Abstract
Background:
Colorectal cancers with deficient DNA mismatch repair (dMMR) are presumed to uniformly have dense lymphocytic infiltration that underlies their favorable prognosis and is critical to their responsiveness to immunotherapy, as compared to MMR-proficient (pMMR) tumors. We examined T-cell densities and their potential heterogeneity in a large cohort of dMMR tumors.
Experimental Design:
CD3+ and CD8+ T-cell densities were quantified at the invasive margin (IM) and tumor core (CT) in 561 stage III colon cancers (dMMR, n=278; pMMR, n=283) from a phase 3 adjuvant trial (N0147). Their association with overall survival (OS) was determined using multivariable Cox analysis.
Results:
While CD3+ and CD8+ T-cell densities in the tumor microenvironment were higher in dMMR vs pMMR tumors overall, inter-tumoral heterogeneity in densities between tumors was significantly higher by 30–88% among dMMR vs pMMR cancers (P<.0001 for all four T-cell subtypes [CD3+IM, CD3+CT, CD8+IM, CD8+CT]). A substantial proportion of dMMR tumors (26% to 35% depending on the T-cell subtype) exhibited T-cell densities as low as that in the bottom half of pMMR tumors. All four T-cell subtypes were prognostic in dMMR with CD3+IM being the most strongly prognostic. Low (vs high) CD3+IM was independently associated with poorer OS among dMMR (HR 4.76 [95% CI 1.43–15.87]; P=.0019) and pMMR tumors (P=.0103).
Conclusions:
Tumor-infiltrating T-cell densities exhibited greater inter-tumoral heterogeneity among dMMR than pMMR colon cancers, with CD3+IM providing robust stratification of both dMMR and pMMR tumors for prognosis. Potentially, lower T-cell densities among dMMR tumors may contribute to immunotherapy resistance.
Keywords: CD3, CD8, tumor-infiltrating lymphocytes, colorectal cancer, colon cancer, prognosis, N0147
INTRODUCTION
Accumulating evidence suggests that tumor progression and recurrence are governed not only by the genetic profile within cancer cells, but by the host anti-tumor immune response. The TNM classification, while valuable (1), provides incomplete prognostic information, given that clinical outcome can vary substantially within the same tumor stage (2). Molecular classifications of colorectal cancer (CRC) have been proposed based upon DNA mismatch repair (MMR) status and gene expression profiles (3, 4). CRCs with deficient MMR (dMMR) and microsatellite instability (MSI) typically show abundant tumor-infiltrating lymphocytes (TILs) and are associated with favorable outcome and decreased likelihood of metastases, as shown by ourselves (5, 6) and others (7, 8). In CRC, dMMR is caused most commonly by epigenetic silencing of the MLH1 MMR gene or by germline mutation in an MMR gene (MLH1, MSH2, MSH6, PMS2 or EpCAM) (conferring Lynch Syndrome) followed by the somatic inactivation of the second allele. This defect leads to accumulation of insertions and deletions in DNA repeat sequences. In genes containing coding repeats, frameshift mutations are a potential source of immunogenic neo-antigens recognized by the immune system which can trigger TILs (9, 10).
The influence of the type, density, and intratumor location of TILs on patient survival has been extensively reported (10–18). The quantification of CD3+ T cells (a pan-T cell marker) and CD8+ T cells (cytotoxic T cell marker) at the invasive margin (IM) (i.e., leading edge) and core of the tumor (CT) has been shown to identify prognostically distinct immune subsets in CRC, as shown by ourselves (14, 19) and others (11, 12, 20–22). TILs may have a dual advantage over TNM staging as a prognostic classifier. First, CD3+ and/or CD8+ TILs appear to be prognostic across tumor stages and treatments (11, 20). Second, the antitumor activity of these naturally infiltrating T cells might be amenable to enhancement by novel immunotherapy approaches (23–25). Data suggest that therapeutic benefit from programmed death receptor-1 (PD-1) blockade is preferentially achieved in patients with a pre-existing intratumoral T-cell response (26, 27). While many MSI tumors demonstrate responsiveness to anti-PD-1 therapy (28, 29), ~60% of patients with dMMR metastatic CRC do not respond to such treatment, indicating that heterogeneity among dMMR tumors exists, and factors underlying response and resistance are poorly understood.
Studies evaluating the association of TILs with survival have been limited to retrospective case series or population-based studies of varying CRC stage, treatment, and follow-up, and generally lacking data on MMR status or other molecular features. To date, the prognostic impact of CD3+ and CD8+ T cells, have not adequately studied dMMR tumors due to their relatively low frequency in CRC (~11% in stage III). In this report, we examined the ability of CD3+ and CD8+ T-cell density at the IM and CT to stratify patients for prognosis in a large cohort of patients with dMMR stage III colon cancer who received adjuvant FOLFOX-based therapy in a phase 3 trial.
METHODS
Study Population
This analysis included participants in the North Central Cancer Treatment Group (NCCTG) (now part of the Alliance for Clinical Trials in Oncology) N0147 adjuvant randomized phase 3 trial whose tumor blocks of resected stage III colon adenocarcinoma were available for analysis. TILs were analyzed in all available dMMR tumors and a similar number of randomly selected pMMR tumors. In the parent trial (enrollment 2004–2009) patients were randomized after surgery to receive 6 months of adjuvant FOLFOX therapy with or without the anti-EGFR antibody cetuximab, as described previously (30). In the present study we pooled data from both arms since adjuvant cetuximab did not improve efficacy (30), and there were no observed interactions between immune markers and treatments (interaction p-values > 0.13).
Immunohistochemistry (IHC) for TILs
IHC staining was performed on whole-slide serial tissue sections from formalin-fixed, paraffin-embedded surgical resection specimens (VENTANA BenchMark ULTRA automated staining instrument at Ventana Medical Systems, Inc.). Briefly, tissue sections were deparaffinized, pretreated with Cell Conditioning 1 for antigen retrieval, followed by inactivation of endogenous peroxidase. Specimens were incubated with CONFIRM anti-CD3 (2GV6) rabbit monoclonal antibody for 20 minutes or anti-CD8 (SP239) rabbit monoclonal antibody (3 μg/ml) for 32 minutes at 36 °C. CD3 and CD8 positive immune cells were visualized using OptiView DAB IHC Detection Kit. Following detection, all samples were counterstained with Hematoxylin II and Bluing Reagent for 4 minutes each, and coverslips were applied.
Digital image analysis for TILs
Hematoxylin and eosin (H&E)-stained sections along with the immunostained slides were scanned on VENTANA iSCAN HT scanner (Figure 1). Three independent pathologists manually annotated H&E sections to outline the entire tumor region containing invasive cancer (i.e., core of the tumor [CT]) using a whole tumor section approach (31). They further demarcated the invasive margin (IM) without knowledge of clinical characteristics or outcome by indicating sections of the tumor outline involved in the invasive process. A registration algorithm (32) automatically transferred pathologist-derived annotations from the H&E onto the adjacent CD3 and CD8 IHC images. From the IM demarcation, an algorithm automatically generated the IM area as 0.5 mm extending into the tumor core and 1.0 mm beyond the tumor. Fully automated computer vision and cell classification (33) captured CD3 and CD8 positive cells in the CT and IM areas with algorithm parameters fixed for all slides in the study. Multiple quality steps were employed to ensure fidelity of tissue slides, digital images, registration, and cell detection. Digital image analysis reports the tissue area and number of detected T-cells in the two observed compartments. CD3+ and CD8+ TIL densities at the IM and CT were quantified by image analysis and normalized to establish semi-continuous density scores (0–100 scale). TIL analysis was performed blinded to patient outcomes. A comparison of manual vs automated counts for both CD3+ and CD8+ T cells in a separate cohort of stage II colon cancers were highly concordant (unpublished data).
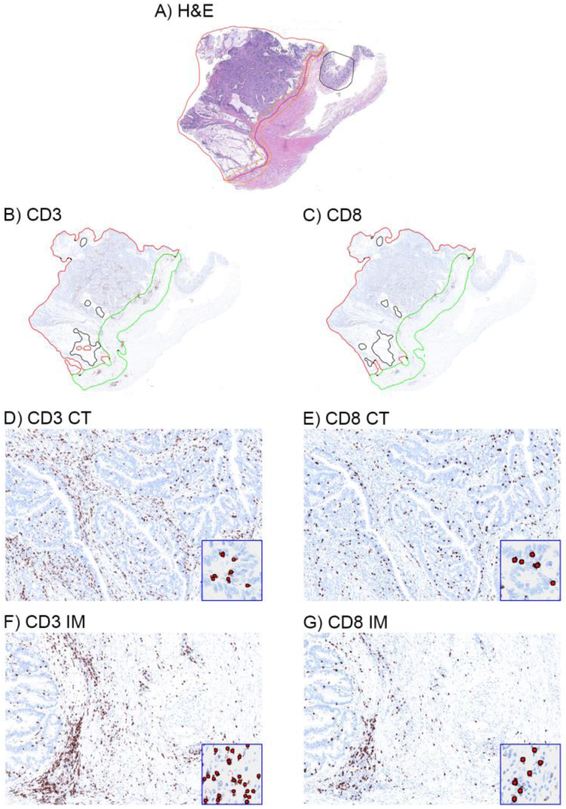
Figure 1. Representative immunohistochemical images of CD3+ and CD8+ T cells in the tumor microenvironment of colon cancers.
H&E-stained whole-tissue sections were manually annotated to outline the entire tumor region (red line) and demarcate the invasive margin (orange line) (a). Annotations were transferred onto adjacent CD3 and CD8 IHC images (b-c) with an algorithm outlining the invasive margin (IM, green line) and core of the tumor (CT, red line) (see Methods). Whole tissue sections (a-c) and magnified regions (40x, d-g) are shown. Insets show algorithmic cell-by-cell classification of CD3 and CD8 T cells (red dots). T-cell densities were quantified (score 0–100) within the entire tumor compartment.
Calculation of Semi-Continuous Density Scores (0–100 scale)
CD3 and CD8 counts were determined at the tumor core and invasive margin. The density of each marker (CD3+IM, CD3+CT, CD8+IM, CD8+CT) was calculated by dividing the count by the area of its tumor compartment. Given the potential right-skew in density distributions, biologically meaningful maximum values were established by truncating large densities, as follows: 1, Density values were categorized starting from zero in incremental steps of 250 cells/mm2; 2, Patients with the highest density values were identified (“edge effect” group) restricted to ~<10% of patients; 3, The density value that represented the cutoff value for the “edge effect” group was identified; 4, The incremental step corresponding to the “edge effect” cutoff value was established as the truncation value. Densities larger than this truncation value were assigned the truncation value. Density values were then standardized to generate a Density Score ranging from 0 to 100:
MMR Status Determination, DNA Mutation Analysis
Mismatch repair (MMR) tumor status was determined by immunohistochemical analysis (IHC) or by MSI testing when IHC findings were indeterminate, as previously described (5). Tumors with a dMMR phenotype were defined as showing loss of expression of 1 or more MMR proteins by IHC or exhibiting high-level tumor DNA MSI on MSI testing by polymerase chain reaction (PCR). Proficient MMR phenotype tumors were defined as showing intact MMR protein expression on IHC or microsatellite-stable or low-level MSI status on MSI testing.
Tumor DNA was extracted from formalin-fixed, paraffin-embedded tissue specimens containing more than 50% tumor cells using the QIAamp DNA Mini Kit (Qiagen). Testing for the BRAF V600E hotspot exon 15 mutation (c.1799T>A/p.V600E) and KRAS exon 2 mutation was performed as described previously(5).
Statistical Analysis
For comparisons of baseline characteristics, categorical factors were analyzed with χ2 tests, and continuous factors were compared with Wilcoxon rank-sum tests. For each T-cell subtype, densities between tumor compartments were compared using median pairwise differences (Wilcoxon Sign Rank tests). A Cox regression model was used to estimate hazard ratios (HRs) and 95% confidence intervals (CIs) and to calculate P values. Analyses were conducted in each MMR group separately. Each immune variable (CD3+IM, CD3+CT, CD8+IM, CD8+CT) was analyzed as a continuous variable with regard to OS in univariate and multivariable analysis. Covariates were prespecified as T3 or T4 (vs T1–2), N2 (vs N1), grade high (vs low), tumor side left (vs right), smoking ever (vs never), and age for each 5 years increase. No interactions were observed in adjusted analysis between any two of the immune markers on OS in either MMR group. Any individual immune variable demonstrating an association with OS at P <.10 adjusting for common clinical-pathological features was then included in a backward selection model. For immune variables with a statistically significant association with OS after backward selection, an optimal cutpoint that distinguished OS was identified using Cox Model Hazard Ratio and Wald p-value methods. Overall survival (OS) was defined as the time between randomization and any-cause death. Time to recurrence (TTR) (i.e., time between randomization and local or metastatic tumor recurrence) was analyzed as a secondary endpoint. Two-sided P values are reported; P <.05 was considered statistically significant. Analyses were performed using SAS software (v9.4, SAS Institute Inc), on a clinical data set locked on 5/8/2015. Data collection and statistical analyses were conducted by the Alliance Statistics and Data Center.
RESULTS
Among N0147 trial participants, 3018 patients received adjuvant mFOLFOX6 with or without cetuximab (30). All patients with dMMR tumors and a similar number of randomly selected patients with pMMR tumors were identified to undergo immune marker testing. A total of 278 dMMR tumors and 283 pMMR tumors had evaluable immune marker data (Figure S1).
Immune Markers and Clinicopathological Variables
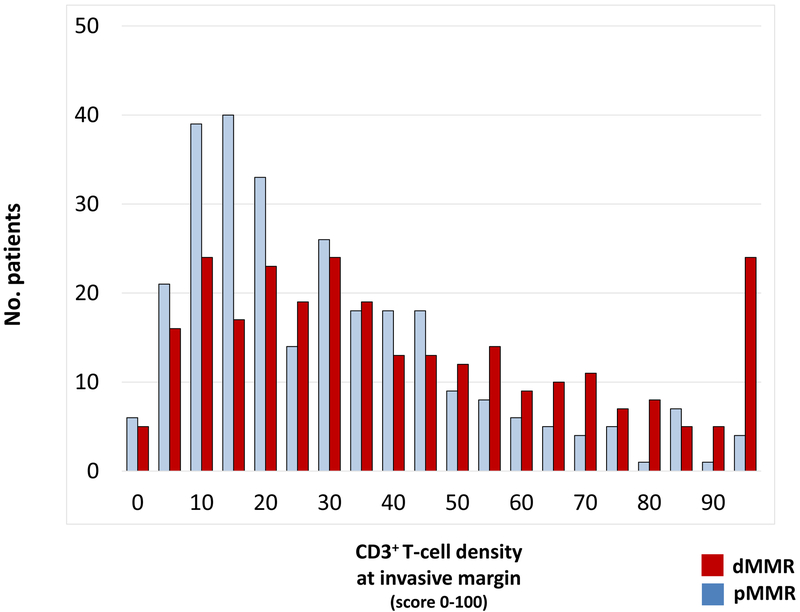
For each T-cell subtype and within each tumor compartment, the continuous density of T-cell infiltration was significantly higher in dMMR vs pMMR tumors (each P <.001). However, considerable inter-tumoral heterogeneity in T-cell subtype densities was observed between patients within each MMR group (Figure 2a, Figure S2). Variance of tumor-infiltrating T-cell densities was significantly higher by 30%−88% in dMMR vs pMMR for all T-cell subtypes: standard deviations were 28.2 vs 21.7 for CD3+IM, 28.8 vs 20.2 for CD3+CT, 28.8 vs 18.8 for CD8+IM, and 29.2 vs 15.5 for CD8+CT, respectively (F-Fold P<.0001 for each subtype; Table S1). A substantial proportion of dMMR tumors (26% to 35% depending on the T-cell subtype) exhibited T-cell densities as low as that in the bottom half of pMMR tumors (Table S1).
Figure 2: Heterogeneity of lymphocytic infiltrate between patients in mismatch repair deficient (dMMR) and proficient (pMMR) tumors.
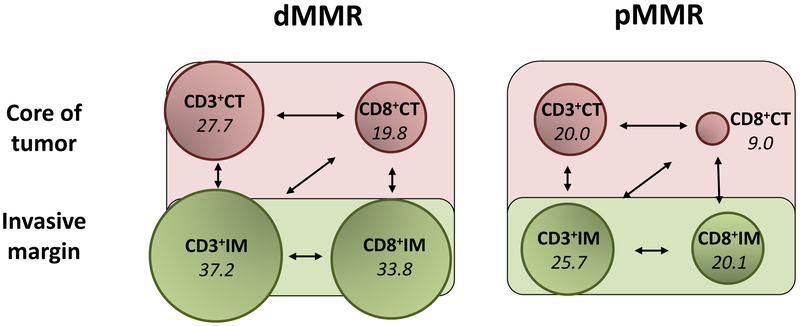
(a) The distribution of CD3+ T-cell density (score 0–100) at the invasive margin is shown across the cohort of colon cancers. For each T-cell subtype, density of tumor infiltration was higher in dMMR vs pMMR tumors (each p <.001). (b) Shown are T-cell subtypes at the invasive margin (IM) and core of the tumor (CT). The density of each T-cell subtype (shown in italics) is reflected in the size of the circles. All associations shown by arrows are statistically different (P <.0001, pair-wise comparisons using Wilcoxon Sign Rank tests).
Each T-cell subtype generally showed excellent-strong correlation with one another (Table S2). Within a given tumor, the density of lymphocytic subtypes differed topographically (Figure 2b). Among dMMR tumors, CD3+ and CD8+ TILs were significantly more abundant at the IM than in the CT: median difference in CD3+ and CD8+ densities at the IM vs CT were 8.6 (IQR 0.0–18.5; P<.0001) and 10.4 (IQR 2.1–20.1; P<.0001), respectively. A similar, though less pronounced, difference was observed among pMMR tumors: median difference in CD3+ and CD8+ densities at the IM vs CT were 5.7 (IQR −1.7–13.8; P<.0001) and 9.1 (IQR 4.1–17.5; P<.0001), respectively.
Among dMMR tumors, a lower (vs higher) CD3+ and CD8+ T-cell density was significantly associated with younger age, never (vs ever) smoking status, and low (vs high) histologic grade within either IM or CT tissue compartments, i.e., CD3+IM, CD3+CT, CD8+IM, and CD8+CT (Table S3). Furthermore, a lower CD3+IM density was significantly associated with higher T and N stage and left-sided tumor location among dMMR tumors (Table 1). Among pMMR tumors, lower densities of CD3+IM, CD3+CT, and CD8+IM were significantly associated with higher T stage (each P <.003), with CD8+CT showing a marginal association (P=.0652). Left-sided (vs right) tumors showed a lower density of most T-cell subtypes (P<.05 for CD3+IM, CD8+IM, and CD8+CT each). TIL densities did not differ significantly according to BRAF/KRAS mutation status in either MMR group. In these pre-treatment tumor specimens, the density of TIL subsets did not differ significantly by treatment arm (i.e., FOLFOX vs FOLFOX + cetuximab). Among dMMR tumors, data for MLH1 methylation and BRAF mutation (the presence of either designating sporadic origin) were available in only a limited number of dMMR tumors (n=97), and no significant differences in TIL densities between tumors with vs without these alterations were observed.
Table 1.
Association of CD3+ T-cell densities at the invasive tumor margin with clinicopathologic characteristics in stage III colon cancers with deficient and proficient DNA mismatch repair
| Deficient mismatch repair tumors | Proficient mismatch repair tumors | ||||||
|---|---|---|---|---|---|---|---|
| N | Median (IQR) | P | N | Median (IQR) | P | ||
| Age | ≤ 59 y > 59 y |
124 154 |
36.1 (19.4, 58.5) 40.0 (24.2, 73.8) |
0.0851 |
161 122 |
25.7 (15.1, 42.3) 25.6 (15.8, 45.5) |
0.8938 |
| Gender | Female | 157 | 39.0 (22.9, 64.0) | 0.5497W | 119 | 26.8 (15.8, 41.8) | 0.9595W |
| Race | Asian | 7 | 20.0 (13.3, 35.6) | 0.0668KW | 15 | 19.3 (11.8, 30.5) | 0.1125KW |
| Smoking | Ever | 72 | 49.6 (24.0, 79.4) | 0.0097W | 88 | 26.9 (15.4, 41.8) | 0.8930W |
| T stage | T1 or T2 | 24 | 57.3 (33.2, 82.7) | 0.0188KW | 35 | 43.3 (30.0, 71.7) | 0.0002KW |
| N stage | N1 | 166 | 41.0 (24.2, 70.5) | 0.0300W | 161 | 28.2 (17.4, 46.3) | 0.0578W |
| T and N Stage | T1–3N1 | 143 | 44.5 (24.9, 73.9) | 0.0059W | 140 | 29.7 (18.4, 47.3) | 0.0138W |
| Tumor Location | Right | 244 | 38.7 (23.1, 67.1) | 0.0369W | 127 | 31.4 (17.4, 48.0) | 0.0315W |
| Grade | High | 145 | 44.9 (26.3, 74.5) | 0.0013W | 67 | 26.5 (14.4, 46.9) | 0.9272W |
| BRAF/KRAS mutation status | Both wild-type | 103 | 38.2 (21.5, 57.5) | 0.9419KW | 165 | 24.1 (14.9, 42.7) | 0.4675KW |
Abbreviations: W, Wilcoxon rank-sum tests; KW, Kruskal Wallis test
Immune markers and prognosis
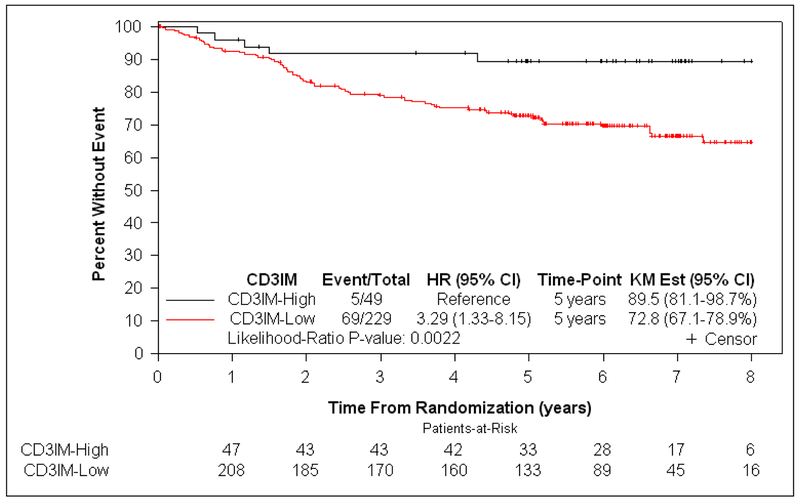
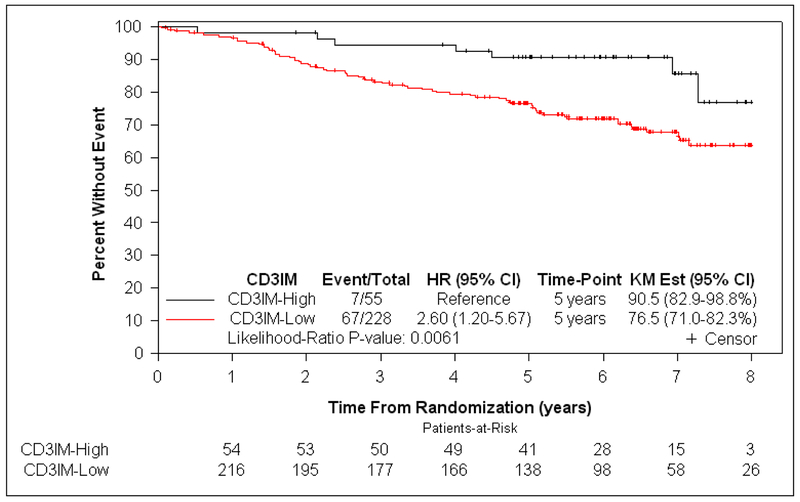
At a median follow-up of 6.3 years, patients with dMMR tumors exhibiting a lower density of CD3+ and CD8+ T cells in CT or IM (CD3+IM, CD3+CT, CD8+IM, and CD8+CT) had significantly shorter OS, independent of covariates (each Padj <.10) (Table 2). Since all associations with OS were P <.10 and all T-cell subtypes were correlated with one another, we sought to determine which T-cell subtype(s) contributed to prognosis independent of the other markers. When all T-cell subtypes were included in a backward selection model using OS as the endpoint (see Methods), all T-cell subtypes were eliminated except for CD3+IM. This finding indicates that the other T-cell subtypes (CD3+CT, CD8+IM, CD8+CT) did not enhance prognostication beyond that contributed by CD3+IM (Table 2, Figure S3). A lower density of CD3+IM among dMMR tumors was associated with shorter OS in univariate (HR for each 10-unit decrease 1.13 [95% CI 1.03–1.24]; P=.0057) and multivariable analysis (HR 1.14 for each 10-unit decrease [95% CI 1.02–1.27]; P=.0151 [Table 2]), with only CD3+IM, T, and N stage having a significant association with OS. A lower density of CD3+IM was also associated with statistically significant shorter TTR in univariate analysis (HR 1.13 for each 10-unit decrease [95% CI 1.03–1.24]; P=.0097), but not in multivariable analysis (HR 1.09 [95% CI 0.96–1.23]; P=.16). When the data were dichotomized at the cutpoint value of CD3+IM density that was intended to optimally distinguish OS (see Methods and Table S4), 82.4% (229/278) and 17.6% (49/278) of dMMR tumors had low and high CD3+IM density, respectively. Patients whose dMMR tumors had low (vs high) CD3+IM demonstrated significantly worse OS (5-year OS rate 89.5% vs 72.8%, respectively; HRadj 4.76 [95% CI 1.43–15.87]; Padj=.0019) independent of covariates (Table 3, Figure 3).
Table 2.
Tumor-infiltrating T-cell densities as continuous variables in multivariable Cox proportional hazards models for overall survival by MMR status in stage III colon cancer patients treated with FOLFOX-based adjuvant therapy
| Variable | MMR-deficient tumors | MMR-proficient tumors | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Four models of individual immune markersa | |||||
| CD3+ at invasive marginb | 1.14 (1.02–1.27) | 0.0151 | 1.20 (1.03–1.41) | 0.0153 | |
| CD3+ at central tumorb | 1.12 (1.00–1.28) | 0.0417 | 1.06 (0.91–1.25) | 0.4255 | |
| CD8+ at invasive marginb | 1.11 (1.00–1.23) | 0.0484 | 1.12 (0.93–1.35) | 0.2279 | |
| CD8+ at central tumorb | 1.11 (0.99–1.27) | 0.0644 | 1.07 (0.83–1.39) | 0.5828 | |
| Backward selection of immune markers a | |||||
| All immune markers eliminated except CD3+ at invasive margin |
Not indicated because only one immune variable (CD3+ at invasive margin) was associated with overall survival at prespecified P <.10 |
||||
In addition to one immune marker, all models included T stage (T3–4 vs T1–2), N stage (N2 vs N1), grade (high vs low), tumor side (left vs right), smoking (ever vs never), BRAF/KRAS mutation (BRAF-mutated vs KRAS-mutated vs both wild-type), age per 5-year increase.
HRs represent 10-unit decrease in immune density.
Abbreviations: HR, hazard ratio; MMR, mismatch repair
Table 3.
Dichotomized CD3+ at invasive margin relative to clinicopathologic variables in multivariable Cox proportional hazards models for overall survival by MMR status in stage III colon cancer patients treated with FOLFOX-based adjuvant therapy a
| Variable | MMR-deficient tumors | MMR-proficient tumors | ||||
|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |||
| CD3+ at invasive margin (low vs high) | 4.76 (1.43–15.87) | 0.0019 | 3.39 (1.16–9.90) | 0.0103 | ||
| T stage | T3 vs T1–2 T4 vs T1–2 |
—b | 0.0086 | 1.29 (0.38–4.37) 2.72 (0.73–10.13) |
0.0994 | |
| N stage | N2 vs N1 | 4.52 (2.49–8.19) | <.0001 | 3.68 (1.98–6.84) | <.0001 | |
| Histologic grade (high vs low) | 0.77 (0.42–1.43) | 0.3751 | 1.64 (0.90–2.98) | 0.1130 | ||
| Tumor side (left vs right) | 1.13 (0.51–2.51) | 0.7658 | 0.70 (0.36–1.37) | 0.2980 | ||
| Mutation status |
KRAS-mutated vs both wild-type BRAF-mutated vs both wild-type |
0.77 (0.28–2.14) 1.17 (0.58–2.36) |
0.7305 | 1.75 (0.93–3.29) 2.68 (1.03–6.98) |
0.0828 | |
| Smoking (ever vs never) | 0.77 (0.42–1.43) | 0.4031 | 0.88 (0.49–1.57) | 0.6620 | ||
| Age (per 5-year increase) | 1.12 (0.98–1.28) | 0.0980 | 1.25 (1.09–1.43) | 0.0010 | ||
CD3+ at invasive margin was dichotomized at an optimized cutpoint value. HRs are adjusted for all variables shown
Precise HR could not be estimated since there were no events in the T1–2 subgroup.
Abbreviations: HR, hazard ratio; MMR, mismatch repair
Figure 3:
Kaplan-Meier curves showing the association of CD3+ T-cell density at the invasive margin with overall survival in stage III colon cancer patients DNA mismatch repair deficiency (a) and proficiency (b).
Among pMMR tumors, CD3+IM was the only T-cell subtype associated with OS at P <.10 (multivariable HR for each 10-unit decrease 1.23 [95% CI 1.04–1.43]; P=.0087) (Table 2). Thus, backward selection was not indicated. A lower density of CD3+IM among pMMR tumors was associated with statistically significant shorter TTR in univariate (HR 1.29 [95% CI 1.13–1.46]; P<.0001) and multivariable analysis (HR 1.19 [95% CI 1.01–1.40]; P=.0236). At the cutpoint value of CD3+IM intended to optimally distinguish OS (Methods, Table S5), 80.6% (228/283) and 19.4% (55/283) of pMMR tumors had low and high CD3+IM density, respectively. Patients with low (vs high) CD3+IM pMMR tumors had significantly worse OS (5-year OS rate 90.5% vs 76.5%, respectively; HRadj 3.39 [95% CI 1.16–9.90]; P adj =.0103) (Table 3, Figure 3).
DISCUSSION
We examined CD3+ and CD8+ T-cell subtypes in a large number of patients with dMMR colon cancers of uniform stage (III) who were treated with FOLFOX-based adjuvant therapy in a clinical trial. We confirmed that dMMR tumors have a significantly higher TIL density compared to pMMR tumors. MMR-deficient tumors have generally been regarded as a relatively homogeneous group with high tumor mutation burden and neoantigen load that trigger abundant TILs (10, 29, 34). We made the novel observation of considerable inter-tumoral heterogeneity in the density of CD3+ and CD8+ T-cell densities between dMMR tumors that was greater than observed between pMMR tumors. A significant proportion (26% to 35%) of dMMR tumors exhibited T-cell densities as low as those observed among pMMR tumors. Moreover, we found that the observed inter-tumoral heterogeneity in TILs impacted patient survival in both dMMR and pMMR colon cancers. A weak pre-existing antitumor T-cell response, as indicated by a low density of CD3+ and CD8+ T cells, was associated with significantly shorter OS independent of confounders. We report for the first time that CD3+ T-cell density can prognosticate within the dMMR population in patients who received protocol-defined therapy. Given that dMMR can predict responsiveness to anti-PD-1 therapy, these new data may have implications for variability in immunotherapy responsiveness in dMMR tumors.
We found that prognostic information provided by immune infiltrates was not the same across T-cell subtypes and topographical location within the tumor microenvironment. Among dMMR tumors, although 3 of the 4 immune variables individually had statistically significant associations with OS, CD3+IM density contributed the most prognostic information, and the other three immune variables (CD8+IM, CD8+CT, CD3+CT) did not add further prognostic value. Similarly, only CD3+IM was prognostic among pMMR tumors. Consistent among MMR groups was the finding that the host anti-tumor immune response was most intense at the IM, presumably to impede further advance of malignant cells. In contrast to the well-described Immunoscore which utilizes all four TIL variables (ie, CD3+IM, CD3+CT, CD8+IM, CD8+CT) (19, 35), we utilized a prognostic model that required fewer immune variables and demonstrated that CD3+IM alone provides robust prognostic information. An advantage of this approach includes a simpler tumor analysis and anticipated lower cost of translating these findings into the clinic.
A number of factors could underlie inter-tumoral heterogeneity of TILs among dMMR tumors including differences in the overall mutation burden, unstable microsatellites, or variability in neoantigen profiles (36). A recent study found that MSI CRCs from patients with Lynch Syndrome (n=85) had a higher density of CD3+ TILs in association with more somatic mutations, higher neoantigen burden, and better survival compared to sporadic-MSI CRCs (n=67) (37). Recent analysis of cancer exomes in 18 cancer types found a correlation between survival outcomes and the overall burden of unstable microsatellites, suggesting that MSI may be more informative when analyzed as a continuous rather than a discrete phenotype (38). Among MSI CRCs, those with WNT/beta-catenin mutations displayed decreased intratumoral CD8+ density and a reduced PD-1+ infiltrate in the central tumor compared to other MSI patients (10). Activated WNT/beta-catenin signaling in melanoma specimens was recently reported to correlate with the absence of a T-cell gene expression signature, and mouse models revealed that activated WNT/beta-catenin signaling led to T-cell exclusion and resistance to anti-PD-L1/anti-cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) monoclonal antibody therapy (27).
Recent reports indicate that anti-PD-1 antibodies yield a response rate of 31–36% in advanced dMMR/MSI-H CRC patients after failure of conventional therapy, with some responses lasting 12 months or longer (39, 40), and with higher response rates seen for the combination of nivolumab plus ipilimumab (41). The observation that a significant portion of dMMR CRC patients do not respond to immune checkpoint inhibition underscores the need to elucidate factors such as immune density or composition that may be contributory. Since our study population was stage III colon cancer, our findings may be relevant to clinical benefit of the anti-PD-L1 antibody, atezolizumab, that is being studied in an ongoing phase 3 trial (ATOMIC) in which stage III dMMR colon cancers are randomized to receive FOLFOX alone or combined with atezolizumab. In contrast to dMMR tumors, anti-PD-1 therapy has not shown a benefit for patients with pMMR/microsatellite stable tumors. Importantly, we found that a subset of pMMR tumors exhibited TIL densities that were similar to those found in dMMR tumors. A dense infiltrate in pMMR tumors also predicted more favorable OS, consistent with prior findings by our group (14, 19) and others (11, 12, 20–22). These findings suggest that the observed inter-tumoral heterogeneity of TILs and its prognostic impact could be associated with responsiveness to immune checkpoint inhibitors, although we emphasize that our study does not provide predictive data for immune markers.
Strengths of our study include the use of a molecularly annotated clinical trial cohort with prospective specimen collection, in which patients of uniform tumor stage received protocol-defined adjuvant therapy. Follow-up data were collected prospectively in a robust and meticulous manner. By contrast, prior evaluations were performed on retrospectively collected specimens from case series or population-based studies of pooled tumor stage, varying treatments, suboptimal follow-up and/or lymphocyte populations that were defined only in H&E-stained sections (10, 18, 21). Limitations include lack of data on other intratumoral immune cell populations and the lack of data on the predictive potential of intratumoral immune infiltration which was not possible in our cohort since all patients received chemotherapy. A recent analysis of CRC samples from non-trial sources of varying disease stages reported that expression of immune checkpoints may act as antagonists to cancel the prognostic relevance of TILs even in MSI-high tumors (42). Such an analysis in our cohort will be the subject of future research.
In conclusion, we report greater inter-tumoral heterogeneity of CD3+ and CD8+ T-cell densities in dMMR vs pMMR stage III colon cancers, as well as the ability of CD3+ TILs to prognosticate among dMMR tumors. Low CD3+ T-cell infiltration at the invasive tumor margin was the most informative T-cell subtype, and identifies both dMMR and pMMR colon cancers with shorter OS. These findings underscore the clinical importance of heterogeneity in T-cells markers in the tumor microenvironment and demonstrate the prognostic utility of measuring T-cell subtype densities within dMMR tumors. Lastly, our data suggest that measurement of CD3+ and CD8+ T-cell densities warrant evaluation as a predictive biomarker for the efficacy of in immunotherapy in patients with CRC.
Supplementary Material
TRANSLATIONAL RELEVANCE.
We examined CD3+ and CD8+ T-cell subtypes in stage III colon cancers from patients who received adjuvant FOLFOX-based chemotherapy in a phase III trial. Mismatch repair-deficient (dMMR) tumors are hypermutated and generally harbor abundant lymphocytic infiltrates which are believed to contribute to their prognostic advantage and may underlie their enhanced responsiveness to immunotherapy. We report that dMMR colon cancers demonstrate significantly greater inter-tumoral heterogeneity in the density of CD3+ and CD8+ tumor-infiltrating T-lymphocytes compared to MMR-proficient (pMMR) tumors. We also report the novel observation that CD3+ and CD8+ lymphocyte density can individually prognosticate within the dMMR group in a clinical trial cohort. The observed inter-tumoral heterogeneity of T-cell densities can account for prognostic differences among observed among dMMR tumors, and may be a relevant factor for the efficacy of adjuvant immunotherapy which is currently being studied in stage III colon cancers.
Acknowledgements
The authors are grateful to Raquel J. Ostby for administrative assistance.
Funding: This work was supported by the National Cancer Institutes of Health R01 CA210509–01A1 (to FAS) and U10CA180790, U10CA180821, U10CA180882 and 1UG1CA189823 (to the Alliance for Clinical Trials in Oncology. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest Disclosure Statement: Azita Djalilvan, Faith Ough, June Clements, Kandavel Shanmugam, Michael Barnes, Rebecca Bowermaster, Andrea Muranyi and Joerg Bredno are employees of Roche Tissue Diagnostics; Azita Djalilvand, Faith Ough, June Clements, Kandavel Shanmugam, Michael Barnes and Andrea Muranyi own stock in Roche Tissue Diagnostics; Azita Djalilvand, Faith Ough, June Clements, Michael Barnes, Rebecca Bowermaster, Andrea Muranyi and Joerg Bredno receive royalties from Roche Tissue Diagnostics; and Andrea Muranyi receives travel reimbursement from Roche Tissue Diagnostics. Wen-Wei Liu is employed by and receives royalties from Ventana Medical Systems, Inc. Harry Yoon has received research funding from Roche/Genentech and Merck. Frank Sinicrope has served in a consulting or advisory role for Ventana Medical Systems, EMD Serono, and Roche/Genentech, has received travel, accommodations and/or expenses from Ventana Medical Systems.
REFERENCES
- 1.Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:5313–27. [DOI] [PubMed] [Google Scholar]
- 2.Nagtegaal ID, Quirke P, Schmoll HJ. Has the new TNM classification for colorectal cancer improved care? Nature reviews Clinical oncology. 2011;9:119–23. [DOI] [PubMed] [Google Scholar]
- 3.Atlas TCG. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinicrope FA, Mahoney MR, Smyrk TC, Thibodeau SN, Warren RS, Bertagnolli MM, et al. Prognostic Impact of Deficient DNA Mismatch Repair in Patients With Stage III Colon Cancer From a Randomized Trial of FOLFOX-Based Adjuvant Chemotherapy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:3664–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaanan A, Shi Q, Taieb J, Alberts SR, Meyers JP, Smyrk TC, et al. Role of Deficient DNA Mismatch Repair Status in Patients With Stage III Colon Cancer Treated With FOLFOX Adjuvant Chemotherapy: A Pooled Analysis From 2 Randomized Clinical Trials. JAMA Oncology. 2018;4:379–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lochhead P, Kuchiba A, Imamura Y, Liao X, Yamauchi M, Nishihara R, et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105:1151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benatti P, Gafa R, Barana D, Marino M, Scarselli A, Pedroni M, et al. Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res. 2005;11:8332–40. [DOI] [PubMed] [Google Scholar]
- 9.Williams DS, Bird MJ, Jorissen RN, Yu YL, Walker F, Zhang HH, et al. Nonsense mediated decay resistant mutations are a source of expressed mutant proteins in colon cancer cell lines with microsatellite instability. PLoS One. 2010;5:e16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity. 2016;44:698–711. [DOI] [PubMed] [Google Scholar]
- 11.Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–66. [DOI] [PubMed] [Google Scholar]
- 12.Pages F, Kirilovsky A, Mlecnik B, Asslaber M, Tosolini M, Bindea G, et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J Clin Oncol. 2009;27:5944–51. [DOI] [PubMed] [Google Scholar]
- 13.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. [DOI] [PubMed] [Google Scholar]
- 14.Yoon HH, Orrock JM, Foster NR, Sargent DJ, Smyrk TC, Sinicrope FA. Prognostic impact of FoxP3+ regulatory T cells in relation to CD8+ T lymphocyte density in human colon carcinomas. PLoS One. 2012;7:e42274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nosho K, Baba Y, Tanaka N, Shima K, Hayashi M, Meyerhardt JA, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222:350–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogino S, Nosho K, Irahara N, Meyerhardt JA, Baba Y, Shima K, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15:6412–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zlobec I, Karamitopoulou E, Terracciano L, Piscuoglio S, Iezzi G, Muraro MG, et al. TIA-1 Cytotoxic Granule-Associated RNA Binding Protein Improves the Prognostic Performance of CD8 in Mismatch Repair-Proficient Colorectal Cancer. PLoS One. 2010;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rozek LS, Schmit SL, Greenson JK, Tomsho LP, Rennert HS, Rennert G, et al. Tumor-Infiltrating Lymphocytes, Crohn’s-Like Lymphoid Reaction, and Survival From Colorectal Cancer. J Natl Cancer Inst. 2016;108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinicrope FA, Shi Q, Hermitte F, Heying EN, Benson AB, Gill S, et al. Association of immune markers and Immunoscore with survival of stage III colon carcinoma (CC) patients (pts) treated with adjuvant FOLFOX: NCCTG N0147 (Alliance). J Clin Oncol. 2017;35:3579-. [Google Scholar]
- 20.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. [DOI] [PubMed] [Google Scholar]
- 21.Galon J, Mlecnik B, Marliot F, Ou F- S, Bifulco CB, Lugli A, et al. Validation of the Immunoscore (IM) as a prognostic marker in stage I/II/III colon cancer: Results of a worldwide consortium-based analysis of 1,336 patients. J Clin Oncol. 2016;34:3500-. [Google Scholar]
- 22.Galon J, Pages F, Marincola FM, Angell HK, Thurin M, Lugli A, et al. Cancer classification using the Immunoscore: a worldwide task force. J Transl Med. 2012;10:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. The New England journal of medicine. 2012;366:2455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Des Guetz G, Schischmanoff O, Nicolas P, Perret GY, Morere JF, Uzzan B. Does microsatellite instability predict the efficacy of adjuvant chemotherapy in colorectal cancer? A systematic review with meta-analysis. Eur J Cancer. 2009;45:1890–6. [DOI] [PubMed] [Google Scholar]
- 25.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. The New England journal of medicine. 2012;366:2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, Carmona M, Kivork C, Seja E, Cherry G, Gutierrez AJ, Grogan TR, Mateus C, Tomasic G, Glaspy JA, Emerson RO, Robins H, Pierce RH, Elashoff DA, Robert C, Ribas A. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014. November 27;515(7528):568–71. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, Alaparthy S, Berman D, Jure-Kunkel M, Siemers NO, Jackson JR, Shahabi V. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother. 2012. July;61(7):1019–31. doi: 10.1007/s00262-011-1172-6. Epub 2011 Dec 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. The New England journal of medicine. 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alberts SR, Sargent DJ, Nair S, Mahoney MR, Mooney M, Thibodeau SN, et al. Effect of oxaliplatin, fluorouracil, and leucovorin with or without cetuximab on survival among patients with resected stage III colon cancer: a randomized trial. JAMA : the journal of the American Medical Association. 2012;307:1383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barnes M, Srinivas C, Bai I, Frederick J, Liu W, Sarkar A, et al. Whole tumor section quantitative image analysis maximizes between-pathologists’ reproducibility for clinical immunohistochemistry-based biomarkers. Laboratory investigation; a journal of technical methods and pathology. 2017;97:1508–15. [DOI] [PubMed] [Google Scholar]
- 32.Sarkar A, Yuan Q, Srinivas C. A robust method for inter-marker whole slide registration of digital pathology images using lines based features. IEEE 11th International Symposium on Biomedical Imaging (ISBI). 2014;April:762–5, doi: 10.1109/ISBI.2014.6867982. [DOI] [Google Scholar]
- 33.Lorsakul A, Bredno J, Ochs RL, Morrison L, Day W. Validation of multiplex immunohistochemistry assays using automated image analysis. International Society for Optics and Photonics Medical Imaging. 2018;Digital Pathology:doi: 10.1117/12.2293168. [DOI] [Google Scholar]
- 34.Maby P, Tougeron D, Hamieh M, Mlecnik B, Kora H, Bindea G, et al. Correlation between Density of CD8+ T-cell Infiltrate in Microsatellite Unstable Colorectal Cancers and Frameshift Mutations: A Rationale for Personalized Immunotherapy. Cancer Res. 2015;75:3446–55. [DOI] [PubMed] [Google Scholar]
- 35.Pages F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391:2128–39. [DOI] [PubMed] [Google Scholar]
- 36.Schwitalle Y, Linnebacher M, Ripberger E, Gebert J, von Knebel Doeberitz M. Immunogenic peptides generated by frameshift mutations in DNA mismatch repair-deficient cancer cells. Cancer Immun. 2004;4:14. [PubMed] [Google Scholar]
- 37.Liu GC, Liu RY, Yan JP, An X, Jiang W, Ling YH, et al. The Heterogeneity Between Lynch-Associated and Sporadic MMR Deficiency in Colorectal Cancers. J Natl Cancer Inst. 2018. [DOI] [PubMed] [Google Scholar]
- 38.Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med. 2016;22:1342–50. [DOI] [PubMed] [Google Scholar]
- 39.Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. The Lancet Oncology. 2017;18:1182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Broderick JM. FDA Approves Pembrolizumab for Microsatellite Instability-High and Mismatch Repair Deficient Cancers. OncLive. 2017:http://www.onclive.com/web-exclusives/fda-approves-pembrolizumab-for-microsatellite-instabilityhigh-and-mismatch-repair-deficient-cancers.
- 41.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, et al. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2018;36:773–9. [DOI] [PubMed] [Google Scholar]
- 42.Marisa L, Svrcek M, Collura A, Becht E, Cervera P, Wanherdrick K, et al. The Balance Between Cytotoxic T-cell Lymphocytes and Immune Checkpoint Expression in the Prognosis of Colon Tumors. J Natl Cancer Inst. 2018;110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.