Significance
Past studies on HIV-1 evolution were mainly at the genetic level. This study provides well-matched genotype and phenotype data and demonstrates the disparate pathogenicity of two major CRF01_AE clusters. While CRF01_AE cluster 4 and cluster 5 are associated with the men-who-have-sex-with-men (MSM) route of transmission, cluster 4 but not cluster 5 causes a low CD4 count, which is associated with the higher prevalence CXCR4 viruses in cluster 4. The higher CXCR4 use tendency in cluster 4 is derived from its unique V3 loop favoring CXCR4 binding. This study demonstrates disparate HIV-1 phenotypes between different phylogenetic clusters. It is important to monitor HIV-1 evolution at both the genotype and phenotype level to identify and control more pathogenic HIV-1 strains.
Keywords: CRF01_AE, Coreceptor tropism, CXCR4, CD4 count, HIV-1 pathogenesis
Abstract
HIV-1 evolved into various genetic subtypes and circulating recombinant forms (CRFs) in the global epidemic. The same subtype or CRF is usually considered to have similar phenotype. Being one of the world’s major CRFs, CRF01_AE infection was reported to associate with higher prevalence of CXCR4 (X4) viruses and faster CD4 decline. However, the underlying mechanisms remain unclear. We identified eight phylogenetic clusters of CRF01_AE in China and hypothesized that they may have different phenotypes. In the National HIV Molecular Epidemiology Survey, we discovered that people infected by CRF01_AE cluster 4 had significantly lower CD4 counts (391 vs. 470, P < 0.0001) and higher prevalence of X4-using viruses (17.1% vs. 4.4%, P < 0.0001) compared with those infected by cluster 5. In an MSM cohort, X4-using viruses were only isolated from seroconvertors in cluster 4, which was associated with low a CD4 count within the first year of infection (141 vs. 440, P = 0.003). Using a coreceptor binding model, we identified unique V3 signatures in cluster 4 that favor CXCR4 use. We demonstrate that the HIV-1 phenotype and pathogenicity can be determined at the phylogenetic cluster level in the same subtype. Since its initial spread to humans from chimpanzees, estimated to be the first half of the 20th century, HIV-1 continues to undergo rapid evolution in larger and more diverse populations. The divergent phenotype evolution of two major CRF01_AE clusters highlights the importance of monitoring the genetic evolution and phenotypic shift of HIV-1 to provide early warning of the appearance of more pathogenic strains.
During global transmission, HIV-1 evolved into various subtypes and their hybrids, the so-called circulating recombinant forms (CRFs) (1). CRF01_AE, one of the major CRFs, spreads mainly in Southeast Asia and China (1, 2). Early studies in Thailand reported faster CD4 loss and shorter survival of CRF01_AE-infected people compared with infections in western countries, where subtype B predominates (3–6). In recent years, faster disease progression of CRF01_AE was also reported in China (7–9). Although past studies reported high prevalence of CXCR4 (X4) viruses and fast disease progression in CRF01_AE infections (7, 8, 10, 11), the data were mainly based on samples without known infection time and on genotypic prediction without phenotypic confirmation. It is still unclear how early X4-using viruses can emerge during natural CRF01_AE infection and whether the genotypic prediction is reliable.
The epidemic of CRF01_AE in China was initiated by multiple phylogenetic clusters introduced from Thailand in the 1990s (2). We earlier identified eight CRF01_AE clusters, which have different geographic distributions and epidemic patterns (2, 12). Therefore, we hypothesized that they may have different phenotypes. We used the National HIV Molecular Epidemiology Survey (NHMES) dataset for screening and a men-who-have-sex-with-men (MSM) seroincidence cohort for in-depth study to determine the time of X4 virus emergence, and phenotypically confirmed viral tropism using matched viral isolates. We observed significantly lower CD4 counts in CRF01_AE cluster 4 compared with cluster 5 in the NHMES dataset. Focusing on the MSM cohort, we demonstrated higher prevalence of the X4 phenotype in cluster 4 among people within the first year of infection. We further determined the genetic and structural basis favoring X4-using coreceptor use in cluster 4. This study demonstrates that different phylogenetic clusters from the same HIV-1 subtype can cause disparate rates of disease progression, and provides the missing link between CRF01_AE genetic makeup, phenotype, and clinical outcomes.
Results
Lower CD4 T Cell Count and Higher Prevalence of the X4-Using Virus in CRF01_AE Cluster 4.
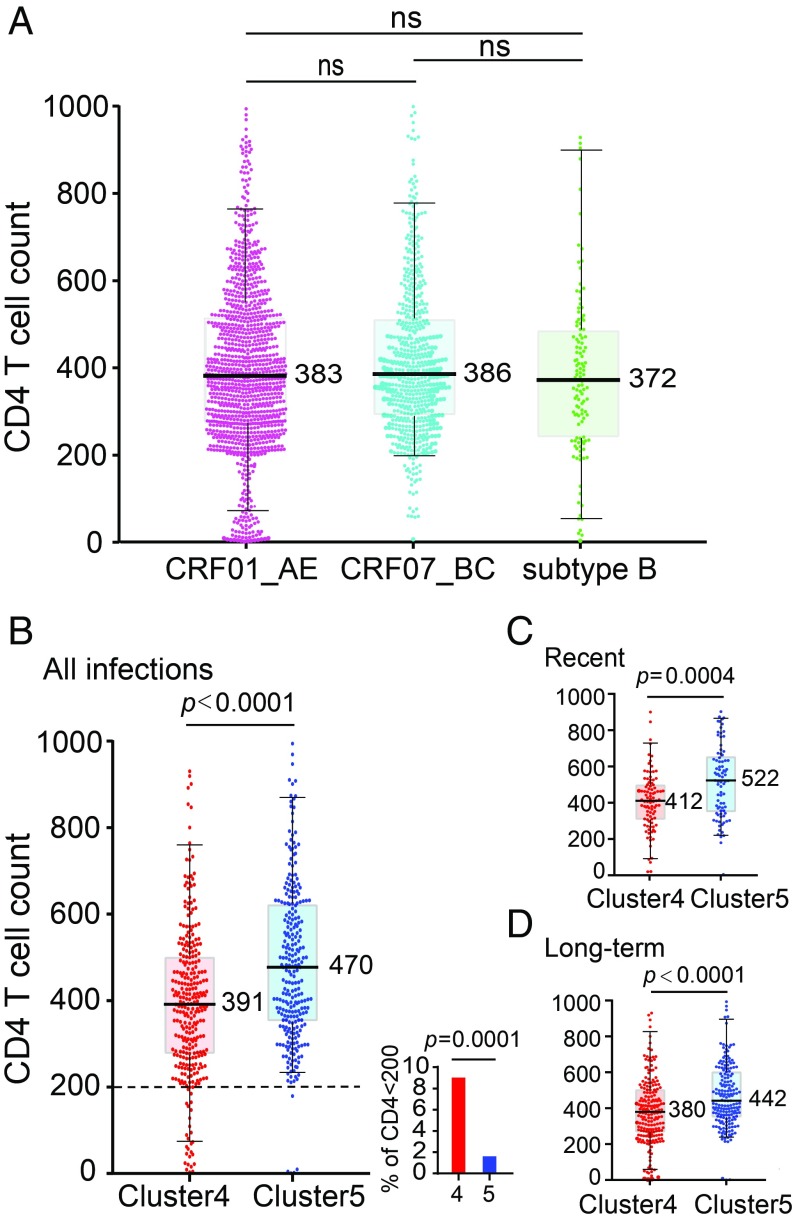
We compared the CD4 T cell counts of 1,118 CRF01_AE, 633 CRF07_BC, and 123 subtype B newly diagnosed HIV-1–positive participants in China’s NHMES study and found no differences (Fig. 1A). When analyzing the two major CRF01_AE clusters which contribute greatly to China’s MSM epidemic, however, the CD4 count in cluster 4 was significantly lower than in cluster 5 (P < 0.0001) (Fig. 1B). Since CD4 counts decline with time during natural infection, we distinguished recent infections from long-term infections by HIV-1 limiting antigen-avidity assay. CRF01_AE clusters 4 and 5 showed equally high rates of recent infection (31.3% and 31.4%), while only 10.1% of recent infections were found in other minor CRF01_AE clusters (SI Appendix, Fig. S1A). Among recent infections, cluster 4 had the lowest CD4 count in comparison with cluster 5 (P = 0.0004) and other minor clusters (P = 0.0037), while cluster 5 and other clusters showed no statistical difference (Fig. 1C and SI Appendix, Fig. S1B). In long-term infections, the CD4 count of cluster 4 remained significantly lower than that of cluster 5 (P < 0.0001), while the CD4 count of other clusters became the lowest (Fig. 1D and SI Appendix, Fig. S1B). Given the current HIV-1 epidemic pattern in China, the lower CD4 count of other clusters in long-term infections is likely due to a longer infection time (see Discussion). Together, the results clearly showed that CRF01_AE cluster 4 but not cluster 5 causes lower CD4 counts during early infection.
Fig. 1.
Comparison of CD4 count between different HIV-1 subtypes and different CRF01_AE clusters. (A) Comparison of CD4 count between CRF01_AE (n = 1,118), CRF07_BC (n = 633), and subtype B (n = 123) infections from the NHMES (B–D) Significantly lower CD4 T cell count in individuals infected by CRF01_AE cluster 4 (n = 308) than those infected by cluster 5 (n = 273) regardless of the stage of infection (B) in recent infection group (C) and in long-term infection group (D). Of the total 308 cluster-4 and 273 cluster-5 infections shown in B, 304 and 271 had plasma samples available for limiting antigen-avidity EIA testing, respectively. The small figure in B shows the percentage of individuals with CD4 below 200. The vertical line, box, and whisker represent the median, upper, and lower quartiles and the 5–95 percentile, respectively. The actual median number of each group is shown. The statistical difference in CD4 count was calculated using the two-tailed Mann–Whitney U test. The percentage of individuals with CD4 below 200 was compared using the two-tailed Fisher’s exact test.
Because CXCR4 tropism is associated with lower CD4 cell counts (13–17), we investigated the prevalence level of X4 viruses in clusters 4 and 5 using genotypic prediction. Geno2pheno prediction showed higher prevalence of the X4 genotype in cluster 4 than in cluster 5 (17.1% vs. 4.4%, P < 0.0001) (SI Appendix, Fig. S2). In cluster 4, the X4 genotype was “concentrated” among patients with low CD4 counts (SI Appendix, Fig. S2). This result indicates a strong association between higher X4 prevalence and lower CD4 counts in CRF01_AE cluster 4.
High Prevalence of the X4-Using Phenotype in CRF01_AE Cluster 4 Among Recently Infected Individuals.
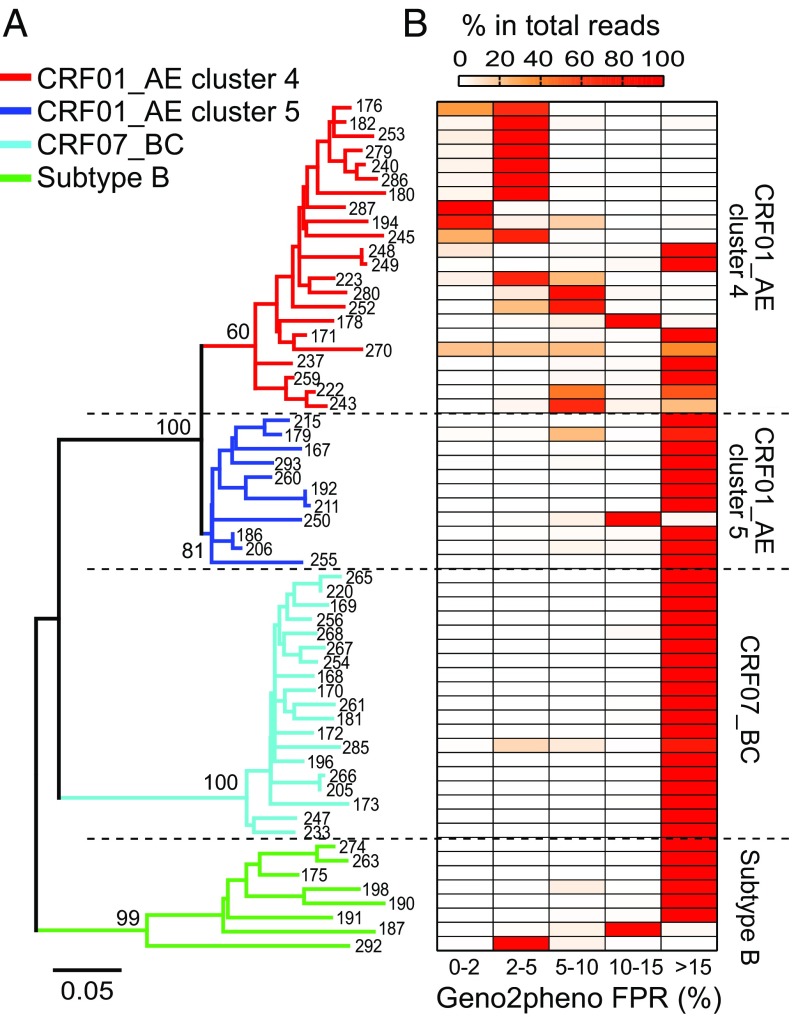
To further characterize the mechanism of fast CD4 cell loss in CRF01_AE cluster 4, we focused on a seroincidence cohort with approximately 2,000 MSMs from Beijing’s Chaoyang District [the Chaoyang MSM (CYM) cohort]. For the 135 MSM seroconvertors, we first performed deep sequencing on all 78 participants with archived first-year blood samples and successfully sequenced 71 of them (SI Appendix, Figs. S3 and S4). Among them, 60 participants purely infected by CRF01_AE, CRF07_BC, or subtype B were further analyzed for coreceptor tropism (with an average of 65,000 sequencing reads per participant) (SI Appendix, Figs. S3 and S4 and Tables S1 and S2). Analysis of Geno2pheno false positive rate (FPR) distribution among the plasma viral quasi-species in each participant again found higher frequency of X4-using variants in people infected by CRF01_AE cluster 4 than by cluster 5, CRF07_BC, or subtype B (Fig. 2 and SI Appendix, Fig. S5 and Table S2). Together, deep sequencing on samples collected within the first year of infection confirmed the observation in a large cross-sectional NHMES study.
Fig. 2.
Higher frequency of predicted X4-using variants in CRF01_AE cluster 4 identified by deep sequencing. (A) Phylogenetic relationship of 60 deep-sequenced individuals from the CYM cohort who were purely infected by CRF01_AE, CRF07_BC, or subtype B HIV-1. The CYM number of each participant is labeled. In each individual, the most frequent haplotype among the deep-sequencing reads was used for phylogenetic inference. The neighbor-joining (NJ) tree was constructed using the Kimura 2-parameter evolutionary model with 1,000 bootstrap replications. The branches for CRF01_AE cluster 4 (n = 22), cluster 5 (n = 11), CRF07_BC (n = 19), and subtype B (n = 8) are color-coded. (B) Heatmap showing the frequency distribution of Geno2pheno FPR values among the deep-sequencing reads in each individual. The samples in the tree and in the heatmap are matched.
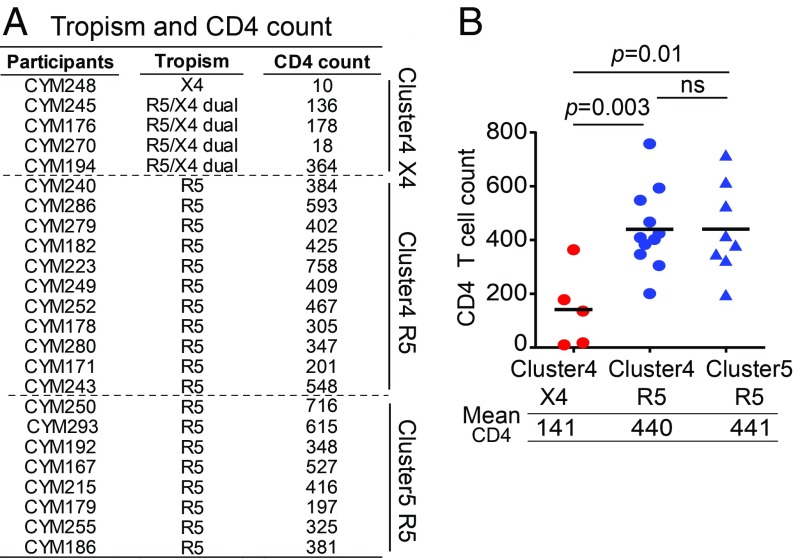
To confirm the genetic prediction phenotypically, we isolated viruses using cryopreserved PBMCs from the deep-sequenced CRF01_AE participants and conducted coreceptor tropism assays using GHOST cell lines. A total of 24 viruses were successfully isolated. The isolation success rate was similar for both clusters: A total of 16 and 8 viruses were isolated from 21 cluster 4 participants (76.2%) and 10 cluster 5 participants (80.0%) who had PBMCs available (Fig. 3A). Five isolates showed the X4-using phenotype, and the remaining 19 were the R5-only phenotype (Fig. 3A). All of the five X4-using isolates, including four dual-tropic and one exclusively X4-tropic, belonged to cluster 4 (Fig. 3A and SI Appendix, Fig. S6). This indicates a much higher prevalence of the X4-using phenotype in cluster 4 than in cluster 5 (31.3% vs. 0%), consistent with genotypic prediction.
Fig. 3.
Significantly lower CD4 count in individuals harboring X4-using viruses in CRF01_AE cluster 4. (A) CD4 count for participants harboring phenotypically confirmed X4 or R5 viruses from the CYM cohort. (B) CD4 count comparison between participants harboring X4 or R5 viruses. The black line represents the mean CD4 count. The statistical difference was calculated using the two-tailed Mann–Whitney u test.
Individuals Harboring X4 Viruses Had Significantly Lower CD4 T Cell Counts.
Previous studies demonstrated the association between X4 tropism and lower CD4 counts, mainly for subtype B HIV-1 (13–17). However, a recent study based on genotypic prediction did not find such an association in CRF01_AE-infected people in China (though there is a trend that people with CD4 < 50 tend to have Geno2pheno FPR < 5) (8). Because genotypic prediction could overestimate actual X4 prevalence, we compared CD4 T cell counts based on virus phenotype (Fig. 3B). Among the 24 phenotype-confirmed participants, those with the X4-using phenotype had significantly lower CD4 counts compared with those with the R5 phenotype in cluster 4 (141 vs. 440, P = 0.003) and in cluster 5 (141 vs. 441, P = 0.01) (Fig. 3B). With well-matched phenotype data, we confirmed that the X4-using phenotype is associated with a significantly lower CD4 count in CRF01_AE. The higher prevalence of the X4 phenotype in cluster 4 during the first year of infection and its association with significantly lower CD4 counts explained why cluster 4 had lower CD4 counts compared with cluster 5 from the early infection stage (Fig. 1 C and D).
Genetic and Structural Determinants for Greater CXCR4 Use Propensity.
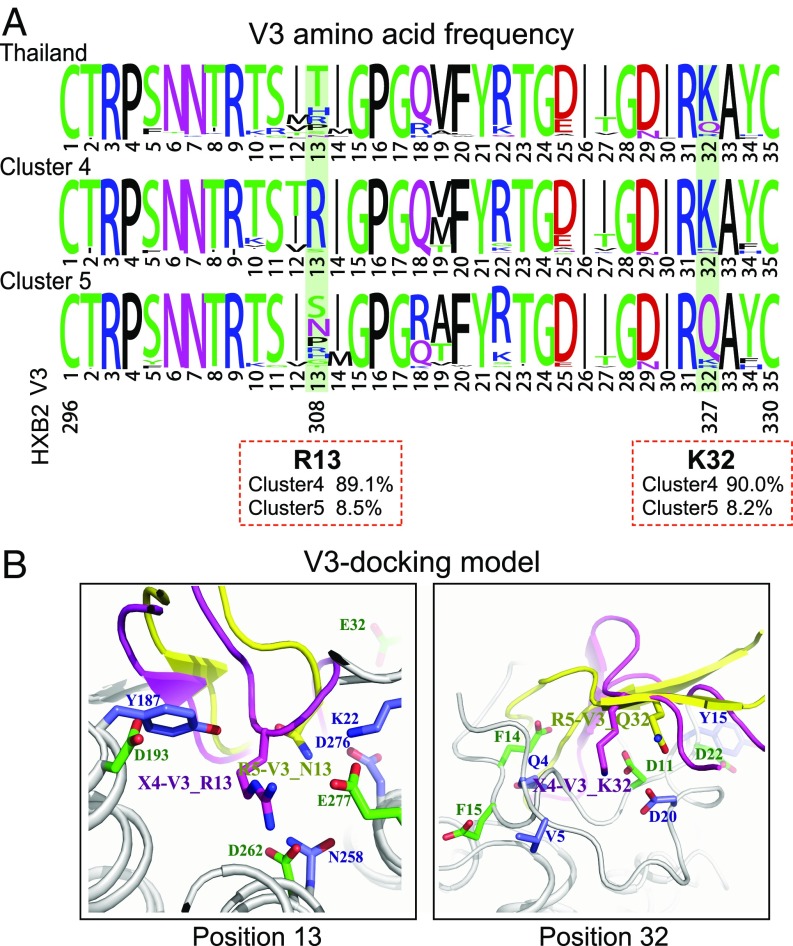
To explore the mechanism for greater X4-using propensity exhibited by cluster 4 viruses, we first compared the V3 sequences between clusters 4 and 5. We found that cluster 4 viruses have two highly conserved basic amino acids at positions 13 and 32 in the V3 loop (R13 and K32, HXB2 nos. R308 and K327, respectively), which were present in only about 8% of the cluster 5 viruses (Fig. 4A). These two highly conserved amino acids confer on cluster 4 viruses a higher positively charged V3 loop. Because the V3 net charge is an important determinant for X4 use, the presence of R13 and K32 may decrease the threshold of R5 to X4 tropism switching in cluster 4, while in cluster 5 the genetic and functional barrier for such switching is higher. Structure analysis using V3-docking models (18) suggested that residue R13 in cluster 4 potentially forms salt bridges with D262 and E277 and a hydrogen bond with the side chain of H281 in the CXCR4 coreceptor, while the corresponding residue in cluster 5 may form hydrogen bonds with K22 and D276 in the CCR5 coreceptor (Fig. 4B). Because the ligand-binding pocket in CXCR4 is more negatively charged than that in CCR5, the positively charged residue R13 in cluster 4 viruses is more favored. The residue K32 in cluster 4 may form salt bridges with CXCR4’s N terminus, which contains more acidic residues than CCR5’s N terminus (Fig. 4B). Therefore, the highly conserved residues R13 and K32 in cluster 4 may be key determinants of the higher tendency toward X4 use. Indeed, the basic amino acid at V3 position 32 has been observed to associate with T cell tropism in subtype B HIV-1 (19). Interestingly, the V3 region of cluster 4 has fewer variations compared with the CRF01_AE ancestor sequences from Thailand—notably the preservation of K32—while cluster 5 is more divergent (Fig. 4A and SI Appendix, Fig. S7).
Fig. 4.
Genetic determinants and structural basis of the higher X4-using tendency in CRF01_AE cluster 4. (A) The frequency of each V3 amino acid was determined using a total of 385 available sequences from cluster 4 and 328 available sequences from cluster 5. The sequences from Thailand (n = 34) were downloaded from the Los Alamos HIV sequence database (before year 2000). The plots were generated using the WebLogo tool (55). The exact frequency of V3 residues R13 and K32 in clusters 4 and 5 is shown in red boxes. (B) Structural analysis for V3 positions 13 and 32 in binding of the CCR5 and CXCR4 coreceptors using the V3-docking model.
Despite the fact that R13 and K32 were present in the vast majority of cluster 4 viruses, only about 30% of the individuals in cluster 4 were phenotypically confirmed to harbor X4 viruses. This indicates the existence of additional determinants to shift to the X4 phenotype. To further identify key amino acids governing the phenotype switch, we analyzed the genetic composition of PBMC isolates at the single-genome level (SI Appendix, Fig. S8 and Table S3). To “sieve out” the X4-using variant(s) from the entire viral population (that is, the exact sequence or sequences accounting for the X4-using phenotype), we further sequenced the viruses released from the GHOST X4 cell culture by single-genome amplification (SGA) for the five X4-using samples. Comparing the genetic composition between the PBMC viral isolates and the concurrent plasma viral population showed two different patterns: In the majority of R5 subjects (15 of 19), the V3 lineages in the PBMC isolates and in plasma were in proportion; that is, the predominant lineage in the PBMC isolates was also the predominant one in plasma. In contrast, in four of the five X4 isolates, the predominant, phenotypically confirmed X4 lineage in the PBMC isolates existed as a minor variant in plasma (SI Appendix, Fig. S9 and Table S3). All V3 lineages detected in the PBMC isolates were present in plasma, and more V3 lineages were detected in plasma than in the isolates (SI Appendix, Fig. S9 and Table S3).
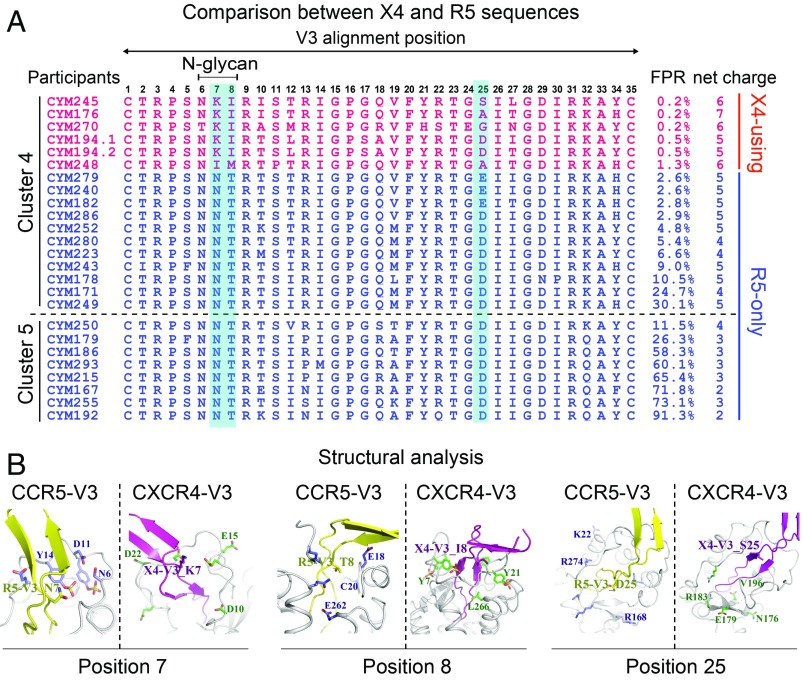
Several V3 alterations were found in the phenotypically confirmed X4 sequences. First, all X4-using sequences lost the N-linked glycan site at the beginning of the V3 loop (V3 positions 6–8, HXB2 nos.301–303), mostly by T to I substitution at position 8 (Fig. 5A). Second, while residue N was invariably found in all R5 sequences at position 7, all but one X4-using sequence had residue K at this position (Fig. 5A). Third, either E or D was found at position 25 in the R5 sequences; however, non-E/D substitutions (S/A/G) were present in the majority of X4-using sequences (Fig. 5A). Interestingly, none of the X4 sequences had positively charged amino acid R or K at V3 position 11 or 25. These two residues are important for X4 use in other HIV-1 subtypes (20–22). This implies a different evolutionary pathway of coreceptor switching in CRF01_AE HIV-1. In genotypic prediction, all X4-using sequences had Geno2pheno FPR values below 2% and a V3 net charge no less than 5. In contrast, all R5 sequences had FPR values higher than 2% and a V3 net charge no more than 5. As expected, cluster 4 had an overall higher V3 net charge than cluster 5 (Fig. 5A). Notably, five sequences in cluster 4 with FPR below 5% were in fact the R5 phenotype (Fig. 5A). Therefore, using FPR 5% as the cutoff may significantly overestimate the prevalence of the X4 phenotype in CRF01_AE. Although determining a coreceptor prediction method was not the focus of the study due to a relatively small sample size, the results showed that three common features were shared by all phenotypically confirmed X4 sequences: The loss of N301 glycan, a V3 net charge no less than 5, and an FPR lower than 2%. Thus, these three features together may strongly indicate an X4 phenotype in CRF01_AE HIV-1.
Fig. 5.
Genetic characteristics of phenotypically confirmed X4 sequences and structural modeling for coreceptor binding. (A) V3 amino acid alignment of SGA-derived sequences from the phenotype-confirmed primary viral isolates. Sequences shown in red and blue are phenotypically confirmed X4 and R5 sequences, respectively. Key V3 positions associated with the X4-using phenotype are shaded in light blue. (B) Structural modeling for V3 positions 7, 8, and 25 in binding of coreceptors CCR5 and CXCR4 using the V3-docking model.
Using the CCR5-V3 and CXCR4-V3 complex models (23, 24), we also investigated the role of V3 positions 7, 8, and 25 in viral tropism from a structural perspective. In the model of the CCR5-V3 complex, residue T8 in the R5 V3 loop is surrounded by hydrophilic amino acids, suggesting that residue T8 is more favored by a hydrophilic environment. However, in the model of the CXCR4-V3 complex, residue I8 in the X4 V3 loop is surrounded by hydrophobic amino acids (Fig. 5B). This could explain why all X4 viruses have T to I/M substitutions at position 8 because residue T8 may not fit the hydrophobic environment within CXCR4’s ligand-binding pocket. In the CXCR4-V3 complex, V3 position 7 is surrounded by negatively charged residues, which favor interaction with positively charged residue K7 in X4 sequences (Fig. 5B). Compared with the corresponding region in the ligand-binding pocket of CXCR4, the ligand-binding pocket in CCR5 around V3 position 25 contains more positively charged residues, which are favored for interacting with negatively charged residues D/E25. This explained why all R5 viruses have D/E at V3 position 25. However, D/E may be less favored in the less positively charged environment in the ligand-binding pocket of CXCR4 (Fig. 5B). Therefore, non-D/E substitutions, as observed in X4-using sequences, would be required for efficient X4 binding (Fig. 5B). Taken together, genetic analysis and structural modeling showed that specific V3 substitutions at positions 7, 8, and 25 may be required to achieve an X4-using phenotype in the context of the CRF01_AE cluster 4 envelope.
Discussion
Since its initial introduction to humans in the early 20th century, HIV-1 evolved genetically and biologically at a faster pace than other viruses, due to the highly error-prone nature of its reverse transcriptase and its diverse transmission routes, such as intravenous drug use (IDU), heterosexual transmission (HT), and MSM activities. The Chinese HIV-1 epidemic, with its multiple subtypes and their genetic clusters circulating at the same time, provides a unique opportunity to monitor virus evolution at both genotype and phenotype levels.
Past studies on HIV-1 evolution were mainly focused on virus genotype, not phenotype, because the latter takes longer observation time and requires large well-matched samples. In this study, we focused on two major CRF01_AE clusters in China. In the large cross-sectional dataset from the NHMES, we discovered a significant difference in CD4 count between people infected by CRF01_AE clusters 4 and 5 at both an early and a later stage of infection. The lower CD4 count in cluster 4 is directly associated with the higher prevalence of the X4 virus based on genotypic prediction. This genotype-based observation in a large population was further confirmed by well-matched genotype and phenotype data from an MSM seroincidence cohort. We observed that, among seroconvertors, those harboring X4 viruses had significantly lower CD4 counts. Although in the NHMES dataset other minor CRF01_AE clusters had the lowest CD4 counts, this could have been mainly due to the higher proportion of chronic infections with longer infection times. Although the exact time of infection was not measurable, it could be estimated that the overall infection time in other clusters is substantially longer than in clusters 4 and 5. This is supported by the following observations: First, China’s HIV-1 epidemic has experienced a transition from IDU and HT to MSM. Second, the current HIV incidence is highest in the MSM population, followed by HT and lastly IDU. Third, the MSM epidemic is mainly caused by clusters 4 and 5, while the HT and IDU epidemics are mainly caused by other clusters. Due to both the different epidemic pattern and the low recent infection rate of other clusters, it was difficult to include them in the current study. However, the pathogenicity of other CRF01_AE clusters will be important to investigate in the future.
It is usually considered that X4 variants emerge during late stages of infection, with the development of immunodeficiency of the host (25). In subtype B infections, around 50% of patients underwent a coreceptor switch, usually after five years of infection, which correlated with rapid CD4 decline and faster progression to AIDS (16, 26–31). The exact times of coreceptor switching in different HIV-1 subtypes are not well understood. A recent study did not detect X4 variants in subtype B-infected people within two years of infection (32). We demonstrated that, in certain HIV-1 subtypes or clusters, coreceptor switching may occur much earlier than previously thought. Investigating the dynamics of CD4 loss in association with coreceptor switching using longitudinal samples will promote better understand of the pathogenicity of the X4 virus. In the current study, we could not rule out the possibility that, in some of individuals harboring X4 viruses, the X4 variants were initially transmitted as the transmitted/founder (T/F) virus. No samples from earlier time points were available to address this possibility. However, nearly all T/F viruses characterized to date were exclusively CCR5-tropic except in some rare cases (33–38). In particular, a study of T/F virus envelopes from another MSM cohort in China showed that all CRF01_AE T/F envelopes tested were CCR5-tropic (38). These all support early coreceptor switching as a more plausible scenario. Nevertheless, it will be important to determine whether X4 transmission more frequently occurs in CRF01_AE than in subtypes B and C, as the existence of X4 variants in up to 30% of recent infections, as observed in cluster 4, may increase the chance of X4 transmission at the population level.
Another interesting finding is the outgrowth of highly replication-competent X4 variants in primary viral isolates from CRF01_AE cluster 4. The low frequency of these X4 variants in plasma may not be explained by their low replication fitness, as they rapidly outcompete other lineages in the in vitro setting. Instead, it is more likely due to the compartmentalization of the R5 and X4 viruses in different cell subsets or tissues in vivo. Due to the differential expressions of CCR5 and CXCR4 coreceptors in memory and naïve CD4 T cell subsets (39, 40), R5 and X4 viruses are considered to preferentially replicate in the memory and naïve CD4 subsets, respectively (41–45). It has been shown that naïve CD4 T cells produce viruses at a lower propagation rate than memory T cells (46, 47), possibly due to the relatively low division rate (48, 49). Therefore, in vivo those minor X4 lineages might compartmentalize in cell subsets or tissues that shunt the viruses less efficiently into the blood. Although the plasma viral population is most widely studied, the relative contribution of different tissues/compartments to plasma viremia and immunopathogenesis remains an unresolved question in the field. The mechanism of CD4 loss caused by such minor X4 lineages in plasma will be an important question to address in the future. Regardless of the mechanism, this observation has a clinical implication: Because conventional sequencing methods may not be sensitive enough to capture those minor X4 variants in plasma, deep sequencing or phenotypic assay will be required to determine the existence of X4 variants in vivo, especially when using treatment regimens with the CCR5 inhibitor.
In summary, we demonstrated that various phylogenetic clusters of the same HIV-1 subtype can have disparate pathogenicities and cause different disease outcomes, providing the missing link between HIV-1 genetic evolution and viral phenotypes. At the phenotype level, CRF01_AE cluster 4 evolved with enhanced X4 tropism and viral pathogenesis, while cluster 5 became more attenuated due to a decreased potential of using CXCR4. Whether the process of “phenotype divergence” occurred as a random founder event from the initial seeding clusters or was due to adaptation to different hosts or transmission routes remains to be studied. Our study emphasizes the importance of monitoring HIV-1 genetic drift and phenotype shift at the phylogenetic cluster level to control the spread of more pathogenic viruses like CRF01_AE cluster 4.
Materials and Methods
Study Participants.
The study participants were from the NHMES and the Beijing Chaoyang District MSM cohort (the CYM cohort) (SI Appendix). Written informed consent was obtained from all study participants. All experiments involving human subjects from the NHMES (project no.: X140617334) and CYM cohort (project no: X080216132) were approved by the Institutional Review Board of the National Center for AIDS/STD Control and Prevention, Chinese Center for Disease Control and Prevention.
Enzyme Immunoassay.
To distinguish recent HIV-1 infections from long-term HIV-1 infections, enzyme immunoassay (EIA) was performed using the Maxim HIV-1 Limiting Antigen Avidity (LAg-Avidity) EIA Kit (Maxim Biomedical). The experiment and the data analysis were performed according to manufacturer’s instructions (Maxim Biomedical).
Viral RNA Extraction and cDNA Synthesis.
Viral RNA was extracted from 200 μL of plasma sample using the QIAamp Viral RNA Mini Kit (Qiagen). RNA was eluted into 50 μL of RNase-free water. A total of 17 μL of viral RNA was used for cDNA synthesis using Invitrogen SuperScript III reverse transcriptase (Thermo Fisher Scientific) with Oligo (dT) primer. The cDNA was immediately used for PCR amplification.
Deep Sequencing.
The Illumina MiSeq library was prepared using a nested PCR approach. The 8-nt Illumina index and adaptors (P5 and P7) were added to both ends of the second-round PCR primers. The pooled DNA library was sequenced on a Illumina MiSeq system using the Reagent Kit V2 (500 cycles) as previously described (50). Data analysis was performed as previously described (51) (SI Appendix).
Genotypic Prediction of Coreceptor Use.
Genotypic prediction of coreceptor use was performed using the Geno2pheno clonal model (52).
Primary Virus Isolation from PBMCs.
Primary viruses were isolated by cocultivation of infected PBMCs with normal PBMCs from healthy donors, after CD8 T cells were depleted from both the infected and normal PBMC samples (SI Appendix).
Coreceptor Tropism Determination.
Coreceptor tropism of the primary viral isolates was determined using the GHOST(3) CCR5 and GHOST(3) CXCR4 cell lines (53). Both GFP expression in the GHOST cell lines and viral p24 production were used to determine coreceptor use (SI Appendix).
Single Genome Amplification.
The SGA was performed as previously described (33). The sequences were aligned using the Los Alamos National Laboratory’s GeneCutter tool followed by manual adjustment to obtain the optimal alignment (SI Appendix).
Statistical Analysis.
All statistical analysis was performed using Prism 7 software (GraphPad). Statistical differences were determined using the two-tailed Mann–Whitney U test or Fisher’s exact test as indicated in the figure legends. The exact P values are provided in the figures.
Data Availability.
Generated nucleic acid sequences were deposited in GenBank (54) (accession nos.: MH672692–MH673032).
Supplementary Material
Acknowledgments
We thank Dr. Cecilia Cheng-Mayer for help editing the manuscript. We thank the staff from the Center for Disease Control and Prevention in China, who participated in the National HIV Molecular Epidemiology Survey and helped collect samples and epidemiology data. This work was supported by the China National Major Project for Infectious Diseases Control and Prevention, the China Key Project of the State Key Laboratory of Infectious Diseases Control and Prevention, and the National Natural Science Foundation of China (Grants NSFC31730027 and NSFC8171101312).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. MH672692–MH673032).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814714116/-/DCSupplemental.
References
- 1.Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med. 2008;358:1590–1602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng Y, et al. The rapidly expanding CRF01_AE epidemic in China is driven by multiple lineages of HIV-1 viruses introduced in the 1990s. AIDS. 2013;27:1793–1802. doi: 10.1097/QAD.0b013e328360db2d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kilmarx PH, et al. Disease progression and survival with human immunodeficiency virus type 1 subtype E infection among female sex workers in Thailand. J Infect Dis. 2000;181:1598–1606. doi: 10.1086/315469. [DOI] [PubMed] [Google Scholar]
- 4.Costello C, et al. HIV-1 subtype E progression among northern Thai couples: Traditional and non-traditional predictors of survival. Int J Epidemiol. 2005;34:577–584. doi: 10.1093/ije/dyi023. [DOI] [PubMed] [Google Scholar]
- 5.Nelson KE, Costello C, Suriyanon V, Sennun S, Duerr A. Survival of blood donors and their spouses with HIV-1 subtype E (CRF01 A_E) infection in northern Thailand, 1992-2007. AIDS. 2007;21(Suppl 6):S47–S54. doi: 10.1097/01.aids.0000299410.37152.17. [DOI] [PubMed] [Google Scholar]
- 6.Rangsin R, et al. The natural history of HIV-1 infection in young Thai men after seroconversion. J Acquir Immune Defic Syndr. 2004;36:622–629. doi: 10.1097/00126334-200405010-00011. [DOI] [PubMed] [Google Scholar]
- 7.Li X, et al. Evidence that HIV-1 CRF01_AE is associated with low CD4+T cell count and CXCR4 co-receptor usage in recently infected young men who have sex with men (MSM) in Shanghai, China. PLoS One. 2014;9:e89462. doi: 10.1371/journal.pone.0089462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, et al. CACT0810 Group CRF01_AE subtype is associated with X4 tropism and fast HIV progression in Chinese patients infected through sexual transmission. AIDS. 2014;28:521–530. doi: 10.1097/QAD.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 9.Chu M, et al. HIV-1 CRF01_AE strain is associated with faster HIV/AIDS progression in Jiangsu Province, China. Sci Rep. 2017;7:1570. doi: 10.1038/s41598-017-01858-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.To SW, et al. Determination of the high prevalence of dual/mixed- or X4-tropism among HIV type 1 CRF01_AE in Hong Kong by genotyping and phenotyping methods. AIDS Res Hum Retroviruses. 2013;29:1123–1128. doi: 10.1089/aid.2013.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng KY, et al. High prevalence of CXCR4 usage among treatment-naive CRF01_AE and CRF51_01B-infected HIV-1 subjects in Singapore. BMC Infect Dis. 2013;13:90. doi: 10.1186/1471-2334-13-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, et al. Tracing the epidemic history of HIV-1 CRF01_AE clusters using near-complete genome sequences. Sci Rep. 2017;7:4024. doi: 10.1038/s41598-017-03820-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melby T, et al. HIV-1 coreceptor use in triple-class treatment-experienced patients: Baseline prevalence, correlates, and relationship to enfuvirtide response. J Infect Dis. 2006;194:238–246. doi: 10.1086/504693. [DOI] [PubMed] [Google Scholar]
- 14.Moyle GJ, et al. Epidemiology and predictive factors for chemokine receptor use in HIV-1 infection. J Infect Dis. 2005;191:866–872. doi: 10.1086/428096. [DOI] [PubMed] [Google Scholar]
- 15.Wilkin TJ, et al. HIV type 1 chemokine coreceptor use among antiretroviral-experienced patients screened for a clinical trial of a CCR5 inhibitor: AIDS Clinical Trial Group A5211. Clin Infect Dis. 2007;44:591–595. doi: 10.1086/511035. [DOI] [PubMed] [Google Scholar]
- 16.Koot M, et al. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 17.Brumme ZL, et al. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J Infect Dis. 2005;192:466–474. doi: 10.1086/431519. [DOI] [PubMed] [Google Scholar]
- 18.Tan Q, et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science. 2013;341:1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milich L, Margolin B, Swanstrom R. V3 loop of the human immunodeficiency virus type 1 env protein: Interpreting sequence variability. J Virol. 1993;67:5623–5634. doi: 10.1128/jvi.67.9.5623-5634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang W, et al. Mutational pathways and genetic barriers to CXCR4-mediated entry by human immunodeficiency virus type 1. Virology. 2011;409:308–318. doi: 10.1016/j.virol.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: Roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 22.Ping LH, et al. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: Underrepresentation of X4 variants. J Virol. 1999;73:6271–6281. doi: 10.1128/jvi.73.8.6271-6281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamamis P, Floudas CA. Molecular recognition of CCR5 by an HIV-1 gp120 V3 loop. PLoS One. 2014;9:e95767. doi: 10.1371/journal.pone.0095767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamamis P, Floudas CA. Molecular recognition of CXCR4 by a dual tropic HIV-1 gp120 V3 loop. Biophys J. 2013;105:1502–1514. doi: 10.1016/j.bpj.2013.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swanstrom R, Coffin J. HIV-1 pathogenesis: The virus. Cold Spring Harb Perspect Med. 2012;2:a007443. doi: 10.1101/cshperspect.a007443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuitemaker H, et al. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: Progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1–infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koot M, et al. Conversion rate towards a syncytium-inducing (SI) phenotype during different stages of human immunodeficiency virus type 1 infection and prognostic value of SI phenotype for survival after AIDS diagnosis. J Infect Dis. 1999;179:254–258. doi: 10.1086/314539. [DOI] [PubMed] [Google Scholar]
- 29.Verhofstede C, Nijhuis M, Vandekerckhove L. Correlation of coreceptor usage and disease progression. Curr Opin HIV AIDS. 2012;7:432–439. doi: 10.1097/COH.0b013e328356f6f2. [DOI] [PubMed] [Google Scholar]
- 30.Moore JP, Kitchen SG, Pugach P, Zack JA. The CCR5 and CXCR4 coreceptors–Central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res Hum Retroviruses. 2004;20:111–126. doi: 10.1089/088922204322749567. [DOI] [PubMed] [Google Scholar]
- 31.Richman DD, Bozzette SA. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J Infect Dis. 1994;169:968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 32.Zhou S, Bednar MM, Sturdevant CB, Hauser BM, Swanstrom R. Deep sequencing of the HIV-1 env gene reveals discrete X4 lineages and linkage disequilibrium between X4 and R5 viruses in the V1/V2 and V3 variable regions. J Virol. 2016;90:7142–7158. doi: 10.1128/JVI.00441-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keele BF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salazar-Gonzalez JF, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isaacman-Beck J, et al. Heterosexual transmission of human immunodeficiency virus type 1 subtype C: Macrophage tropism, alternative coreceptor use, and the molecular anatomy of CCR5 utilization. J Virol. 2009;83:8208–8220. doi: 10.1128/JVI.00296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rademeyer C, et al. Features of recently transmitted HIV-1 clade C viruses that impact antibody recognition: Implications for active and passive immunization. PLoS Pathog. 2016;12:e1005742. doi: 10.1371/journal.ppat.1005742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang C, et al. Primary infection by a human immunodeficiency virus with atypical coreceptor tropism. J Virol. 2011;85:10669–10681. doi: 10.1128/JVI.05249-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, et al. Comprehensive characterization of the transmitted/founder env genes from a single MSM cohort in China. J Acquir Immune Defic Syndr. 2015;69:403–412. doi: 10.1097/QAI.0000000000000649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci USA. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blaak H, et al. In vivo HIV-1 infection of CD45RA(+)CD4(+) T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4(+) T cell decline. Proc Natl Acad Sci USA. 2000;97:1269–1274. doi: 10.1073/pnas.97.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Rij RP, et al. Differential coreceptor expression allows for independent evolution of non-syncytium-inducing and syncytium-inducing HIV-1. J Clin Invest. 2000;106:1569. doi: 10.1172/jci7953c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimura Y, et al. Resting naive CD4+ T cells are massively infected and eliminated by X4-tropic simian-human immunodeficiency viruses in macaques. Proc Natl Acad Sci USA. 2005;102:8000–8005. doi: 10.1073/pnas.0503233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ribeiro RM, Hazenberg MD, Perelson AS, Davenport MP. Naïve and memory cell turnover as drivers of CCR5-to-CXCR4 tropism switch in human immunodeficiency virus type 1: Implications for therapy. J Virol. 2006;80:802–809. doi: 10.1128/JVI.80.2.802-809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Council OD, Joseph SB. Evolution of host target cell specificity during HIV-1 infection. Curr HIV Res. 2018;16:13–20. doi: 10.2174/1570162X16666171222105721. [DOI] [PubMed] [Google Scholar]
- 46.Eckstein DA, et al. HIV-1 actively replicates in naive CD4(+) T cells residing within human lymphoid tissues. Immunity. 2001;15:671–682. doi: 10.1016/s1074-7613(01)00217-5. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z, et al. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science. 1999;286:1353–1357. doi: 10.1126/science.286.5443.1353. [DOI] [PubMed] [Google Scholar]
- 48.Mclean AR, Michie CA. In vivo estimates of division and death rates of human T lymphocytes. Proc Natl Acad Sci USA. 1995;92:3707–3711. doi: 10.1073/pnas.92.9.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCune JM, et al. Factors influencing T-cell turnover in HIV-1-seropositive patients. J Clin Invest. 2000;105:R1–R8. doi: 10.1172/JCI8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams WB, et al. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science. 2015;349:aab1253. doi: 10.1126/science.aab1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J, et al. Extensive recombination due to heteroduplexes generates large amounts of artificial gene fragments during PCR. PLoS One. 2014;9:e106658. doi: 10.1371/journal.pone.0106658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol. 2007;25:1407–1410. doi: 10.1038/nbt1371. [DOI] [PubMed] [Google Scholar]
- 53.Vödrös D, et al. Quantitative evaluation of HIV-1 coreceptor use in the GHOST3 cell assay. Virology. 2001;291:1–11. doi: 10.1006/viro.2001.1163. [DOI] [PubMed] [Google Scholar]
- 54. Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2005) GenBank. Nucleic Acids Res 33:D34–D38. Deposited July 25, 2018. [DOI] [PMC free article] [PubMed]
- 55.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Generated nucleic acid sequences were deposited in GenBank (54) (accession nos.: MH672692–MH673032).