Significance
Tumor cells reprogram their metabolism to support cell growth, proliferation, and differentiation, thus driving cancer progression. Profiling of the metabolic signatures in heterogeneous tumors facilitates the understanding of tumor metabolism and introduces potential metabolic vulnerabilities that might be targeted therapeutically. We proposed a spatially resolved metabolomics method for high-throughput discovery of tumor-associated metabolite and enzyme alterations using ambient mass spectrometry imaging. Metabolic pathway-related metabolites and metabolic enzymes that are associated with tumor metabolism were efficiently discovered and visualized in heterogeneous esophageal cancer tissues. Spatially resolved metabolic alterations hold the key to defining the dependencies of metabolism that are most limiting for cancer growth and exploring metabolic targeted strategies for better cancer treatment.
Keywords: mass spectrometry imaging, metabolomics, esophageal cancer, metabolic alterations, airflow-assisted ionization
Abstract
Characterization of tumor metabolism with spatial information contributes to our understanding of complex cancer metabolic reprogramming, facilitating the discovery of potential metabolic vulnerabilities that might be targeted for tumor therapy. However, given the metabolic variability and flexibility of tumors, it is still challenging to characterize global metabolic alterations in heterogeneous cancer. Here, we propose a spatially resolved metabolomics approach to discover tumor-associated metabolites and metabolic enzymes directly in their native state. A variety of metabolites localized in different metabolic pathways were mapped by airflow-assisted desorption electrospray ionization mass spectrometry imaging (AFADESI-MSI) in tissues from 256 esophageal cancer patients. In combination with in situ metabolomics analysis, this method provided clues into tumor-associated metabolic pathways, including proline biosynthesis, glutamine metabolism, uridine metabolism, histidine metabolism, fatty acid biosynthesis, and polyamine biosynthesis. Six abnormally expressed metabolic enzymes that are closely associated with the altered metabolic pathways were further discovered in esophageal squamous cell carcinoma (ESCC). Notably, pyrroline-5-carboxylate reductase 2 (PYCR2) and uridine phosphorylase 1 (UPase1) were found to be altered in ESCC. The spatially resolved metabolomics reveal what occurs in cancer at the molecular level, from metabolites to enzymes, and thus provide insights into the understanding of cancer metabolic reprogramming.
Metabolic reprogramming represents cancer-associated metabolic changes during tumorigenesis and has been recognized as a new hallmark of cancer (1). Profiling the differential metabolic dependencies of cancer at the highest achievable coverage is significant for understanding the complex molecular processes taking place in tumors and will yield important insights into how to target tumor metabolism (2). Metabolites and metabolic enzymes are the two most important constituents of biological metabolic networks. Metabolic enzymes, as important nodes in biological metabolic networks, can regulate and control the flux of metabolites to maintain metabolic homeostasis (3). Emerging work suggests that the dysregulation of metabolic pathways contribute to the pathogenesis of cancer, and this altered metabolism introduces metabolic liabilities that can be exploited for cancer therapy (4–6). However, high-throughput discovery of tumor-associated metabolic alterations at both the metabolite and metabolic enzyme levels is still a great challenge.
A technique that enables a direct view of altered enzyme features from cancer tissues will facilitate the exploitation of metabolic enzyme driven tumor-targeting therapy. Liquid chromatography–mass spectrometry (LC-MS)-based proteomic profiling of cancer tissue reveals differentially expressed proteins; however, the spatial distribution discrepancy of heterogeneous tissue samples is lost over the course of elaborate sample pretreatment (7). Matrix-assisted laser desorption ionization mass spectrometry imaging (MALDI-MSI) is an effective tool to visualize the spatial location of proteins and metabolites with high spatial resolution, but high-vacuum environments and matrix assistance are usually needed for ionization (8–11). Immunohistochemistry (IHC) is a traditional tool to characterize the distribution and intensity of specific enzymes in tissue based on antibody–antigen interactions. However, its dependency on targeted antibodies and its time-consuming procedure make it difficult to investigate multiple unknown features in tissues for tumor-associated enzyme discovery (12).
Metabolites are the direct products or substrates of metabolic enzymes, and the level of metabolites in tissue may reflect enzyme capacities (13). Characterization of the metabolic pathway-related metabolites in heterogeneous cancer tissue provides important insights into tumor-associated metabolic enzyme discovery. Ambient MSI (AMSI) technology, represented by desorption electrospray ionization (DESI)-MSI can directly extract and visualize numerous molecular features in tissue sections (9, 14–20). Untargeted AMSI has been regarded as a rational approach for monitoring lipid dysregulation in cancer tissues for biological and clinical studies (21–25). Airflow-assisted desorption electrospray ionization (AFADESI)-MSI is a high-coverage ambient molecular imaging technique that was developed by our group, and it can map numerous functional metabolites located in different metabolic pathways (26). However, the chemical noise arising from tissue biomatrix makes the directly mapping of proteins using AMSI techniques challenging, especially for the low-content functional metabolic enzymes.
In this study, we propose a strategy for high-throughput discovery of cancer-associated metabolites and metabolic enzymes in their native state. The design of this strategy is shown in SI Appendix, Fig. S1. High-coverage airflow-assisted desorption electrospray ionization (AFADESI)-MSI was first applied to acquire region-specific tissue metabolite profiles in 256 esophageal squamous cell carcinoma (ESCC) patients. Then, MSI-based metabolomics combined with multivariate statistical analysis was applied to screen discriminating metabolites between cancerous and normal tissues, and metabolic pathway matching analysis of the screened metabolites was conducted to reveal potential tumor-associated metabolic enzymes. Afterward, specific IHC staining was performed on adjacent tissue sections to validate the spatial expression of the potential tumor-associated enzymes. We report a method for the high-throughput discovery of tumor-associated metabolite and enzyme alterations based on a spatially resolved MSI metabolomics approach. Using this approach, the proline biosynthesis, glutamine metabolism, uridine metabolism, histidine metabolism, fatty acid (FA) biosynthesis, and polyamine biosynthesis pathways were found to be altered in ESCC. Six abnormally expressed metabolic enzymes, including pyrroline-5-carboxylate reductase 2 (PYCR2), glutaminase (GLS), uridine phosphorylase 1 (UPase1), histidine decarboxylase (HDC), FA synthase (FASN), and ornithine decarboxylase (ODC), which are directly associated with the altered metabolites in pathways, were further discovered. Notably, the expression levels of PYCR2 and UPase1 were found to be altered in ESCC. The integration of spatially resolved enzyme information and the corresponding downstream metabolite information will expand our understanding of tumor metabolism and could further facilitate the discovery of altered metabolic pathways. Furthermore, extensively altered tumor metabolism provides insights for developing novel drugs to target multiple metabolic abnormalities.
Results and Discussion
Region-Specific Molecule Profiling.
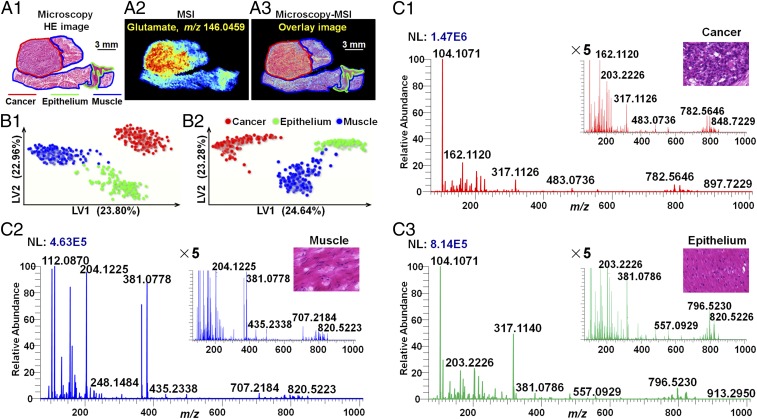
Postoperative ESCC tissue sections were divided into three histologic types based on the cell type and components: cancer tissue, epithelial tissue, and muscular tissue. After tissue MSI, microscopy was integrated with the MS image to form a microscopy-MSI overlay image, which delivers both the spatial resolution of microscopy and the chemical signatures of MSI in one integrated whole (Fig. 1A). However, it is worth noting that the combined image is just an overlay of the histology and the MS image, and the discrimination of different tissue region still depends on the pathologist. Based on microscopy-MSI overlay image, cancer, epithelium, and muscle specific mass spectra were precisely extracted and are illustrated in Fig. 1C and SI Appendix, Fig. S2. As shown in the figures, the significant difference in the mass profiles among cancer tissue, epithelial tissue, and muscular tissue is obvious. In addition, the ion intensities of different molecules varied over a dynamic range that spanned more than three orders of magnitude (SI Appendix, Fig. S3).
Fig. 1.
The strategy to extract region-specific MS spectra in heterogeneous ESCC tissue. (A) Example of a microscopy-MSI overlay. (A1) H&E image of ESCC tissue section. (A2) MS image of glutamate (m/z 146.0459) in ESCC tissue section. (A3) Microscopy-MSI overlay image. (B) PLS-DA models based on positive (B1) and negative (B2) ion mode AFADESI-MSI data. (C) Representative mass spectra of cancer tissue (C1), muscular tissue (C2), and epithelial tissue (C3) in positive ion mode.
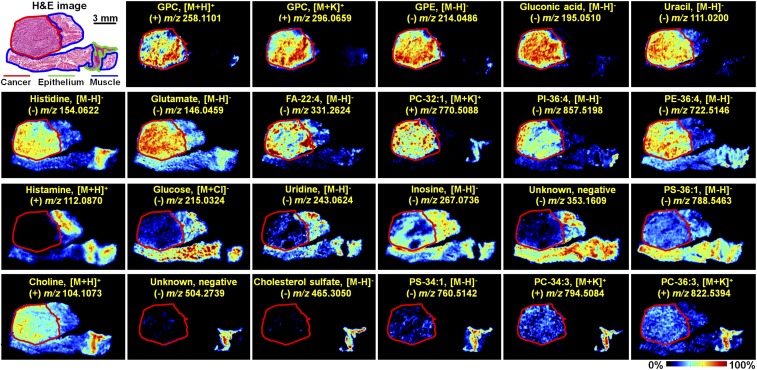
To explore the global discriminating molecules between different tissue types, a partial least squares discriminant analysis (PLS-DA) model based on the MS image pixel point was built to screen region-specific biomarkers. Because the ion signals were measured on the same scale in the raw MSI data, ions that typically exhibit stronger intensities have a greater influence on multivariate statistical analysis and potential region-specific biomarker screening, resulting in the neglect of low-abundance functional metabolites (27). To alleviate the dependency of heteroscedasticity on the ion signal intensity, metabolite peaks from all pixel points were picked, underwent log transformation, and then were subjected to PLS-DA analysis. As shown in Fig. 1B, the PLS-DA models based on (±) AFADESI-MSI data achieved great separation among cancer tissue, epithelial tissue, and muscular tissue. Region-specific metabolite biomarkers were first screened based on their respective classification loadings. Then, independent t tests were carried out to validate the significance of the discriminated metabolites between cancer and normal tissues. A multitude of region-specific small molecule metabolites and lipids were screened and visualized in ESCC tissues (Fig. 2).
Fig. 2.
Region-specific MS images of ESCC tissue section. GPC, glycerophosphorylcholine; GPE, glycerophosphorylethanolamine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine.
Tumor-Associated Metabolic Pathway Discovery.
Tissue is the lesion location of cancer and contains global biological metabolic information at both the metabolic enzyme and metabolite levels. As important nodes in biological metabolic networks, metabolic enzymes connect and regulate complex metabolic reactions and have always been recognized as potential anticancer drug targets. Here, MSI data combined with PLS-DA analysis enabled the determination of region-specific discriminating metabolites. Then, the discriminating metabolites were imported into Kyoto Encyclopedia of Genes and Genomes (www.kegg.jp) to perform metabolic pathway matching analysis, facilitating the discovery of altered metabolic pathways (28). This analysis suggested that arginine and proline metabolism; FA biosynthesis; alanine, aspartate, and glutamate metabolism; pyrimidine metabolism; and histidine metabolism were significantly dysregulated in ESCC (SI Appendix, Fig. S4). Six crucial metabolic enzymes that are directly associated with the altered metabolites in pathways were chosen as potential tumor-associated metabolic enzymes. The detailed metabolic enzymes and related metabolite information are illustrated in SI Appendix, Tables S1 and S2: PYCR2 catalyzes the biosynthesis of proline, GLS catalyzes the first reaction in the primary pathway for the catabolism of glutamine, UPase1 catalyzes the reversible phosphorolytic cleavage of uridine to uracil, HDC stimulates the decarboxylation of histidine to form histamine, FASN catalyzes the formation of long-chain FAs, and ODC regulates amine and polyamine biosynthesis.
In Situ Validation of Crucial Metabolites and Metabolic Enzymes in Tumor-Associated Metabolic Pathways.
In this study, MSI-based in situ metabolomics combined with metabolic pathway analysis contributed to the discovery of potential tumor-associated metabolic enzymes in ESCC tissue. Then, targeted IHC testing of the suspected metabolic enzymes was performed on successive tissue sections (adjacent to the tissue section analyzed by AFADESI-MSI) to validate our discovery.
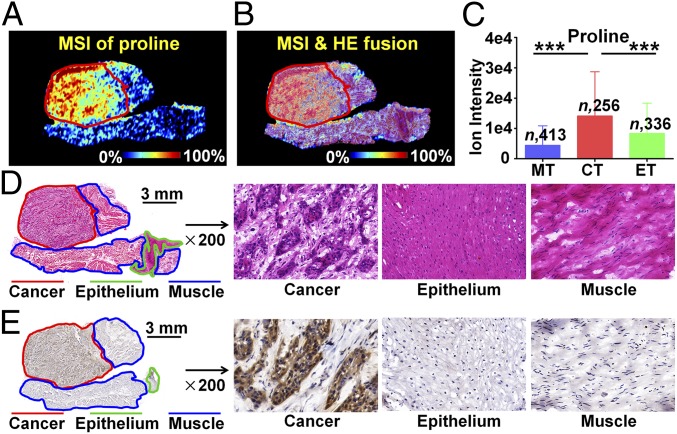
Proline, as an important amino acid in the cellular microenvironment, participates in apoptosis and autophagy, and it is drawing increasing attention for its crucial role in cancer metabolism (29, 30). The MS image indicated that proline was significantly up-regulated in the cancer region compared with the normal epithelium and muscle region (Fig. 3A). Microscopy-MSI overlay image facilitates the extraction of region-specific metabolite profiles (Fig. 3B), which suggested that the ion intensity of proline in cancer regions is significantly higher than that in epithelium and muscle regions according to the statistical data of 256 ESCC tissue samples (P < 0.001, Fig. 3C). Based on the proline biosynthesis metabolic pathway, PYCR2 is an essential rate-limiting enzyme for the biosynthesis of proline. Emerging studies have shown that PYCR2 is indispensable for cancer cell proliferation and progression (31). We speculated that the up-regulated expression of proline in ESCC tissue may be attributed to elevated proline biosynthesis. IHC staining was performed to explore the spatial expression of PYCR2 in ESCC tissue sections and to evaluate the spatial matching of PYCR2 and proline (Fig. 3E). Interestingly, IHC analysis indicated that PYCR2 was mainly expressed in the cancer region, which was consistent with the spatial distribution of proline in ESCC tissue sections. The spatial expression of proline and PYCR2 in other ESCC tissue section is illustrated in SI Appendix, Fig. S5. Notably, we identified dysregulated PYCR2 in ESCC. In addition, we analyzed the PYCR2 expression in a tumor section that did not present changes in proline level, and the results suggest that there was no difference of PYCR2 (SI Appendix, Fig. S6).
Fig. 3.
In situ visualization of crucial metabolite and metabolic enzyme in the proline biosynthesis pathway. (A) MS image of proline. (B) MSI and H&E overlay image. (C) Proline levels in cancer and paired epithelium and muscle tissues from 256 ESCC patients (means ± SD). ***P < 0.001. (D) H&E image of ESCC tissue section. (E) Expression of PYCR2 in different regions of ESCC tissue section. CT, cancer tissue; ET, epithelial tissue; MT, muscular tissue.
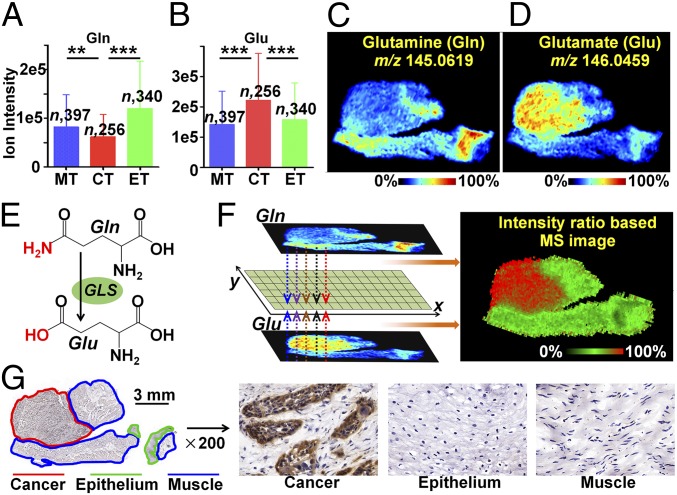
Glutamine (Gln) is indispensable to the maintenance of cell energy metabolism, nucleotide and amino acid biosynthesis, and redox homeostasis (32). Extensive studies have demonstrated that cancerous cells display a strong addiction to Gln, which makes glutamine metabolism an appealing target for cancer diagnosis and treatment (33). The catabolism of Gln is mediated by GLS through the hydrolysis of Gln to glutamate (Glu). Here, the elevated glutamine consumption in cancer tissue was further proved by our MSI results. As shown in Fig. 4 A and C, Gln is down-regulated in cancer tissues compared with normal muscle and epithelial tissues. Instead, Glu as the hydrolysis product of Gln was dramatically increased in cancer tissues (Fig. 4 B and D). The MS image is composed of consecutive pixels, each of which can reflect the relative content of the metabolites in the region. Here, the pixel-by-pixel intensity ratio of Glu to Gln was calculated to construct an intensity ratio-based MS image (Fig. 4F), and it suggested that the cancer tissue (red region) possessed a higher ion-intensity ratio than normal tissue (green region). Moreover, the ion-intensity ratio-based MSI offers an approach to the diagnosis of esophageal cancer. The altered intensity ratio across different tissue regions may reflect the in situ Gln hydrolysis rate, which is mediated by GLS. The subsequent IHC assay showed that GLS was remarkably up-regulated in cancer tissue compared with normal tissue, in good agreement with the intensity ratio-based MS image (Fig. 4G). The spatial expressions of Gln, Glu, and GLS in other tissue sections are demonstrated in SI Appendix, Fig. S7.
Fig. 4.
In situ visualization of crucial metabolites and metabolic enzyme in the glutamine metabolism pathway. (A and B) Glutamine (Gln) and glutamate (Glu) levels in cancer and paired epithelium and muscle tissues from 256 ESCC patients (means ± SD). **P < 0.01; ***P < 0.001. (C and D) MS images of Gln and Glu in ESCC tissue section. (E) GLS-mediated metabolic process of converting Gln to Glu. (F) The newly constructed MS image based on the ion-intensity ratio of Glu to Gln. (G) Expression of GLS in different regions of the ESCC tissue section. CT, cancer tissue; ET, epithelial tissue; MT, muscular tissue.
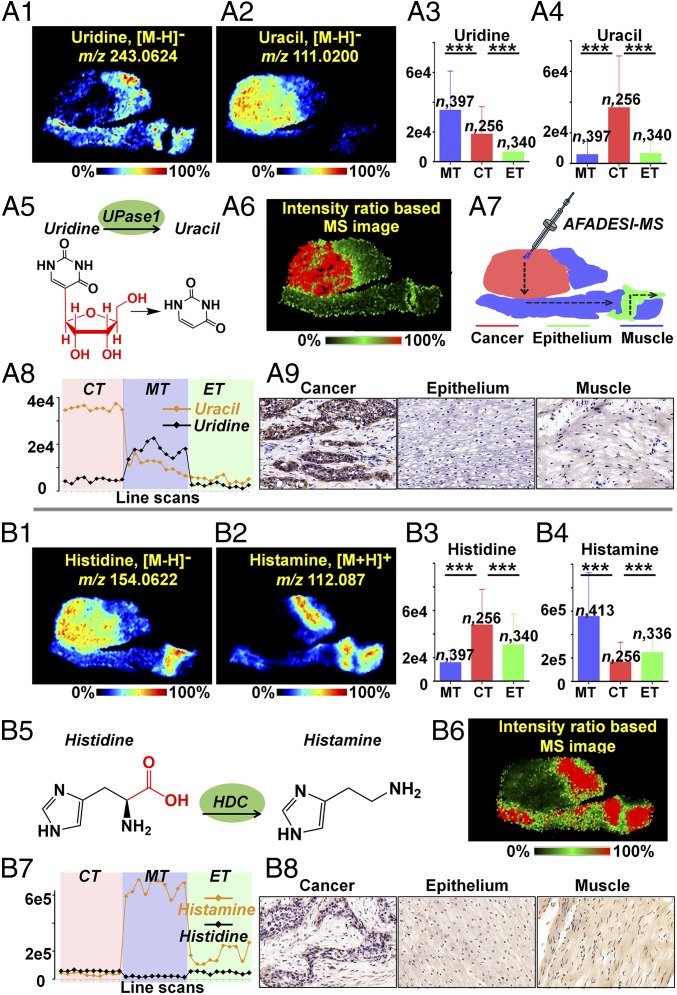
Uridine, an important nucleoside precursor for the synthesis of RNA, also participates in the regulation of purine nucleotide biosynthesis and carbohydrate metabolism (34). Moreover, the level of uridine in tissue is critical to pyrimidine antimetabolite-based anticancer treatment. A recent study indicated that uridine homeostatic disorder can trigger p53-mediated DNA damage and lead to tumorigenesis (35). Our MSI and statistical data suggested that uridine metabolism was severely dysregulated in ESCC tissues: although the ion intensity of uridine in cancer regions is higher than that in paired epithelium regions, it is lower than that in muscle regions (P < 0.001; Fig. 5 A1 and A3). Meanwhile, the uracil level was dramatically up-regulated in cancer tissue (P < 0.001, Fig. 5 A2 and A4). Based on the pyrimidine metabolism pathway, UPase1, which reversibly catalyzes the phosphorolysis of uridine into uracil, is the key enzyme of pyrimidine nucleoside metabolism (Fig. 5A5). Thus, UPase1 is regarded as a potential tumor-associated metabolic enzyme in ESCC. The pixel-by-pixel intensity ratio of uracil to uridine was calculated to map the intensity ratio-based MS image (Fig. 5A6). Furthermore, we extracted the mass spectra of consecutive image pixels along the illustrated path over an ESCC tissue section (Fig. 5A7). The dramatic intensity difference of uracil and uridine in cancer, muscle, and epithelium are illustrated in Fig. 5A8. These data suggest that the intensity ratio of uracil to uridine was significantly increased in the cancer region and can serve as a biomarker to distinguish cancer from paracancerous normal tissue. Considering all of the above findings, we predicted that the UPase1-mediated phosphorolytic cleavage of uridine would be stronger in cancer tissue than in paracancerous normal tissue. Excitingly, the IHC data on the ESCC tissue section confirm our prediction that UPase1 was up-regulated in the cancer region (Fig. 5A9). Furthermore, we report differentially expressed UPase1 in ESCC.
Fig. 5.
In situ visualization of crucial metabolites and metabolic enzyme in the uridine metabolism pathway (A) and histidine metabolism pathway (B). (A1 and A2) MS images of uridine and uracil. (A3 and A4) Uridine and uracil levels in cancer and paired epithelium and muscle tissues from 256 ESCC patients (means ± SD). ***P < 0.001. (A5) UPase1-mediated metabolic process of converting uridine to uracil. (A6) The newly constructed MS image based on the ion-intensity ratio of uracil to uridine. (A7) Scanning path of AFADESI-MSI. (A8) Plot of the intensity changes of uridine and uracil occurring during the transition from cancer, muscle, to epithelial tissue. (A9) Expression of UPase1 in different regions of an ESCC tissue section. (B1 and B2) MS images of histidine and histamine. (B3 and B4) Histidine and histamine levels in cancer and paired epithelium and muscle tissues from 256 ESCC patients. (B5) The HDC-mediated metabolic process of converting histidine to histamine. (B6) The newly constructed MS image based on the ion-intensity ratio of histamine to histidine. (B7) Plot of the intensity changes of histidine and histamine occurring during the transition from cancer, muscle, to epithelial tissue. (B8) Expression of HDC in different regions of an ESCC tissue section. CT, cancer tissue; ET, epithelial tissue; MT, muscular tissue.
Histamine is derived from the decarboxylation of histidine, which is exclusively catalyzed by HDC (36). There is growing evidence suggesting that histamine is directly involved in carcinogenesis and may serve as a potential cytoprotective agent to improve cancer therapy (37). According to some investigators, histamine-based therapies facilitate DNA damage, apoptosis, and senescence in carcinoma cells and remarkably increase the survival of tumor-bearing animals (38). In this study, histidine and histamine presented totally opposite spatial distributions. Histidine was significantly up-regulated in cancer according to the MSI and statistical data of 256 ESCC tissue samples (P < 0.001; Fig. 5 B1 and B3), while histamine was dramatically down-regulated in cancer tissue (P < 0.001; Fig. 5 B2 and B4). Meanwhile, methylhistamine, the metabolic product of histamine, did not exhibit obvious dysregulation in cancer tissue (SI Appendix, Fig. S9). The dramatic intensity difference of histamine and histidine is illustrated in Fig. 5B7. The pixel-by-pixel intensity ratio of histamine to histidine was calculated and then imaged to investigate the HDC-mediated decarboxylation of histidine (Fig. 5B6), and the decarboxylation rate was found to be relatively weaker in cancer tissue than in muscular and epithelial tissue. IHC validation was then applied to evaluate the expression of HDC in ESCC tissue (Fig. 5B8). As predicted by the intensity ratio-based MS image, cancer tissue demonstrated a lower level of HDC expression than muscular and epithelial tissues.
FAs are important endogenous molecules for cellular energy metabolism and biological signal transmission. SI Appendix, Fig. S11 demonstrates that the ion intensities of representative FAs were stronger in cancer and epithelial tissues than in muscle tissue. Overall, the FA ion intensities demonstrated an increasing trend from muscle to epithelium to cancer tissue in ESCC. It has been reported that cancer cells rapidly produce FAs to meet the urgent need for membrane biosynthesis, cellular signaling, and energy consumption (3, 39). In the FA biosynthesis pathway, FASN is a key metabolic enzyme for the de novo synthesis of FAs. Therefore, FASN was selected as a potential tumor-associated enzyme according to the significantly increased levels of FAs in the cancer tissue. Then, IHC staining of FASN was performed on adjacent tissue sections. Notably, the spatial expression of FASN was consistent with the distribution of FAs. FASN was mainly expressed in cancer tissue, followed by epithelial tissue and muscular tissue, which means that region-specific FAs may be able to predict the expression of FASN in ESCC tissue. The MS images and statistical data of other representative FAs in ESCC tissue are illustrated in SI Appendix, Fig. S12.
Polyamines, including spermine and spermidine, have long been recognized as indispensable components for cell growth, especially for unwanted cancer cell proliferation (40). As demonstrated in SI Appendix, Fig. S13, spermine and spermidine were highly expressed in cancer tissue, which is consistent with the stronger proliferation ability of cancer cells. Metabolic pathway analysis suggested that ODC, which converts ornithine into putrescine to form spermidine and spermine, is a rate-limiting enzyme in polyamine biosynthesis. Therefore, ODC was selected as another potential tumor-associated metabolic enzyme. IHC testing verified our discovery that the expression of ODC in cancer is higher than that in paired normal tissues (SI Appendix, Fig. S13A9). Quantification of IHC signals of the six enzymes are shown in SI Appendix, Fig. S14. Targeted inhibition/inducement of the dysregulated enzymes or altering the levels of downstream metabolites may shed light on metabolism-based therapy. However, what we offer is only potential metabolic vulnerabilities. Further study of the roles of the altered metabolic pathway in tumor progression is needed. Other kinds of detectable metabolites are shown in SI Appendix, Figs. S15 and S16.
Diagnostic Features of Unknown Samples.
To verify the diagnostic ability of the MSI-based PLS-DA model on untested esophageal cancer, we further analyzed 36 newly collected samples. The spatially resolved metabolite profiles were extracted and then imported into the PLS-DA classifier for automatic class identification and recognition. As shown in SI Appendix, Fig. S17, most of the newly collected samples were correctly classified with few exceptions, and the overall accuracy was 94.4% for identifying the three different tissue types. The altered metabolites screened by spatially resolved metabolomics still present tissue-specific distributions in newly collected samples (SI Appendix, Fig. S18).
Conclusions
In summary, we have developed a spatially resolved metabolomics approach for the high-throughput characterization of tumor-associated metabolic alterations at both the metabolite and enzyme levels. Differentially expressed metabolic enzymes that are closely associated with tumors were efficiently discovered based on region-specific and pathway-related metabolites. A major advantage of this approach is its direct predictive applicability to large sets of candidate metabolites and metabolic enzymes without a priori definition of specific targets of interest. Moreover, the tumor-associated metabolic enzymes and corresponding metabolites information expand our understanding of the complex tumor metabolic reprogramming. However, it is worth noting that this approach provides potentially altered metabolic enzymes based on spatially resolved metabolomics, and the subsequent IHC validation is still needed. In addition, the screened tumor-associated metabolic pathways have numerous potential metabolic enzymes; we paid more attention to key enzymes that are directly related to the remarkably dysregulated metabolites in certain pathways.
This MSI-based metabolomics study of 256 cases of cancer and matched normal tissues suggests that the proline biosynthesis, glutamine metabolism, uridine metabolism, histidine metabolism, FA biosynthesis, and polyamine biosynthesis pathway were significantly altered in ESCC. Six abnormally expressed metabolic enzymes, including PYCR2, GLS, UPase1, HDC, FASN, and ODC, which are extensively involved in ESCC carcinogenesis, were discovered. Most importantly, PYCR2 and UPase1 were found to be differentially expressed in ESCC tissue. Furthermore, this spatially resolved tumor metabolic information in ESCC offers insights for understanding the complex cancer metabolic reprogramming.
Materials and Methods
Sample Preparation and Process.
All cohort patients provided written informed consent. Approval to perform metabolic analysis on tissue samples was obtained from the local Ethical Review Board of Linzhou Esophageal Cancer Hospital. A total of 256 pairs of matched human ESCC tissue samples, including cancer tissues, adjacent noncancerous tissues (collected at 0–2 cm surrounding the cancer tissue), and distal noncancerous tissue (collected at 5 cm away from the cancer tissue) were collected. The esophageal cancer tissues were flash-frozen in liquid nitrogen for 10 s after resection, then were transferred to cryogenic vials, and were stored at −80 °C until sectioned at 10-μm thickness using a CM 1860 UV cryostat microtome (Leica). The tissue sections were thaw-mounted onto microscope slide and stored in closed containers at −80 °C. Before AFADESI-MSI analysis, the microscope slides were dried in a vacuum for ∼15 min. The typical H&E images of different tissues are illustrated in SI Appendix, Fig. S20, and it suggests that adjacent noncancerous tissue include adjacent muscle and epithelium, while distal noncancerous tissue include distal muscle and epithelium.
AFADESI-MSI Analysis.
AFADESI-MSI analysis was carried out in both positive- and negative-ion mode on a Q-Exactive mass spectrometer (Thermo Scientific) over an m/z range of 70–1,000 at a nominal mass resolution of 70,000. A mixture of acetonitrile and water (8:2, vol/vol) was used as the spray solvent at a flow rate of 5 μL/min. The sprayer and transport tube voltages were set at 7,500 and 2,000 V in positive-ion mode and at −5,500 and −1,500 V in negative-ion mode. The extracting gas flow was 45 L/min, and the capillary temperature was 350 °C. The MSI experiments were performed by continuously scanning the tissue surface in the x direction at a constant rate of 200 μm/s, with a 200-μm vertical step separating the adjacent lines in the y direction.
Data Processing.
The collected .raw files were converted into .cdf format and then imported into custom-developed imaging software (MassImager, a dedicated imaging software based on the C++ programming language) for ion image reconstructions and multivariate statistical analysis (41). After background subtraction, region-specific MS profiles were precisely extracted by matching high-spatial resolution H&E images. The discriminating endogenous molecules of different tissue microregions were screened by a supervised statistical analytical method: PLS-DA. The detailed process of screening discriminant m/z features was illustrated in SI Appendix, Fig. S21. Two-tailed t test and analysis of variance were performed using SPSS statistical software (SPSS21.0).
Immunohistochemistry.
The IHC characterization of PYCR2, GLS, FASN, UPase1, HDC, and ODC in ESCC tissues are provided in SI Appendix.
Analyte Identification.
Extracted adducted ions were compared with the free databases the Human Metabolome Database (www.hmdb.ca), Metlin (https://metlin.scripps.edu), and LIPID MAPS (www.lipidmaps.org) using exact molecular weights and a mass accuracy of less than 5 ppm, combining the isotope abundance from high-resolution MS help to give the elemental composition and possible list of endogenous metabolites (42). Then, the metabolites were performed high-resolution tandem MS directly from tissue sections. Additional details are provided in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China Grants 21335007 and 81773678; the Leading Talent Support Program of State Ethnic Affairs Commission, Program of Collaborative Innovation Center for Ethnic Minority Development of Minzu University of China (Grant 017004040501); and National Instrumentation Program Grant 2016YFF0100304.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.G.V.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808950116/-/DCSupplemental.
References
- 1.Ward PS, Thompson CB. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vander Heiden MG, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luengo A, Gui DY, Vander Heiden MG. Targeting metabolism for cancer therapy. Cell Chem Biol. 2017;24:1161–1180. doi: 10.1016/j.chembiol.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez-Outschoorn UE, Peiris-Pagés M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: A therapeutic perspective. Nat Rev Clin Oncol. 2017;14:11–31. doi: 10.1038/nrclinonc.2016.60. [DOI] [PubMed] [Google Scholar]
- 5.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Longuespée R, et al. MALDI mass spectrometry imaging: A cutting-edge tool for fundamental and clinical histopathology. Proteomics Clin Appl. 2016;10:701–719. doi: 10.1002/prca.201500140. [DOI] [PubMed] [Google Scholar]
- 8.Caprioli RM. Imaging mass spectrometry: Molecular microscopy for the new age of biology and medicine. Proteomics. 2016;16:1607–1612. doi: 10.1002/pmic.201600133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kompauer M, Heiles S, Spengler B. Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-μm lateral resolution. Nat Methods. 2017;14:90–96. doi: 10.1038/nmeth.4071. [DOI] [PubMed] [Google Scholar]
- 10.Maier SK, et al. Comprehensive identification of proteins from MALDI imaging. Mol Cell Proteomics. 2013;12:2901–2910. doi: 10.1074/mcp.M113.027599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Q, Chu JL, Rubakhin SS, Gillette MU, Sweedler JV. Dopamine-modified TiO2 monolith-assisted LDI MS imaging for simultaneous localization of small metabolites and lipids in mouse brain tissue with enhanced detection selectivity and sensitivity. Chem Sci (Camb) 2017;8:3926–3938. doi: 10.1039/c7sc00937b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swanson PE. Immunohistochemistry as a surrogate for molecular testing: A review. Appl Immunohistochem Mol Morphol. 2015;23:81–96. doi: 10.1097/PAI.0000000000000181. [DOI] [PubMed] [Google Scholar]
- 13.Fendt SM, et al. Tradeoff between enzyme and metabolite efficiency maintains metabolic homeostasis upon perturbations in enzyme capacity. Mol Syst Biol. 2010;6:356. doi: 10.1038/msb.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takáts Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 15.Nemes P, Vertes A. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal Chem. 2007;79:8098–8106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 16.Wu C, Dill AL, Eberlin LS, Cooks RG, Ifa DR. Mass spectrometry imaging under ambient conditions. Mass Spectrom Rev. 2013;32:218–243. doi: 10.1002/mas.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eberlin LS, et al. Molecular assessment of surgical-resection margins of gastric cancer by mass-spectrometric imaging. Proc Natl Acad Sci USA. 2014;111:2436–2441. doi: 10.1073/pnas.1400274111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Römpp A, et al. Histology by mass spectrometry: Label-free tissue characterization obtained from high-accuracy bioanalytical imaging. Angew Chem Int Ed Engl. 2010;49:3834–3838. doi: 10.1002/anie.200905559. [DOI] [PubMed] [Google Scholar]
- 19.Balog J, et al. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci Transl Med. 2013;5:194ra93. doi: 10.1126/scitranslmed.3005623. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee S, et al. Diagnosis of prostate cancer by desorption electrospray ionization mass spectrometric imaging of small metabolites and lipids. Proc Natl Acad Sci USA. 2017;114:3334–3339. doi: 10.1073/pnas.1700677114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eberlin LS, et al. Alteration of the lipid profile in lymphomas induced by MYC overexpression. Proc Natl Acad Sci USA. 2014;111:10450–10455. doi: 10.1073/pnas.1409778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eberlin LS, et al. Pancreatic cancer surgical resection margins: Molecular assessment by mass spectrometry imaging. PLoS Med. 2016;13:e1002108. doi: 10.1371/journal.pmed.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guenther S, et al. Spatially resolved metabolic phenotyping of breast cancer by desorption electrospray ionization mass spectrometry. Cancer Res. 2015;75:1828–1837. doi: 10.1158/0008-5472.CAN-14-2258. [DOI] [PubMed] [Google Scholar]
- 24.Jarmusch AK, et al. Lipid and metabolite profiles of human brain tumors by desorption electrospray ionization-MS. Proc Natl Acad Sci USA. 2016;113:1486–1491. doi: 10.1073/pnas.1523306113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calligaris D, et al. Application of desorption electrospray ionization mass spectrometry imaging in breast cancer margin analysis. Proc Natl Acad Sci USA. 2014;111:15184–15189. doi: 10.1073/pnas.1408129111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He J, et al. A sensitive and wide coverage ambient mass spectrometry imaging method for functional metabolites based molecular histology. Adv Sci. 2018;5:1800250. doi: 10.1002/advs.201800250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enot DP, et al. Preprocessing, classification modeling and feature selection using flow injection electrospray mass spectrometry metabolite fingerprint data. Nat Protoc. 2008;3:446–470. doi: 10.1038/nprot.2007.511. [DOI] [PubMed] [Google Scholar]
- 28.Kanehisa M, et al. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W, et al. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci USA. 2012;109:8983–8988. doi: 10.1073/pnas.1203244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phang JM, Liu W, Hancock CN, Fischer JW. Proline metabolism and cancer: Emerging links to glutamine and collagen. Curr Opin Clin Nutr Metab Care. 2015;18:71–77. doi: 10.1097/MCO.0000000000000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu W, Hancock CN, Fischer JW, Harman M, Phang JM. Proline biosynthesis augments tumor cell growth and aerobic glycolysis: Involvement of pyridine nucleotides. Sci Rep. 2015;5:17206. doi: 10.1038/srep17206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith B, et al. Addiction to coupling of the Warburg effect with glutamine catabolism in cancer cells. Cell Rep. 2016;17:821–836. doi: 10.1016/j.celrep.2016.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, et al. Metabolomics profiles delineate uridine deficiency contributes to mitochondria-mediated apoptosis induced by celastrol in human acute promyelocytic leukemia cells. Oncotarget. 2016;7:46557–46572. doi: 10.18632/oncotarget.10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao Z, et al. Uridine homeostatic disorder leads to DNA damage and tumorigenesis. Cancer Lett. 2016;372:219–225. doi: 10.1016/j.canlet.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 36.Ruffell B, Coussens LM. Histamine restricts cancer: Nothing to sneeze at. Nat Med. 2011;17:43–44. doi: 10.1038/nm0111-43. [DOI] [PubMed] [Google Scholar]
- 37.Martinel Lamas DJ, Nicoud MB, Sterle HA, Cremaschi GA, Medina VA. Histamine: A potential cytoprotective agent to improve cancer therapy? Cell Death Dis. 2015;6:e2029. doi: 10.1038/cddis.2015.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinel Lamas DJ, et al. Enhancement of ionizing radiation response by histamine in vitro and in vivo in human breast cancer. Cancer Biol Ther. 2015;16:137–148. doi: 10.4161/15384047.2014.987091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen T, Li H. Fatty acid metabolism and prospects for targeted therapy of cancer. Eur J Lipid Sci Technol. 2017;119:1600366. [Google Scholar]
- 40.Murray-Stewart TR, Woster PM, Casero RA., Jr Targeting polyamine metabolism for cancer therapy and prevention. Biochem J. 2016;473:2937–2953. doi: 10.1042/BCJ20160383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He J, et al. MassImager: A software for interactive and in-depth analysis of mass spectrometry imaging data. Anal Chim Acta. 2018;1015:50–57. doi: 10.1016/j.aca.2018.02.030. [DOI] [PubMed] [Google Scholar]
- 42.Palmer A, et al. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat Methods. 2017;14:57–60. doi: 10.1038/nmeth.4072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.