genes in two biological replicates as measured by Bru-seq (Fig. 6A, S8A, B) as well as accumulation of mature hypoxia-inducible mRNAs as measured by qRT-PCR (Fig. 6B), but had only minor effects on basal mRNA expression levels (Fig. S8C). TFIISDN did not reduce transcription of HIF1A or its main binding partner, ARNT (Fig. S8D).
Figure 6. Pol II cleavage activity is required to rapidly activate gene expression.
(A) TFIISDN impairs transcriptional activation of hypoxia inducible genes in response to DMOG. Metaplots of pulse-labelled nascent RNA-seq (Bru-seq) signals for cells treated +/−2 mM DMOG for 16 hours. Data were plotted for genes with increased signal +DMOG (p-value < 0.01). The mean signal and SEM was plotted for two biological replicates as in Fig. 2A. Negative values correspond to anti-sense signal.
(B) qRT-PCR showing the fold change in mRNA abundance for hypoxia-responsive transcripts in cells treated with 2 mM DMOG for indicated times. Error bars represent the SEM for at least two biological replicates.
(C) Lactate/pyruvate ratios were determined by UHPLC-MS in uninduced and TFIISDNexpressing cells +/−2 mM DMOG for 16 hrs. ** p < 0.01, *** p < 0.001 ANOVA.
(D) qRT-PCR showing the fold change in LDHA mRNA as in B.
(E) qRT-PCR showing the fold change in heat shock mRNA after incubation at 42° as in B.
To determine if the reduced transcriptional response to DMOG caused by TFIISDN is physiologically significant, we assayed the metabolic response of the cells by ultra high-pressure liquid chromatography-mass spectrometry (UHPLC-MS). The hypoxic response involves elevated conversion of pyruvate to lactate by the HIF inducible enzyme lactate dehydrogenase encoded by the LDHA gene (Firth et al., 1995). Expression of TFIISDN prior to DMOG treatment reduced the accumulation of LDHA transcripts (Fig. 6C) and importantly decreased lactate/pyruvate ratios relative to control cells (Fig. 6D). The results therefore suggest that rescue of backtracked pol II complexes is required for cells to mount a normal metabolic response to hypoxic stress.
We also tested how inhibition of pol II cleavage activity alters the heat shock response. Expression of TFIISDN prior to heat shock at 42° significantly reduced the accumulation of heat shock responsive transcripts, but only had minor effects on basal mRNA levels (Fig. 6E, S8E). Consistent with a role in activation of stress-inducible genes, TFIIS can be detected with paused pol II at the 5’ ends of these genes prior to induction (Fig. S4C, D). Overall the results in Figure 6 show that the ability of cells to rescue backtracked polymerases is important for rapid gene activation in response to stress stimuli.
Discussion
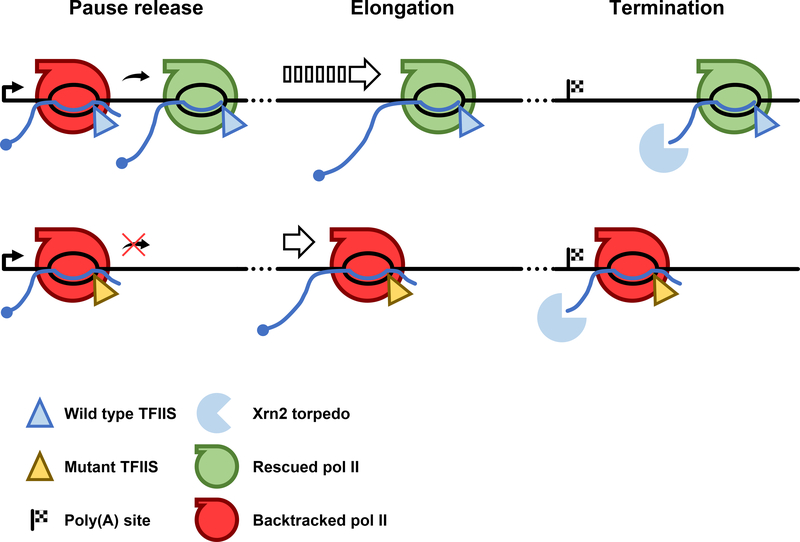
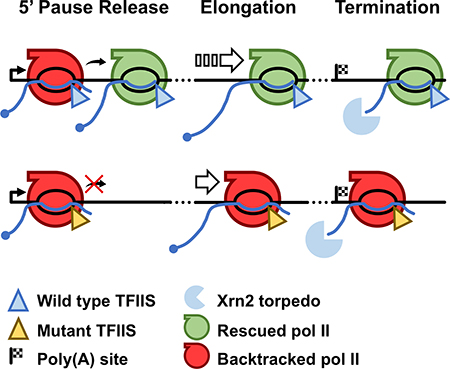
In this report, we used a dominant-negative mutant TFIIS that inhibits the RNA cleavage activity of pol II (Sigurdsson et al., 2010) to identify where pol II backtracks on human genes. The results revealed important effects of RNA cleavage-mediated rescue of backtracked complexes at multiple stages of the transcription cycle (Fig. 7). Inhibition of RNA cleavage by TFIISDN increased production of 3’ extended nascent RNAs within 15 bp downstream of pause sites (Fig. 4E, S7A) as predicted for backtracked complexes. mNET-seq revealed approximately 180,000 pause sites and 11,000 high confidence backtracking sites (Fig. 4, 5). Compared to yeast genes (Churchman and Weissman, 2011), which are relatively short, backtracking is less uniformly distributed along human genes which have defined regions of high pausing and backtracking frequency within 2 kb downstream of the TSS and in the termination zone downstream of poly(A) sites (Fig. 4A, B, 5C). We did not detect elevated pausing in the vicinity of splice sites, however. The functional significance of elevated backtracking at 5’ and 3’ ends of genes is supported by our finding that TFIISDN altered both escape from the 5’ pause and the position where termination occurs (Fig. 1A, 2, S3A, F, G).
Figure 7. Expression of TFIISDN causes widespread effects throughout the transcription cycle.
Model showing the effect TFIISDN expression has on the release of promoter proximally paused pol II, elongation through the gene, and transcription termination.
At 5’ ends, inhibition of RNA cleavage activity impaired the ability of 5’ paused polymerases to resume elongation in the run-on (GRO-seq) assay without inhibiting recruitment of pol II to 5’ ends as measured by anti-pol II ChIP-seq and mNET-seq (Fig. 1, S3). These results indicate that pol II cleavage activity is widely required to prepare promoter proximally paused pol II for release into the gene body by maintaining it in an elongation-competent state. This idea is supported by a recent structural study showing that NELF and TFIIS occupy overlapping sites within the polymerase, suggesting that NELF could promote 5’ pausing by competing with TFIIS and preventing the rescue of backtracked pol II (Vos et al., 2018).
Following release from the promoter proximal pause, elongation efficiency improves over the following 2 kb as indicated by gradually reduced levels of pausing and backtracking (Fig. 4B, 5C). The high frequency of pausing and backtracking within the first part of the gene body provides a likely explanation for the finding that transcription elongation is slower within more proximal gene regions than further downstream (Danko et al., 2013; Jonkers et al., 2014). This effect could be due to a local property of the template in the first part of the gene that promotes pausing and backtracking. Alternatively, the pol II transcription complex may become intrinsically more resistant to pausing and backtracking later during elongation as a result of an unknown maturation process possibly caused by modification of pol II or associated factors. For example it is possible that backtracking is suppressed by changes in phosphorylation of the CTD (Buratowski, 2009; Heidemann et al., 2013) or the elongation factor Spt5 (Bourgeois et al., 2002; Yamada et al., 2006) that occur as pol II traverses the first part of the gene.
Pausing and backtracking as well as TFIIS occupancy are elevated downstream of poly(A) sites and furthermore, stabilization of backtracked pol II by TFIISDN shifted the termination zone more proximally (Fig. 2, 4B, 5C). Backtracking is therefore common at 3’ termination sites where it facilitates early termination. The position where termination occurs is dictated by a kinetic competition between two enzymes moving 5’−3’; the Xrn2 RNA exonuclease “torpedo”, and the pol II molecule that it is chasing (Fong et al., 2015). The implication of proximal termination caused by TFIISDN is that backtracked pol II complexes are susceptible to Xrn2 mediated termination. This conclusion is consistent with previous work showing that an A/U-rich sequence element predicted to promote backtracking by forming an unstable RNA-DNA hybrid coincides with transcription termination (Schwalb et al., 2016). Early termination like that induced by TFIISDN is also associated with TFIIS deletion in S. pombe (Lemay et al., 2014). The susceptibility of backtracked pol II to termination is suggested by the fact that backtracking by more than one base induces a conformation with an unfolded trigger loop (Cheung and Cramer, 2011; Zhang et al., 2010) that may be more prone to opening of the clamp that stabilizes the ternary complex (Sekine et al., 2015). It is also possible that frequent backtracking near promoter proximal pause sites could sensitize pol II to premature termination leading to turnover of paused complexes (Erickson et al., 2018; Steurer et al., 2018). This idea is suggested by the unexpected observation that inhibition of elongation at the promoter proximal pause by TFIISDN did not increase pol II density there (Fig. 1B-D, S3B-D). Suppression of premature termination by TFIIS is also suggested by the fact that it is specifically required for completion of long transcripts as shown by the observation that TFIISDN preferentially inhibited transcription of the 3’ ends of the longest genes (Fig. 3D, E, S5B, C).
Remarkably inhibition of pol II cleavage activity by TFIISDN caused a ~50% reduction in the elongation rate within numerous genes (Fig. 3A-C, S5A). The implication of this finding is that even when the pol II complex is expected to be at its most productive within the body of genes, it still relies heavily on its RNA cleavage activity to move at its normal speed of ~2 kb/minute. These results suggest that TFIIS-stimulated rescue of backtracked pol II has a remarkably profound effect on average elongation rates throughout the genome under normal growth conditions. The slowing of elongation by mutant TFIIS is most likely caused by inhibition of the strong TFIIS-activated RNA cleavage activity of pol II, but inhibition of the weak intrinsic TFIIS-independent cleavage activity (Sigurdsson et al., 2010) might also contribute. We cannot completely eliminate the possibility that in vivo the TFIIS D282A, E283A double mutant slows elongation by an additional unknown mechanism but this scenario seems unlikely because: 1) the Metal ion B Mg2+ coordinated by D282 and E283 does not participate in the nucleotide addition reaction 2) neither WT yeast nor human TFIIS affects the pause-free rate of elongation in vitro (Ishibashi et al., 2014; Zhang and Burton, 2004) and 3) yeast TFIISDN does not affect the elongation reaction in vitro (Sigurdsson et al., 2010).
The major influence of TFIIS-stimulated RNA cleavage on elongation under normal conditions suggests that physiological modulation of TFIIS activity could regulate elongation rates. In this context it is of interest that Ccr4-Not and DSIF/NELF can positively and negatively regulate TFIIS activity in vitro (Dutta et al., 2015; Palangat et al., 2005) and NELF likely competes with TFIIS for a common binding site on pol II (Vos et al., 2018). In summary, the widespread effects of TFIISDN on multiple steps in the transcription cycle show that rescue of backtracked pol II complexes is essential for the success of most rounds of transcription in vivo under normal conditions rather than being a specialized adaptation to stress conditions. Our results also suggest that expression of TFIISDN may be of value more generally as a means of experimentally or therapeutically manipulating pol II elongation rate.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David Bentley (david.bentley@ucdenver.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human cell lines
Flp-In-293 TREX cells (Female, Invitrogen) expressing inducible dominant negative mouse TFIIS (TCEA1) (D282A, E283A) with a N-terminal Avitag (TFIISDN) were generated through integration of a pcDNA5/FRT/Avi-TFIISDN construct using Flp recombinase-mediated sitespecific recombination. The cells were maintained in DMEM supplemented with 10% FBS, 200 μg/mL hygromycin B, 6.5 μg/mL blasticidin, and penicillin/streptomycin. All experiments were performed after induction with 2 μg/mL doxycycline for 24 hr.
HMLE-RAS cells (Elenbaas et al., 2001) expressing doxycycline inducible mouse Avi-TFIISDN and Avi-TFIISWT were made by lentiviral infection and blasticidin selection.
METHOD DETAILS
Antibodies
Rabbit anti-TFIIS was raised against recombinant full-length mouse GST-tagged TFIIS and affinity purified. Mouse TFIIS is 96.7% identical to its human homolog and this antibody is therefore expected to react equally well with both. Rabbit anti-pol II pan CTD, rabbit antiCstF77, 8WG16 anti-pol II CTD, and B44 anti-BrdU have been described (Glover-Cutter et al., 2008; Gratzner, 1982; Thompson et al., 1990). Rabbit anti Avi-tag was purchased from GenScript (A00674). Rat anti-CTD Ser2-P (3E10) was purchased from Chromotek.
Immunoblotting
Immunoblots were developed with HRP conjugated swine anti-rabbit secondary antibody (DAKO, P0217) and ECL Plus (Perkin Elmer, NEL103E001EA).
Flow cytometry
HEK293 TFIISDN cells were collected + and - 24 hr induction with doxycycline (2 μg/ml) and were incubated in Krishan stain for 72 hours before performing flow cytometry using a Beckman Coulter Gallios 561. ModFit LT software (Verity Software House, Topsham, ME) was used for cell cycle analysis.
ChIP-seq
ChIP of human extracts has been described previously (Glover-Cutter et al., 2008). 1 mg of each sonicated lysate was pre-cleared with 30 μl protein A sepharose beads (Life technologies, 101042) for 1 hr at 4°. Pre-cleared lysates were mixed with 30 μl protein A sepharose beads (blocked with 1 mg/ml BSA and 0.5 mg/ml E. coli tRNA) and 15μl of rabbit anti-total pol II CTD serum, 5 μg anti-Avi tag antibody, or 25 μl rat anti-Ser2-PO4-CTD antibody. For Ser2-PO4 IPs, used 4.5 μl rabbit anti-rat IgG secondary (Jackson labs, 312–005-003).
Libraries were sequenced on an Illumina Hi-seq 4000 (1X50) or NovaSeq 6000 (2×150). Adapters were trimmed using cutadapt version 1.16 and reads were mapped to the hg19 UCSC human genome (Feb. 2009) or sacCer3 UCSC yeast genome (April 2011), where appropriate with Bowtie2 version 2.3.2. PCR duplicates were removed using Picard Tools version 2.18.7. Read coordinates were collapsed to a single basepair and centered assuming a fragment size of 180 bp.
For Figs 1E, F, and S4, 100 μg crosslinked S. cerevisiae lysate expressing Avi-tagged Rpb3 was mixed with 1 mg cross linked human lysate prior to sonication. Each lysate was divided into two IPs using either 15μl of rabbit anti-total pol II CTD serum or 5 ug anti-Avi-tag antibody (GenScript, A00674). For metaplots, read counts in each bin were divided by the size of the bin and the number of mapped sacCer3 spike-in reads. To calculate TFIIS/pol II ratios for each gene, the normalized Avi-tag ChIP-seq signal was divided by the normalized pol II ChIPseq signal for each bin. For bins that lacked pol II ChIP-seq signal, a pseudo count (smallest pol II ChIP-seq value for the gene / 2) was used. The TFIIS/pol II ratios for each bin were then averaged across the plotted genes. The 5’ metaplots included genes >2 kb long, separated by >2 kb, containing at least one mapped read in the region, and were in common between all ChIP-seq datasets. The 3’ metaplots included genes >2 kb long, separated by >5 kb, containing at least one mapped read in the region, and were in common between all ChIP-seq datasets.
Bru-seq
Bru-seq was performed as described (Paulsen et al., 2014) with minor modifications. Cells were incubated with 2 mM Bromouridine for 30 min and labeled RNA was immunopurified with B44 monoclonal anti-BrdU (Gratzner, 1982). All libraries were generated using the KAPA biosciences stranded RNA-seq kit (KK8401) and sequenced on an Illumina Hi-Seq 4000 (1×50 or 1×150). After filtering out rRNA, reads were mapped to the hg19 UCSC human genome (Feb. 2009) with Bowtie2 version 2.3.2. Adapters were trimmed using cutadapt version 1.16 and PCR duplicates were removed using Picard Tools version 2.18.7. Read coordinates were collapsed to a single basepair and centered assuming a fragment size of 180 bp. For all metaplots, read counts were normalized by the size of the region and the library size. For metaplots in Fig. 3E, 6A, and S5B-D, signal was plotted for 200 bp bins for the regions from −5 kb to the TSS and from the pAS to +5 kb, and 30 bins of variable length for region from the TSS to the pAS. For Fig. 3D, fold changes in Bru-seq signal were calculated by comparing the number of reads mapping to each gene using DESeq2 version 1.10.1 (Love et al., 2014). For Fig. 6A, genes upregulated after treatment with 2 mM DMOG for 16 hrs were identified using DESeq2 version 1.10.1 (p < 0.01, N = 288).
Calculation of elongation rates
Cells were treated with 100 μM DRB for 3 hrs. For DRB Bru-seq experiments, the cells were washed twice with PBS and incubated in -DRB medium containing 2 mM bromouridine for 5, 10, or 20 min, before harvesting in with Trizol (Thermo Fisher). For DRB pol II ChIP-seq experiments (Fig. 2C, D), the cells were washed twice with PBS and incubated in -DRB media for 5, 10, or 20 min before crosslinking. The pol II wave was identified by first calculating the mean signal for the region from +70 kb to +75 kb downstream of the TSS, which we defined as background. We then identified the position where signal rose 8 standard deviations above the mean background signal. We only included genes where the front edge of the wave was called at a position downstream of the wave for the previous timepoint. For Bru-seq DRB washout metaplots, signal was plotted for 500bp bins for non-overlapping genes >75 kb long. For pol II ChIP-seq DRB washout metaplots, signal was plotted for 100 bp bins for the region from the TSS to +500 bp and 500 bp bins for the region from +500 bp to +60 kb for non-overlapping genes >75 kb long.
mNET-seq
mNET-seq was performed as previously described using the 8WG16 anti-pol II CTD antibody (Nojima et al., 2016). Each mNET-seq library was made from 8–12 15 cm dishes of cells grown to 80% confluency. Nuclei were extracted with NUN1 and NUN2 buffers as described (Nojima et al., 2016) and washed pellets were digested in 100 μl with 2 μl MNase (NEB M0247) for 2 min at 37° in a thermomixer. Reactions were stopped by adding10 μl 250 mM EGTA and pooled supernatants (400–600 μl) were diluted 10X with NET-2 buffer and immunoprecipitated with 400–600 μl antibody-conjugated protein A/G dynabeads (Invitrogen 10015D). Beads were washed 8x with 1 ml of NET-2 buffer. RNA was extracted with Trizol, treated with T4 PNK (3’ phosphatase minus, NET M0236) and DNaseI (NEB, M0303) and libraries were generated using the Lexogen small RNA-seq kit (052). After filtering out rRNA, reads were then mapped to the hg19 UCSC human genome (Feb. 2009) with Bowtie2 version 2.3.2. Adapters were trimmed using cutadapt version 1.16 and read coordinates were collapsed to a single basepair coordinate corresponding to the RNA 3’ end. PCR duplicates were filtered using a custom script that removes reads having the same 5’ and 3’ ends. 64–75% of reads were between 20 and 30 bases long and were assumed to correspond to pol II footprints. We allowed up to 5 copies of reads <30 bp in length to be kept, for reads >30 bp in length only a single copy of each read was kept.
Identification of pol II pause sites
Pol II pause sites were located using a custom script to identify positions where there were at least 5 mapped reads and mNET-seq signal was >3 standard deviations above the mean signal (not including pauses) for the surrounding 200 bp. This was accomplished by performing successive rounds of “searching” where the pause sites identified during the previous rounds were excluded from the calculations for the current round. This was performed until no additional pause sites were identified. Pauses that mapped to splice sites or snoRNAs were excluded from all additional analysis except for Fig. 4C. For analysis of signals and sequences surrounding pause sites (Fig. 4D, E, S6C, D, S7) and backtracking analysis (Fig. 5B, S7B) pauses were only included that were separated by >30 bp and contained signal within 15 bp upstream or downstream of the pause site. Pause density and backtrack density (Fig. 4B, 5C) were calculated by binning the sites into 100 bp windows and normalizing by window size.
Pause sites were only used that were shared between the two uninduced datasets (179,605 pauses) and that mapped between the TSS and +5 kb downstream of the poly(A) site. To normalize the site density, the mNET-seq signal within each bin for each gene was averaged for the biological replicates. The site density along each gene was then divided by the mNET-seq signal within the bin. For bins with no mapped mNET-seq reads, a pseudo count (smallest mNET-seq value for the gene / 2) was used. The normalized density was then averaged across non-overlapping genes >5 kb long for the region downstream of the TSS or it was averaged across genes >2 kb long and >5 kb separated for the region around the poly(A) site. Genes were only included that contained at least one pause or backtrack site in the region plotted. For Fig. 4D nucleotide frequency was calculated for the region +/−5 bp around the indicated sites and sequence logos were generated using the R package, ggseqlogo (https://github.com/omarwagih/ggseqlogo). For Fig. S6C, D, E, S7C, sequence logos were generated using WebLogo 3 (http://weblogo.threeplusone.com/create.cgi).
Cross correlation analysis
The mNET-seq signal at each bp was computed for each gene. R was then used to calculate the Pearson’s correlation coefficient for each gene after shifting one of the replicate datasets by the amount indicated on the x-axis. The correlation coefficients were then averaged over the entire gene list. Genes were only included if they contained at least one of the pause sites used in Fig. 4E.
GRO-seq
GRO-seq was performed as previously described with BrUTP labeling for 5 minutes (Core et al., 2008). Libraries were generated using the Lexogen small RNA-seq kit (052) and sequenced on an Illumina HiSeq 4000 (1×50 or 1×150). After filtering out rRNA, reads were mapped to the hg19 UCSC human genome (Feb. 2009) with Bowtie2 version 2.3.2. Adapters were trimmed using cutadapt version 1.16 and PCR duplicates were removed using Picard Tools version 2.18.7. Read coordinates were collapsed to a single basepair and centered assuming a fragment size of 80 bp.
Quantifying fold change in TSS signal
GRO-seq, pol II ChIP-seq, and mNET-seq TSS signal was calculated as the number of reads (normalized for library size) that mapped between the TSS to +300bp for all genes longer than 1 kb, separated by >2 kb, contained at least one read in the region, and were in common between all ChIP-seq, GRO-seq, and mNET-seq datasets. Fold changes were calculated after expression of TFIISDN for each biological replicate. The fold changes for the replicates were averaged in Fig. 1D.
5’ Metaplots
GRO-seq, pol II ChIP-seq, and mNET-seq 5’ metaplots from Fig. 1A-C, F and Fig. S3A-C include all genes longer than 5 kb, separated by >2 kb, containing at least one read in the region, and were in common between all ChIP-seq, GRO-seq and mNET-seq datasets. Read counts were normalized by the size of the bin. To calculate the relative signal for each gene, the signal for each bin was divided by the total signal within the region plotted. The mean relative signal was then calculated by averaging the relative signal across the genes plotted for each individual replicate. The replicates were then averaged with the shaded region representing the standard error of the means (SEM) for two (GRO-seq and mNET-seq) or three (pol II ChIP-seq) replicates.
3’ Metaplots
Pol II ChIP-seq and mNET-seq 3’ metaplots from Fig. 2 and Fig. S3F, G include all genes longer than 1 kb, separated by >5 kb, containing at least one read in the region and were in common between all ChIP-seq and mNET-seq datasets. Read counts for each bin were normalized by the size of the bin and the total number of mapped reads. The mean signal was calculated by averaging the normalized read counts across the genes plotted for each individual replicate. The replicates were then averaged with the shaded region representing the standard error of the means (SEM) for two (mNET-seq) or three (pol II ChIP-seq) replicates.
qRT-PCR
Quantitative RT-PCR was performed on 2–3 independent biological replicates of uninduced and TFIISDN-expressing cells (induced with 2 μg/ml doxycycline for 24 hrs) treated +/− 2mM DMOG for 2, 8, or 16 hrs or incubated at 42° for 30 min or 1 hr. Random primed cDNA was synthesized with SuperScript III (Invitrogen) and amplified using primers shown in Table S1. Fold change in mRNA abundance was calculated using the ΔΔCt method using β-Actin as a reference gene.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical details including the number of replicates (N) are provided in figure legends. Significance was defined by p-values determined by using the Welch t-test. When appropriate, pvalues were corrected for multiple testing using the Bonferroni-Holm method.
Supplementary Material
Dominant negative TFIIS stabilizes backtracked pol II throughout human genes
RNA cleavage by pol II is essential for rapid activation of stress-inducible genes
Rescue from backtracking is important for escape from promoter-proximal pause sites
Rescue from backtracking is a major stimulus of rapid transcriptional elongation
Acknowledgments
We thank T. Blumenthal, J. Hesselberth, S. Ramachandran, N. Mukherjee, R. Landick and members of our lab for helpful discussions and S. Bustos and R. Cohrs for help with extract sonication and K. Helm and the U. Colorado Cancer Center Support grant (P30CA046934) for flow cytometry. We thank K. Diener, B. Gao and the UC Denver Sequencing facility for sequencing. R.S is a scholar of the UC Denver RNA Bioscience Initiative. This work was supported by a Boettcher Webb-Waring early career award to A.D. and NIH grant R35GM118051 to D.B.
Footnotes
Declaration of interests
Authors declare no competing interests.
DATA AND SOFTWARE AVAILABILITY
ChIP-Seq, Gro-seq, mNET-seq, and Bru-seq datasets have been deposited in GEO under GSE120201. All custom software is available at https://github.com/sheridar/TFIISdn_paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, and Lis JT (2005). Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell 17, 103112. [DOI] [PubMed] [Google Scholar]
- Anamika K, Gyenis À, Poidevin L, Poch O, and Tora L (2012). RNA polymerase II pausing downstream of core histone genes is different from genes producing polyadenylated transcripts. PLoS ONE 7, e38769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL (2014). Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15, 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, and Studitsky VM (2006). Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell 24, 469–479. [DOI] [PubMed] [Google Scholar]
- Bourgeois C, Kim Y, Churcher M, West M, and Karn J (2002). Spt5 cooperates with Human Immunodeficiency virus Type 1 Tat by Preventing Premature Release at Terminator Sequences. Mol. Cell. Biol 22, 1079–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S (2009). Progression through the RNA polymerase II CTD cycle. Mol Cell 36, 541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrière L, Graziani S, Alibert O, Ghavi-Helm Y, Boussouar F, Humbertclaude H, Jounier S, Aude J-C, Keime C, Murvai J, et al. (2012). Genomic binding of Pol III transcription machinery and relationship with TFIIS transcription factor distribution in mouse embryonic stem cells. Nucleic Acids Research 40, 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AC, and Cramer P (2011). Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471, 249–253. [DOI] [PubMed] [Google Scholar]
- Churchman LS, and Weissman JS (2011). Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469, 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S, and Manley JL (1988). A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev 2, 440–452. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, and Lis JT (2008). Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, and Kornberg RD (2001). Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292, 1863–1876. [DOI] [PubMed] [Google Scholar]
- Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, and Kraus WL (2013). Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell 50, 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Babbarwal V, Fu J, Brunke-Reese D, Libert DM, Willis J, and Reese JC (2015). Ccr4Not and TFIIS Function Cooperatively To Rescue Arrested RNA Polymerase II. Mol Cell Biol 35, 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, Popescu NC, Hahn WC, and Weinberg RA (2001). Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev 15, 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson B, Sheridan RM, Cortazar MA, and Bentley DL (2018). Dynamic turnover of paused pol II complexes at human promoters. Genes Dev 32, 1215–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exinger F, and Lacroute F (1992). 6-azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Current Genetics 22, 9–11. [DOI] [PubMed] [Google Scholar]
- Firth JD, Ebert BL, and Ratcliffe PJ (1995). Hypoxic Regulation of Lactate Dehydrogenase A. J Biol Chem 270, 21021–21027. [DOI] [PubMed] [Google Scholar]
- Fong N, Brannan K, Erickson B, Kim H, Cortazar MA, Sheridan RM, Nguyen T, Karp S, and Bentley DL (2015). Effects of Transcription Elongation Rate and Xrn2 Exonuclease Activity on RNA Polymerase II Termination Suggest Widespread Kinetic Competition. Mol Cell 60, 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Voichek Y, Benjamin S, Gilad S, Amit I, and Oren M (2014). 4sUDRB-seq: measuring genomewide transcriptional elongation rates and initiation frequencies within cells. Genome Biol 15, R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, and Bentley DL (2008). RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol 15, 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Herreros F, de Miguel-Jimenez L, Morillo-Huesca M, Delgado-Ramos L, Munoz-Centeno MC, and Chavez S (2012). TFIIS is required for the balanced expression of the genes encoding ribosomal components under transcriptional stress. Nucleic Acids Res 40, 6508–6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratzner HG (1982). Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: A new reagent for detection of DNA replication. Science 218, 474–475. [DOI] [PubMed] [Google Scholar]
- Hazelbaker DZ, Marquardt S, Wlotzka W, and Buratowski S (2013). Kinetic competition between RNA Polymerase II and Sen1-dependent transcription termination. Mol Cell 49, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann M, Hintermair C, Voss K, and Eick D (2013). Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta 1829, 55–62. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Dangkulwanich M, Coello Y, Lionberger TA, Lubkowska L, Ponticelli AS, Kashlev M, and Bustamante C (2014). Transcription factors IIS and IIF enhance transcription efficiency by differentially modifying RNA polymerase pausing dynamics. Proc Natl Acad Sci U S A 111, 3419–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban MG, and Luse DS (1992). The RNA polymerase II ternary complex cleaves the nascent transcript in a 3’--−−5’ direction in the presence of elongation factor SII. Genes Dev 6, 1342–1356. [DOI] [PubMed] [Google Scholar]
- Izban MG, and Luse DS (1993). The increment of SII-facilitated transcript cleavage varies dramatically between elongation competent and incompetent RNA polymerase II ternary complexes. J Biol Chem 268, 12874–12885. [PubMed] [Google Scholar]
- James K, Gamba P, Cockell SJ, and Zenkin N (2017). Misincorporation by RNA polymerase is a major source of transcription pausing in vivo. Nucleic Acids Res 45, 1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon C, Yoon H, and Agarwal K (1994). The transcription factor TFIIS zinc ribbon dipeptide Asp-Glu is critical for stimulation of elongation and RNA cleavage by RNA polymerase II. Proc Natl Acad Sci 91, 9106–9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, and Lis JT (2014). Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife 3, e02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, and Lis JT (2015). Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 16, 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenberger H, Armache K-J, and Cramer P (2003). Architecture of the RNA Polymerase II-TFIIS Complex and Implications for mRNA Cleavage. Cell 114, 347–357. [DOI] [PubMed] [Google Scholar]
- Kettenberger H, Armache KJ, and Cramer P (2004). Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell 16, 955–965. [DOI] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, and Buratowski S (2004). The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432, 517–522. [DOI] [PubMed] [Google Scholar]
- Lemay JF, Larochelle M, Marguerat S, Atkinson S, Bahler J, and Bachand F (2014). The RNA exosome promotes transcription termination of backtracked RNA polymerase II. Nat Struct Mol Biol 21, 919–926. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik I, Qiu C, Snavely T, and Kaplan CD (2017). Wide-ranging and unexpected consequences of altered Pol II catalytic activity in vivo. Nucleic Acids Res, 45, 4431–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Landry HM, and Churchman LS (2017). Pause and go: from the discovery of RNA polymerase pausing to its functional implications. Curr Opin Cell Biol 46, 72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naftelberg S, Schor IE, Ast G, and Kornblihtt AR (2015). Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem 84, 165–198. [DOI] [PubMed] [Google Scholar]
- Nechaev S, and Adelman K (2011). Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta 1809, 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo D, Dos Santos G, Liu L, Gao Y, and Adelman K (2010). Global Analysis of Short RNAs Reveals Widespread Promoter-Proximal Stalling and Arrest of Pol II in Drosophila. Science 327, 335–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Gomes T, Carmo-Fonseca M, and Proudfoot NJ (2016). Mammalian NET-seq analysis defines nascent RNA profiles and associated RNA processing genome-wide. Nat Protoc 11, 413–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Gomes T, Grosso AR, Kimura H, Dye MJ, Dhir S, Carmo-Fonseca M, and Proudfoot NJ (2015). Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing. Cell 161, 526–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palangat M, Renner DB, Price DH, and Landick R (2005). A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc Natl Acad Sci 102, 15036–15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen MT, Veloso A, Prasad J, Bedi K, Ljungman EA, Magnuson B, Wilson TE, and Ljungman M (2014). Use of Bru-Seq and BruChase-Seq for genome-wide assessment of the synthesis and stability of RNA. Methods 67, 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ (2016). Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science 352, aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D (1992). Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J Biol Chem 267, 3795–3800. [PMC free article] [PubMed] [Google Scholar]
- Schwalb B, Michel M, Zacher B, Frühauf K, Demel C, Tresch A, Gagneur J, and Cramer P (2016). TT-seq maps the human transient transcriptome. Science 352, 1225–1228. [DOI] [PubMed] [Google Scholar]
- Sekine S. i., Murayama Y, Svetlov V, Nudler E, and Yokoyama S (2015). The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol Cell 57, 408–421. [DOI] [PubMed] [Google Scholar]
- Semenza GL (2011). Regulation of metabolism by hypoxia-inducible factor 1. Cold Spring Harb Symp Quant Biol 76, 347–353. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, and Reines D (2000). Saccharomyces cerevisiae Transcription Elongation Mutants Are Defective in PUR5 Induction in Response to Nucleotide Depletion.Mol Cell Biol 20, 7427–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S, Dirac-Svejstrup AB, and Svejstrup JQ (2010). Evidence that Transcript Cleavage Is Essential for RNA Polymerase II Transcription and Cell Viability. Mol Cell 38, 202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, and Padgett RA (2009). Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol 16, 1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosunov V, Sosunova E, Mustaev A, Bass I, Nikiforov V, and Goldfarb A (2003). Unified twometal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J 22, 2234–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosunova E, Sosunov V, Kozlov M, Nikiforov V, Goldfarb A, and Mustaev A (2003). Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc Natl Acad Sci 100, 15469–15474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steurer B, Janssens RC, Geverts B, Geijer ME, Wienholz F, Theil AF, Chang J, Dealy S, Pothof J, van Cappellen WA, et al. (2018). Live-cell analysis of endogenous GFP-RPB1 uncovers rapid turnover of initiating and promoter-paused RNA Polymerase II. Proc Natl Acad Sci 67, 201717920–201717929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MJ, Platas AA, and Hawley DK (1998). Transcriptional fidelity and proofreading by RNA polymerase II. Cell 93, 627–637. [DOI] [PubMed] [Google Scholar]
- Thompson N, Aronson D, and Burgess R (1990). Purification of Eukaryortic RNA polymerase II by Immunoaffinity Chromatography. J. Biol. Chem 265, 7069–7077. [PubMed] [Google Scholar]
- Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, and Ljungman M (2014). Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res 24, 896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Urlaub H, and Cramer P (2018). Structure of paused transcription complex Pol II-DSIF-NELF. Nature 560, 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CM, Ramachandran S, and Henikoff S (2014). Nucleosomes are context-specific, H2A.Zmodulated barriers to RNA polymerase. Mol Cell 53, 819–830. [DOI] [PubMed] [Google Scholar]
- West S, Gromak N, and Proudfoot NJ (2004). Human 5’ --> 3’ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432, 522–525. [DOI] [PubMed] [Google Scholar]
- Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, and Handa H (2006). P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell 21, 227–237. [DOI] [PubMed] [Google Scholar]
- Zhang C, and Burton ZF (2004). Transcription factors IIF and IIS and nucleoside triphosphate substrates as dynamic probes of the human RNA polymerase II mechanism. J Mol Biol 342, 1085–1099. [DOI] [PubMed] [Google Scholar]
- Zhang J, Palangat M, and Landick R (2010). Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol 17, 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.