Significance
Mitochondria are intracellular hubs of metabolism that play critical roles in mammals, but knowledge of the metabolic landscape within mitochondria in mammalian tissues remains incomplete. Prior studies have often relied on interrogating metabolites within whole tissue, but this approach does not reveal the behavior of metabolites within specific organelles and cell types. To address this, we generated a transgenic mouse that can express a mitochondrially localized epitope tag (MITO-Tag) with spatiotemporal control to allow for rapid, cell-type-specific immunoisolation of mitochondria from tissues. We demonstrate that these MITO-Tag Mice can be utilized for profiling a variety of molecular components of mitochondria, such as proteins, lipids, and polar metabolites, and thus believe that MITO-Tag Mice will be useful for studying mitochondrial physiology.
Keywords: mitochondria, MITO-Tag Mice, metabolomics, proteomics, lipidomics
Abstract
Mitochondria are metabolic organelles that are essential for mammalian life, but the dynamics of mitochondrial metabolism within mammalian tissues in vivo remains incompletely understood. While whole-tissue metabolite profiling has been useful for studying metabolism in vivo, such an approach lacks resolution at the cellular and subcellular level. In vivo methods for interrogating organellar metabolites in specific cell types within mammalian tissues have been limited. To address this, we built on prior work in which we exploited a mitochondrially localized 3XHA epitope tag (MITO-Tag) for the fast isolation of mitochondria from cultured cells to generate MITO-Tag Mice. Affording spatiotemporal control over MITO-Tag expression, these transgenic animals enable the rapid, cell-type-specific immunoisolation of mitochondria from tissues, which we verified using a combination of proteomic and metabolomic approaches. Using MITO-Tag Mice and targeted and untargeted metabolite profiling, we identified changes during fasted and refed conditions in a diverse array of mitochondrial metabolites in hepatocytes and found metabolites that behaved differently at the mitochondrial versus whole-tissue level. MITO-Tag Mice should have utility for studying mitochondrial physiology, and our strategy should be generally applicable for studying other mammalian organelles in specific cell types in vivo.
Mitochondria are membrane-bound organelles that house metabolic pathways that play critical roles in mammalian physiology. The mitochondrial matrix, the innermost mitochondrial compartment, is a unique metabolic space within the cell because of the presence of a distinct array of metabolic processes and the selective permeability of the inner mitochondrial membrane (1). Because of the unique metabolic landscape within the mitochondrial matrix, metabolomic approaches with organellar-level resolution can provide important insights into mitochondrial and cellular physiology that otherwise may be missed using whole-cell or whole-tissue analyses (2). Importantly, given that mitochondria have been implicated in a wide variety of pathologies, such as myocardial infarction and Parkinson’s disease (1, 3), a complete understanding of how mitochondrial dysfunction contributes to these various disease states will likely require the study of mitochondrial metabolites in vivo.
We have previously developed a mitochondrial metabolomics workflow for cultured mammalian cells that utilizes a mitochondrially localized 3XHA epitope tag (MITO-Tag) to achieve the rapid isolation of mitochondria followed by metabolite profiling by liquid chromatography and mass spectrometry (LC/MS) (2, 4). Utilizing this methodology, we were able to observe changes in matrix metabolism that were not appreciated using whole-cell metabolomics. Similar approaches from our laboratory for studying lysosomes in cultured cells have also revealed the dynamics of lysosomal metabolism during different cellular states (5, 6). Because mammalian physiology can often be better modeled in vivo rather than in cultured cells, we wanted to implement our workflow for studying mitochondria in mice. Of particular importance, mitochondrially targeted epitope tags would allow for the rapid isolation of mitochondria from specific cell types in complex tissues without the need for cell sorting, thus improving the speed of the workflow and reducing distortion of the mitochondrial metabolite profile (7). This would be useful for cell types such as neurons, as the state of these cells can be particularly vulnerable to distortions during traditional cell-sorting methods due to their frailty and sprawling cellular architecture (8). Of note, there is a paucity of methodologies for rapidly isolating mitochondria with cell-type specificity from mammalian tissues in vivo (9–14).
To that end, we generated MITO-Tag Mice, Rosa26 knock-in mice that express the MITO-Tag only in cells that express Cre recombinase, as a result of a loxP-STOP-loxP (LSL) cassette. This strategy allows for rapid isolation of mitochondria with cell-type specificity from mammalian tissues as long as one has an appropriate promoter driving expression of Cre recombinase (15). Note that the MITO-Tag is the 3XHA-EGFP-OMP25 or HA-MITO construct described previously (2, 4). Of note, the HA-MITO protein lacks the N-terminal PDZ domain of OMP25 and contains only the OMP25 C-terminal transmembrane and outer mitochondrial membrane localization sequences, which should render the HA-MITO protein generally innocuous (16, 17). We show that these MITO-Tag Mice allow for the rapid immunopurification and proteomic, lipidomic, and polar metabolomic analyses of mitochondria from hepatocytes in vivo. Using both targeted and untargeted metabolomics, we uncovered changes in a diverse array of mitochondrial metabolites during fasted and refed conditions, thus shedding light on how hepatocyte mitochondria behave during these two important nutritional states. We also identified metabolites that behave differently when assessed at the mitochondrial versus whole-tissue level, thus underscoring the importance of mitochondrial metabolomics. Taken together, our work demonstrates the utility of MITO-Tag Mice for studying mitochondrial biology in vivo and offers a framework for studying other mammalian organelles in specific cell types within complex tissues.
Results
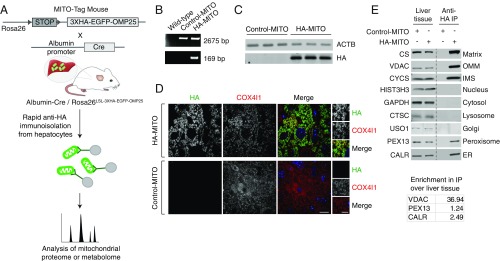
To enable rapid isolation of mitochondria from specific cell types in vivo, we generated MITO-Tag Mice by knocking our HA-MITO construct and an upstream LSL cassette into the Rosa26 locus (Fig. 1A). In the absence of Cre recombinase, the LSL cassette will prevent the production of the HA-MITO protein, and anti-HA immunopurifications will not yield mitochondria. However, once present, Cre recombinase will excise the LSL cassette and thus enable epitope tagging and rapid isolation of mitochondria. Importantly, by selecting the appropriate promoters for controlling the expression of Cre recombinase, one can restrict epitope tagging of mitochondria to specific cell types and consequently immunoisolate mitochondria with cell-type specificity from a piece of tissue without the need for cell sorting.
Fig. 1.
Characterization of MITO-Tag Mice and associated in vivo workflow. (A) Schematic demonstrating the design of the MITO-Tag Mice and how they can be utilized for rapid, cell-type–specific isolation and multimodal analysis of mitochondria in vivo. MITO-Tag Mice contain the 3XHA-EGFP-OMP25 (HA-MITO) gene knocked into the Rosa26 locus. The expression of this gene is dependent on the presence of Cre recombinase due to an upstream LSL cassette. Mating MITO-Tag Mice with mice expressing Cre under the control of certain promoters, such as the Albumin promoter as in this study, allows for cell-type-specific expression of the HA-MITO gene and, consequently, allows for the rapid, cell-type-specific isolation of mitochondria in vivo (mitochondrial isolation takes ∼10 min once liver tissue has been homogenized). Because KPBS, the mitochondrial isolation buffer utilized in our workflow, is compatible with mass spectrometric analyses, the isolated mitochondria can subsequently be interrogated with either proteomics or metabolomics. The actual immunopurification workflow used here overall is similar to that used in our prior work (2, 4); however, it is important to note that the material inputs in this work are liver tissue from Rosa26LSL-3XHA-EGFP-OMP25/+ mice (Control-MITO mice) and Alb-Cre+/−, Rosa26LSL-3XHA-EGFP-OMP25/+ mice (HA-MITO mice) and that no measurements of mitochondrial volume were done in this study because we performed relative rather than absolute quantification of metabolites. Arrowheads indicate loxP sites. (B) Genomic PCR analysis of liver tissue taken from mice with the indicated genotypes. Upper (2,675 bp) and Lower (169 bp) bands indicate the presence of the desired knock-in at the Rosa26 locus and successful Cre-mediated excision of the floxed region, respectively. The presence of nonexcised versions of the knock-in cassette in the HA-MITO liver tissue could be secondary to heterogeneity of Albumin promoter activity across hepatocytes and there being a significant number of cells in liver that are not hepatocytes and thus not expected to express Cre recombinase (35). (C) Immunoblot analysis of liver tissue taken from mice with the indicated genotypes. ACTB was used as a loading control. Control-MITO mice (n = 3); HA-MITO mice (n = 3). (D) Immunofluorescence of liver tissue taken from mice with the indicated genotypes. HA (green), COX4l1 (red), and Hoescht (blue) signals are shown. (Scale bars in the large and small images represent 100 and 50 μm, respectively.) (E) Immunoblot analysis of liver tissue and the anti-HA immunoprecipitates (anti-HA IP) from mice with the indicated genotypes. The line indicates that two parts of the same membrane have been brought together. The names of the protein markers used and the corresponding subcellular compartments appear to the left and right of the immunoblots, respectively. ER, endoplasmic reticulum; Golgi, Golgi complex; IMS, mitochondrial intermembrane space; Matrix, mitochondrial matrix; OMM, outer mitochondrial membrane. The enrichment in the HA-MITO IP, relative to the HA-MITO liver tissue, of the markers VDAC, PEX13, and CALR was quantified and is shown in the table below. All mice used for these experiments were fed ad libitum. For A–E, the following abbreviations are used: Control-MITO, Control-MITO mice; HA-MITO, HA-MITO mice.
To begin our studies utilizing the MITO-Tag Mice, we chose to examine mitochondria within hepatocytes, as these cells respond to a range of interesting physiological stimuli and possess mitochondria that house a diverse array of metabolic reactions. To specifically isolate mitochondria from hepatocytes, we mated MITO-Tag Mice to transgenic animals expressing Cre recombinase under the control of the Albumin promoter (Fig. 1A). Subsequent progeny could then be used for the rapid immunopurification of mitochondria from hepatocytes, with mitochondrial isolation taking ∼10 min once the liver tissue had been homogenized. Because the mitochondrial isolation buffer used in our workflow (i.e., KPBS) is compatible with mass spectrometric analyses, the isolated mitochondria could then be characterized using either proteomic or metabolomic analyses. The immunopurification workflow and methodology was generally similar to that used in our prior studies with two primary differences (2, 4): first, the material inputs in this work were liver tissue from Rosa26LSL-3XHA-EGFP-OMP25/+ mice (Control-MITO mice) and Alb-Cre+/−, Rosa26LSL-3XHA-EGFP-OMP25/+ mice (HA-MITO mice); and second, no measurement of matrix volume was done because we performed relative rather than absolute quantification of metabolites.
Importantly, MITO-Tag Mice behaved as expected when assessed using several orthogonal methods. Utilizing genomic PCR and immunoblot analyses of liver tissue, we observed that excision of the LSL cassette and production of the HA-MITO protein occurred only in the presence of Cre recombinase, respectively (Fig. 1 B and C). Immunofluorescent examination also revealed that the HA-MITO protein colocalized with COX4l1, a mitochondrial marker (Fig. 1D). Cells lacking HA-MITO protein in the images were either not hepatocytes and thus lacked Albumin promoter activity or were a hepatocyte that did not express the HA-MITO gene, which we observed occasionally. Immunoblot analysis of the HA-MITO immunoprecipitate (HA-MITO IP) revealed not only substantial enrichment of mitochondria relative to multiple nonmitochondrial, subcellular compartments, but also minimal organellar contamination of the Control-MITO immunoprecipitate (Control-MITO IP) (Fig. 1E). Consistent with the fact that mitochondria can form contacts with the endoplasmic reticulum and peroxisomes in certain cell types (18, 19), it is not surprising that our rapid, gentle workflow did not remove all traces of these two organelles, although mitochondrial markers were still highly enriched in the immunoprecipitate (IP) by comparison (see table in Fig. 1E).
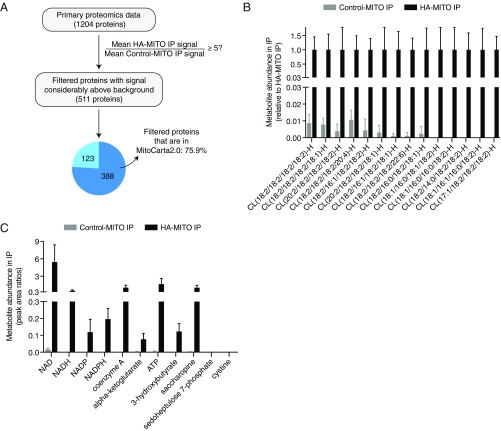
We next performed proteomic and metabolomic analyses on the IP material from our in vivo workflow. From the 1,204 proteins in our primary proteomics data, we identified 511 proteins that met a stringent enrichment criterion of having a mean protein signal in the HA-MITO IPs (i.e., isolated mitochondria) that was at least five times greater than that in the Control-MITO IPs (i.e., background); of these 511 proteins with signal considerably above background, 75.9% of them were found in MitoCarta2.0 (Fig. 2A and Dataset S1) (20). Our proteomic analysis also provided evidence supporting the cell-type specificity of our in vivo workflow (Dataset S1). We were able to detect BDH1, an enzyme required for synthesis of the ketone body 3-hydroxybutyrate and known to be present within hepatocyte mitochondria (21) at ∼15-fold signal above background. In addition, BCL2, a mitochondrial protein present in lymphocytes but much less so in hepatocytes (22, 23), was absent from our primary proteomics data. We chose to look for a mitochondrial protein in lymphocytes because circulating lymphocytes in principle can be present within the liver tissue as not all blood is washed out of the tissue before homogenization, but we would not expect to isolate lymphocyte mitochondria since lymphocytes should not contain Cre recombinase when its expression is controlled by the Albumin promoter. We also carried out metabolomic analyses of the IP material from our in vivo workflow. Using lipidomics, we were able to detect numerous species of cardiolipin, a well-known class of mitochondrial lipids, in the HA-MITO IPs at levels that were greater than background and met our criteria for being mitochondrial (Fig. 2B and Dataset S2). Likewise, using polar metabolomics, we found that the HA-MITO IPs had known mitochondrial metabolites, such as NAD, NADH, NADP, NADPH, CoA, alpha-ketoglutarate, ATP, 3-hydroxybutyrate, and saccharopine, at levels above background and met our criteria for being mitochondrial (Fig. 2C and Dataset S3). We also found that metabolites not expected to be in mitochondria, such as cystine (lysosome) and sedoheptulose 7-phosphate (cytosol), were undetectable in both Control-MITO and HA-MITO IPs. Collectively, these data demonstrate that our in vivo workflow successfully isolates mitochondria and can be utilized for proteomic, lipidomic, and polar metabolomic analyses.
Fig. 2.
Mitochondria isolated with MITO-Tag Mice can be used for multiple downstream analyses and possess both protein and metabolite features characteristic of mitochondria. (A) Flowchart of how proteomics analysis was done. In brief, the 1,204 proteins in our primary proteomics data were filtered based on a stringent enrichment criterion of having a mean pseudonumbered protein signal in the HA-MITO IPs that was at least five times greater than that in the Control-MITO IPs. Of these 511 proteins with signal considerably above that of the Control-MITO IPs (i.e., the background), 75.9% of them were found in MitoCarta2.0 by cross-referencing the NCBI gene IDs, with the respective numerical breakdown indicated in the pie chart wedges (20). Control-MITO mice (n = 3); HA-MITO mice (n = 3). Mice used for these experiments were fed ad libitum (Dataset S1). (B) Lipidomic analysis of IPs for cardiolipin species. Negative lipid ion peak area signals for various cardiolipins (CLs) were used to generate this graph. Control-MITO IPs (n = 4 mice); HA-MITO IPs (n = 4 mice). Mice were fasted for 12 h and then refed for 4 h. Data are shown as means with SDs. All CLs presented here were categorized as grade A and satisfy the criteria of being considered mitochondrial, as defined in Materials and Methods (see also Dataset S2). (C) Targeted metabolomic analysis of IPs for various polar metabolites. Shown are metabolite peak area ratios, which are calculated by dividing the metabolite peak areas by internal standard peak areas, for the Control-MITO and HA-MITO IPs. NAD, NADH, NADP, NADPH, CoA, alpha-ketoglutarate, ATP, 3-hydroxybutyrate, and saccharopine are known mitochondrial metabolites, whereas cystine (lysosome) and sedoheptulose 7-phosphate (cytosol) are markers for extramitochondrial compartments. Control-MITO IPs (n = 4 mice); HA-MITO IPs (n = 4 mice). Mice were fasted for 12 h and then refed for 4 h. Data are shown as means with SDs. With the exception of cystine and sedoheptulose 7-phosphate, all metabolites shown here meet the criteria of being considered mitochondrial, as defined in Materials and Methods (see also Dataset S3).
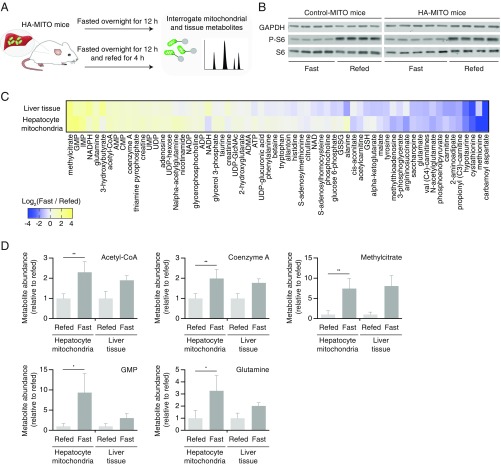
Because the polar metabolome of mitochondria is generally more prone to distortion during mitochondrial isolation than the proteome and lipidome are, we next leveraged the rapidity of our cell-type-specific workflow to study how polar mitochondrial metabolites behave in hepatocytes during fasted and refed conditions, two important nutritional states. Control-MITO and HA-MITO mice were fasted for 12 h overnight and then either fasted or refed for 4 h, after which we interrogated polar metabolites from hepatocyte mitochondria and whole liver tissue (Fig. 3A). To verify that our fasting and refeeding conditions were having the intended effects, we confirmed that there was greater phosphorylation of ribosomal protein S6 at Ser240/244 in the livers of refed Control-MITO and HA-MITO mice (Fig. 3B). Immunoblot analyses revealed that the immunopurifications generally behaved similarly under fasting and refeeding conditions (SI Appendix, Fig. S1), with similar patterns of mitochondrial enrichment as immunopurifications performed with mice that were fed ad libitum (Fig. 1E). Targeted, polar metabolomic analyses of liver tissue and hepatocyte mitochondria revealed changes across a diverse range of metabolites (Fig. 3C and Dataset S3). We observed increased acetyl-CoA levels in hepatocyte mitochondria isolated from animals that were fasted but not refed, which is consistent with the fact that there is increased mitochondrial fatty acid oxidation during fasted conditions (Fig. 3D and Dataset S3) (24). Mitochondrial CoA levels were elevated in fasting conditions as well, potentially reflecting increased rates of ketogenesis within mitochondria. We also observed that mitochondrial methylcitrate was considerably elevated when mice were fasted but not refed, which may be secondary to increased amounts of mitochondrial propionyl CoA. In the fasted state, there were also considerably elevated levels of mitochondrial GMP, which may indicate altered energy charge dynamics within the matrix, and increased amounts of mitochondrial glutamine, which may reflect decreased matrix glutaminolysis. Taken together, our data demonstrate the utility of our in vivo workflow for interrogating changes in polar mitochondrial metabolites during different physiological states.
Fig. 3.
In vivo dynamics of hepatocyte mitochondrial metabolites during fasting and refeeding. (A) Schematic demonstrating general design of fasting and refeeding experiment. In brief, Control-MITO and HA-MITO mice were fasted overnight for 12 h and then either fasted or refed for another 4 h before liver tissue was harvested for mitochondrial or whole-tissue polar metabolomics. (B) Immunoblot analysis of liver tissue taken from Control-MITO and HA-MITO mice during fasted and refed conditions. Protein names appear to the left of the membrane. P-S6, phospho-S6 [Ser240/244]. GAPDH was used as a loading control. (C) Heat map of changes in metabolites as assessed at the mitochondrial and whole-tissue level during fasted and refed conditions. The data are presented as the log2-transformed mean fold difference (fast/refed). For a metabolite to be included in this heat map, it had to be considered mitochondrial in at least the fasted or refed state. See Materials and Methods for criteria for being considered mitochondrial. See also Dataset S3 for more detail. (D) Comparison of select metabolites during fasted and refed conditions. Metabolite peak area ratios, which are calculated by dividing the metabolite peak areas by internal standard peak areas, are used to generate these graphs. See also Dataset S3. Data are presented as means with SDs. *P < 0.05 and **P < 0.01 as determined by unpaired, parametric, two-tailed Student’s t test. For A–D, fasted Control-MITO mice (n = 4), fasted HA-MITO mice (n = 5), refed Control-MITO mice (n = 4), and refed HA-MITO mice (n = 4).
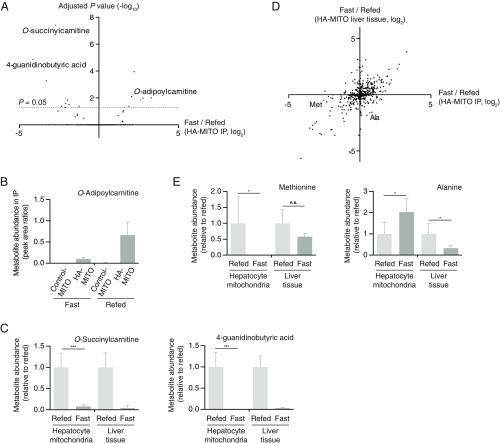
To complement our targeted studies on polar mitochondrial metabolites, we also incorporated untargeted metabolomics into our in vivo workflow. From this untargeted analysis, we found several hits that had indeterminate chemical identities and significantly different abundances within the HA-MITO IPs during fasted and refed conditions (Fig. 4A and Dataset S4). Using authentic chemical standards that were commercially available, we successfully identified several of these hits to be O-adipoylcarnitine, O-succinylcarnitine, and 4-guanidinobutyric acid. Although the differences in O-adipoylcarnitine levels within mitochondria during fasted and refed conditions did not reach statistical significance when peak areas were manually integrated and appropriately adjusted, the untargeted analysis was still fruitful as it allowed us to determine that, under both nutritional states, O-adipoylcarnitine was present in the HA-MITO IPs above background and at levels meeting our criteria for being considered mitochondrial (Fig. 4B and Dataset S4). We did find, however, that the differences in mitochondrial O-succinylcarnitine and 4-guanidinobutyric acid during fasted and refed conditions remained statistically significant after manual integration and appropriate adjustment of peak areas (Fig. 4C and Dataset S4), and it will be interesting to see future work decipher the mechanisms underlying these metabolic changes. Taken together, these data demonstrate how untargeted metabolomics can be used with our in vivo workflow to study mitochondrial metabolites without knowledge of their molecular identities beforehand.
Fig. 4.
Interrogation of mitochondrial metabolites during fasting and refeeding using untargeted metabolomics. (A) Scatter plot of compounds in the HA-MITO IPs (i.e., isolated mitochondria) as generated from the untargeted analysis. The data presented here are the log2-transformed fold differences (median fasted HA-MITO IP signal/median refed HA-MITO IP signal). Ad hoc adjusted P values (Benjamini–Hochberg method) are presented here, and the line indicates where the adjusted P value = 0.05. All points above the line are considered significant. Compounds that were subsequently validated using authentic chemical standards are highlighted in red and indicated by their metabolite name. Note that many compounds appear along the x axis as a result of their adjusted P values. See also Dataset S4 for more detail. (B) Metabolomic analysis of IPs for O-adipoylcarnitine. Shown are metabolite peak area ratios, which are calculated by dividing the metabolite peak areas by internal standard peak areas, for the Control-MITO and HA-MITO IPs during fasted and refed conditions. O-adipoylcarnitine meets the criteria of being mitochondrial in both fasted and refed conditions, as described in Materials and Methods. Data are shown as means with SDs. See also Dataset S4. (C) Metabolomic interrogation of O-succinylcarnitine and 4-guanidinobutyric acid during fasted and refed conditions. Metabolite peak area ratios, which are calculated by dividing the metabolite peak areas by internal standard peak areas, were used to generate these graphs. See also Dataset S4. Data are shown as means with SDs. ***P < 0.001 as determined by unpaired, parametric, two-tailed Student’s t test. (D) Scatter plot of compounds from untargeted analysis when assessed on the mitochondrial versus whole-tissue level. The data presented here are the log2-transformed fold differences for the HA-MITO IP samples (median fasted HA-MITO IP signal/median refed HA-MITO IP signal) and the HA-MITO whole-tissue samples (median fasted HA-MITO whole-tissue signal/median refed HA-MITO whole-tissue signal). Methionine and alanine are highlighted in red as the behavior of these two metabolites was confirmed in our targeted analysis. See Datasets S3 and S4 for more detail. (E) Metabolomic interrogation of methionine and alanine during fasted and refed conditions. Metabolite peak area ratios, which are calculated by dividing the metabolite peak areas by internal standard peak areas, are used to generate these graphs. See also Dataset S3. Data are shown as means with SDs. *P < 0.05 as determined by unpaired, parametric, two-tailed Student’s t test. n.s., not statistically significant. For A–E, fasted Control-MITO mice (n = 4), fasted HA-MITO mice (n = 5), refed Control-MITO mice (n = 4), and refed HA-MITO mice (n = 4).
Perhaps the most interesting metabolites identified by direct profiling of organelles are those that have significant discordance in their behaviors when assessed using organellar versus whole-tissue metabolomics. To that end, we also examined which compounds from our untargeted analysis behaved differently in the HA-MITO IPs versus in liver tissue during fasted and refed conditions. Our untargeted analysis identified methionine and alanine as two metabolites that exhibited the desired behavior (Fig. 4D and Dataset S4). This was reassuring as the changes seen for these two metabolites were validated during our targeted analysis (Fig. 4E and Dataset S3). Interestingly, there were considerably reduced levels of methionine within mitochondria when mice were fasted but not refed, but methionine levels were not significantly different at the whole-tissue level. This may reflect that mitochondrial pools of methionine are less well-buffered during fasting than those in other compartments that contribute to the whole-tissue signal, such as the cytosol. With regards to alanine, we observed that fasted mice had elevated alanine within mitochondria but decreased levels of the amino acid at the whole-tissue level. During fasting, the increase in mitochondrial alanine could be a result of decreased pyruvate dehydrogenase activity, increased mitochondrial pyruvate, and subsequent equilibration between pyruvate and alanine pools. The decreased amounts of whole-tissue alanine could be a reflection of an increased gluconeogenic drive to convert alanine to pyruvate within the cytosol. Collectively, these data demonstrate the value of our in vivo workflow, when used in conjunction with untargeted metabolomics, for taking a broad approach to studying mitochondrial metabolites during different physiological states.
Discussion
Methods for rapidly isolating mitochondria and studying mitochondrial metabolites with cell-type specificity in vivo have thus far been limited in mammalian systems (9–14). During the preparation of this manuscript, a similar strategy using a different immunocapture handle and different strain of knock-in mice was reported, although in that study the workflow was slower (e.g., 60–90 min of incubating mitochondria-containing suspensions with antibody-coated magnetic beads versus 3.5 min in our workflow) and no lipidomic or polar metabolomic analyses were performed (25). In our study, building on our prior methodology using epitope tags for fast isolations of mitochondria from cultured cells (2, 4), we have generated MITO-Tag Mice that allow for the rapid, cell-type-specific immunopurification and proteomic, lipidomic, and polar metabolomic analysis of mammalian mitochondria in vivo. By combining our in vivo workflow for rapid isolation of mitochondria with metabolomics, we uncovered changes in a diverse array of hepatocyte mitochondrial metabolites during fasted and refed conditions via targeted and untargeted approaches. Interestingly, many metabolites behaved similarly when assessed at the mitochondrial and whole-tissue level, which may be a reflection of multiple factors, such as the nature of the fasting and refeeding perturbations and the time points at which liver tissue were taken. Of note though, we were still able to identify metabolites, such as methionine and alanine, that behaved differently during fasted and refed conditions when assessed using mitochondrial versus whole-tissue metabolomics, thus underscoring the importance of examining metabolites at the organellar level.
We believe that, in addition to their utility for assessing mitochondrial metabolism in vivo, these MITO-Tag Mice will be a widely useful tool for studying mitochondrial physiology in mammalian systems. Multimodal analysis of mitochondria from specific cell types will be important for trying to dissect mitochondrial heterogeneity in complex tissues, such as the brain and kidney. Furthermore, the ability to rapidly isolate mitochondria from specific cell types without the need for cell sorting and long centrifugation steps can reduce the magnitude of distortion of the original mitochondrial state, as well as significantly decrease the time and effort needed to acquire the desired mitochondrial material, which can be useful for not just metabolomic but also proteomic work by better preserving the native proteomic mitochondrial profile and the interactions of proteins that peripherally associate with mitochondrial membranes (26). Of note, the design of our in vivo workflow is such that it can also be combined with other downstream studies aside from those utilized here, such as genomic and transcriptomic analyses. We thus believe that these MITO-Tag Mice will have utility in the study of mitochondria and feel that this work can serve as a general strategy for the cell-type-specific interrogation of other mammalian organelles in vivo.
Materials and Methods
Reagents and Equipment.
The following items were used: Pierce Anti-HA magnetic beads (88837); Lucigen QuickExtract DNA Extraction Solution (QE09050); Miltex Biopsy Punch with Plunger, inner diameter 4.0 mm, outer diameter 4.36 mm (15110–40; Ted Pella); Self Healing Cutting Mat, 3.5″ × 5.5″ (15085; Ted Pella); VWR PowerMax Dual Shaft Laboratory Mixer (14215-266); Pierce BCA Protein Assay (23224); Roche cOmplete Mini EDTA-free protease inhibitor (11836170001); Roche PhosSTOP phosphatase inhibitor (4906845001); Omni International Bead Ruptor 24 Bead Mill Homogenizer (10032-376); Omni BR-Cryo Cooling Unit (19-8005); Cambridge Isotope Laboratories Metabolomics Amino Acid Mix Standard (MSK-A2-1.2); Thermo Fisher Scientific Hoescht 33342 (H3570); Triton lysis buffer (50 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and cOmplete EDTA-free protease inhibitor); Nonidet P-40 lysis buffer (50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40 and cOmplete EDTA-free protease inhibitor), used for proteomics due to incompatibility of Triton detergent with downstream proteomics; and Odyssey Nitrocellulose Membranes (926-31092).
Antibodies.
For immunoblotting, the following antibodies were used: antibodies against HA (2367), CS (14309), VDAC (4661), HIST3H3 (4499), CALR (12238), S6 (2217), and P-S6 [Ser240/244] (2215) (Cell Signaling Technology); the antibody against CYCS (MA511674) (Thermo Fisher Scientific); the antibodies against GAPDH (GTX627408) and ACTB (GTX109639) (GeneTex); the antibody against CTSC (sc-74590) (Santa Cruz Biotechnology); the antibody against USO1 (612260) (BD Biosciences); the antibody against PEX13 (ABC 143) (MilliporeSigma). The IRDye 800CW donkey anti-rabbit IgG (925-32213) and IRDye 680LT donkey anti-mouse IgG (925-68022) were purchased from LI-COR. For immunofluorescence, the following antibodies were used: antibody against the HA epitope (3724) (Cell Signaling Technology); the antibody against COX4I1 (ab14744) (Abcam); and Alexa Fluor 488-conjugated donkey anti-rabbit IgG (A-21206) and the Alexa Fluor 568-conjugated donkey anti-mouse IgG (A10037) (Thermo Fisher Scientific).
MITO-Tag Mice.
MITO-Tag Mice (Rosa26LSL-3XHA-EGFP-OMP25 mice) were generated by first constructing a gene targeting plasmid using a CAG-driven R26TV LSL backbone that was a gift from Thales Papagiannakopoulos, New York University, New York (64812; Addgene). Gene blocks with the HA-MITO gene and homologous arms of 1 kb to the target site in the Rosa26 locus were cloned into the vector with Gibson assembly. The HA-MITO gene was placed downstream of the LSL cassette. The construct was electroporated into Cy2.4 ES cells [B6(Cg)-Tyr <c2J> genetic background]. Positive clones were identified by Southern blot analysis of the neomycin cassette. ES cells were injected into blastocysts, chimeras were screened based on white/black coat-color selection, and the strain was maintained as heterozygous. MITO-Tag Mice were generated in the K.B. laboratory with the assistance of the Transgenic Services Laboratory at The Rockefeller University.
For the purposes of this study, we designated Rosa26LSL-3XHA-EGFP-OMP25/+ mice as Control-MITO mice and Alb-Cre+/−, Rosa26LSL-3XHA-EGFP-OMP25/+ mice as HA-MITO mice. We generated HA-MITO mice by mating MITO-Tag Mice with mice expressing Cre recombinase under the control of the Albumin promoter [B6.Cg-Speer6-ps1Tg(Alb-Cre)21Mgn/J mice, stock no: 003574, The Jackson Laboratory] (27). PCR genotyping for alleles containing the HA-MITO and Alb-Cre genes was performed on genomic DNA isolated from mouse tails with the following primers: MITO-Tag_F—GGTCACCACACTCACCTATGGCGTACAGT; MITO-Tag_R—CCTTGCTCAGTGCAGATTGCGTGCTCAG; Alb-Cre_F—CTGCCACGACCAAGTGACAGCAATGC; and Alb-Cre_R—GACGGAAATCCATCGCTCGACCAGTTTAGT. Excision of the floxed site was analyzed using the following primers: Excision_F—TCTGCTAACCATGTTCATGCCTTC; and Excision_R—ATCGCACAGCTAGCGTTCGAG.
Genomic PCR analysis, such as in Fig. 1B, was done using ∼25 mg of liver tissue.
MITO-Tag Mice Accessibility.
MITO-Tag Mice [B6.Cg-Gt(ROSA)26Sortm1(CAG-EGFP)Brsy/J, stock no. 032290, The Jackson Laboratory] have been deposited in The Jackson Laboratory for use by the research community.
Mouse Studies.
C57BL/6J mice were obtained from The Jackson Laboratory (stock no. 000664). Animal studies and procedures were conducted according to a protocol approved by the Institutional Animal Care and Use Committee at The Rockefeller University. All mice were maintained on a standard light–dark cycle with food and water ad libitum. For all experiments used in this study, mice were 8–12 wk old and male. Control-MITO mice were Rosa26LSL-3XHA-EGFP-OMP25/+ mice and HA-MITO mice were Alb-Cre+/−, Rosa26LSL-3XHA-EGFP-OMP25/+ mice. For the fasting and refeeding experiment, 18 mice (10 HA-MITO mice, 8 Control-MITO mice) were randomly allocated into either the fasted or refed groups, with five HA-MITO mice and four Control-MITO mice per group. The fasting and refeeding experiments described in this study were performed as follows: mice were fasted overnight for 12 h and then either refed or fasted for the next 4 h. Following cervical dislocation, liver tissue was immediately harvested for further processing. Note that one of the HA-MITO mice from the refed group was excluded from all analyses as this mouse did not exhibit the expected elevation in liver P-S6 signal upon refeeding, perhaps due to low food intake during the refeeding period (see Fig. 3B for expected P-S6 dynamics during fasted and refed conditions). The same mice were used for the S6 signaling immunoblot in Fig. 3B, the organellar marker immunoblot in SI Appendix, Fig. S1, the lipidomic analysis, and the polar targeted and untargeted analyses. To avoid decreasing the speed of the workflow, we conducted immunopurifications intended for different downstream analyses in parallel for a given animal. Aside from the aforementioned analyses, all other experiments were performed on mice that were fed ad libitum.
Protein Extraction from Liver Tissue.
Approximately 25 mg of liver tissue was excised using a punch biopsy tool and was washed three times in ice-cold PBS before treatment with Triton lysis buffer supplemented with phosphatase inhibitors. Homogenization was performed with a Bead Ruptor, and cooling was achieved via liquid nitrogen. The protein concentration was determined by using the Pierce BCA Protein Assay kit with BSA as a protein standard. Samples were subsequently used for immunoblotting.
Immunofluorescence.
Following cervical dislocation and dissection, 4- × 4-mm pieces of liver were cut and washed in ice-cold PBS. Next, the pieces were fixed in 10% neutral buffered formalin for 8 h, followed by washing three times in 70% ethanol. The histological preparation of the specimens in paraffin blocks was done at the Center of Comparative Medicine and Pathology, Memorial Sloan Kettering Cancer Center, New York. Liver sections were deparaffinized, rehydrated, and blocked (5% normal donkey serum, 0.3% Triton X-100 in PBS) for 2 h at room temperature. The sections were then incubated with primary antibodies [anti-HA, 3724 (1:100); Cell Signaling Technology; anti-COX4l1, ab14744 (1:100); Abcam] for 48 h at 4 °C. After washing, the sections were incubated with Alexa Fluor-conjugated secondary antibodies [Alexa Fluor 488-conjugated donkey anti-rabbit IgG (1:500); Alexa Fluor 568-conjugated donkey anti-mouse IgG (1:500)] for 2 h at room temperature, washed, and counterstained with Hoechst 33342 to label nuclei. Images were acquired on an RPI spinning disk confocal microscope using MetaMorph acquisition software.
Rapid Isolation of Hepatocyte Mitochondria from Liver Tissue.
Isolation of mitochondria was performed as previously described with modifications (2, 4). All procedures were performed on ice. The liver tissue was excised with the punch biopsy tool on the cutting mat and was immediately washed three times in ice-cold PBS before extraction. Two pieces of liver tissue (∼50 mg total) were homogenized in 5 mL of KPBS (136 mM KCl, 10 mM KH2PO4, pH 7.25, in Optima LC/MS water) with a pestle attached to the mixer to rotate at 220 rpm in a 4 °C cold room. Five microliters of this homogenate was taken as a sample of liver tissue and was extracted in the appropriate reagent depending on the downstream analysis. The remaining homogenate was spun down at 1,000 × g for 2 min at 4 °C. The supernatant was subjected to immunoprecipitation with prewashed anti-HA beads for 3.5 min, followed by three rounds of washing in KPBS. In the final wash, if desired, 20% of the suspension of beads was set aside and used for protein extraction and immunoblotting by aspirating the KPBS and then mixing the beads with 50 μL of Triton lysis buffer. The remainder of the beads were then extracted using a reagent appropriate for the desired downstream analysis.
Immunoblotting.
For immunoblot analyses of material obtained from the mitochondrial isolation workflow, the IP material or 5 μL of homogenized liver tissue was mixed with 50 μL of Triton lysis buffer. Afterward, the samples containing homogenized liver tissue or IP material were placed on an end-over-end rotator for 10 min in a 4 °C cold room. For the IP samples, beads were magnetized, and the extract was transferred to a clean tube. Magnetization and transfer to a clean tube was repeated once more to make sure that no magnetic beads were recovered in the final extract.
In general, all Triton lysates were mixed with 5× Laemmli buffer after protein quantification with BCA and then heated at 95 °C for 5 min before loading onto gels. All samples were resolved on 8 or 12% SDS/PAGE gels and analyzed by immunoblotting as described previously (28). In brief, gels were run at 125 V for 1.5 h using Tris–glycine SDS running buffer and transferred onto Odyssey Nitrocellulose Membranes (pore size 0.22 μm) at 150 mA for 2 h in Tris–glycine transfer buffer. Membranes were blocked for 1 h using 5% BSA in TBST (Tris-buffered saline with Tween 20) and then incubated with primary antibodies diluted in 5% BSA in TBST overnight at 4 °C. All primary antibodies were used at a final dilution of 1:1,000, except for the HA antibody (2367; Cell Signaling Technology), which was used at 1:2,000. Membranes were then washed four times in TBST and incubated with IRDye secondary antibodies diluted 1:10,000 in 5% BSA in TBST for 1 h at room temperature. The membranes were washed four times in TBST and then imaged using Odyssey CLx. All band quantifications were performed on Image Studio Lite.
Proteomics.
For proteomic analyses, mitochondria were isolated as described above. In the final wash, 20% of the bead suspension was set aside and extracted in 50 μL Triton lysis buffer and processed for immunoblotting to examine the purity of the IPs. The remainder of the beads were then extracted using 50 μL of Nonidet P-40 lysis buffer, and samples were placed on an end-over-end rotator for 10 min in a 4 °C cold room. Beads were magnetized, and the extract was transferred to a clean tube. Magnetization and transfer to a clean tube was repeated once more to make sure that no magnetic beads were recovered in the final extract. Proteins from the resulting Nonidet P-40 lysates were purified by trichloroacetic acid precipitation, and the resulting pellet was resuspended in a 100-μL buffer of 6 M urea and 100 mM Tris at pH 7.8. Reduction and alkylation of disulfide bonds were then carried out by adding 5 μL of 200 mM DTT and incubating for 60 min to reduce disulfide bonds. The resulting free cysteine residues were subjected to an alkylation reaction by adding 20 μL of 200 mM iodoacetamide and incubating for 60 min to form carbamidomethyl cysteine. This solution was brought to a volume of 900 μL to reduce the urea concentration. One hundred microliters of 20 ng/μL of trypsin was added for a final volume of 1,000 μL and allowed to digest overnight at 37 °C with gentle shaking. The resulting peptides were washed, extracted, and concentrated by solid phase extraction using Waters Sep-Pak Plus C18 cartridges. For subsequent analyses, organic solvent was removed, and the volumes were reduced to 20 μL using a speed vac.
For chromatographic separation, the digestion extracts were analyzed by reversed phase high-performance liquid chromatography (HPLC) using Thermo EASY-nLC 1200 pumps and an autosampler and a Thermo Q Exactive HF-X Hybrid Quadrupole-Orbitrap mass spectrometer using a nano-flow configuration. Samples were loaded on a 6-cm × 100-μm column packed with 10 μm ODS-A C18 material (YMC) and washed with 4 μL of total volume to trap and wash peptides. These were then eluted onto the analytical column, which was self-packed with 1.7 μm of Aeris C18 material (Phenomenex) in a fritted 14-cm × 75-μm fused silica tubing pulled to a 5-μm tip. The gradient was isocratic 1% Buffer A for 1 min at a rate of 250 nL/min with increasing buffer B concentrations to 19% buffer B at 52 min, 34% buffer B at 77.5 min, and 50% buffer B at 103 min. The column was washed with high-percentage buffer B and re-equilibrated between analytical runs for a total cycle time of ∼127 min. Buffer A consisted of 1% formic acid in water and buffer B consisted of 1% formic acid in acetonitrile. The mass spectrometer was operated in a data-dependent acquisition mode where the 20 most abundant peptides detected in the Orbitrap using full scan mode with a resolution of 60,000 were subjected to daughter ion fragmentation in the linear ion trap. A running list of parent ions was tabulated to an exclusion list to increase the number of peptides analyzed throughout the chromatographic run.
Peptides were identified from the mass spectrometry (MS) data using PEAKS Studio 8.5. The Mus musculus Refseq protein FASTA entries were downloaded from NIH/National Center for Biotechnology Information (NCBI) and concatenated to a database of common contaminants (keratin, trypsin, etc). A false discovery rate threshold of 1% for identification of peptides was used. For each protein, areas were determined for each IP replicate, mean areas were calculated for the Control-MITO IPs and HA-MITO IPs, and mean areas were then pseudonumbered by replacing values of 0 with 0.01 to facilitate calculating the degree of protein enrichment in the HA-MITO IPs relative to the Control-MITO IPs. Proteins were then filtered based on a stringent enrichment criterion of having a pseudonumbered mean protein signal in the HA-MITO IPs that was at least five times greater than that in the Control-MITO IPs (i.e., the background). For the proteins that satisfied this criterion, we then determined whether they were present in MitoCarta2.0 using the corresponding NCBI gene IDs (20).
Lipidomics.
To extract lipids, 333 μL of 100% methanol was added to the IP material or 5 μL of homogenized liver tissue, followed by 667 μL of chloroform. Five microliters of KPBS was also processed as a quality control measure. Samples were vortexed at maximum speed in a 4 °C cold room for 15 min, followed by the addition of 200 μL of 0.9% (wt/vol) NaCl. Samples were vortexed at maximum speed at 4 °C for 15 min and were spun down at 3,000 × g for 15 min at 4 °C. The bottom layer containing the lipids was taken and briefly dried under a cold stream of nitrogen gas.
Lipids were separated on an Ascentis Express C18 2.1- × 150-mm, 2.7-μm column (MilliporeSigma) connected to a Dionex UltiMate 3000 UPLC system and a QExactive benchtop orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with a heated electrospray ionization (HESI) probe. Dried lipid extracts were reconstituted in 50 μL 65:30:5 acetonitrile: isopropanol:water (vol/vol/vol). Five microliters of sample were injected onto the column, with separate injections for positive and negative ionization modes. Mobile phase A in the chromatographic method consisted of 60:40 water:acetonitrile with 10 mM ammonium formate and 0.1% formic acid, and mobile phase B consisted of 90:10 isopropanol:acetonitrile, with 10 mM ammonium formate and 0.1% formic acid. The chromatographic gradient was described previously (29). The column oven and autosampler were held at 55 °C and 4 °C, respectively. The mass spectrometer parameters were described previously (30) and were modified as described in ref. 31. The spray voltage was set to 4.2 kV, and the heated capillary and the HESI were held at 320 °C and 300 °C, respectively. The S-lens RF level was set to 50, and the sheath and auxillary gas were set to 35 and 3 units, respectively. These conditions were held constant for both positive and negative ionization mode acquisitions. External mass calibration was performed every 7 d using the standard calibration mixture.
Mass spectra were acquired in both full-scan and data-dependent MS/MS mode. For the full-scan acquisition, the resolution was set to 70,000, the automatic gain control (AGC) target was 1 × 106, the maximum injection time was 50 ms, and the scan range was m/z = 133.4–2,000. For data-dependent MS/MS, the top 10 ions in each full scan were isolated with a 1.0-Da window, fragmented with a step-wise collision energy of 15, 25, and 35 units and analyzed at a resolution of 17,500 with an AGC target of 2 × 105 and a maximum injection time of 100 ms. The underfill ratio was set to 0. The selection of the top 10 ions was set to isotopic exclusion, a dynamic exclusion window of 5.0 s, and an exclusion list of background ions based on a solvent blank.
High-throughput identification and relative quantification of lipids was performed separately for positive and negative ionization mode data, using LipidSearch software (Thermo Fisher Scientific/Mitsui Knowledge Industries) (32, 33) with the default parameters for QExactive product search and alignment. After alignment, raw peak areas for all identified lipids were exported to Microsoft Excel and filtered according to the following predetermined quality control criteria: Rej (“Reject” parameter calculated by LipidSearch) equal to 0; PQ (“Peak Quality” parameter calculated by LipidSearch software) greater than 0.85; CV [SD/mean peak area across triplicate injections of a representative (pooled) biological sample] below 0.4; R [linear correlation across a three-point dilution series of the representative (pooled) biological sample] greater than 0.9. Typically, ∼70% of identified lipids passed all four quality control criteria. Cardiolipins were validated by manually checking the peak alignment and matching the MS/MS spectra to the characteristic fragmentation patterns found in the LIPIDMAPS database (http:www.lipidmaps.org), as well as those described previously (34). Validated cardiolipin species were reanalyzed using Xcalibur Quanbrowser 4.1 (Thermo Fisher Scientific) and graded based on the quality of MS/MS spectra collected: Grade A—CL annotation is correct, and all four acyl chains are correctly annotated; Grade B—3/4 acyl chains are correctly annotated; Grade C—2/4 acyl chains are correctly annotated; and Grade D—1/4 acyl chains are correctly annotated or CL species were confirmed through the presence of head group, but acyl chain identification is ambiguous.
For a lipid species to be examined for mitochondrial abundance, it had to pass the aforementioned four quality control criteria. For cardiolipin analysis, only grade A cardiolipins were used. A lipid species was considered mitochondrial within a given dietary state only if (i) the mean Control-MITO IP peak area was 0 and the peak areas of each HA-MITO IP replicate were greater than 0 or (ii) the mean Control-MITO IP peak area was greater than 0 but the peak areas of each HA-MITO IP replicate were at least 1.5-fold or greater than the mean Control-MITO IP peak area.
Polar Metabolomics.
For polar metabolites, the extraction of the IP material or the 5 μL of homogenized liver tissue was performed using 50 μL of ice-cold 80% methanol in water with internal standards. Five microliters of KPBS was also processed as a quality control measure. Afterward, the samples were placed on an end-over-end rotator for 10 min in a 4 °C cold room. For the IP samples, beads were magnetized and the extract was transferred to a clean tube. Magnetization and transfer to a clean tube was repeated once more to make sure that no magnetic beads were recovered in the final extract. Afterward, all samples were spun down at 21,000 × g for 10 min at 4 °C, and the supernatant was transferred into a clean tube.
Metabolite profiling was conducted on a QExactive bench top orbitrap mass spectrometer equipped with an Ion Max source and a HESI II probe, which was coupled to a Dionex UltiMate 3000 HPLC system (Thermo Fisher Scientific). External mass calibration was performed using the standard calibration mixture every 7 d. Typically, 5 μL of each sample was injected onto a SeQuant ZIC-pHILIC 150- × 2.1-mm analytical column equipped with a 2.1- × 20-mm guard column (both 5-mm particle size; EMD Millipore). Buffer A was 20 mM ammonium carbonate, 0.1% ammonium hydroxide; Buffer B was acetonitrile. The column oven and autosampler tray were held at 25 °C and 4 °C, respectively. The chromatographic gradient was run at a flow rate of 0.150 mL/min as follows: 0–20 min—linear gradient from 80 to 20% Buffer B; 20–20.5 min—linear gradient from 20 to 80% Buffer B; and 20.5–28 min—hold at 80% Buffer B. The mass spectrometer was operated in full-scan, polarity-switching mode, with the spray voltage set to 3.0 kV, the heated capillary held at 275 °C, and the HESI probe held at 350 °C. The sheath gas flow was set to 40 units, the auxiliary gas flow was set to 15 units, and the sweep gas flow was set to 1 unit. MS data acquisition was performed in a range of m/z = 70–1,000, with the resolution set at 70,000, the AGC target at 1 × 106, and the maximum injection time at 20 ms. A sample of KPBS in 80% methanol with internal standards was included in each polar metabolomics run to monitor for signs of buffer contamination. Relative quantitation of polar metabolites was performed with XCalibur QuanBrowser 4.1 (Thermo Fisher Scientific) using a 5-ppm mass tolerance and referencing an in-house library of chemical standards. Peak area ratios were calculated using the peak area of each metabolite and the peak area of an internal standard. No adjustment for KPBS matrix effects was done for polar metabolomics as we performed only relative quantification, and all biological samples had residual KPBS in them.
Determination of mitochondrial abundances of polar metabolites within hepatocytes during fasted and refed conditions was done as follows. Within a given dietary state, metabolites were classified as mitochondrial if (i) the mean Control-MITO IP peak area ratio was 0 and the peak area ratio of each HA-MITO IP replicate was greater than 0 or (ii) the mean Control-MITO IP peak area ratio was greater than 0, but the peak area ratio of each HA-MITO IP replicate was at least 1.5-fold or greater than the mean Control-MITO IP peak area ratio. Only metabolites that were deemed mitochondrial were then used for further calculations. Within a given dietary state, background correction was done for each mitochondrial metabolite by subtracting the mean Control-MITO IP peak area ratio from the peak area ratio of each HA-MITO IP replicate. Background-corrected HA-MITO IP metabolite values were then normalized based on the amount of isolated mitochondria in each HA-MITO IP, which can be assessed via immunoblotting for citrate synthase and quantitation of band intensities. (Note that, in this work, we found that the background-corrected HA-MITO IP peak area ratio of the mitochondrial metabolite FAD actually performed better than citrate synthase for estimating the amount of isolated mitochondria in each HA-MITO IP and was thus used for normalizing samples.) Because of the reproducibility of the punch biopsy tool, equal amounts of liver tissue were homogenized and then taken for metabolite extraction, so no normalization was done for the liver tissue samples.
Untargeted Polar Metabolomics.
Data were acquired as described above, with additional data-dependent MS/MS collected on pooled samples to aid with unknown metabolite identification. For data-dependent MS/MS, the top 10 ions in each full scan were isolated with a 1.0-Da window, fragmented with a step-wise collision energy of 15, 30, and 45 units, and analyzed at a resolution of 17,500 with an AGC target of 2 × 105 and a maximum injection time of 100 ms. The underfill ratio was set to 0. The selection of the top 10 ions was set to isotopic exclusion, a dynamic exclusion window of 5.0 s, and an exclusion list of background ions based on a solvent bank. Data were analyzed using Compound Discoverer 2.1 (Thermo Fisher Scientific) and by including an in-house mass-list. Compounds were excluded if they were present in samples containing only KPBS. P values were adjusted according to the Benjamini–Hochberg method. O-adipoylcarnitine, O-succinylcarnitine, 4-guanidinobutyric acid, methionine, and alanine were validated using chemical standards. These metabolites were also analyzed in a targeted fashion. All analyses and calculations for these metabolites were done as described previously for polar mitochondrial metabolites within hepatocytes (Polar Metabolomics).
Statistics and Analysis.
All P values were calculated using an unpaired, two-tailed, parametric Student’s t test using Graph Pad Prism 7, except for Fig. 4A and Dataset S4, P values were generated using Compound Discoverer 2.1, and then ad hoc adjusted P values were calculated using the Benjamini–Hochberg method. Heatmaps were generated using Graph Pad Prism 7.
Supplementary Material
Acknowledgments
We thank all members of the K.B. and D.M.S. laboratories and Elizaveta Freinkman and Hoi See Tsao for helpful suggestions; and Eli Freiman, Shyam Deshpande, Amanda Gomez, Laura Chiel, Katherine Nash, and all other members of the Boston Combined Residency Program for their assistance. C4- and C5-carnitines were synthesized by Rajan Pragani and provided by Jared Mayers. E.C.B. is supported by Robertson Therapeutic Funds of The Rockefeller University. A.L.C. is supported by NIH F31 National Research Service Award Fellowship F31 DK113665. K.B. is supported by NIH Director’s New Innovator Award DP2 OD024174-01, the Irma-Hirschl Trust, and an American Association for Cancer Research NextGen grant; K.B. is a Searle Scholar, Sidney Kimmel Scholar, Pew-Stewart Scholar, and Basil O’Connor Scholar of the March of Dimes. This work was supported by NIH Grants R01CA103866, R01CA129105, and R37AI047389, and by Department of Defense Grant W81XWH-15-1-0230 (to D.M.S.). D.M.S. is an investigator of the Howard Hughes Medical Institute and an American Cancer Society Research Professor. W.W.C. is supported by the Boston Combined Residency Program and Boston Children’s Hospital.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 20.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816656115/-/DCSupplemental.
References
- 1.Wallace DC. A mitochondrial bioenergetic etiology of disease. J Clin Invest. 2013;123:1405–1412. doi: 10.1172/JCI61398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen WW, Freinkman E, Wang T, Birsoy K, Sabatini DM. Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell. 2016;166:1324–1337.e11. doi: 10.1016/j.cell.2016.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chouchani ET, et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen WW, Freinkman E, Sabatini DM. Rapid immunopurification of mitochondria for metabolite profiling and absolute quantification of matrix metabolites. Nat Protoc. 2017;12:2215–2231. doi: 10.1038/nprot.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abu-Remaileh M, et al. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science. 2017;358:807–813. doi: 10.1126/science.aan6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyant GA, et al. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell. 2017;171:642–654.e12. doi: 10.1016/j.cell.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Llufrio EM, Wang L, Naser FJ, Patti GJ. Sorting cells alters their redox state and cellular metabolome. Redox Biol. 2018;16:381–387. doi: 10.1016/j.redox.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahier A, et al. Affinity purification of cell-specific mitochondria from whole animals resolves patterns of genetic mosaicism. Nat Cell Biol. 2018;20:352–360. doi: 10.1038/s41556-017-0023-x. [DOI] [PubMed] [Google Scholar]
- 10.Roede JR, Park Y, Li S, Strobel FH, Jones DP. Detailed mitochondrial phenotyping by high resolution metabolomics. PLoS One. 2012;7:e33020. doi: 10.1371/journal.pone.0033020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross-Inta C, Tsai C-Y, Giulivi C. The mitochondrial pool of free amino acids reflects the composition of mitochondrial DNA-encoded proteins: Indication of a post-translational quality control for protein synthesis. Biosci Rep. 2008;28:239–249. doi: 10.1042/BSR20080090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tischler ME, Hecht P, Williamson JR. Determination of mitochondrial/cytosolic metabolite gradients in isolated rat liver cells by cell disruption. Arch Biochem Biophys. 1977;181:278–293. doi: 10.1016/0003-9861(77)90506-9. [DOI] [PubMed] [Google Scholar]
- 13.Zuurendonk PF, Tager JM. Rapid separation of particulate components and soluble cytoplasm of isolated rat-liver cells. Biochim Biophys Acta. 1974;333:393–399. doi: 10.1016/0005-2728(74)90022-x. [DOI] [PubMed] [Google Scholar]
- 14.Franko A, et al. Efficient isolation of pure and functional mitochondria from mouse tissues using automated tissue disruption and enrichment with anti-TOM22 magnetic beads. PLoS One. 2013;8:e82392. doi: 10.1371/journal.pone.0082392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gierut JJ, Jacks TE, Haigis KM. Strategies to achieve conditional gene mutation in mice. Cold Spring Harb Protoc. 2014;2014:339–349. doi: 10.1101/pdb.top069807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung V, et al. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. eLife. 2017;6:e24463. doi: 10.7554/eLife.24463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nemoto Y, De Camilli P. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 1999;18:2991–3006. doi: 10.1093/emboj/18.11.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis SC, Uchiyama LF, Nunnari J. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science. 2016;353:aaf5549. doi: 10.1126/science.aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schrader M, Grille S, Fahimi HD, Islinger M. Peroxisome interactions and cross-talk with other subcellular compartments in animal cells. Subcell Biochem. 2013;69:1–22. doi: 10.1007/978-94-007-6889-5_1. [DOI] [PubMed] [Google Scholar]
- 20.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grabacka M, Pierzchalska M, Dean M, Reiss K. Regulation of ketone body metabolism and the role of PPARα. Int J Mol Sci. 2016;17:E2093. doi: 10.3390/ijms17122093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt T, et al. ProteomicsDB. Nucleic Acids Res. 2018;46:D1271–D1281. doi: 10.1093/nar/gkx1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilhelm M, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 24.Rui L. Energy metabolism in the liver. Compr Physiol. 2014;4:177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fecher C, et al. 2018. Profound functional and molecular diversity of mitochondria revealed by cell type-specific profiling in vivo. bioRxiv:403774. Preprint, posted August 29, 2018.
- 26.Wyant GA, et al. NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science. 2018;360:751–758. doi: 10.1126/science.aar2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 28.Birsoy K, et al. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature. 2014;508:108–112. doi: 10.1038/nature13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu C, et al. RPLC-ion-trap-FTMS method for lipid profiling of plasma: Method validation and application to p53 mutant mouse model. J Proteome Res. 2008;7:4982–4991. doi: 10.1021/pr800373m. [DOI] [PubMed] [Google Scholar]
- 30.Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Serum lipidomics profiling using LC-MS and high-energy collisional dissociation fragmentation: Focus on triglyceride detection and characterization. Anal Chem. 2011;83:6648–6657. doi: 10.1021/ac201195d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruzicka J, McHale KJ, Peake DA. 2014 Data acquisition parameters optimization of quadrupole orbitrap for global lipidomics on LC-MS/MS time frame (Thermo Scientific, Waltham, MA). Available at https://pdfs.semanticscholar.org/1c5e/d1ce779ebeef7012c4be850d868c12506fb4.pdf. Accessed June 12, 2018.
- 32.Taguchi R, Ishikawa M. Precise and global identification of phospholipid molecular species by an orbitrap mass spectrometer and automated search engine lipid search. J Chromatogr A. 2010;1217:4229–4239. doi: 10.1016/j.chroma.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 33.Yamada T, et al. Development of a lipid profiling system using reverse-phase liquid chromatography coupled to high-resolution mass spectrometry with rapid polarity switching and an automated lipid identification software. J Chromatogr A. 2013;1292:211–218. doi: 10.1016/j.chroma.2013.01.078. [DOI] [PubMed] [Google Scholar]
- 34.Minkler PE, Hoppel CL. Separation and characterization of cardiolipin molecular species by reverse-phase ion pair high-performance liquid chromatography-mass spectrometry. J Lipid Res. 2010;51:856–865. doi: 10.1194/jlr.D002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansel MC, et al. The history and use of human hepatocytes for the treatment of liver diseases: The first 100 patients. Curr Protoc Toxicol. 2014;62:14.12.1–14.12.23. doi: 10.1002/0471140856.tx1412s62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.