Abstract
Purpose:
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive disease, affecting children and adults. Chemotherapy treatments show high response rates but have debilitating effects and carry risk of relapse. Previous work implicated NOTCH1 and other oncogenes. However, direct inhibition of these pathways affects healthy tissues and cancer alike. Our goal in this work has been to identify enzymes active in T-ALL whose activity could be targeted for therapeutic purposes.
Experimental Design:
To identify and characterize new NOTCH1 druggable partners in T-ALL, we coupled studies of the NOTCH1 interactome to expression analysis and a series of functional analyses in cell lines, patient samples and xenograft models.
Results:
We demonstrate that ubiquitin-specific protease 7 (USP7) interacts with NOTCH1 and controls leukemia growth by stabilizing the levels of NOTCH1 and JMJD3 histone demethylase. USP7 is highly expressed in T-ALL and is transcriptionally regulated by NOTCH1. In turn, USP7 controls NOTCH1 levels through deubiquitination. USP7 binds oncogenic targets and controls gene expression through stabilization of NOTCH1 and JMJD3 and ultimately H3K27me3 changes. We also show that USP7 and NOTCH1 bind T-ALL superenhancers, and inhibition of USP7 leads to a decrease of the transcriptional levels of NOTCH1 targets and significantly blocks T-ALL cell growth in vitro and in vivo.
Conclusions:
These results provide a new model for USP7 deubiquitinase activity through recruitment to oncogenic chromatin loci and regulation of both oncogenic transcription factors and chromatin marks to promote leukemia. Our studies also show that targeting USP7 inhibition could be a therapeutic strategy in aggressive leukemia.
Keywords: leukemia, epigenetics, post-translational regulation, targeted therapy, xenograft
Introduction
Acute lymphoblastic leukemia (ALL) represents 25% of childhood tumors1,2. T cell ALL (T-ALL)1–7 is the most aggressive subtype of ALL and these tumors exhibit a significant risk of recurrence due to tumor cells that are refractory to chemotherapy, a common characteristic of high-risk T-ALL. In this regard, 20% of pediatric, and more than 50% of adult patients, with T-ALL fail to achieve a complete remission after chemotherapy, making resistance to therapy the most substantial challenge in treatment5,8,9. Immunotherapy using engineered T cells appears promising for some patients, but these experimental therapies also appear fraught with unacceptable toxicities. ALL, thus, represents a high unmet need. To this end, improvement in therapy, as well as personalized cancer medicine, should identify subgroups of patients, based on the molecular mechanism of resistance, which will benefit patients through the development of novel therapeutic strategies, including targeted agents in high risk T-ALL patients.
The NOTCH11,2,10–26 signaling pathway is activated in more than 50% of T-ALL cases. NOTCH1 mutations were initially identified in the form of chromosomal translocation t(7;9)(q34;q34.3), which leads to the expression of a truncated and constitutively active form of NOTCH110. In most T-ALL cases NOTCH1 is activated via mutations that disrupt specific domains responsible for the nuclear levels of NOTCH1. NOTCH1-positive leukemia presents with infiltration of bone marrow, spleen and liver with leukemic blasts and lead to a fulminant terminal disease that, if untreated, can lead to death over the course of months. Seminal work has described the major oncogenic pathways (i.e. NOTCH1) in T cell ALL (T-ALL)1,2,10–21. Chemical inhibitors or antibodies against NOTCH1 have been unsuccessful in the clinic, due to high levels of on-target toxicity, given the involvement of this protein in multiple physiological processes11,27.
Others and we have previously shown that loss of the repressive mark trimethylation of lysine 27 on histone H3 (H3K27me3) is important for leukemia initiation and progression15, and NOTCH1 recruitment to target genes and loss of H3K27me3 correlate during leukemogenesis15. We also identified and characterized the pro-oncogenic role of the epigenetic modulator Jumonji D3 (JMJD3)28–31, which mediates NOTCH1 oncogenic function in T-ALL16. JMJD3 is a NOTCH1 interactor and is recruited by NOTCH1 to oncogenic targets16.
Here we sought out to identify additional NOTCH1 co-factor proteins with pro-oncogenic functions that can be targeted in T-ALL contexts without causing toxicity. In this study, we used a number of patient samples and cell lines of T-ALL, as well as preclinical models, including patient-derived xenografts. We identified a novel member of the NOTCH1 transcriptional complex in T-ALL, the first deubiquitinase (DUB) protein with critical role in leukemia, referred to as ubiquitin-specific peptidase 7 (USP7)32–43. We have shown that: (1) the NOTCH1 complex interacts with and is stabilized by USP7; (2) USP7 levels are significantly increased in T-ALL tumor samples compared to normal T cells via a positive feedback loop with NOTCH1; (3) a small chemical molecule that inhibits USP7 (USP7i), leads to a blockade in T-ALL cell growth, without associated toxicity in healthy animals. Thus, it is proposed to determine the mechanisms employed by USP7/NOTCH1 that leads to a T-ALL phenotype and most importantly, can targeting USP7 reverse this phenotype in patients.
Materials and Methods
Cell lines and primary cells
The human T-ALL cell lines CUTLL1 (gift from Iannis Aifantis, New York University), LOUCY (gift from Pieter Van Vlierberghe, Cancer Research Institute, Belgium), CCRF-CEM (American Type Culture Collection (ATCC), Manassas, VA, #CCL-119), JURKAT (ATCC), HPB (HPB-ALL) and KOPTK1 (Iannis Aifantis’ group) were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated FBS (Sigma-Aldrich, St. Louis, MO), 2% penicillin/streptomycin (Gibco, Fisher Scientific, Hampton, NH), and 1% GlutaMAX (Gibco, Fisher Scientific). 293T cells (ATCC, #CRL-11268) were maintained in DMEM medium supplemented with 10% heat-inactivated FBS, 2% penicillin/streptomycin, and 1% GlutaMAX. Human pan T cells were purchased from AllCells.com (Alameda, CA). Primary human samples were collected by collaborating institutions with informed consent and analyzed under the supervision of the Institutional Review Board of Ghent University. Informed consent to use leftover material for research purposes was obtained from all of the patients at trial entry in accordance with the Declaration of Helsinki.
Antibodies and reagents
The following antibodies were used for western blot: mouse anti-Actin (Millipore, Billerica, MA, clone C4), rabbit anti-JMJD3 (Cell Signaling Technology, Danvers, MA, #3457), rabbit anti-USP7 (Bethyl Laboratories, Montgomery, TX, #A300–033A-7), rabbit anti-Cleaved NOTCH1 (Val1744) (Cell Signaling Technology, #4147), rabbit anti-K48 specific ubiquitin (Millipore, Billerica, MA, 05–1307), rabbit anti-Lamin B1(Proteintech, 12987–1-AP), rabbit anti-USP9X (Bethyl Laboratories, Montgomery, TX, #A301–351A), anti-Flag (Sigma, M2 clone), anti-HA (Abcam, Cambridge, MA, ab18181) and rabbit anti-USP24 (Bethyl Laboratories, Montgomery, TX, #A300–938A).
The following antibodies were used for ChIP: rabbit anti-H3K27Ac (Cell Signaling Technology, #8173S), rabbit anti-H3K27me3 (Cell Signaling Technology, #9733S), rabbit anti-H2BK120ub (Cell Signaling Technology, #5546S), rabbit anti-H2AK119ub (Cell Signaling Technology, #8240S), rabbit anti-H3K79me2 (made by Ali Shilatifard’s laboratory, Northwestern University), rabbit anti-H3K4me3 (Ali Shilatifard’s laboratory), normal rabbit IgG (Millipore, #12–370), anti-Flag (Sigma, M2 clone) and rabbit anti-USP7 (Bethyl Laboratories, Montgomery, TX, #A300–033A-7), The rabbit anti-C-MYC:(N-262, Santa Cruz Biotechnology) and goat anti-RUNX1:(Santa Cruz Biotech, SC- 8563) antibodies were used in published ChIP-seq studies we use in our analysis. A RUNX1 antibody from Cell Signaling Technology was used for our western blot and ChIP-qPCR studies (AML1 antibody #4334). Secondary antibodies for Western blots were HRP-conjugated anti-rabbit and anti-mouse IgG (GE Healthcare, Chicago, IL).
Quick Start Bovine Gamma Globulin (BGG) Standard Set (protein standards) were purchased from Bio-Rad (Hercules, CA); benzonase, RNase A, dithiothreitol (DTT), EZview Red Anti-HA Affinity Gel (HA beads), and Influenza Hemagglutinin (HA) peptide were purchased from Sigma-Aldrich; NaV and NaF were purchased from New England BioLabs (Ipswich, MA); Protein G Dynabeads were purchased from Life Technologies (Carlsbad, CA); IgG-free BSA was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA); phenol chloroform was purchased from ThermoScientific (Waltham, MA); and proteinase K, Tousimis formaldehyde, and Peptide International Z-Leu-Leu-Leu-H aldehyde (MG132) were purchased from Fisher Scientific. The latest generation of Progenra inhibitors (USP7i), P217564, was a kind gift from Progenra (Malvern, PA). Several additional inhibitors from Progenra were used (P5091 and P22077) yielding results similar to P217564. All USP7 inhibitors used in the manuscript have been published previously as referred to in the manuscript. GSKJ4 was purchased from Cayman Chemical, Ann Arbor, MI, #12073).
Immunoprecipitation (IP)
100 million T-ALL cells were collected and washed with chilled PBS. Cells were resuspended in 5 volumes of Buffer A (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 1:100 protease inhibitor (Sigma-Aldrich, P8340), 1 mM NaV, 1 mM NaF, and 0.5 mM DTT in H2O), incubated on ice for 10 min, and lysed using a Dounce homogenizer. Nuclear pellets were resuspended in 1.5 ml of TENT buffer (50 mM Tris pH 7.5, 5 mM EDTA, 150 mM NaCl, 0.05% v/v Tween 20, 1:100 protease inhibitor (Sigma-Aldrich, P8340), 1 mM NaV, 1 mM NaF, and 0.5 mM DTT in H2O) containing 5 mM MgCl2 and 100 units benzonase, and incubated at 4°C for 30 min, rotating. Lysates were passed through a 251/2G needle/syringe 5 times, and spun down at 4°C, 2000 RPM, for 7 min to remove debris. Protein G magnetic beads were added to the lysates to decrease non-specific binding and incubated at 4°C for 30 min, rotating. Precleared lysates were then incubated with the appropriate antibody-conjugated beads (5 μg antibody per 100 million cells) at 4°C overnight, rotating. Beads were washed 4 times in TENT buffer at 4°C for 3 min, and protein complexes were eluted in 200 μl 0.1M glycine pH 2.5 for 10 min at 25°C, shaking. 20 μl of 1M Tris pH 8.0 was then added to the supernatants. For IP of HA-JMJD3, precleared lysates were incubated with HA-coated beads overnight, and protein complexes were eluted in 200 μl TENT buffer containing 400 μg/ml HA peptide (Sigma-Aldrich) at 4°C overnight, rotating.
Mass spectrometry
Histone epiproteomics analysis was performed as previously described44,45. For the JMJD3 mass spectrometry the affinity-purified proteins were reduced, alkylated, and loaded onto an SDS-PAGE gel to remove any detergents and LCMS incompatible reagents. The gel plugs were excised, destained, and subjected to proteolytic digestion with trypsin. The resulting peptides were extracted and desalted as previously described46. An aliquot of the peptides was analyzed with LCMS coupled to a ThermoFisher Scientific Orbitrap QExactive Mass Spectrometer operated in data dependent mode. The data was searched against a UniProt human database, using Sequest within Proteome Discoverer. The results were filtered with a 1% FDR searched against a decoy database and for proteins with at least two unique peptides. Word clouds were generated using the statistical program R, and gene ontology was performed using the Gene Ontology Consortium (http://www.geneontology.org/).
In more detail:
As previously described47 the affinity purified proteins were resuspended in NuPAGE® LDS Sample Buffer (Novex). The samples were reduced with 2μl of 0.2M dithiothreitol (Sigma) for one hour at 57 ⁰C at pH 7.5. Next the samples were alkylated with 2μl of 0.5M iodoacetamide (Sigma) for 45 minutes at room temperature in the dark. The samples were loaded on a NuPAGE® 4–12% Bis-Tris Gel 1.0 mm (Life Technologies) and run for 8 minutes at 200V. The gel was stained with GelCode Blue Stain Reagent (Thermo). The gel plugs were excised and destained for 15 minutes in a 1:1 (v/v) solution of methanol and 100mM ammonium bicarbonate. The buffer was exchanged and the samples were destained for another 15 minutes. This was repeated for another 3 cycles. The gel plugs were dehydrated by washing with acetonitrile, and further dried by placing them in a SpeedVac for 20 minutes. 300ng of sequencing grade modified trypsin (Promega) was added directly to the dried gel plugs followed by enough 100mM ammonium bicarbonate to cover the gel pieces. The gel plugs were allowed to shake at room temperature and digestion proceeded overnight. The digestion was halted by adding a slurry of R2 50 μm Poros beads (Applied Biosystems) in 5% formic acid and 0.2% trifluoroacetic acid (TFA) to each sample at a volume equal to that of the ammonium bicarbonate added for digestion. The samples were allowed to shake at 4⁰C for 120 mins. The beads were loaded onto C18 ziptips (Millipore), equilibrated with 0.1% TFA, using a microcentrifuge for 30 s at 6,000 rpm. The beads were washed with 0.5% acetic acid. Peptides were eluted with 40% acetonitrile in 0.5% acetic acid followed by 80% acetonitrile in 0.5% acetic acid. The organic solvent was removed using a SpeedVac concentrator and the sample reconstituted in 0.5% acetic acid.
Analysis--Mass Spectrometry
An aliquot of each sample was loaded onto an Acclaim PepMap100 C18 75-μm x 15-cm column with 3μm bead size coupled to an EASY-Spray 75-μm x 50-cm PepMap C18 analytical HPLC column with a 2μm bead size using the auto sampler of an EASY-nLC 1000 HPLC (ThermoFisher) and solvent A (2% acetonitrile, 0.5% acetic acid). The peptides were eluted into a Thermo Fisher Scientific Orbitrap Q Exactive Mass Spectrometer increasing from 2% to 30% solvent B (90% acetonitrile, 0.5% acetic acid) in 60 minutes, followed by an increase from 30% to 40% solvent B in 10 minutes and 40–100% solvent B in another 10 minutes.
High resolution full MS spectra were obtained with a resolution of 70,000, an AGC target of 1e6, with a maximum ion time of 120ms, and a scan range from 400 to 1500m/z. Following each full MS scan, twenty data-dependent MS/MS spectra were acquired. The MS/MS spectra were collected with a resolution of 17,500 an AGC target of 5e4, maximum ion time of 120ms, one microscan, 2m/z isolation window, fixed first mass of 150 m/z, dynamic exclusion of 30 sec, and Normalized Collision Energy (NCE) of 27.
Data Processing—Mass spectrometry
All acquired MS2 spectra were searched against a UniProt human database using Sequest within Proteome Discoverer (ThermoScientific). The search parameters were as follows: precursor mass tolerance ±10 ppm, fragment mass tolerance ± 0.02 Da, digestion parameters trypsin allowing 2 missed cleavages, fixed modification of carbamidomethyl on cysteine, variable modification of oxidation on methionine, and variable modification of deamidation on glutamine and asparagine. The results were filtered with a 1% FDR searched against a decoy database and for proteins with at least two unique peptides. Proteins enriched in the sample affinity purification over the empty vector control were further interrogated.
Reverse phase protein array (RPPA)
RPPA was performed as described in Milani G. et al., Oncotarget 2014, and Serafin V. et al., Leukemia 2017. Briefly, cells were lysed in an appropriate lysis buffer with proteases and phosphatases inhibitors, serially diluted into four-points dilution curves and printed on nitrocellulose-coated glass slides with the 2470 Aushon Arrayer (Aushon Biosystems).
Western blot
To make total cell extracts, up to 10 million cells were collected and resuspended in 20 μl RIPA buffer (50 mM Tris HCl pH 8.0, 150 mM NaCl, 1% NP-40/IGEPAL, 0.5% sodium deoxycholate, 0.1% SDS, 1:100 protease inhibitor (Sigma-Aldrich, P8340), 1 mM NaV, and 1 mM NaF in H2O) per 1 million cells. Cells were lysed on ice for 20 min, and spun down at 4°C, max speed, for 10 min to remove debris.
Protein concentrations were determined via Bradford assay. Samples and buffer were diluted 1:10 in H2O. 2 μl of protein standards, H2O, or diluted sample were added to wells of a 96-well plate in duplicate. Then, 2 μl of diluted buffer and 100 μl Quick Start Bradford 1X Dye Reagent (Bio-Rad) were added to each well, and absorbance was measured at 600nm using the GloMax-Multi Detection System (Promega, Madison, WI).
Up to 50 μg sample was boiled in 1X SDS loading dye (Bio-Rad) at 95°C for 10 min prior to loading into 4–15% Tris-glycine polyacrylamide gels (Bio-Rad). 8 μl of PageRuler Plus Prestained Protein Ladder (10–250kD; Fisher Scientific) was also loaded. Gels were run at 100V until samples reached the separating part of the gel, and then were run at 130V. Gels were transferred for 1.5h at 80V or overnight at 35–40V, and membranes were blocked in 5% milk in TBST (0.1% Tween 20 in 1X TBS) for 1h. Membranes were incubated at 4°C overnight with the appropriate antibody in TBST. Then, the membranes were washed 3 times for 10 min with TBST, incubated for 2h at 4°C with the appropriate secondary antibody, washed 3 times for 10 min with TBST, and developed using Clarity Western ECL Substrate (Bio-Rad), or SuperSignal West Femto Maximum Sensitivity Substrate (ThermoScientific) as needed, on a Bio-Rad ChemiDoc Touch Imaging System. Analysis was performed using Image Lab software (Bio-Rad).
Chromatin immunoprecipitation (ChIP)
10 million T-ALL cells were cross-linked in 1 ml/million cells fixation buffer (1% formaldehyde, 1X PBS, and 1% FBS in H2O) for 10 min at 25°C. Then, 1:12.5 glycine [2.5M] was added for 5 min. Pelleted cells were then lysed according to the type of ChIP performed.
For histone ChIPs, cells were lysed in 375 μl of Nuclei Incubation Buffer (15 mM Tris pH 7.5, 60 mM KCl, 150 mM NaCl, 15 mM MgCl2, 1 mM CaCl2, 250 mM Sucrose, 0.3% NP-40, 1 mM NaV, 1 mM NaF, and 1 EDTA-free protease inhibitor tablet (Roche, Pleasanton, CA)/10 ml in H2O) for 10 min on ice. Nuclei were washed once with Digest Buffer (10 mM NaCl, 10 mM Tris pH 7.5, 3 mM MgCl2, 1 mM CaCl2, 1 mM NaV, 1 mM NaF, and 1 EDTA-free protease inhibitor tablet (Roche)/10 ml in H2O) and resuspended in 57 μl Digest Buffer containing 4.5 units MNase (USB, Cleveland, OH) for 1h at 37°C. MNase activity was quenched for 10 min on ice upon the addition of EDTA to a final concentration of 20 mM. Pelleted nuclei were lysed in 300 μl Nuclei Lysis Buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA pH 8.0, 1% SDS, 1 mM NaV, 1 mM NaF, and 1 EDTA-free protease inhibitor tablet (Roche)/10 ml in H2O) using a Bioruptor Pico (Diagenode, Denville, NJ) for 5 min (30 sec on, 30 sec off). Lysate was centrifuged at max speed for 5 min to remove debris, and 9 volumes of IP Dilution Buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA pH 8.0, 16.7 mM Tris-HCl pH 8.0, 167 mM NaCl, 1 mM NaV, 1 mM NaF, and 1 EDTA-free protease inhibitor tablet (Roche)/10 ml in H2O) were added to the supernatant. 50 μl protein G magnetic beads, blocked with IgG-free BSA, were added to the sample and incubated at 4°C for 30 min, rotating. 1% of the precleared sample was kept out as input, and the remaining sample was split into 3 tubes. 50 μl protein G magnetic beads conjugated to 15 μl of the appropriate antibody were added to each tube, and incubated at 4°C overnight, rotating. Bead-bound complexes were washed for 5 min each in 1 ml of Low Salt Buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% w/v Triton X-100, and 0.1% w/v SDS in H2O), High Salt Buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 2 mM EDTA, 1% w/v Triton X-100, and 0.1% w/v SDS in H2O), LiCl Buffer (10 mM Tris-HCl pH8.0, 250 mM LiCl, 1 mM EDTA, 1% w/v NP-40, and 1% w/v deoxycholic acid in H2O), and TE.
For epigenetic regulator and transcription factor ChIPs, cells were lysed in 1 ml LB1 Buffer (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% Glycerol, 0.5% NP-40, 0.25% Triton X-100, 1:100 protease inhibitor (Sigma-Aldrich, P8340), 1 mM NaV, and 1 mM NaF in H2O) for 10 min at 4°C, rotating. Nuclei pellets were resuspended in LB2 Buffer (10 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1:100 protease inhibitor (Sigma-Aldrich, P8340), 1 mM NaV, and 1 mM NaF in H2O) and incubated for 10 min at 25°C, rotating. Finally, nuclei pellets were lysed in 300 μl LB3 Buffer (10 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 0.1% sodium deoxycholate, 0.5% N-lauroylsarcosine, 1:100 protease inhibitor (Sigma-Aldrich, P8340), 1 mM NaV, and 1 mM NaF in H2O) using a Bioruptor Pico (Diagenode) for 5 min (30 sec on, 30 sec off). 1% Triton X-100 was added, and samples were centrifuged at max speed to remove debris. 50 μl protein G magnetic beads, blocked with IgG-free BSA, were added to the sample and incubated at 4°C for 30 min, rotating. 1% of the precleared sample was kept out as input, and the remaining sample was used for IP. 50 μl protein G magnetic beads conjugated to 5–10 μg of the appropriate antibody were added to each tube, and incubated at 4°C overnight, rotating. Bead-bound complexes were washed for 5 min each in 1 ml of RIPA Wash Buffer (50 mM HEPES-KOH pH 7.6, 300 mM LiCl, 1 mM EDTA, 1% NP-40, and 0.7% sodium deoxycholate in H2O), 5 times, and 1 time in 1 ml TE Wash Buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, and 50 mM NaCl in H2O).
To elute bead-bound complexes, 50 μl of Elution Buffer (for 10 ml total volume, 7.5 ml H2O, 0.5 ml 20% SDS, and 2 ml 0.5M sodium bicarbonate) was added to each sample, and samples were incubated at 65°C for 15 min, shaking at 1000 RPM on a thermomixer (ThermoScientific). Elution was repeated a second time, and then 100 μl RNase Buffer (12 μl of 5M NaCl, 0.2 μl 30 mg/ml RNase, and 88 μl TE) was added to each ChIP and input sample. Samples were incubated at 37°C for 20 min, followed by the addition of PK Buffer (2.5 μl 20 mg/ml proteinase K, 5 μl 20% SDS, and 92.5 μl TE) overnight at 65°C. An equal volume of phenol chloroform solution was added to the samples, which were vortexed for 1 min and transferred to MaXtract High Density tubes (Qiagen). Samples were centrifuged for 8 min at max speed, and the upper phase was transferred to new tubes containing 1.5 μl 20 mg/ml glycogen. Then, 30 μl sodium acetate and 800 μl 100% ethanol were added, and tubes were incubated on dry ice to 30–60 min. DNA pellets were washed in 70% ethanol, air-dried, and resuspended in 30 μl H2O.
SRM method development and mass spectral data analysis
SRMs capable of discriminating 19 acetylation sites from 13 different histone peptides were developed with two to four fragment ions for each species, as described previously44. Various histone peptides with different modifications were purchased or custom synthesized from Anaspec or Genescript to assist the method development. Data were analysed in Skyline with Savitzky–Golay smoothing34. All labelled and unlabelled peaks were grouped as a whole for manual peak determination to avoid bias. Total peak areas from SRMs were used for quantification (as listed in ftp://PASS01134:HG6634ca@ftp.peptideatlas.org/). To access files via FTP, use credentials:
Servername: ftp.peptideatlas.org
Username: PASS01134
Password: HG6634ca
ChIP-Seq
Libraries were prepared using Agencourt AMPure XP beads (Beckman Coulter, Brea, CA) and the KAPA HTP Library Preparation Kit (KAPA Biosystems, Wilmington, MA), according to the manufacturer’s protocol (v4.15). DNA fragment size was determined using High Sensitivity DNA Chips read on a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Libraries were sequenced on the Illumina NextSeq 500 (San Diego, CA; 50bp single reads). FASTQ reads were first trimmed from the 3’ end using Trimmomatic48 version 0.33, such that the Phred33 score of all the nucleotides was above 30 and all reads shorter than 20bp were discarded. The resulting reads were then aligned to the hg19 genome using bowtie version 1.1.2 with the following parameters: bowtie -p 5 -m 1 -v 2 -S49. Next, the bam file was sorted and bigwig tracks were created by extending each read by 150 bp and using the GenomicAlignments50 package in R in order to calculate the coverage of reads in counts per million (CPM) normalized to the total number of reads for each sample in the library.
Peak calling
Peaks were called using MACS51 version 1.4.2 with the default parameters, using the specific aligned ChIP-Seq data for a particular antibody as the treatment sample and an aligned bam file of the unprecipitated input as a control sample. Only the top 10,000 peaks ordered by peak score (which is proportional to the FDR of the peak) were chosen for further analysis.
Calculation of overlaps and statistical significance
Overlap between two ChIP-Seq peak datasets were calculated using the mergePeaks tool in the HOMER52 tools. In brief, the intersection between two ChIP-Seq peak datasets was determined by calculating the number of peaks in one set that overlap with the peaks in the second set. Statistical significance of the overlaps between ChIP-Seq datasets was done by using reservoir sampling53 with a set of 161 transcription factor peak binding sites downloaded fromhttp://hgdownload.cse.ucsc.edu/goldenpath/hg19/encodeDCC/wgEncodeRegTfbsClustered/(wgEncodeRegTfbsClusteredV3.bed.gz, ENCODE data). The tool for implementing the sampling schema is called poverlaps and can be downloaded from github at https://github.com/brentp/poverlap. A total of 100 permutations per pair-wise combination of transcription factor overlaps was used. Venn diagrams of overlaps were generated using an online Venn diagram generator (http://jura.wi.mit.edu/bioc/tools/venn.php).
Motif analysis and pathway enrichment analysis
HOMER, with the standard default parameters, was used to determine the enrichment for known and unknown motifs in the USP7 ChIP-Seq peaks. Pathway enrichment analysis of USP7-bound genes were determined using GREAT (Genomic Regions Enrichment of Annotations Tool)54.
Generation of heatmaps
Heatmap representation of log2 fold-changes in epigenetic marks between DMSO and USP7 inhibition were visualized using ngs.plot55. In brief, log2 fold-changes of H3K27me3, H2Aub, H2Bub, and H3K79me2 upon USP7 inhibition versus DMSO treatment at the top 10,000 USP7 peaks were visualized. A k-means clustering algorithm option was used with the number of clusters set to 5. Correlated gene expression changes were calculated by determining the log2 fold-change in gene expression upon USP7 or JMJD3 inhibition of the nearest protein-coding gene to the USP7 peak. Gene expression changes were visualized using Java TreeView56.
Quantitative polymerase chain reaction (qPCR)
RNA was isolated from T-ALL cells using the Aurum Total RNA Mini Kit (Bio-Rad), and was quantified on a Qubit 3.0 Fluorometer (Life Technologies) using the Qubit RNA HS (High Sensitivity) Assay Kit (Life Technologies) according to the manufacturers’ instructions. cDNA was made using the High Capacity cDNA RT Kit (Applied Biosystems, Foster City, CA), according to the manufacturer’s protocol. qPCR reactions were carried out using iTaq Universal SYBR Green Supermix (Bio-Rad) and the following primers: USP7 – 5’-CGGTGTTGTGTCCATCACTC-3’ (forward) and 5’-AGTTGAGCGAGCCCGAG-3’ (reverse), NOTCH3 – 5’-GTAGAGGGCATGGTGGAAGA-3’ (forward) and 5’-AAGTGGTCCAACAGCAGCTT-3’ (reverse), DTX1 – 5’-CTCGCCACTGCTATCTACCC-3’ (forward) and 5’-CGTGCCGATAGTGAAGATGA-3’ (reverse), and HES1 – 5’-GCAGATGACGGCTGCGCTGA-3’ (forward) and 5’-AAGCGGGTCACCTCGTTCATGC-3’ (reverse). Samples were run on a CFX Connect Real-Time System (Bio-Rad) under the following conditions: 95°C for 10 sec (denaturation), 60°C for 30 sec (annealing), and 72°C for 30 sec (elongation), for 40 cycles. Analyses were performed using GraphPad Prism software (GraphPad Software, La Jolla, CA). Statistical comparisons were made using the Student’s unpaired, two-sided t-test. p values <0.05 were considered statistically significant. Relative mRNA abundance refers to the levels of the gene of interest normalized to housekeeping genes GAPDH or G6PD. All values were then normalized to the control sample.
Analysis of data from publically available databases
Analysis of microarray data from GEO was done using the NCBI GEO2R online tool for microarray analysis. Adjusted p-value calculations were done using Benjamini & Hochberg (False discovery rate or FDR) option. A FDR of <0.05 was considered to be statistically significant.
Ubiquitin competition assay
This assay was performed as previously described, using recombinant enzymes and a panel of USP7 inhibitors, including P21756457. In brief, in vitro recombinant enzymes in 20 mM Tris-HCl (pH 8.0), 2 mM CaCl2, and 2 mM b-mercaptoethanol were incubated with dose ranges of P5091 for 30 min in a 96-well plate before the addition of Ub-PLA2 and NBD C6-HPC (Invitrogen Carlsbad, CA, USA) or Ub-EKL and EKL substrate, as previously described58. The liberation of a fluorescent product within the linear range of the assay was monitored using a Perkin Elmer Envision fluorescence plate reader. Activity-based DUB probes were used for the evaluation of the in vivo activity of DUBs. These are based on the sequence of Ub as the DUB-targeting motif and comprise a reactive C-terminal warhead such as vinyl methyl ester (VME), and an N-terminal epitope tag59.
Cell transfection and virus production
293T cells that reach up to 70% confluency were used for transfection. 48 hours after transfection, 293T cells were collected for the subsequent experiment as required. The following USP7-specific shRNA (Sigma-Aldrich, MISSION system) was used: shUSP7.1:5’-CCGGCCTGGATTTGTGGTTACGTTACTCGAGTAACGTAACCACAAATCCAGGTTTTT-3’ and shUSP7.2: 5’-CCGGCCAGCTAAGTATCAAAGGAAACTCGAGTTTCCTTTGATACTTAGCTGGTTTTT-3’. Retroviral NOTCH1 plasmid (gift from Iannis Aifantis, New York University) and retroviral JMJD3 plasmids (from addgene and Paul Khavari’s group, wild-type #21212 and mutant #21214) were used for retrovirus production. Retrovirus was used to infect T-ALL cells as previously described60.
Viability assays, apoptosis, and cell cycle analysis
500,000 cells were plated in wells of a 24-well plate, and treated with DMSO or USP7i as indicated in the Figure Legends. Cells were counted each day via trypan blue, and inhibitor/medium was changed. When cells reached a confluency of >1 million cells/ml, cells were diluted 1:2, and this dilution was factored into the cell numbers for viability assays. For apoptosis analysis, cells were stained with LIVE/DEAD Fixable Near-IR Dead Cell Stain (Life Technologies) according to the manufacturer’s protocol, except that cells were stained for 20 min at 4°C, prior to staining with PE-conjugated Annexin V (Life Technologies) in Annexin V Binding Buffer (BD Biosciences, San Jose, CA), according to the manufacturers’ instructions. To measure cell cycle, cells were fixed in 100 μl Fix and Perm Medium A (Life Technologies) for 15 min, washed with PBS, and incubated with 100 μl Fix and Perm Medium B (Life Technologies) + 1 μg/ml DAPI (Invitrogen, Carlsbad, CA) for 1h at 4°C. Flow cytometry was performed on an LSR II (BD, Franklin Lakes, NJ) and analyses were performed using FlowJo software (Tree Star, Ashland, OR). Statistical analyses were performed using GraphPad Prism software (GraphPad Software). Comparisons were made using the Student’s unpaired, two-sided t-test. p values <0.05 were considered statistically significant.
TUBE and ubiquitination assays
200 million JURKAT/CUTLL1 T-ALL cells were treated in duplicate with either DMSO or 5 μM USP7i for 8h. Cell pellets were collected, washed with PBS, and frozen. Ubiquitinated substrates were pulled down using agarose-TUBE beads, eluted, and treated with the deubiquitinase USP2 to remove ubiquitin prior to western blotting as previously described57. The wild-type and mutant USP7-expressing plasmids are purchased by addgene (#46751 and 46752 correspondingly, from the Maertens and Peters labs).
RNA-Seq
RNA was extracted from up to 10 million T-ALL cells using the Aurum Total RNA Mini Kit (Bio-Rad) according to the manufacturer’s instructions. RNA was quantified on a Qubit 3.0 Fluorometer (Life Technologies) using the Qubit RNA HS (High Sensitivity) Assay Kit (Life Technologies) according to the manufacturer’s instructions. RNA integrity and DNA fragment size were determined using RNA Nano Chips and High Sensitivity DNA Chips read on a 2100 Bioanalyzer (Agilent Technologies). Libraries were prepared using Agencourt AMPure XP beads (Beckman Coulter) and the TruSeq RNA Library Prep Kit v2 (Illumina), according to the manufacturer’s Low Sample (LS) protocol, and sequenced on the Illumina NextSeq 500 (50bp single reads). FASTQ reads were aligned to the hg19 genome using tophat version 2.1.061 using the following options --no-novel-juncs --read-mismatches 2 --read-edit-dist 2 --max-multihits 5. The generated bam files were then used to count the reads only at the exons of genes using htseq-count62 with the following parameters -q -m intersection-nonempty -s reverse -t exon. Differential expression analysis was done using the R package edgeR63. Bigwig tracks of RNA-Seq expression were generated by using the GenomicAlignments package in R in order to calculate the coverage of reads in counts per million (CPM) normalized to the total number of uniquely mapped reads for each sample in the library.
Superenhancer analysis
Superenhancers were determined using a H3K27Ac ChIP-Seq dataset in JURKAT cells treated with DMSO or USP7 inhibitor. Aligned bam files of H3K27Ac ChIP-Seq signal and input control were analyzed using the Rank Ordering of Super-Enhancers algorithm (ROSE)64,65. A stitching distance of 12.5Kb were used to stitch together enhancer regions, and regions within 2.5Kb of a transcriptional start site were ignored in order to prevent promoter bias. The initial set of enhancer regions used in the analysis was determined by merging the peaks of H3K4me1 and H3K27Ac in CUTLL1 cells (ChIP-Seq dataset for H3K4me1 signal and H3K27Ac was downloaded from GEO with the accession numbers GSM732910 and GSM1252938 for H3K4me1 and H3K27Ac, respectively).
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was done using the Broad Institute GSEA software (http://software.broadinstitute.org/gsea/index.jsp)66,67. RNA-Seq gene expression data were used to create a pre-ranked order of genes, where genes were sorted using the p-value of the differential expression between USP7 inhibition and DMSO treatment, and gene up or downregulation upon USP7 inhibition was determined. In brief, the genes were ranked by calculating –log(p-value)*sign(logFC), and a PreRanked GSEA analysis was done using the Hallmark database and the standard weighted enrichment statistic with 5,000 permutations. Enriched datasets with a FDR < 0.05 were considered statistically significant.
Intravenous and subcutaneous xenograft studies
All mice were housed in a barrier facility, and procedures were performed as approved by the Northwestern University Institutional Animal Care and Use Committee (protocol Ntziachristos #IS00002058 and Mazar #IS00000556) and the Ghent University Animal Ethics Committee (protocol #ECD16/56).
For CUTLL1 T-ALL subcutaneous studies, 0.7 million (PLKO.1 and shUSP7.1) cells were injected subcutaneously into the right flank of 8-week-old NOD.Cg-Prkdcscid male mice (#005557, Jackson Laboratories, Portage, MI) with an equal volume of BD Matrigel, at 50 μl of cells to 50 μl Matrigel. Body weight and tumor size (via calipers) were measured 3 times per week.
For JURKAT T-ALL subcutaneous studies, 0.7 million cells were injected subcutaneously into the right flank of 8-week-old NOD.Cg-Prkdcscid male mice (#005557, Jackson Laboratories, Portage, MI) with an equal volume of BD Matrigel, at 50 μl of cells to 50 μl Matrigel. The following day, mice were randomly assigned to treatment groups (7 mice/group), and were treated with either DMSO or 10 mg/kg USP7i 3 times per week i.p. for 8 doses. One week later, mice were treated 5 times per week i.p. for 17 days. Body weight and tumor size (via calipers) were measured 3 times per week.
For JURKAT T-ALL transplant studies, 1 million cells were injected via tail vein into 8-week-old NOD.Cg-Prkdcscid male mice (#005557, Jackson Laboratories) in 100 μl PBS. Animals were monitored by IVIS every 3 days until luciferase signal was detected, and then animals were randomly assigned to treatment groups (9 mice/group). Mice were treated 3 times per week i.p. with DMSO or 10 mg/kg USP7i (in 10% DMSO + 2% Tween 80) for up to 10 doses. IVIS images were taken twice per week (Perkin Elmer) and animal weight was measured 3 times per week to ensure accurate dosing.
For primagraft studies, 1.2 million primary human T-ALL cells (from the spleen of leukemic primary xenograft engrafted with cells of T-ALL patient FV, N16/0267) in 150 μl PBS were injected via tail vein into 1.5-month-old female NGS mice (Jackson Laboratories). Two weeks after transplantation, blood was collected via tail nick for analysis of hCD45 expression (engraftment) using PE-conjugated mouse anti-human CD45 antibody (Miltenyi Biotec, Auburn, CA; clone 5B1). Flow cytometry was performed on and analyzed using software FlowJo. One week later, mice were randomly divided into two groups (7 mice/group) and treated with either DMSO or 10 mg/kg USP7i in 2% Tween 80 + 10% DMSO in PBS. Treatment was administered on days 1–5 and 8–11 i.v., and on days 12 and 15–22 i.p. Blood was collected via tail nick on days 17 and 22 to measure tumor burden by staining for hCD45.
For toxicity studies, 8-week-old NOD.Cg-Prkdcscid male mice (#005557, Jackson Laboratories) were randomly assigned to treatment groups (3 mice/group). Mice were treated with either vehicle (10% DMSO, 2% Tween-80, and 10% captisol in PBS), 10 mg/kg USP7i, 10 mg/kg USP7i + 10 mg/kg GSKJ4, 10 mg/kg USP7i + 20 mg/kg GSKJ4, or 10 mg/kg USP7i + 50 mg/kg GSKJ4 i.v. daily for 5 days to evaluate treatment toxicity.
For combination treatment studies, 1 million JURKAT T-ALL cells were injected subcutaneously into the right flank of 8-week-old NOD.Cg-Prkdcscid male mice (#005557, Jackson Laboratories) with an equal volume of BD Matrigel, at 50 μl of cells to 50 μl Matrigel. After tumors reached 150–200 mm3, mice were randomly assigned to treatment groups and treated i.v. with vehicle (10% DMSO, 2% Tween 80, and 10% captisol in PBS; n=10), 10 mg/kg USP7i (n=6 per inhibitor), or 10 mg/kg USP7i + 50 mg/kg GSKJ4 (n=10) 5 times the first week, and then 3 times per week until tumors reached 1.5 cm3.
Analyses were performed using GraphPad Prism software (GraphPad Software). Statistical comparisons were made using the Student’s unpaired, two-sided t-test. P values <0.05 were considered statistically significant.
Complete blood cell count analysis
Mouse blood samples were collected via cardiac puncture and run on a Hemavet 950 FS (Drew Scientific, Inc., Miami Lakes, FL) to obtain blood cell counts.
Mouse Histology
This was performed as described in the past by Milano et al., 200468, and Real et al., 200911.
Drug synergism
3,000 cells per well were seeded using a microplate Dispenser (MultiFlo™, BioTek) in 384-well clear bottom, black wall plates (Corning). Drugs were added using the Tecan D300e digital dispenser (Tecan). After 72-h incubation, alamarBlue™ cell viability reagent (ThermoFisher) was added and viability was quantified by measuring fluorescence in a plate reader (Tecan Infinite m1000 pro, Ex: 530 nm; Eλ: 590). Synergy analysis was conducted using Compusyn software69.
Ub-VME based DUB activity assay
The studies were performed as previously described110. JURKAT cells were treated with compound or DMSO for 5 hours. Cells were washed once with PBS, harvested and lysed in 1% NP40 lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP40, 10% Glycerol, 1 mM PMSF, 2 mM β-ME). 20 μg of lysate per sample was incubated at at 37 °C for 30 min with Ub-VME at a final concentration of 400 nM in a total reaction volume of 10 μl. The reaction was stopped by adding 2 μl of 6X SDS sample buffer and boiling for 5 min, followed by SDS-PAGE electrophoresis, transfer to PVDF membranes. The blots were probed with antibodies against USP7 and USP24 to determine the formation of DUB-Ub-VME complex.
Results
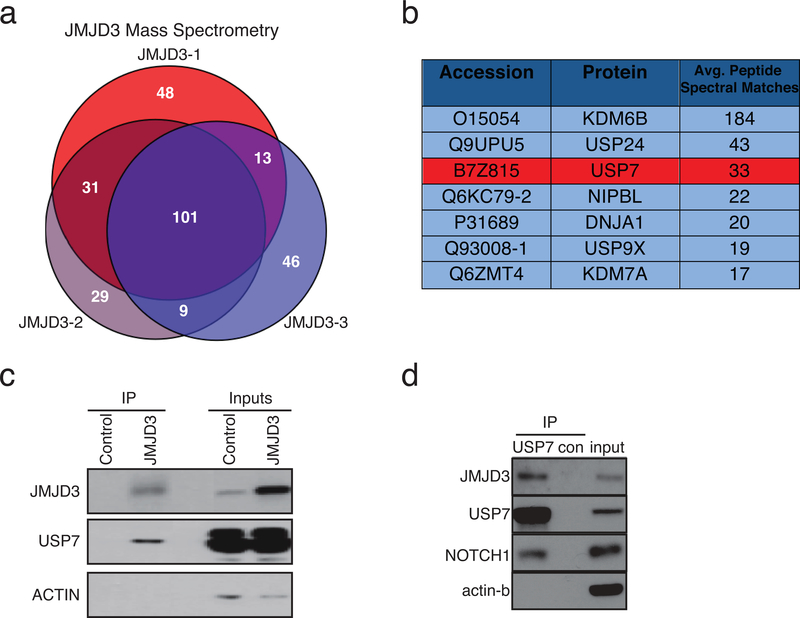
To identify potential NOTCH1/JMJD3-associated proteins and understand NOTCH1 complex stability in leukemia, we retrovirally expressed hemaglutinin (HA) tagged-JMJD3 in CCRF-CEM T-ALL cells. We performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) following immunoprecipitation (IP) of HA-JMJD3 (Fig. 1a). 101 proteins were uniquely associated with HA-JMJD3, and these proteins were enriched in members of the ubiquitin specific protease (USP) family of deubiquitinases (DUBs), including USP7, USP9X, and USP24 (Fig. 1b, Supplementary Table 1, and Supplementary Fig. 1a). Indeed, gene ontology analysis of the JMJD3 interactome showed enrichment in proteins involved in various metabolic processes, cell cycle progression and members of the proteasome complex (Supplementary Fig. 1b).
Figure 1. USP7 is a member of the JMJD3/NOTCH1 complex in T-ALL.
a, HA immunoprecipitation (for HA-tagged JMJD3) followed by mass spectrometry in CCRF-CEM T-ALL cells. Shown is the overlap of HA-JMJD3-asssociated proteins across 3 biological replicates, revealing 101 common proteins associated with HA-JMJD3. b, Top interactors for JMJD3, showing the average peptide spectral matches. Members of the USP family are among these top interacting proteins. c, Representative western blot studies following immunoprecipitation of HA-JMJD3 in CCRF-CEM T-ALL cells. Control cells express the HA-tagged vector. d, Western blot studies following immunoprecipitation of USP7 in CUTLL1 T-ALL cells, showing interactions of JMJD3 and NOTCH1 with USP7. For all immune-blots presented here, one of three representative biological replicates is shown.
Dynamic NOTCH1 (de)ubiquitination plays a pivotal role in the regulation of NOTCH1 protein levels in leukemia, and ubiquitin ligases such as FBXW7 have been demonstrated to regulate NOTCH1 degradation and are tumor suppressors in T-ALL70,71. To this end, we hypothesized that USPs might be acting as oncogenic co-factors for the NOTCH1 complex, controlling complex stability and, ultimately, transcriptional potency. Ubiquitination occurs through a sequence of enzymatic reactions involving the activity of two E1 ubiquitin-activating enzymes, multiple E2 enzymes and hundreds (~700) of E3 enzymes in humans. E3 ligases determine complex specificity72. In contrast, DUBs73,74 mainly include the ubiquitin-specific protease superfamily (USP/UBP, 58 members), the ovarian tumor (OUT, 14) superfamily, the Machado-Josephin domain (MJD, 5) superfamily, the ubiquitin C-terminal hydrolase (UCH, 4) superfamily (all the aforementioned categories are cysteine proteases) and the Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM, 14) domain superfamily proteins that binds zinc (metalloproteases).
USP732–43 was a top interacting partner of JMJD3. USP7 might also interact with NOTCH175 and has a demonstrated role in other hematological malignancies, in multiple myeloma in particular76. We confirmed the interaction of NOTCH1 and JMJD3 with USP7 in reciprocal IP experiments (Fig. 1c, d). USP24 and USP9x77, in contrast, have not been found to interact with NOTCH175. Indeed, our pull-down studies for NOTCH1, followed by detection of USP24 and USP9x in western blot experiments, showed that neither USP9x nor USP24 interact with NOTCH1 (Supplementary Fig. 1c, d). Furthermore, our studies show that, in contrast to USP7 and NOTCH1, both USP9x and USP24 are mainly localized in the cytoplasm (Supplementary Fig. 1e).
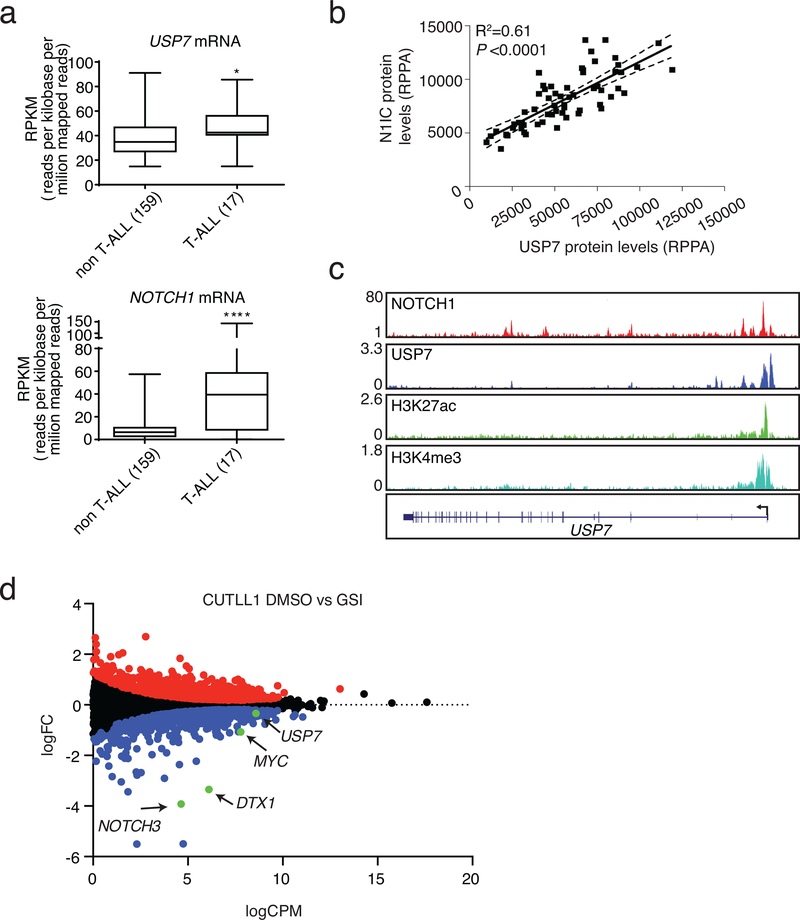
Given the potential participation of USP7 in NOTCH1 protein complexes, we studied the levels of USP7 mRNA in cancer using data from 176 cancer cell lines (cancer cell line encyclopedia). Our analysis showed that USP7, like NOTCH1, is significantly overexpressed in T-ALL compared to other hematologic and solid tumors (Fig. 2a). We then used reverse phase protein array (RPPA) analysis78,79 to quantify NOTCH1 and USP7 protein levels in a panel of 64 pediatric leukemia patient samples. We noticed that USP7 and intracellular or total NOTCH1 levels significantly correlate in T-ALL (Fig. 2b and Supplementary Fig. 2a). We then hypothesized that NOTCH1 might directly bind and control USP7 gene locus. Analysis of ChIP-Seq data in CUTLL1 T-ALL cells, showed that NOTCH1 directly binds the promoter of USP7 gene (Fig. 2c). We also demonstrated that inhibition of NOTCH1, using an established method for NOTCH1 inhibition, using gamma secretase inhibitors (GSI)11,15, decreased NOTCH1 binding enrichment on USP7 gene locus as well as other well-characterized NOTCH1 targets, DTX1 and NOTCH3 (Supplementary Fig. 2b). Furthermore, GSI treatment or NOTCH1 silencing using short hairpins against NOTCH1 (shNOTCH1), led to downregulation of USP7 mRNA and protein levels and other well-characterized NOTCH1 targets (Fig. 2d, Supplementary Fig. 2c-e). In contrast, NOTCH1 over-expression leads to a transcriptional upregulation of USP7 (Supplementary Fig. 2f).
Figure 2. USP7 is highly expressed in T-ALL and is transcriptionally regulated by NOTCH1.
a, RPKM values for NOTCH1 and USP7 in 176 blood cancer cell lines were obtained fromhttps://software.broadinstitute.org/morpheus/, using the CCLE RNAseq data. This data is available for download at https://portals.broadinstitute.org/ccle_legacy/. These include several T-ALL cell lines, ALLSIL, DND41, HPBALL, HUT78, JURKAT, KE37, LOUCY, MOLT13, MOLT16, MOLT3, P12ICHIKAWA, PEER, PF382, RPMI8402, SUPT1, SUPT11 and TALL1. All other cell lines were analyzed against these T-ALL cell lines. Two-tailed unpaired t-test was conducted using the RPKM values (P < 0.05 *, P < 0.0001 ****). b, USP7 protein levels correlate with NOTCH1 intracellular (N1IC) levels in primary T-ALL (n=64). c, Tracks showing NOTCH1, USP7, and the activating histone marks H3K27Ac and H3K4me3 ChIP-Seq signal enrichment in T-ALL cells (CUTLL1), at the USP7 locus. d, Gene expression analysis from CUTLL1 RNA-Seq showing up- and downregulated genes (red and blue dots, respectively) upon GSI treatment, as well as genes with no statistically significant expression changes (black dots, P<0.05). NOTCH1 target genes showing statistically significant down regulation upon GSI treatment are indicated in the plot. One of three representative biological replicates is shown.
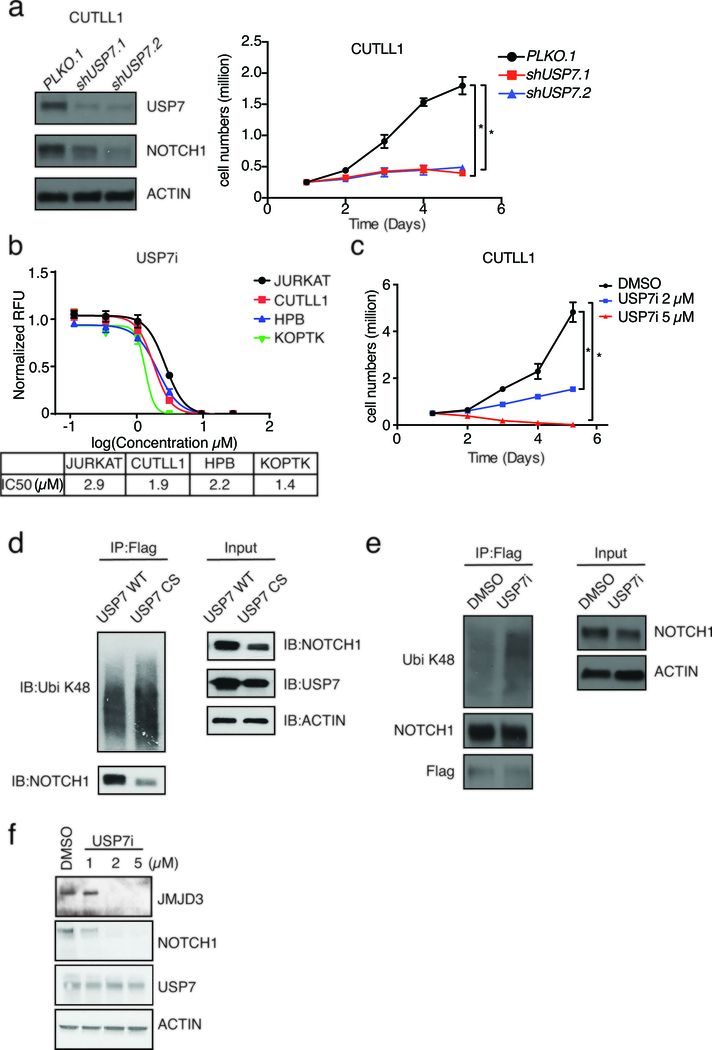
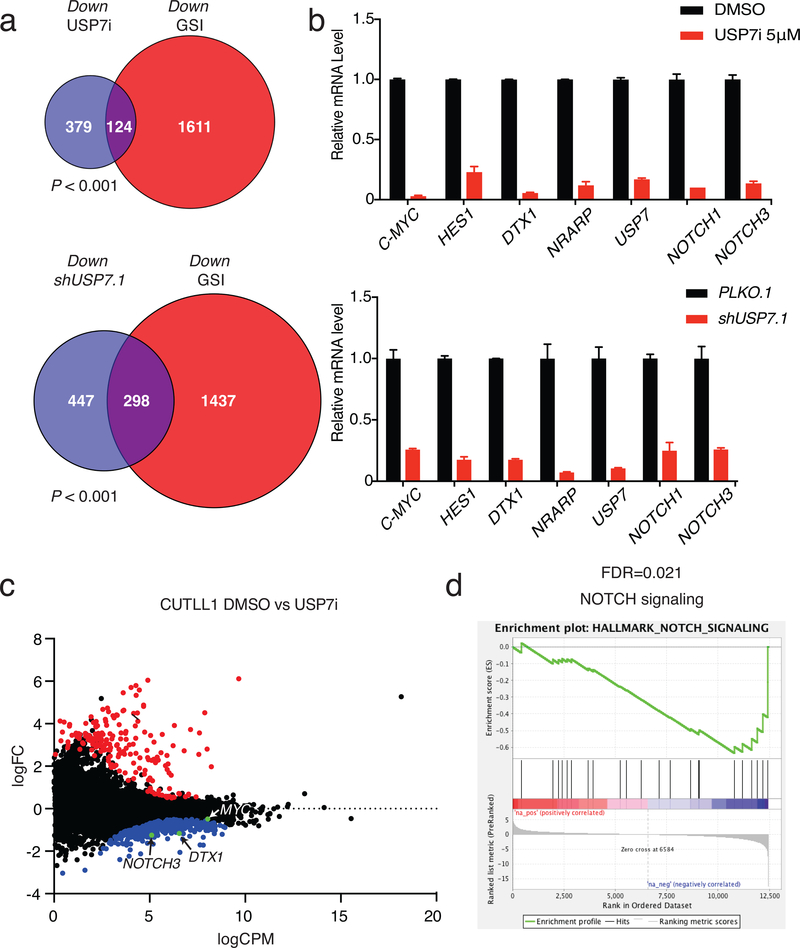
As USP7 has a characterized enzymatic activity as deubiquitinating enzyme, we hypothesized that it might control NOTCH1 protein levels through active deubiquitination. We silenced USP7 in T-ALL using short hairpin RNAs (shUSP7) and examined NOTCH1 protein levels, to observe a decrease in the levels of NOTCH1, suggesting that USP7 might control NOTCH1 protein stability (Fig. 3a). We also noticed a marked inhibition of T-ALL growth upon shUSP7 compared to the control (Fig. 3a, Supplementary Fig. 3a, b). USP7 silencing led to increased apoptosis and a decrease in S phase of T-ALL cells, as demonstrated using shUSP7 (Supplementary Fig. 3c, d). We then took advantage of established USP7-small molecule inhibitors (USP7i, see also methods), developed by Progenra Inc. and successfully used in preclinical models of multiple myeloma and other types of cancer and in immunological contexts57,76,80–84, to block USP7 activity in leukemia cells. The efficacy and specificity of those compounds has been previously demonstrated in several systems57,76,80–84. USP7i significantly inhibited the growth of various T-ALL lines, including CUTLL1, JURKAT and CEM, in the micromolar (μM) range of concentrations, similarly to shUSP7 (Fig. 3b, c, and Supplementary Fig. 4a, 5a). The compound inhibits USP7 activity in a dose-dependent manner from 1 to 5 μM (Supplementary 4b), without significantly blocking total DUB activity in T-ALL, even at 5 μM concentrations (Supplementary Fig. 4c). Treatment of T-ALL cells with USP7i for one and 3 days coupled to apoptosis and cell cycle analysis, using Annexin V and DAPI staining respectively, led to a significant increase in apoptosis and a decrease in S phase (Supplementary Fig. 5b, c), similarly to the shUSP7.
Figure 3. USP7 plays a critical role in T-ALL maintenance via regulation of NOTCH1 protein levels.
a, CUTLL1 T-ALL cells were transduced with a control retroviral vector and two different shUSP7 (shUSP7.1 and shUSP7.2). Left panel: protein levels of USP7 and NOTCH1 in CUTLL1 T-ALL cells (control, shUSP7.1 and shUSP7.2). Right panel: Growth study was performed after knocking down of USP7 in CUTLL1 cells. Data are shown as mean ± SD derived from 3 independent biological replicates, * P≤0.05. b, IC50 curve of USP7i in several T-ALL cell lines (JURKAT, CUTLL1, HPB and KOPTK). c, Growth curve of CUTLL1 T-ALL cells upon daily treatment with increasing concentrations of USP7i. Data are shown as mean ± SD derived from 3 independent experiments, * P≤0.05. d, Representative western blot studies following immunoprecipitation of Flag-NOTCH1 in 293T cells at denaturing condition. 293T cells were transfected with Flag-NOTCH1, ubiquitin and FBXW7 associated with USP7 WT or USP7 CS as indicated. e, A representative western blot following immunoprecipitation of Flag-NOTCH1, conducted in three biological replicates, is shown. Flag-NOTCH1 overexpressed CUTLL1 T-ALL cells were treated with DMSO (control) or USP7i (5μM) for 8h, whole cell extracts were isolated under denaturing conditions to disrupt protein-protein interactions, followed by Flag-NOTCH1 immunoprecipitation. f, Western blot analysis of JMJD3, NOTCH1, and USP7 protein levels upon treatment of CUTLL1 T-ALL cells with increasing concentrations of USP7i for 24h. For all immune-blots presented, one of three representative biological replicates is shown.
To confirm the role of USP7 in NOTCH1 stabilization, we expressed wild-type or a catalytic mutant of USP7 together with NOTCH1, and measured NOTCH1 expression in 293T cells. Indeed, NOTCH1 was stabilized in the presence of wild-type USP7 (Fig. 3d, right panel). Similarly, we show that only the wild-type but not the catalytically inactive USP7 can efficiently deubiquitinate NOTCH1 (Fig. 3d, left panel). These results show that USP7 and its catalytic activity are critical for NOTCH1 deubiquitination and stabilization. To evaluate the role of USP7 in NOTCH1 ubiquitination in T-ALL, we inhibited USP7 catalytic activity, pulled down ubiquitinated proteins using tandem ubiquitin binding entity (TUBE) technology85, and measured NOTCH1 levels (Supplementary Fig. 6a). We then performed similar cell treatments followed by Flag-tagged NOTCH1 pull-down and detection of ubiquitination using antibodies against lysine 48 (K48)-linked poly-ubiquitin chains, a type of polyubiquitination coupled to protein degradation through proteasomal activity86. We demonstrated an increase of NOTCH1 poly-ubiquitination upon USP7i treatment (Fig. 3e). NOTCH1 is a critical target of USP7, as NOTCH1 overexpression partially rescues USP7i-associated blockade in T-ALL growth and expression of NOTCH1 targets in T-ALL cells (Supplementary Fig. 7a-c). Similar to NOTCH1, we demonstrated a significant decrease in the levels of JMJD3 upon silencing of USP7 (Supplementary Fig. 6b). Treatment with USP7i led to a similar decrease in NOTCH1 and JMJD3 protein levels, starting at 1uM USP7i (Fig. 3f). Likewise, inhibition of USP7 in T-ALL cells leads to an increasing of K48-linked ubiquitination of JMJD3 (Supplementary Fig. 6c). JMJD3 overexpression using a retroviral system could not rescue the effect of the drug on leukemic cells (Supplementary Fig. 7d), potentially due to the simultaneous requirement for high levels of NOTCH1 expression for leukemia maintenance.
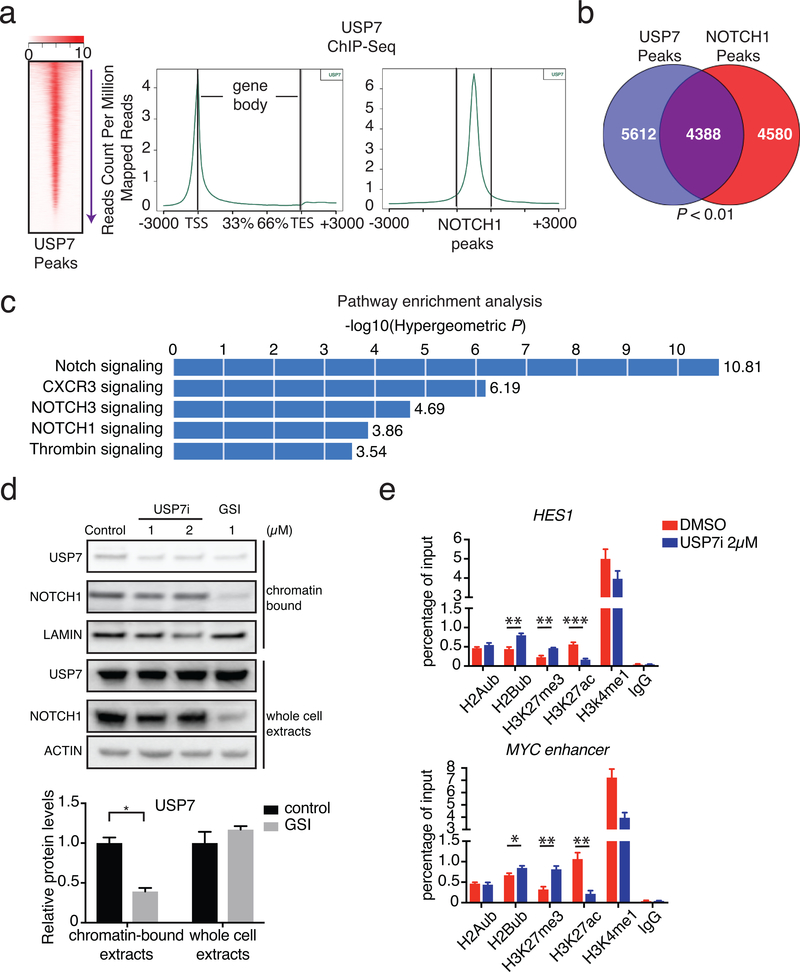
Due to the activity of intracellular NOTCH1 (N1-IC) on chromatin, we evaluated genome-wide USP7 binding via chromatin immunoprecipitation combined with sequencing (ChIP-Seq). We identified ~10,000 high-confidence peaks for USP7 that localized near promoters (Fig. 4a) and had a strong overlap with NOTCH1 binding sites (Fig. 4b). Both USP7 and NOTCH1 bound to classical NOTCH1 targets, such as DTX1 and NOTCH3, as well as NOTCH1 itself (Supplementary Fig. 8a). Pathway analysis of the ChIP-Seq data showed a striking enrichment for NOTCH1 signaling (Fig. 4c). Furthermore, NOTCH1 recruited USP7 to chromatin, as treatment with GSI for depletion of intracellular NOTCH1 levels significantly decreased chromatin-bound USP7 levels (Fig. 4d). USP7 controls NOTCH1 stability and inhibition of USP7 led to a reduction of global NOTCH1 levels and depletion of NOTCH1 from chromatin (Fig. 4d). We further performed USP7 and NOTCH1 ChIP studies focusing on USP7 targets upon USP7 inhibition. Our results show that NOTCH1 and USP7 binding to chromatin is significantly affected upon USP7i treatment whereas binding of another T-ALL transcription factor, RUNX1 (used as control) remained largely unaffected (Supplementary Fig. 8b). Global cellular levels of USP7 did not change over the period of treatment (Fig. 4d). These findings suggest that USP7 and NOTCH1 form a positive feedback loop (see also our discussion), where NOTCH1 induces USP7 gene expression and then recruits USP7 to chromatin, resulting in increased expression of NOTCH1 targets, through stabilization of the NOTCH1 complex.
Figure 4. USP7 and NOTCH1 co-bind oncogenic targets.
a, Heatmap of USP7 peak intensity (left) and metaplots showing the average counts per million of USP7 ChIP-Seq signal across all genes (middle), and NOTCH1 peaks (right). One of three representative biological replicates is shown. b, Analysis of ChIP-Seq data showing the overlap of USP7 peaks with NOTCH1 peaks. c, GREAT (Genomic Regions Enrichment of Annotations Tool) analysis showing enrichment for specific gene functions amongst the USP7-bound peaks used in panel b. d, Upper panel: representative protein levels of USP7 and NOTCH1 in the chromatin-bound fraction and whole cell extracts of JURKAT T-ALL cells upon treatment with DMSO (control), USP7i, or GSI. One of three biological replicates is shown. Lower panel: quantification of USP7 protein levels in upper panel. e, CUTLL1 T-ALL cells were treated with DMSO or USP7i 2μM for 2 days. Chromatin immunoprecipitation (ChIP) of indicative histone marker coupled to detection by quantitative real-time PCR at indicative loci were presented. Data are shown as mean ± SD derived from 3 independent experiments, * P≤0.05, * * P≤0.01, * * * P≤0.001.
We and others have shown that NOTCH1 activity leads to eviction of the repressive mark H3K27me3 from the promoters of oncogenic targets15,16,87. We hypothesized that inhibition of USP7 would lead to an increase in H3K27me3 levels through reduction of JMJD3 protein levels. We assessed the levels of H3K27me3 on USP7 targets upon USP7i treatment for 24h. Indeed, USP7 and NOTCH1 target loci presented a significant gain of H3K27me3 upon USP7i treatment (Fig. 4e and Supplementary Fig. 8c). As USP7 has been shown to control histone ubiquitination88,89, we also examined histone H2B lysine 120 ubiquitination (H2Bub) levels following USP7 inhibition. We showed that USP7 colocalized with genes exhibiting a gain of H2B ubiquitination upon USP7 inhibition (Fig. 4e and Supplementary Figs. 8c and 9)90,91. These changes in H2Bub and H3K27ac/me3 seemed to localize to USP7 targets and not a genome-wide phenomenon, as global levels of these marks were not significantly altered following treatment with USP7i as we demonstrated using global bottom-up proteomics92 (Supplementary Fig. 10). Changes in H2A lysine 119 ubiquitination (H2Aub) on down-regulated loci were minimal but detectable, potentially associated to H3K27me3 changes (Fig. 4e and Supplementary Figs. 8c and 9)93.
Our data suggest that USP7 regulates gene expression of critical NOTCH1 targets. To delineate the molecular effects of USP7 activity, we performed global expression analysis using RNA-Seq upon treatment with USP7i, and comparison with GSI (NOTCH1 inhibitor), and GSKJ4 (JMJD3 inhibitor). The transcriptional signatures of NOTCH1, USP7, and JMJD3 chemical inhibition overlapped significantly, both with regards to downregulated as well as upregulated genes (Fig. 5a, b upper panels and Supplementary Fig. 11a-c). Specifically, inhibition of USP7 led to downregulation of NOTCH1 targets, including NOTCH3 and DTX1 (Fig. 5b, c and Supplementary Fig. 11c). Similar to enzymatic inhibition, USP7 silencing led to downregulation of NOTCH1 targets (Fig. 5a, b lower panels) and significant overlap with USP7i treatment (Supplementary Fig. 11d). Gene set enrichment analysis upon USP7 inhibition showed that major oncogenic pathways (NOTCH1, MYC, DNA damage response, and metabolism) were downregulated (Fig. 5d and Supplementary Fig. 12). Together, these data show that USP7 enzymatic activity is important for sustaining oncogenic activity in T-ALL.
Figure 5. USP7 and NOTCH1 control the expression of oncogenic targets.
a, Analysis of RNA-Seq data showing the overlap of downregulated genes upon USP7 inhibition (1μM) and GSI (1μM) treatment in CUTLL1 T-ALL cells (upper panel). Analysis of RNA-Seq data showing the overlap of downregulated genes upon GSI (1μM) and knockdown of USP7 in CUTLL1 T-ALL cells (lower panel). RNA-Seq data are the average of three biological replicates. b, Upper panel: CUTLL1 T-ALL cells were treated with DMSO or 5μM of USP7i for 3 days. NOTCH1 targets were determined by RT-PCR at mRNA level. Lower panel: RT-PCR of NOTCH1 targets upon treatment of CUTLL1 T-ALL cells with shUSP7.1 and a control retroviral vector. Data are shown as mean ± SD derived from 3 independent experiments. c, Gene expression data from CUTLL1 RNA-Seq showing up- and downregulated genes (red and blue dots, respectively) upon USP7i treatment, as well as genes with no statistically significant expression changes (black dots, P<0.05). NOTCH1 target genes are indicated in the plot. One of three representative biological replicates is shown. d, Gene set enrichment analysis for downregulated genes upon USP7 inhibition. Shown is the NOTCH1 signaling pathway that is significantly downregulated in USP7i versus control cells (FDR=0.021).
It is well known that NOTCH1 regulates oncogenic enhancers94,95. Analysis of USP7 ChIP-Seq data showed that leukemia-specific superenhancers (SEs)65,96, as determined by H3K27 acetylation signal intensity (Supplementary Fig. 13a), were enriched for binding of USP7 and NOTCH1: 192 out of 254 leukemia SEs were co-bound by USP7 and NOTCH1. In agreement with the association of SEs with transcriptional activation, we found that the USP7-bound genes were more highly expressed compared to the rest of the transcriptome (Supplementary Fig. 13b). We hypothesized that combinatorial inhibition of USP7 and the SE component BRD4 using JQ1 inhibitors might be an additive or synergistic effect in inhibiting T-ALL growth. Indeed, our studies using combinatorial inhibition of the bromodomain proteins that recognize acetylated histones (JQ1)97,98 and USP7 showed a synergistic effect (Supplementary Fig. 13c) on T-ALL inhibition. This was further underlined by our targeted and genome wide analysis of USP7 interactions showing that USP7 interacts with Mediator 1, a main component of SEs (Ntziachristos group, unpublished), suggesting that USP7 may regulate gene activation via interactions with the T-ALL transcriptional machinery in SE areas.
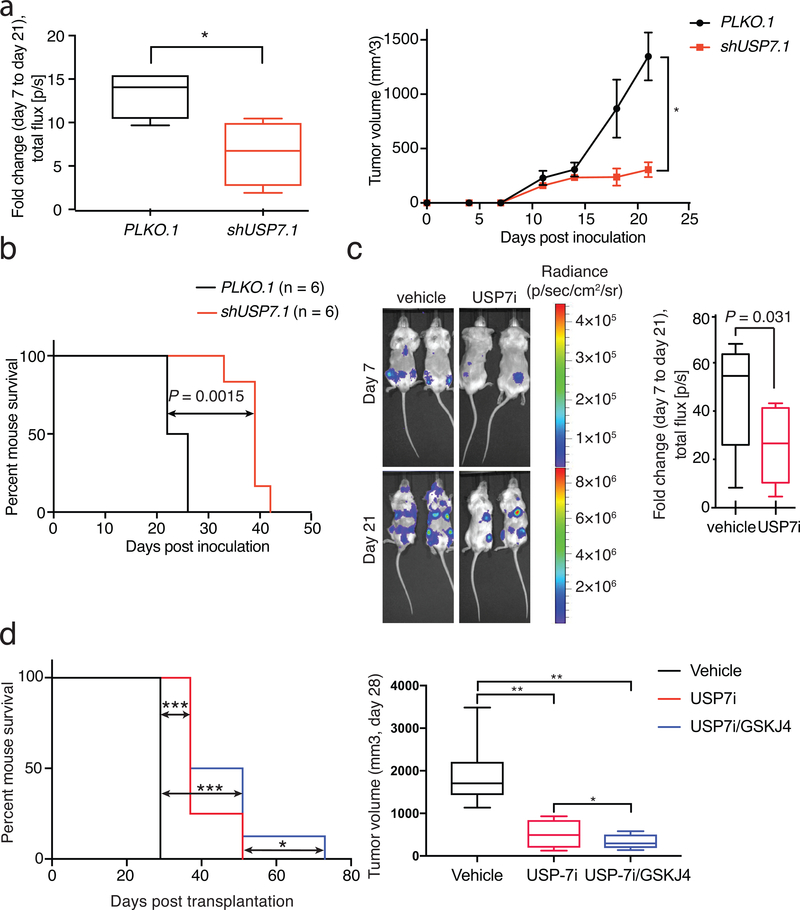
These findings show that USP7 inhibition is a valid therapeutic tool in high-risk leukemia, where other treatments have failed to lead to significant disease regression. Silencing of USP7 in human: mouse xenograft models, using CUTLL1 T-ALL cells, led to a significant inhibition of tumor growth in animals (Fig. 6a) coupled to an extension of mouse survival (Fig. 6b). This significant finding led us to assess the potential of chemical inhibition of USP7 alone or in combination with JMJD3, using the GSKJ4 compound, to block leukemia growth in vivo. Due to the role of H3K27me3 epigenetic mark in the regulation of NOTCH1 targets in T-ALL progression, we and others have successfully used GSKJ4 to target JMJD3 in T-ALL in the past with decent effect on leukemia inhibition16,99. We initially evaluated inhibitor toxicity by injecting mice intravenously (i.v.) with vehicle, USP7i alone [10mg/Kg body mass], or USP7i with increasing concentrations of GSKJ4 [up to 50mg/Kg body mass]. The GSKJ4 concentrations used have been previously shown to successfully inhibit tumor growth99,100. Immunocompromised mice treated daily for five days showed no signs of treatment-associated toxicity, as determined by complete blood cell count, body weight and analysis of spleen and liver (Supplementary Fig. 14a and data not shown). We also examined USP7i-treated animals for gastrointestinal (GI) toxicity as GSI treatment for direct NOTCH1inhibition leads to goblet cell metaplasia in animals. Our studies show no signs of toxicity associated to USP7i inhibition in the GI track (Supplementary Fig. 14b). We then transplanted luciferase-expressing JURKAT T-ALL cells either intravenously (i.v.) or subcutaneously (SubQ) into immunocompromised mice15,16. Upon tumor detection using bioluminescence imaging (IVIS), mice were randomized into different groups that were treated with vehicle or USP7i [10 mg/kg]. In both models, tumors in USP7i-treated animals showed a significant growth disadvantage compared to the control group (Fig. 6c and Supplementary Fig. 15a). Similar results were obtained using a primagraft i.v. model using a patient sample with mutations affecting the gene of the main ubiquitin ligase for NOTCH1, FBXW7, and high NOTCH1 expression levels as well as CCND3 mutation, Supplementary Table 2), where T-ALL progression was detected in the peripheral blood using the marker human CD45 (Supplementary Fig. 15b, c). To further potentiate the therapeutic impact of USP7i in T-ALL, we investigated the effect of combinatorial USP7 and JMJD3 inhibition on leukemia growth by treating mice with USP7i [10mg/Kg body mass] and GSKJ4 [50mg/Kg body mass] in a subcutaneous model of T-ALL and monitored animal survival (Fig. 6d). Similar to our previous studies (Fig. 6c and Supplementary Fig. 15b) we found that USP7i as a single therapy significantly inhibits tumor growth (Fig. 6d, right panel) and extends mouse survival (Fig. 6d, left panel). Moreover, combinatorial inhibition significantly decreased tumor growth, more effectively than vehicle or USP7 inhibition alone (Fig. 6d,right panel) without any associated toxicity based on our analysis of mouse weight (Supplementary Fig. 15d) and histology studies (not shown). Moreover, we demonstrated significant survival differences between mice injected with vehicle or those injected with a combination of USP7 and JMJD3 inhibitors (Fig. 6d, left panel). USP7i/GSKJ4 treatment yielded a prolonged mouse survival compared to vehicle or USP7i therapy (Fig. 6d left panel and data not shown), lending rationale to the use of combinatorial drug treatments against epigenetic regulators in T cell leukemia.
Figure 6. USP7 inhibition blocks leukemia growth in preclinical models of T-ALL.
a, Luciferase-expressing CUTLL1 cells were transduced with a control or shUSP7.1 retroviral vectors. CUTLL1 T-ALL cells (PLKO.1 and shUSP7.1) were injected subcutaneously into immunocompromised mice. Left panel: Fold change in total flux from day 7 to day 21 is shown (PLKO.1, n=6; shUSP7.1, n=6). Right panel: tumor volume for a period of 21 days is shown. * P≤0.05. b, Mouse survival for the PLKO.1 and shUSP7.1 groups. c, JURKAT T-ALL cells were injected intravenously (i.v.) into immunocompromised mice. Upon detection of leukemic blasts using bioluminescence (with IVIS), mice were treated i.p. with 10 mg/kg USP7i 3 days/week for 3 weeks. Relative luminescence intensity is shown for 2 representative mice per treatment group on days 7 and 21 of treatment. The fold change in total flux from day 7 to day 21 is shown on the right (vehicle, n=8; USP7i, n=9). d, Immunocompromised mice were injected subcutaneously with JURKAT T-ALL cells. Once tumors became visible by IVIS, mice were administered USP7i [10mg/Kg body mass], alone or in combination with GSKJ4 [50mg/Kg body mass], i.v., 3 times/week till termination of the study. Shown is mouse survival (lower left panel) and the mean tumor volume ± SD (right panel) for the different groups (vehicle, n=7; USP7i, n=5; and USP7i/GSKJ4, n=8). * P≤0.05, ** P≤0.01, *** P≤0.001.
Discussion
Therapeutic targeting of high-risk acute lymphoblastic leukemia (ALL) has been challenging, rendering this disease an unmet clinical need. In this study, we performed a series of epigenetic, biochemical, functional and pharmacological studies to show that the deubiquitinase USP7 functionally and physically interacts with and controls NOTCH1 pathway and ultimately the oncogenic transcriptional circuitry in T cell ALL. USP7 inhibition of its enzymatic activity could be a valid therapeutic target in this disease.
Given the previously unrecognized importance of deubiquitination in acute leukemia progression, we characterized a novel role for USP7 in controlling NOTCH1 and USP7 stability T-ALL, providing the first evidence that the ubiquitination-methylation axis can serve as an oncogenic switch and therapeutic target in T-ALL. USP7 is the first deubiquitinase identified and thoroughly characterized to regulate the NOTCH oncogenic pathway. Our studies showed that USP7, NOTCH1, and JMJD3 act in a positive feedback loop (Supplementary Fig. 16), where the NOTCH1/JMJD3 complex induces expression of USP7, and it subsequently results in USP7 recruitment to target genes. Ultimately, USP7 recruitment leads to stabilization of the oncogenic complex and activation of targets through demethylation of H3K27 and deubiquitination of H2B, as well as changes in H3K27Ac at SEs. Past and ongoing studies in our group aim at mapping NOTCH1 areas that interact with USP7. NOTCH1 molecules interact with and controlled by the E3 ubiquitin ligase FBXW7 via their PEST domain and engineered PEST mutants (theonine 2512 to alanine, T2512A) for NOTCH1, are impervious to FBXW7 regulation70,71. In contrast, we demonstrate that this NOTCH1 mutant interacts with USP7, suggesting that other parts of NOTCH1 intracellular fragment might control its binding to and regulation via USP7 (Supplementary Fig.17a). To this end, we generated a series of NOTCH1 truncations to demonstrate that the ankyrin domain NOTCH1 (amino acids 1850–2123) mediates interaction with USP7 (Supplementary Fig.17b). Although further studies might be required to further map the exact interacting amino acid regions on USP7 and NOTCH1, our findings potentially suggest a novel mode of post-translational regulation of NOTCH1 independently of the PEST domain and FBXW7 ligase. Of note, there are no hitherto identified ankyrin mutants for NOTCH1 in T-ALL10. We mined 264 T-ALL patient data available through the pediatric cancer genome project (PeCan) data portal (https://pecan.stjude.cloud/home), in our search for NOTCH1 mutations. No mutations in the ankyrin domain of NOTCH1 were identified, in agreement with published evidence.
A better understanding of H3K27me3 demethylation and H2B deubiquitination changes and whether levels of those two marks are intertwined provide us with information useful in designing epigenetic therapies. Intriguingly, and in contrast to the previously characterized pro-oncogenic role of USP7 in T-ALL and multiple myeloma76, neuroblastoma81, chronic lymphocytic leukemia101 and other solid tumors102–104, there are mutations affecting USP7 in pediatric cancers, TAL1-positive leukemias in particular105–107. Moreover, it is known that USP7 can have different substrates, including PTEN35, and the p53-HDM2 axis36–38,108,109. Thus, similarly to what we and others have shown for another epigenetic modulator, the histone demethylase UTX, which can play dual roles (oncogene or tumor suppressor) in NOTCH1- and TAL1-postitive contexts of T-ALL 16,99,110, the role of USP7 might also be context-specific. These context-specific roles of the epigenetic modulators are another intriguing aspect of their biology that we can exploit to develop elegant targeted therapeutic approaches in cancer. Further research is needed to study the role of USP7 in leukemia contexts other than NOTCH1-positive T-ALL, such as TAL1-positive leukemia.
Our findings strongly suggest that USP proteins, and USP7, in particular, may be exploited for pharmacological inhibition in certain T-ALL patients. Previous studies using the same USP7 inhibitor backbone in mouse models showed that the drug inhibits USP7 in specific physiological mouse systems and in mouse tumor cells57,81,111. This is not surprising as USP7 gene/protein is significantly conserved between human and mouse. Although further studies are needed to directly compare the effect of the drug on human and mouse tissues, these published and our findings suggest that USP7 compounds might have significant efficacy with minimal toxicity in clinical trials against patients with T-ALL. The therapeutic window for USP7 inhibition in NOTCH1+ T-ALL might be explained by the significantly higher levels of USP7 in this disease compared to physiological tissues. Combinations of targeted therapies are hailed as the future of cancer therapy, with hematological malignancies leading the way (i.e. bortezomib, lenalidomide, and dexamethasone for multiple myeloma, and rituximab and ibrutinib for chronic lymphocytic leukemia). As epigenetic modulators are one of the main gene families controlling tumor biology, we feel our study can provide insights in the use of deubiquitinases inhibitors and combination with chemotherapy or other epigenetics inhibitors for therapy in leukemia. Three recent studies characterized small molecule inhibitors with efficacy against USP7 in different cancer contexts, including colorectal carcinoma, osteosarcoma and prostate cancer112–114. Thus, combined inhibition of USP7 and JMJD3 may represent an attractive therapeutic approach for T-ALL.
Supplementary Material
Statement of Translational Relevance.
Current therapeutic options and drugs in clinical trials for T-cell acute lymphoblastic leukemia (T-ALL) are rather limited and mainly restricted to chemotherapy, unlike other hematological malignancies, such as B-ALL. Additionally, T-ALL presents with higher rates of relapse compared to B-ALL rendering this disease an unmet clinical need. Here, we report a novel targeted therapy in T-ALL, against a protein (USP7) that is upregulated and transcriptionally regulated by oncogenic NOTCH1 in T-ALL. Inhibition of USP7 significantly blocks disease progression and extends survival, without associated toxicities, in preclinical models of T-ALL. We speculate that this targeted therapy (USP7 inhibition), could be used in combination with systemic chemoradiation, to increase frontline response rates, especially in patients with high risk of relapse/non-response, or potentially as a replacement for systemic cytotoxics in frail patients, likely to suffer from extensive adverse events associated with frontline intensive chemotherapy. USP inhibition could be also used as an addition to immunotherapy approaches.
Acknowledgments:
We want to thank all members of the Ntziachristos laboratory for critical review of the manuscript and their comments. We want to thank Miklos Bekes and Tony Huang (New York University) for providing USP7-expressing plasmids.
Financial Support: This work was supported by the NIH T32CA080621–11 Oncogenesis and Developmental Biology Training Grant and Cancer Smashers Postdoctoral Fellowship (to K.M.A.), and by the National Cancer Institute (R00CA188293–02), the American Society of Hematology, the Leukemia Research Foundation, the St. Baldrick’s Foundation, the H Foundation, the Gabrielle’s Angel Foundation, and the Zell Foundation (to P.N.). This work is also supported by AIRC IG 19186 grant to G.B. High-throughput sequencing data have been deposited into Gene Expression Omnibus, accession number GSE97435. Proteomics services were performed by the Northwestern proteomics core, generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center and the National Resource for Translational and Developmental Proteomics supported by P41 GM108569. For some of the in vivo studies we collaborated with the Developmental Therapeutics Core (DTC), also supported by Cancer Center Support Grant P30 CA060553 from the National Cancer Institute.
Footnotes
Conflict of Interest Statement: Feng Wang, Jian Wu, Hui Wang, Ivan Sokirniy, Joseph Weinstock, and Suresh Kumar are employees of Progenra, Inc., developers of the USP7i compounds used in this paper. The remaining authors have no conflicts of interest.
Data availability:
The raw files and the Proteome discoverer results are now accessible at MassIVE accession: MSV000081771 and ProteomeXchange accession: PXD008330. For now, the log in information for reviewers at MassIVE is:
Username: MSV000081771_reviewer
Password: reviewersonly
The raw MS and skyline files for the epiproteomics analysis are deposited to PeptideAtlas (http://www.peptideatlas.org/)
Official URL for this dataset: ftp://PASS01134:HG6634ca@ftp.peptideatlas.org/
To access files via FTP, use credentials:
Servername: ftp.peptideatlas.org
Username: PASS01134
Password: HG6634ca
References
- 1.Hunger SP & Mullighan CG Acute Lymphoblastic Leukemia in Children. The New England journal of medicine 373, 1541–1552, doi: 10.1056/NEJMra1400972 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Relling MV & Downing JR Acute lymphoblastic leukemia. The New England journal of medicine 350, 1535–1548, doi: 10.1056/NEJMra023001 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Pui CH, Mullighan CG, Evans WE & Relling MV Pediatric acute lymphoblastic leukemia: where are we going and how do we get there? Blood 120, 1165–1174, doi: 10.1182/blood-2012-05-378943 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pui CH & Evans WE Treatment of acute lymphoblastic leukemia. The New England journal of medicine 354, 166–178, doi: 10.1056/NEJMra052603 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Aifantis I, Raetz E & Buonamici S Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol 8, 380–390, doi:nri2304 [pii], 10.1038/nri2304 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Pui CH, Robison LL & Look AT Acute lymphoblastic leukaemia. Lancet 371, 1030–1043, doi: 10.1016/S0140-6736(08)60457-2 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Belver L & Ferrando A The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nature reviews. Cancer 16, 494–507, doi: 10.1038/nrc.2016.63 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Haydu JE & Ferrando AA Early T-cell precursor acute lymphoblastic leukaemia. Curr Opin Hematol 20, 369–373, doi: 10.1097/MOH.0b013e3283623c61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coustan-Smith E et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol 10, 147–156, doi: 10.1016/S1470-2045(08)70314-0 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weng AP et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271, doi: 10.1126/science.1102160 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Real PJ et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nature medicine 15, 50–58, doi:nm.1900 [pii], 10.1038/nm.1900 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tosello V et al. WT1 mutations in T-ALL. Blood 114, 1038–1045, doi:blood-2008–12-192039 [pii], 10.1182/blood-2008-12-192039 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Keersmaecker K & Ferrando AA TLX1-induced T-cell acute lymphoblastic leukemia. Clin Cancer Res 17, 6381–6386, doi: 10.1158/1078-0432.CCR-10-3037 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Zenatti PP et al. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nature genetics 43, 932–939, doi: 10.1038/ng.924 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ntziachristos P et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nature medicine 18, 298–301, doi: 10.1038/nm.2651 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ntziachristos P et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature, doi: 10.1038/nature13605 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trimarchi T et al. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell 158, 593–606, doi: 10.1016/j.cell.2014.05.049 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buonamici S et al. CCR7 signalling as an essential regulator of CNS infiltration in T-cell leukaemia. Nature 459, 1000–1004, doi:nature08020 [pii], 10.1038/nature08020 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullighan CG & Downing JR Global genomic characterization of acute lymphoblastic leukemia. Seminars in hematology 46, 3–15, doi: 10.1053/j.seminhematol.2008.09.005 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481, 157–163, doi: 10.1038/nature10725 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts KG & Mullighan CG Genomics in acute lymphoblastic leukaemia: insights and treatment implications. Nat Rev Clin Oncol 12, 344–357, doi: 10.1038/nrclinonc.2015.38 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Arcipowski KM, Martinez CA & Ntziachristos P Histone demethylases in physiology and cancer: a tale of two enzymes, JMJD3 and UTX. Current opinion in genetics & development 36, 59–67, doi: 10.1016/j.gde.2016.03.010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King B et al. The Ubiquitin Ligase FBXW7 Modulates Leukemia-Initiating Cell Activity by Regulating MYC Stability. Cell 153, 1552–1566, doi: 10.1016/j.cell.2013.05.041 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ntziachristos P, Abdel-Wahab O & Aifantis I Emerging concepts of epigenetic dysregulation in hematological malignancies. Nature immunology 17, 1016–1024, doi: 10.1038/ni.3517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ntziachristos P, Lim JS, Sage J & Aifantis I From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell 25, 318–334, doi: 10.1016/j.ccr.2014.02.018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ntziachristos P, Mullenders J, Trimarchi T & Aifantis I Mechanisms of epigenetic regulation of leukemia onset and progression. Advances in immunology 117, 1–38, doi: 10.1016/B978-0-12-410524-9.00001-3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Real PJ & Ferrando AA NOTCH inhibition and glucocorticoid therapy in T-cell acute lymphoblastic leukemia. Leukemia 23, 1374–1377, doi:leu200975 [pii], 10.1038/leu.2009.75 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agger K et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731–734, doi: 10.1038/nature06145 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Agger K et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes & development 23, 1171–1176, doi: 10.1101/gad.510809 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Santa F et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130, 1083–1094, doi: 10.1016/j.cell.2007.08.019 (2007). [DOI] [PubMed] [Google Scholar]
- 31.De Santa F et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. The EMBO journal 28, 3341–3352, doi: 10.1038/emboj.2009.271 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sowa ME, Bennett EJ, Gygi SP & Harper JW Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403, doi: 10.1016/j.cell.2009.04.042 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lecona E, Narendra V & Reinberg D USP7 cooperates with SCML2 to regulate the activity of PRC1. Molecular and cellular biology 35, 1157–1168, doi: 10.1128/MCB.01197-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang ZM et al. An Allosteric Interaction Links USP7 to Deubiquitination and Chromatin Targeting of UHRF1. Cell reports 12, 1400–1406, doi: 10.1016/j.celrep.2015.07.046 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song MS et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455, 813–817, doi: 10.1038/nature07290 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M, Brooks CL, Kon N & Gu W A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell 13, 879–886 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Li M et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416, 648–653, doi: 10.1038/nature737 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Kon N et al. Inactivation of HAUSP in vivo modulates p53 function. Oncogene 29, 1270–1279, doi: 10.1038/onc.2009.427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Horst A et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol 8, 1064–1073, doi: 10.1038/ncb1469 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Du Z et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal 3, ra80, doi: 10.1126/scisignal.2001462 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Espinosa JM Histone H2B ubiquitination: the cancer connection. Genes & development 22, 2743–2749, doi: 10.1101/gad.1732108 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen ST et al. The Deubiquitinating Enzyme USP7 Regulates Androgen Receptor Activity by Modulating Its Binding to Chromatin. J Biol Chem 290, 21713–21723, doi: 10.1074/jbc.M114.628255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liefke R, Karwacki-Neisius V & Shi Y EPOP Interacts with Elongin BC and USP7 to Modulate the Chromatin Landscape. Mol Cell 64, 659–672, doi: 10.1016/j.molcel.2016.10.019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng Y, Thomas PM & Kelleher NL Measurement of acetylation turnover at distinct lysines in human histones identifies long-lived acetylation sites. Nat Commun 4, 2203, doi: 10.1038/ncomms3203 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng Y et al. Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proceedings of the National Academy of Sciences of the United States of America 109, 13549–13554, doi: 10.1073/pnas.1205707109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cotto-Rios XM, Bekes M, Chapman J, Ueberheide B & Huang TT Deubiquitinases as a signaling target of oxidative stress. Cell Rep 2, 1475–1484, doi: 10.1016/j.celrep.2012.11.011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chirco KR et al. Preparation and evaluation of human choroid extracellular matrix scaffolds for the study of cell replacement strategies. Acta Biomater 57, 293–303, doi: 10.1016/j.actbio.2017.05.011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120, doi: 10.1093/bioinformatics/btu170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25, doi: 10.1186/gb-2009-10-3-r25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawrence M et al. Software for computing and annotating genomic ranges. PLoS Comput Biol 9, e1003118, doi: 10.1371/journal.pcbi.1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome biology 9, R137, doi: 10.1186/gb-2008-9-9-r137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589, doi: 10.1016/j.molcel.2010.05.004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haiminen N, Mannila H & Terzi E Determining significance of pairwise co-occurrences of events in bursty sequences. BMC Bioinformatics 9, 336, doi: 10.1186/1471-2105-9-336 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLean CY et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28, 495–501, doi: 10.1038/nbt.1630 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen L, Shao N, Liu X & Nestler E ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284, doi: 10.1186/1471-2164-15-284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saldanha AJ Java Treeview--extensible visualization of microarray data. Bioinformatics 20, 3246–3248, doi: 10.1093/bioinformatics/bth349 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Wang L et al. Ubiquitin-specific Protease-7 Inhibition Impairs Tip60-dependent Foxp3+ T-regulatory Cell Function and Promotes Antitumor Immunity. EBioMedicine 13, 99–112, doi: 10.1016/j.ebiom.2016.10.018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicholson B et al. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci 17, 1035–1043, doi: 10.1110/ps.083450408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Borodovsky A et al. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol 9, 1149–1159 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Ntziachristos P et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature 514, 513–517, doi: 10.1038/nature13605 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trapnell C, Pachter L & Salzberg SL TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111, doi: 10.1093/bioinformatics/btp120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, doi: 10.1093/bioinformatics/btu638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, doi: 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loven J et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334, doi: 10.1016/j.cell.2013.03.036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whyte WA et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319, doi: 10.1016/j.cell.2013.03.035 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mootha VK et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273, doi: 10.1038/ng1180 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Milano J et al. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol Sci 82, 341–358, doi: 10.1093/toxsci/kfh254 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Chou TC Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 70, 440–446, doi: 10.1158/0008-5472.CAN-09-1947 (2010). [DOI] [PubMed] [Google Scholar]
- 70.Thompson BJ et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med 204, 1825–1835, doi:jem.20070872 [pii], 10.1084/jem.20070872 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson BJ et al. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J Exp Med 205, 1395–1408, doi:jem.20080277 [pii], 10.1084/jem.20080277 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Skaar JR, Pagan JK & Pagano M SCF ubiquitin ligase-targeted therapies. Nat Rev Drug Discov 13, 889–903, doi: 10.1038/nrd4432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nijman SM et al. A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786, doi: 10.1016/j.cell.2005.11.007 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Komander D, Clague MJ & Urbe S Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10, 550–563, doi: 10.1038/nrm2731 (2009). [DOI] [PubMed] [Google Scholar]
- 75.Yatim A et al. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol Cell 48, 445–458, doi: 10.1016/j.molcel.2012.08.022 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chauhan D et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell 22, 345–358, doi: 10.1016/j.ccr.2012.08.007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peterson LF et al. Targeting deubiquitinase activity with a novel small-molecule inhibitor as therapy for B-cell malignancies. Blood 125, 3588–3597, doi: 10.1182/blood-2014-10-605584 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Milani G et al. Low PKCalpha expression within the MRD-HR stratum defines a new subgroup of childhood T-ALL with very poor outcome. Oncotarget 5, 5234–5245, doi: 10.18632/oncotarget.2062 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Serafin V et al. Phosphoproteomic analysis reveals hyperactivation of mTOR/STAT3 and LCK/Calcineurin axes in pediatric early T-cell precursor ALL. Leukemia 31, 1007–1011, doi: 10.1038/leu.2017.13 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Fan YH et al. USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis 4, e867, doi: 10.1038/cddis.2013.400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tavana O et al. HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nature medicine 22, 1180–1186, doi: 10.1038/nm.4180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Das DS et al. A novel hypoxia-selective epigenetic agent RRx-001 triggers apoptosis and overcomes drug resistance in multiple myeloma cells. Leukemia 30, 2187–2197, doi: 10.1038/leu.2016.96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pozhidaeva A et al. USP7-Specific Inhibitors Target and Modify the Enzyme’s Active Site via Distinct Chemical Mechanisms. Cell Chem Biol 24, 1501–1512 e1505, doi: 10.1016/j.chembiol.2017.09.004 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Dar A, Shibata E & Dutta A Deubiquitination of Tip60 by USP7 determines the activity of the p53-dependent apoptotic pathway. Molecular and cellular biology 33, 3309–3320, doi: 10.1128/MCB.00358-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hjerpe R et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep 10, 1250–1258, doi: 10.1038/embor.2009.192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Komander D & Rape M The ubiquitin code. Annu Rev Biochem 81, 203–229, doi: 10.1146/annurev-biochem-060310-170328 (2012). [DOI] [PubMed] [Google Scholar]
- 87.Simon C et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes & development 26, 651–656, doi: 10.1101/gad.186411.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van der Knaap JA et al. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell 17, 695–707, doi: 10.1016/j.molcel.2005.02.013 (2005). [DOI] [PubMed] [Google Scholar]
- 89.van der Knaap JA, Kozhevnikova E, Langenberg K, Moshkin YM & Verrijzer CP Biosynthetic enzyme GMP synthetase cooperates with ubiquitin-specific protease 7 in transcriptional regulation of ecdysteroid target genes. Molecular and cellular biology 30, 736–744, doi:MCB.01121–09 [pii], 10.1128/MCB.01121-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Strikoudis A et al. Regulation of transcriptional elongation in pluripotency and cell differentiation by the PHD-finger protein Phf5a. Nat Cell Biol 18, 1127–1138, doi: 10.1038/ncb3424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strikoudis A, Lazaris C, Ntziachristos P, Tsirigos A & Aifantis I Opposing functions of H2BK120 ubiquitylation and H3K79 methylation in the regulation of pluripotency by the Paf1 complex. Cell cycle, 0, doi: 10.1080/15384101.2017.1295194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tran JC et al. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature 480, 254–258, doi: 10.1038/nature10575 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang H et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878, doi: 10.1038/nature02985 (2004). [DOI] [PubMed] [Google Scholar]
- 94.Herranz D et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nature medicine, doi: 10.1038/nm.3665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang H et al. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proceedings of the National Academy of Sciences of the United States of America 111, 705–710, doi: 10.1073/pnas.1315023111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hnisz D et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947, doi: 10.1016/j.cell.2013.09.053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Filippakopoulos P et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073, doi: 10.1038/nature09504 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Delmore JE et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917, doi: 10.1016/j.cell.2011.08.017 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Benyoucef A et al. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes & development 30, 508–521, doi: 10.1101/gad.276790.115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hashizume R et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nature medicine 20, 1394–1396, doi: 10.1038/nm.3716 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Carra G et al. Therapeutic inhibition of USP7-PTEN network in chronic lymphocytic leukemia: a strategy to overcome TP53 mutated/deleted clones. Oncotarget, doi: 10.18632/oncotarget.16348 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.An T et al. USP7 inhibitor P5091 inhibits Wnt signaling and colorectal tumor growth. Biochem Pharmacol 131, 29–39, doi: 10.1016/j.bcp.2017.02.011 (2017). [DOI] [PubMed] [Google Scholar]
- 103.Malapelle U et al. USP7 inhibitors, downregulating CCDC6, sensitize lung neuroendocrine cancer cells to PARP-inhibitor drugs. Lung Cancer 107, 41–49, doi: 10.1016/j.lungcan.2016.06.015 (2017). [DOI] [PubMed] [Google Scholar]
- 104.Morra F et al. The combined effect of USP7 inhibitors and PARP inhibitors in hormone-sensitive and castration-resistant prostate cancer cells. Oncotarget, doi: 10.18632/oncotarget.16463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huether R et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat Commun 5, 3630, doi: 10.1038/ncomms4630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Richter-Pechanska P et al. Identification of a genetically defined ultra-high-risk group in relapsed pediatric T-lymphoblastic leukemia. Blood Cancer J 7, e523, doi: 10.1038/bcj.2017.3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu Y et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nature genetics 49, 1211–1218, doi: 10.1038/ng.3909 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kon N et al. Roles of HAUSP-mediated p53 regulation in central nervous system development. Cell Death Differ 18, 1366–1375, doi: 10.1038/cdd.2011.12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stolte B et al. Genome-scale CRISPR-Cas9 screen identifies druggable dependencies in TP53 wild-type Ewing sarcoma. J Exp Med, doi: 10.1084/jem.20171066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Van der Meulen J et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood 125, 13–21, doi: 10.1182/blood-2014-05-577270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang F et al. Active site-targeted covalent irreversible inhibitors of USP7 impair the functions of Foxp3+ T-regulatory cells by promoting ubiquitination of Tip60. PloS one 12, e0189744, doi: 10.1371/journal.pone.0189744 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kategaya L et al. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature 550, 534–538, doi: 10.1038/nature24006 (2017). [DOI] [PubMed] [Google Scholar]
- 113.Turnbull AP et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature 550, 481–486, doi: 10.1038/nature24451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gavory G et al. Discovery and characterization of highly potent and selective allosteric USP7 inhibitors. Nat Chem Biol 14, 118–125, doi: 10.1038/nchembio.2528 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.