Abstract
Objective/Hypothesis
Gene expression analyses of head and neck cancer have revealed four molecular subtypes: basal (BA), mesenchymal (MS), atypical (AT), and classical (CL). We evaluate whether gene expression subtypes in oral cavity (OCSCC) and laryngeal (LSCC) can be used to predict nodal metastasis and prognosticate survival.
Level of Evidence
2b
Study Design
Retrospective cohort study and genomic analysis
Methods
OCSCC and LSCC cases were identified from the TCGA head and neck cancer cohort. RNA-Seq by Expected Maximization (RSEM) was used to quantify gene expression levels from TCGA RNA-seq data and to assign each case to one of four subtypes. Descriptive statistics were used to describe patient, disease and treatment characteristics in each subtype. Cox regression and Kaplan Meier analyses were used to determine associations with survival.
Results
OCSCC cases were comprised primarily of the MS and BA subtypes, while LSCC was comprised primarily of CL and AT subtypes. In OCSCC, the MS subtype was significantly associated with higher risk of nodal metastasis. In a subset analysis of clinically T1-2N0M0 OCSCC, we demonstrate that the MS subtype was predictive of occult nodal metastasis (RR=3.38, 95% CI 1.08–10.69). In LSCC, the CL subtype was associated with significantly worse overall survival (HR=4.32, 95% CI 1.77–10.54, p=0.001).
Conclusions
Gene expression analysis reveals potential novel markers of nodal metastasis and survival in HPV (−) head and neck cancer. Future studies will continue to refine and validate these markers, with the goal of providing molecular risk assessments that guide therapy and improve patient outcomes.
Introduction
Head and neck squamous cell carcinoma (HNSCC) - including cancers of the oral cavity, oropharynx, nasopharynx, hypopharynx, and larynx - is one of the six most common cancers worldwide.1 In the United States, it was estimated that there were approximately 60,000 new cases and 12,000 deaths in 2017.2 The majority of HNSCC are associated with heavy tobacco and alcohol use, although over the last thirty years there has been an increase in the incidence of human papillomavirus (HPV)-related cancer, primarily in the oropharynx.3 While the treatment of HNSCC depends on multiple tumor and patient-related factors, the three main modalities used in the management of HNSCC are surgical resection, radiation therapy, and chemotherapy. Patients with early stage tumors are generally treated with a single modality therapy while those with advanced stage tumors often require multiple modalities. Oncologic outcomes in HNSCC are driven largely by stage at presentation: The 5-year overall survival for Stage I–II and III–IV HNSCC is approximately 70–90% and 40–60%, respectively.4,5
HPV-negative, tobacco-associated cases continue to comprise the vast majority of head and neck cancer cases. Oncologic outcomes in HPV-negative head and neck cancer remain poor and have not improved significantly in 60 years.4 Oral cavity squamous cell carcinoma (OCSCC) is the most common head and neck cancer, comprising 1/3 of all cases. Dependent on clinical staging, OCSCC treatment involves surgical excision of the primary tumor with or without neck dissection, followed by radiation with or without chemotherapy.6–8 Laryngeal squamous cell carcinoma (LSCC) is the second most common HPV-negative cancer of the head and neck and is almost exclusively tobacco-associated. Primary radiation-based treatments are common for early and intermediate stage cancers of the larynx in order to preserve function, with surgical resection often reserved for locally advanced tumors or salvage after failed radiation therapy.
The advent of next generation sequencing and modern bioinformatics has allowed investigators to more clearly understand tumor heterogeneity and its impact on clinical outcomes. Gene expression studies have identified previously unrecognized variation in squamous cell carcinoma, specifically of the lung and head and neck. Four mRNA expression patterns (primitive, classical, secretory and basal) demonstrating unique genomic features and prognostic significance were discovered in lung cancer.9 This line of research was extended into head and neck squamous cell carcinoma with remarkably similar observations.
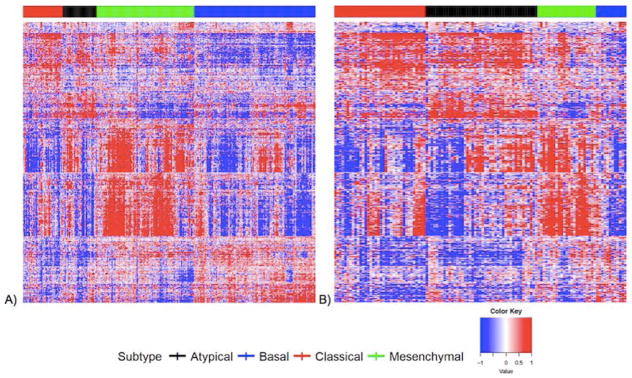
Pioneering work by Chung et al.10 and Walter et al.11 demonstrated four distinct molecular classes in HNSCC based on gene expression patterns: basal, mesenchymal, atypical, and classical. HNSCC subtypes based on gene expression show varied mutational profiles and may complement risk stratification in head and neck cancer based on HPV status, stage, anatomic site, and other characteristics (Figure 1).10,11 The basal subtype in head and neck squamous cell carcinoma is similar to the basal subtype in lung cancer, and is characterized by over-expression of genes functioning in cell adhesion including COL17A1, and growth factor and receptor TGFA and EGFR. The basal subtype is also associated with the highest expression of transcription factor TP63.11 The mesenchymal subtype displays over-expression of genes involved in immune response,12,13 and is characterized by expression of genes associated with epithelial to mesenchymal transition including vimentin, desmin, TWIST1, and HGF.11 It has been suggested previously that epithelial to mesenchymal transition pathways are important in the initiation of nodal metastasis.9,11 The classical subtype is characterized by over-expression of genes related to oxidative stress response and xenobiotic metabolism, and is most strongly associated with tobacco exposure. Deregulation of the KEAP1/NRF2 oxidative stress pathway appears to be a critical element of carcinogenesis in the classical subtype, and there is growing evidence to suggest that the KEAP1-NRF2 mediated oxidative stress response plays a role in resistance to radiation in several human cancers [21–24]. The atypical subtype is characterized by elevated expression of CDKN2A, LIG1, and RPA2, and has also been associated with low EGFR expression.9,11,12
Figure 1.
Gene expression heat maps including 728 reduced gene set for A) OCSCC and B) LSCC.
The discovery of four distinct gene expression subtypes in head and neck cancer provides important insight into the biologic heterogeneity of this disease. What remains unknown is whether these molecular signatures have prognostic or predictive significance in head and neck squamous cell carcinoma. In this study, we undertake a gene expression subtyping analysis of oral cavity and laryngeal squamous cell carcinoma within The Cancer Genome Atlas (TCGA) head and neck cancer cohort.12 We focus deliberately on HPV-negative head and neck cancer in an attempt to establish novel molecular markers of treatment response and survival for a subset of tumors with persistently poor oncologic outcomes. The aims of this study are 1) to compare the distribution and prognostic significance of gene expression subtypes in oral cavity (OCSCC) and laryngeal (LSCC) squamous cell carcinoma, and 2) to determine the association between gene expression subtype, nodal metastasis, and survival in these groups. We hypothesize that the distribution of gene expression subtypes will differ between laryngeal and oral cavity squamous cell carcinoma, reflecting different drivers of carcinogenesis in HPV-negative head and neck cancer across anatomic sites. Furthermore, we hypothesize that gene expression subtypes can be used to predict nodal metastasis and prognosticate survival in head and neck cancer.
Methods
OCSCC and LSCC cases were identified within the TCGA head and neck cancer dataset. The TCGA12 is a comprehensive cancer genomic data repository sponsored by The Cancer Genome Atlas Research Network of the National Cancer Institute, and includes DNA sequencing, RNA sequencing, and protein expression data on 33 cancer types. The TCGA head and neck cancer dataset includes 517 cases across all anatomic sites. Clinical, tumor, and treatment data are also available for each case.12 For this analysis, we chose to focus only on HPV-negative head and neck cancer. In order to avoid the potential of including HPV-positive cases, we chose to exclude oropharyngeal cancers and limit the analysis to LSCC and OCSCC.
RNA Sequencing Analysis
RNA-Seq by Expected Maximization (RSEM)14 was used to quantify gene expression levels from TCGA RNA-seq data. The RSEM gene expression measurements for n = 517 head and neck cancer cases were transformed using log2 (RSEM + 1) and subsequently median centered by gene, and LSCC (n=125) and OCSCC (n=309) cases were selected for further analysis. The centroids in the gene expression subtype classifier originally presented by Walter et al.11 (2013) were reduced from 838 genes to 728 genes, as described in the TCGA genomic characterization of head and neck cancer cohort.12 Each subject was then assigned to one of the four subtypes (basal, mesenchymal, atypical, or classical) by identifying the nearest centroid using a correlation-based similarity metric. A total of 267 of the 279 subjects (95.7%) profiled in the original TCGA head and neck cancer cohort12 received the same subtype classification in both analyses.
Gene expression heat maps including the reduced 728 gene set as well as including 14 genes relevant to head and neck squamous cell carcinoma were generated using ConsensusCluster-Plus as described previously.11,15 In order to facilitate comparisons between OCSCC and LSCC expression, the 728-gene list was ordered by combining expression data for the OCSCC and LSCC samples, clustering the rows and genes, then retaining the ordering for separate OCSCC and LSCC heat maps. The 14 gene lists were also ordered identically.
Statistical Analysis
Descriptive statistics were used to describe patient, disease, and treatment characteristics between each gene expression subtype. P-values were calculated with a chi-square test. Overall survival (OS) was measured from baseline diagnosis to death obtained from the National Death Index. Cases were censored at 3 years. Kaplan-Meier curves and log-rank values were calculated. Unadjusted hazard ratios were calculated with Cox proportional hazards model. Proportional hazards assumption was tested and satisfied. Statistical analysis was performed using R version 3.1.4.
Results
Descriptive Statistics
We first describe the distribution and gene expression characteristics of each subtype in the OCSCC and LSCC cohorts. Of the 309 OCSCC cases, 128 (41.4%) demonstrated a basal subtype, 103 (33.3%) mesenchymal, 43 (14%) classical, and 35 (11.3%) atypical. Of the 125 LSCC cases, 43 (34.4%) expressed an atypical subtype, 38 (30.4%) classical, 27 (21.6%) mesenchymal, and 12 (9.6%) basal. The demographic, tumor, and treatment characteristics of the OCSCC and LSCC cases by subtype are found in Table I. There was no significant difference with respect to clinical TNM stage between OCSCC subtypes. Overall, mesenchymal tumors were significantly more likely to be pathologically node positive (65.4% node positive) compared to the other groups. While the classical OCSCC cases were more likely to be smokers, no statistically significant difference is duration or pack year history of tobacco use was noted between the groups. Among LSCC cases, there was no significant difference with respect to race, gender, smoking status, clinical TNM stage, pathologic TNM stage, or adjuvant radiation therapy by gene expression subtypes.
Table I.
Descriptive statistics of clinical and demographic variables by subtype for each cancer site.
| Oral Cavity Cancer | Laryngeal Cancer | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| Atypical n = 35 | Basal n = 128 | Classical n = 43 | Mesenchymal n = 103 | p-value | Atypical n = 48 | Basal n = 12 | Classical n = 38 | Mesenchymal n = 27 | p-value | |
|
|
|
|||||||||
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |||
| Pathologic N Stage | ||||||||||
| N0 | 13 (41.9) | 55 (50.9) | 21 (50.0) | 28 (33.3) | 0.019 | 17 (44.7) | 5 (55.6) | 10 (35.7) | 9 (34.6) | 0.88 |
| N1 | 7 (22.6) | 17 (15.7) | 6 (14.3) | 16 (19.0) | 5 (13.2) | 0 (0.0) | 3 (10.7) | 4 (15.4) | ||
| N2 | 9 (29.0) | 36 (33.3) | 15 (35.7) | 39 (46.4) | 16 (42.1) | 4 (44.4) | 14 (50.0) | 12 (46.2) | ||
| N3 | 4 | 20 | 1 | 19 | 0 (0.0) | 0 (0.0) | 1 (3.6) | 1 (3.8) | ||
| Missing | 10 | 3 | 10 | 1 | ||||||
| Pathologic T Stage | ||||||||||
| T1 | 5 (15.2) | 12 (10.1) | 1 (2.3) | 11 (11.5) | 0.178 | 3 (7.7) | 1 (10.0) | 2 (6.2) | 1 (3.7) | 0.91 |
| T2 | 10 (30.3) | 37 (31.1) | 10 (23.3) | 37 (38.5) | 3 (7.7) | 1 (10.0) | 5 (15.6) | 3 (11.1) | ||
| T3 | 3 (9.1) | 28 (23.5) | 10 (23.3) | 18 (18.8) | 12 (30.8) | 2 (20.0) | 10 (31.2) | 5 (18.5) | ||
| T4 | 15 (45.5) | 42 (35.3) | 22 (51.2) | 30 (31.2) | 21 (53.8) | 6 (60.0) | 15 (46.9) | 18 (66.7) | ||
| Missing | 2 | 9 | 0 | 7 | 9 | 2 | 6 | 0 | ||
| Race | ||||||||||
| American Indian | 0 (0.0) | 1 (0.8) | 0 (0.0) | 0 (0.0) | 0.028 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (3.8) | 0.458 |
| Asian | 0 (0.0) | 9 (7.3) | 0 (0.0) | 1 (1.0) | 0 (0.0) | 0 (0.0) | 1 (2.8) | 0 (0.0) | ||
| Black | 3 (9.1) | 6 (4.8) | 7 (16.3) | 5 (5.0) | 9 (18.8) | 2 (18.2) | 3 (8.3) | 6 (23.1) | ||
| White | 30 (90.9) | 108 (87.1) | 36 (83.7) | 94 (94.0) | 39 (81.2) | 9 (81.8) | 32 (88.9) | 19 (73.1) | ||
| Missing | 2 | 4 | 0 | 3 | 0 | 1 | 2 | 1 | ||
| Smoking | ||||||||||
| Current | 10 (30.3) | 38 (29.7) | 18 (42.9) | 27 (27.6) | 0.273 | 29 (61.7) | 5 (41.7) | 11 (29.7) | 14 (53.8) | 0.084 |
| Former | 13 (39.4) | 47 (36.7) | 18 (42.9) | 44 (44.9) | 17 (36.2) | 6 (50.0) | 23 (62.2) | 9 (34.6) | ||
| Never | 10 (30.3) | 43 (33.6) | 6 (14.3) | 27 (27.6) | 1 (2.1) | 1 (8.3) | 3 (8.1) | 3 (11.5) | ||
| Missing | 2 | 0 | 1 | 5 | 1 | 0 | 1 | 1 | ||
| Radiation | ||||||||||
| No | 6 (46.2) | 19 (43.2) | 6 (31.6) | 11 (35.5) | 0.753 | 3 (16.7) | 0 (0.0) | 2 (28.6) | 3 (37.5) | 0.431 |
| Yes | 7 (53.8) | 25 (56.8) | 13 (68.4) | 20 (64.5) | 15 (83.3) | 4 (100.0) | 5 (71.4) | 5 (62.5) | ||
| Missing | 22 | 84 | 24 | 72 | 30 | 8 | 31 | 19 | ||
| Clinical N Stage | ||||||||||
| N0 | 21 (61.8) | 64 (53.3) | 23 (54.8) | 56 (55.4) | 0.762 | 19 (42.2) | 5 (55.6) | 22 (57.9) | 12 (46.2) | 0.665 |
| N1 | 4 (11.8) | 25 (20.8) | 7 (16.7) | 17 (16.8) | 9 (20.0) | 1 (11.1) | 3 (7.9) | 5 (19.2) | ||
| N2 | 8 (23.5) | 31 (25.8) | 12 (28.6) | 27 (26.7) | 17 (37.8) | 3 (33.3) | 11 (28.9) | 8 (30.8) | ||
| N3 | 1 (2.9) | 0 (0.0) | 0 (0.0) | 1 (1.0) | 0 (0.0) | 0 (0.0) | 2 (5.3) | 1 (3.8) | ||
| Missing | 1 | 8 | 1 | 2 | 3 | 3 | 0 | 1 | ||
| Clinical T Stage | ||||||||||
| T1 | 3 (8.8) | 5 (4.1) | 2 (4.8) | 6 (5.9) | 0.509 | 1 (2.1) | 0 (0.0) | 1 (2.6) | 1 (3.8) | 0.504 |
| T2 | 9 (26.5) | 40 (32.5) | 10 (23.8) | 39 (38.2) | 7 (14.9) | 1 (10.0) | 10 (26.3) | 2 (7.7) | ||
| T3 | 7 (20.6) | 36 (29.3) | 9 (21.4) | 23 (22.5) | 20 (42.6) | 3 (30.0) | 10 (26.3) | 7 (26.9) | ||
| T4 | 15 (44.1) | 42 (34.1) | 21 (50.0) | 34 (33.3) | 19 (40.4) | 6 (60.0) | 17 (44.7) | 16 (61.5) | ||
| Gender | 1 | 5 | 1 | 1 | ||||||
| Female | 7 (20.0) | 46 (35.9) | 10 (23.3) | 38 (36.9) | 0.125 | 7 (20.0) | 46 (35.9) | 10 (23.3) | 38 (36.9) | 0.125 |
| Male | 28 (80.0) | 82 (64.1) | 33 (76.7) | 65 (63.1) | 28 (80.0) | 82 (64.1) | 33 (76.7) | 65 (63.1) | ||
Gene expression profiles
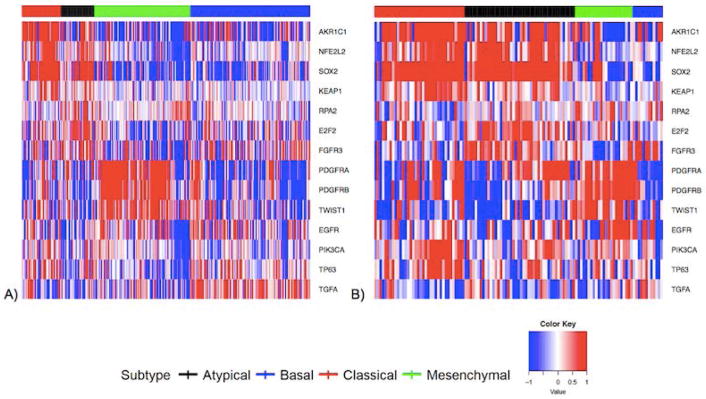
OCSCC and LSCC gene expression heat maps for the 728-gene set are found in Figure 1A and 1B, respectively. The 14 gene expression heat-maps for OCSCC and LSCC are found in Figure 2A and 2B, respectively. We demonstrate clustering of cases into the four subtypes based on gene expression signatures among both OCSCC and LSCC cases, with differences in subtype distribution by anatomic site.
Figure 2.
Gene expression heat maps including 14 reduced gene set for A) OCSCC and B) LSCC.
Survival Analysis
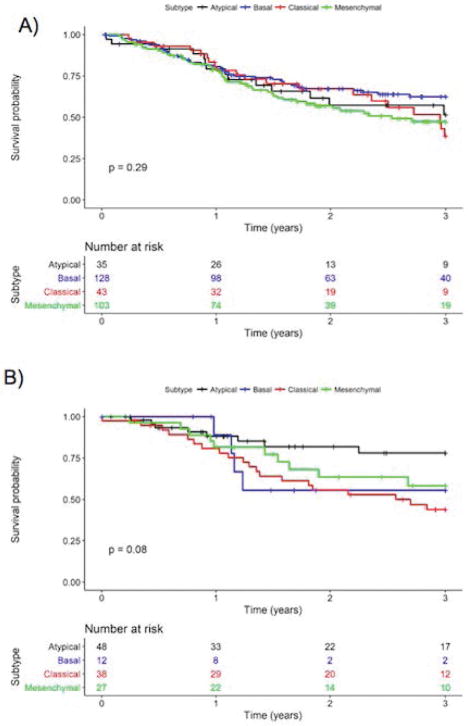
Kaplan Meier survival curves by subtype for OCSCC and LSCC are found in Figure 3A and 3B, respectively. Among OCSCC cases, the basal subtype had the best 3-year survival (62.5%, 95% CI: 54.0%–72.4%) followed by the atypical (51.5%, 95% CI: 35.2% – 75.2%) and mesenchymal (47.3%, 95% CI: 37.5% – 59.8%) subtypes. The classical subtype demonstrated the worst 3-year survival (38.7%, 95% CI: 24.1% – 62.1%). Among LSCC cases, the classical subtype was also associated with the worst 3-year overall survival (43.7%, 95% CI: 30.0 – 63.7%) while the atypical subtype had the best overall survival (78.05%, 95% CI: 65.2% – 93.2%). The basal and mesenchymal subtypes had similar 3-year survival (55.6%, 95% CI: 31.0% – 99.7% and 58.3%, 95% CI: 41.1 – 82.5%, respectively).
Figure 3.
Kaplan Meier survival curves for A) OCSCC and B) LSCC by subtype.
The results of a multivariate regression analysis for factors associated with risk of death in OCSCC and LSCC are found in Table II. In OCSCC, gene expression subtype was not statistically associated with an increased risk of death. In LSCC, the classical subtype was associated with an increased risk of death (HR=4.32, 95% CI 1.77–010.54, p=0.001). Female gender was associated with significantly worse survival compared to male (HR=4.2, 95% CI 1.99–8.90, p<0.001).
Table II.
Adjusted hazard ratios for oral cavity and laryngeal cancers
| Oral Cavity | Laryngeal | |||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Subtype | ||||
| Atypical | 1.00 | 1.00 | ||
| Basal | 0.70 (0.37, 1.32) | 0.265 | 0.93 (0.18, 4.83) | 0.935 |
| Classical | 0.91 (0.44, 1.86) | 0.793 | 4.32 (1.77, 10.54) | 0.001 |
| Mesenchymal | 1.05 (0.56, 1.96) | 0.888 | 2.51 (0.91, 6.91) | 0.076 |
| Stage | ||||
| IV | 1.00 | 1.00 | ||
| I–II | 0.83 (0.53, 1.3) | 0.415 | 0.89 (0.25, 3.17) | 0.864 |
| III | 1.03 (0.64, 1.68) | 0.893 | 0.98 (0.41, 2.38) | 0.973 |
| Gender | ||||
| Male | 1.00 | 1.00 | ||
| Female | 1.13 (0.75, 1.69) | 0.558 | 4.2 (1.99, 8.90) | <0.001 |
| Race | ||||
| White | 1.00 | 1.00 | ||
| Non-White | 1.36 (0.73, 2.52) | 0.328 | 1.87 (0.82, 4.25) | 0.135 |
| Smoking | ||||
| Current | 1.00 | 1.00 | ||
| Never/Former | 0.74 (0.50, 1.11) | 0.148 | 0.52 (0.26, 1.04) | 0.064 |
CI: Confidence interval; HR: Hazards ratio
Occult Nodal Metastasis in OCSCC
Given the association demonstrated between the OCSCC mesenchymal subtype and nodal metastasis, we conducted a subset analysis of T1/T2, clinically node-negative OCSCC cases in order to test the predictive value of gene expression subtypes in detecting occult nodal metastasis. Of the 67 cases identified that fit criteria for inclusion, 24 (35.8%) expressed a basal subtype, 26 (38.8%) a mesenchymal subtype, 8 (12%) a classical subtype, and 9 (13.4%) an atypical subtype. No significant difference in gender, clinical T-stage, or adjuvant therapy use was noted between the groups. Non-Hispanic Whites were significantly more likely to express a mesenchymal subtype compared to African-Americans and Asians. When risk of occult nodal metastasis was considered, mesenchymal subtype tumors were significantly more likely to have pathologically positive lymph nodes at the time of neck dissection (RR=3.38, 95% CI 1.08–10.69) compared to the other subtypes.
Discussion
In this study, we examine the distribution and prognostic significance of gene expression subtypes in TCGA OCSCC and LSCC cases. We demonstrate substantive differences in subtype distribution by site; OCSCC cases were comprised primarily of mesenchymal and basal tumors, while LCSCC of classical and atypical tumors. We also demonstrate an association between the OCSCC mesenchymal subtype and lymph node metastasis. Finally, our findings suggest a significant survival disadvantage associated with the LSCC classical subtype. This analysis provides important insight into tumor heterogeneity in HPV-negative head and neck cancer. If further validated in prospective studies or clinical trials, gene expression subtyping may have a role in prognostication and therapeutic decision-making for HPV-negative head and neck cancer.
In concordance with previous observations by Walter et al11, the vast majority of OCSCC tumors in this series have a basal or mesenchymal gene expression signature. Similar findings were also reported in an integrative genomic analysis of OCSCC by Pickering et al.16, in which unsupervised clustering revealed two gene clusters similar in composition to the basal and mesenchymal groups. In the present study, the basal and mesenchymal subtype comprised over 70% of the OCSCC cohort. We demonstrate that the mesenchymal subtype, characterized by epithelial to mesenchymal transition, is associated with nodal metastasis in OCSCC. Epithelial to Mesenchymal transition is a complex multistep process by which epithelial malignancies undergo loss of cell adhesion, loss of polarity and cohesion, increased motility, and acquire a mesenchymal phenotype.17 Previous studies have explored the role of epithelial to mesenchymal transition in tumor invasiveness and lymph node metastasis in head and neck cancer. El Naggar et al.17 examined several mesenchymal biomarkers in 11 head and neck cancer cell lines and 50 primary tumors. They demonstrated a strong association between decreased E-cadherin expression, increased p-Src, Vimentin expression and lymph node metastasis. Another recent analysis of epithelial to mesenchymal transition markers found that high expression of Vimentin was associated with poor disease-specific survival in oral tongue squamous cell carcinoma.18
Several transcription factors have been identified that act as inducers of epithelial to mesenchymal transition in head and neck squamous cell carcinoma, including Slug, Snail, and Twist1.19 As demonstrated in the present study, Twist1 overexpression is characteristic of the OCSCC mesenchymal subtype. Previous studies have examined Twist1 expression as a potential prognostic and predictive indicator in OCSCC.19–22 In a microarray RNA expression analysis of 74 OCSCC cases, da Silva et al.23 noted that Twist1 upregulation was associated with advanced stage tumors, lymph node and distant metastasis, and poor survival. Immunohistochemical studies of Twist1 expression have also been conducted in OCSCC, confirming the potential role of Twist1 expression as a possible marker of lymph node metastasis and the importance of epithelial to mesenchymal transition in OCSCC.20,21 A recently published meta-analysis of 15 studies in head and neck cancer further supports the importance of Twist1 as a potential prognosticator in head and neck cancer; overall, Twist1 overexpression was associated with a nearly two-fold increased risk of death compared to those without overexpression. (HR= 1.92, 95% CI 1.13–3.25).19
In contrast to OCSCC, LSCC cases were comprised primarily of the classical and atypical subtypes. Furthermore, the LSCC classical subtype was significantly associated with worse overall survival. As demonstrated in this study, a hallmark of the classical subtype is overexpression of KEAP1 and NRF2. The KEAP1/NRF2 pathway, an essential regulator of oxidative stress from reactive oxygen species and xenobiotics, has been identified as a possible mechanism of chemoradiation resistance in multiple cancers including head and neck squamous cell carcinoma.25–28 Loss of function mutations in the KEAP1 tumor suppressor gene and activating mutations in the KEAP1 binding domain of NFE2L2 have been described in multiple cancers, and result in the constitutive activation of NRF2.25,29,30 Constitutive activation of NRF2 in turn has pro-tumorigenic effects, including inhibition of apoptosis, promotion of cell proliferation, and chemoresistance.27,28,31 Kawasaki et al demonstrated that NRF2 immunohistochemical expression was an independent predictor of response to chemotherapy and radiation in a cohort of esophageal squamous cell carcinoma patients.32 The same group and others have demonstrated a similar association between NRF2 expression and chemoresistance in gastric cancer,33,34 while Bao et al demonstrated an association between NRF2 expression and cisplatin resistance in ovarian cancer.35 In head and neck squamous cell carcinoma, a recent analysis of The Cancer Genome Atlas (TCGA) data by Martinez et al. noted that 64% of head and neck cancer cases had disruption of the NRF2 pathway, and that NRF2 activation was associated with worse survival.31 We build on these previous findings by demonstrating a specific phenotype in LSCC, characterized by KEAP/NRF2 over-expression as well as other genetic alterations in oxidative stress pathways, is associated with poor outcomes.
The identification of distinct molecular signatures associated with OCSCC nodal metastasis and LSCC survival has potential therapeutic and prognostic implications. In OCSCC, previous investigators have sought to establish predictive gene expression markers of lymph node metastasis.36,37 We expand on this work by demonstrating the potential use of the mesenchymal gene expression signature to predict occult nodal metastasis in early stage, clinically N0 OCSCC. If further validated, this signature could be used alongside clinical and pathologic characteristics to determine novel criteria for neck dissection. In LSCC, this subtyping analysis has the potential to guide decision-making between primary surgery and radiation therapy for early to intermediate stage cancers. KEAP1/NRF2 pathway alterations have been associated with radiation resistance, and may be contributing to the poor survival noted in classical subtype tumors. If further validated, LSCC gene expression subtype could be used to identify patients at higher risk for radiation therapy failure. This would allow clinicians to suggest alternate therapeutic approaches, including surgical resection as the primary treatment modality or the addition of adjuvant therapy.
Recently, single-cell RNA sequencing technology has provided the potential to refine our understanding of intratumoral heterogeneity in head and neck cancer beyond the limitations of bulk RNA sequencing techniques. By independently sequencing malignant, stromal, and immune cells derived from tumors, Puram et al24 demonstrated that the mesenchymal OCSCC subtype may represent a subset of the basal subtype. They propose that the mesenchymal subtype may actually represent high stromal representation in bulk RNA samples rather than a distinct malignant phenotype. Based on this analysis, they also describe a partial EMT signature that predicts nodal metastasis and other adverse tumor features.24 Through broad application across cancer sites and in larger samples sets, single cell sequencing other novel techniques will continue to provide new insight into intratumoral heterogeneity.
This study has several strengths and limitations that should be considered. An important strength is the use of data from the TCGA head and neck cancer cohort, a large multi-institutional cancer sequencing effort with well-annotated and comprehensive RNA sequencing data. These data are publicly available, allowing other investigators to validate the results presented herein as well as further refine or expand on our findings.12 This study also has several limitations. While TCGA includes a breadth of genomic data, no data are available on recurrence or disease-specific survival. Detailed pathologic characteristics, including depth of invasion, perineural invasion, or lymphovascular invasion were also unavailable through TCGA. Finally, while the TCGA data include a large head and neck cancer cohort drawn from multiple institutions throughout the United States, it may not be reflective of the population as a whole.
This analysis of gene expression subtypes in OCSCC and LCSCC demonstrates potential novel markers of nodal metastasis and survival in HPV-negative head and neck cancer, and highlights the biologic heterogeneity of this disease across anatomic sites. Future studies will continue to refine and validate these gene expression subtypes, with the goal of providing molecular risk assessments that improve treatment response and patient outcomes.
Footnotes
Financial Disclosures: N/A
Conflicts of Interest: JPZ and DNH hold a provisional patent application based on the findings presented in this manuscript
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dahlstrom KR, Calzada G, Hanby JD, et al. An evolution in demographics, treatment, and outcomes of oropharyngeal cancer at a major cancer center: a staging system in need of repair. Cancer. 2013;119(1):81–89. doi: 10.1002/cncr.27727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pulte D, Brenner H. Changes in survival in head and neck cancers in the late 20th and early 21st century: a period analysis. The oncologist. 2010;15(9):994–1001. doi: 10.1634/theoncologist.2009-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Cruz AK, Dandekar MR. Elective versus therapeutic neck dissection in the clinically node negative neck in early oral cavity cancers: do we have the answer yet? Oral Oncol. 2011;47(9):780–782. doi: 10.1016/j.oraloncology.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 7.D’Cruz AK, Vaish R, Kapre N, et al. Elective versus Therapeutic Neck Dissection in Node-Negative Oral Cancer. N Engl J Med. 2015;373(6):521–529. doi: 10.1056/NEJMoa1506007. [DOI] [PubMed] [Google Scholar]
- 8.Dias FL, Kligerman J, Matos de Sa G, et al. Elective neck dissection versus observation in stage I squamous cell carcinomas of the tongue and floor of the mouth. Otolaryngol Head Neck Surg. 2001;125(1):23–29. doi: 10.1067/mhn.2001.116188. [DOI] [PubMed] [Google Scholar]
- 9.Wilkerson MD, Yin X, Hoadley KA, et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res. 2010;16(19):4864–4875. doi: 10.1158/1078-0432.CCR-10-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung CH, Parker JS, Karaca G, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5(5):489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 11.Walter V, Yin X, Wilkerson MD, et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS One. 2013;8(2):e56823. doi: 10.1371/journal.pone.0056823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keck MK, Zuo Z, Khattri A, et al. Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin Cancer Res. 2015;21(4):870–881. doi: 10.1158/1078-0432.CCR-14-2481. [DOI] [PubMed] [Google Scholar]
- 14.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pickering CR, Zhang J, Yoo SY, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer discovery. 2013;3(7):770–781. doi: 10.1158/2159-8290.CD-12-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandal M, Myers JN, Lippman SM, et al. Epithelial to mesenchymal transition in head and neck squamous carcinoma: association of Src activation with E-cadherin down-regulation, vimentin expression, and aggressive tumor features. Cancer. 2008;112(9):2088–2100. doi: 10.1002/cncr.23410. [DOI] [PubMed] [Google Scholar]
- 18.Liu PF, Kang BH, Wu YM, et al. Vimentin is a potential prognostic factor for tongue squamous cell carcinoma among five epithelial-mesenchymal transition-related proteins. PLoS One. 2017;12(6):e0178581. doi: 10.1371/journal.pone.0178581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhuo X, Luo H, Chang A, Li D, Zhao H, Zhou Q. Is overexpression of TWIST, a transcriptional factor, a prognostic biomarker of head and neck carcinoma? Evidence from fifteen studies. Sci Rep. 2015;5:18073. doi: 10.1038/srep18073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wushou A, Hou J, Zhao YJ, Shao ZM. Twist-1 up-regulation in carcinoma correlates to poor survival. International journal of molecular sciences. 2014;15(12):21621–21630. doi: 10.3390/ijms151221621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wushou A, Pan HY, Liu W, et al. Correlation of increased twist with lymph node metastasis in patients with oral squamous cell carcinoma. Journal of oral and maxillofacial surgery : official journal of the American Association of Oral and Maxillofacial Surgeons. 2012;70(6):1473–1479. doi: 10.1016/j.joms.2011.06.212. [DOI] [PubMed] [Google Scholar]
- 22.da Silva SD, Morand GB, Alobaid FA, et al. Epithelial-mesenchymal transition (EMT) markers have prognostic impact in multiple primary oral squamous cell carcinoma. Clinical & experimental metastasis. 2015;32(1):55–63. doi: 10.1007/s10585-014-9690-1. [DOI] [PubMed] [Google Scholar]
- 23.da Silva SD, Alaoui-Jamali MA, Soares FA, et al. TWIST1 is a molecular marker for a poor prognosis in oral cancer and represents a potential therapeutic target. Cancer. 2014;120(3):352–362. doi: 10.1002/cncr.28404. [DOI] [PubMed] [Google Scholar]
- 24.Puram SV, Tirosh I, Parikh AS, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 2017;171(7):1611–1624. e1624. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox biology. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends in biochemical sciences. 2009;34(4):176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Ohta T, Iijima K, Miyamoto M, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer research. 2008;68(5):1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 28.Shibata T, Kokubu A, Gotoh M, et al. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135(4):1358–1368. 1368 e1351–1354. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- 29.Hast BE, Cloer EW, Goldfarb D, et al. Cancer-derived mutations in KEAP1 impair NRF2 degradation but not ubiquitination. Cancer research. 2014;74(3):808–817. doi: 10.1158/0008-5472.CAN-13-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hast BE, Goldfarb D, Mulvaney KM, et al. Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer research. 2013;73(7):2199–2210. doi: 10.1158/0008-5472.CAN-12-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez VD, Vucic EA, Thu KL, Pikor LA, Lam S, Lam WL. Disruption of KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex components by multiple genetic mechanisms: Association with poor prognosis in head and neck cancer. Head & neck. 2015;37(5):727–734. doi: 10.1002/hed.23663. [DOI] [PubMed] [Google Scholar]
- 32.Kawasaki Y, Okumura H, Uchikado Y, et al. Nrf2 is useful for predicting the effect of chemoradiation therapy on esophageal squamous cell carcinoma. Annals of surgical oncology. 2014;21(7):2347–2352. doi: 10.1245/s10434-014-3600-2. [DOI] [PubMed] [Google Scholar]
- 33.Kawasaki Y, Ishigami S, Arigami T, et al. Clinicopathological significance of nuclear factor (erythroid-2)-related factor 2 (Nrf2) expression in gastric cancer. BMC cancer. 2015;15:5. doi: 10.1186/s12885-015-1008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bao J, Li J, Li D, Li Z. Correlation between expression of NF-E2-related factor 2 and progression of gastric cancer. International journal of clinical and experimental medicine. 2015;8(8):13235–13242. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Bao LJ, Jaramillo MC, Zhang ZB, et al. Nrf2 induces cisplatin resistance through activation of autophagy in ovarian carcinoma. International journal of clinical and experimental pathology. 2014;7(4):1502–1513. [PMC free article] [PubMed] [Google Scholar]
- 36.O’Donnell RK, Kupferman M, Wei SJ, et al. Gene expression signature predicts lymphatic metastasis in squamous cell carcinoma of the oral cavity. Oncogene. 2005;24(7):1244–1251. doi: 10.1038/sj.onc.1208285. [DOI] [PubMed] [Google Scholar]
- 37.Enokida T, Fujii S, Takahashi M, et al. Gene expression profiling to predict recurrence of advanced squamous cell carcinoma of the tongue: discovery and external validation. Oncotarget. 2017 doi: 10.18632/oncotarget.18692. [DOI] [PMC free article] [PubMed] [Google Scholar]