Significance
Human activities alter and reduce phenotypic variation in many species, but the long-term consequences (e.g., ability of previous variation to reemerge), and thus the need for conservation action, are unclear. Here we show that dramatic, human-induced changes in adult migration characteristics of wild Chinook salmon are explained by rapid evolution at a single locus and can lead to loss of a critical adaptive allele. The decline and loss of this allele will likely hinder current and future restoration efforts, as well as compromise resilience and evolutionary potential. Thus, human-induced phenotypic change can result in rapid loss of important adaptive variation, and conservation action to address human impacts on phenotypic variation will sometimes be necessary to preserve evolutionarily significant biodiversity.
Keywords: conservation, evolution, genetics, biodiversity, salmon
Abstract
Phenotypic variation is critical for the long-term persistence of species and populations. Anthropogenic activities have caused substantial shifts and reductions in phenotypic variation across diverse taxa, but the underlying mechanism(s) (i.e., phenotypic plasticity and/or genetic evolution) and long-term consequences (e.g., ability to recover phenotypic variation) are unclear. Here we investigate the widespread and dramatic changes in adult migration characteristics of wild Chinook salmon caused by dam construction and other anthropogenic activities. Strikingly, we find an extremely robust association between migration phenotype (i.e., spring-run or fall-run) and a single locus, and that the rapid phenotypic shift observed after a recent dam construction is explained by dramatic allele frequency change at this locus. Furthermore, modeling demonstrates that continued selection against the spring-run phenotype could rapidly lead to complete loss of the spring-run allele, and an empirical analysis of populations that have already lost the spring-run phenotype reveals they are not acting as sustainable reservoirs of the allele. Finally, ancient DNA analysis suggests the spring-run allele was abundant in historical habitat that will soon become accessible through a large-scale restoration (i.e., dam removal) project, but our findings suggest that widespread declines and extirpation of the spring-run phenotype and allele will challenge reestablishment of the spring-run phenotype in this and future restoration projects. These results reveal the mechanisms and consequences of human-induced phenotypic change and highlight the need to conserve and restore critical adaptive variation before the potential for recovery is lost.
Phenotypic variation buffers species and populations against environmental variability and is important for long-term persistence (1–7). In phenotypically diverse populations, environmental fluctuations that negatively impact one phenotype may have a neutral or positive impact on another (5, 8). This decreases variance in population size across time and reduces vulnerability to extirpation or extinction. Furthermore, phenotypic variation increases the potential for species to persist through long-term environmental changes (e.g., climate change) by serving as the substrate upon which evolution can act. Thus, maintaining intraspecific phenotypic variation is an important component of biodiversity conservation.
Anthropogenic activities have major effects on phenotypic variation across a broad array of species and traits, often producing substantial phenotypic shifts and reductions in overall variation (5, 6, 9–12). Despite the recognized importance of intraspecific variation, the urgency of addressing human-driven phenotypic change through conservation policy and action is unclear because the ability of affected populations and/or species to recover previous characteristics (e.g., variation) is not well understood (5, 13, 14). If previous variation can quickly reemerge, human-induced phenotypic change may have limited impact on long-term persistence and evolutionary potential. However, permanent changes and reductions in variation could have severe consequences such as limiting potential response to future environmental fluctuations, constraining the ability to colonize new habitat that may become available and curtailing evolutionary potential (15–17). Thus, in cases where anthropogenic activities threaten the potential to recover previous characteristics, immediate steps to reduce human impacts on intraspecific phenotypic variation are warranted.
The mechanisms that underlie human-induced phenotypic change (i.e., phenotypic plasticity and/or genetic evolution) will influence the potential for previous characteristics to reemerge. For example, if phenotypic changes are due to plasticity (i.e., the ability of the same genotype to produce different phenotypes when exposed to different environments), previous characteristics may rapidly reemerge if environmental conditions become favorable (e.g., habitat is restored or new habitat becomes accessible) (18, 19). However, phenotypic change due to genetic evolution (i.e., changes in allele and genotype frequencies across generations) may severely impact the ability to recover previous characteristics (5, 12, 20). In the case of genetic evolution, the ability to recover previous phenotypic characteristics will depend on factors such as the genetic architecture of the affected trait (21). Unfortunately, understanding the genetic basis of phenotypic variation, and thus the potential consequences of human-driven phenotypic change, can be challenging because the genes that influence specific traits in natural populations are usually unknown (22, 23).
The adult migration characteristics of Chinook salmon (Oncorhynchus tshawytscha) are a clear example of adaptive phenotypic variation that has been impacted by anthropogenic activities (11, 24, 25). Across the southern part of their coastal (i.e., noninterior) range in North America, Chinook display two primary phenotypes in the characteristics of their spawning migration (26). Premature-migrating Chinook enter freshwater from the ocean in a sexually immature state during the spring, migrate high into watersheds to near their spawning grounds, and hold over the summer in a fasted state while their gonads develop before spawning in the fall. In contrast, mature-migrating Chinook enter freshwater in a sexually mature state in the fall and migrate directly to their spawning grounds to spawn immediately (26). Although a suite of characteristics distinguishes premature- and mature-migrating Chinook (e.g., gamete maturation state and body fat content at freshwater entry, time between freshwater entry and spawning, etc.), freshwater entry date is commonly used as a proxy when more comprehensive measurements are not available (26, 27). Thus, the premature and mature migration phenotypes are commonly referred to as “spring-run” and “fall-run,” respectively, which will be the nomenclature used here. The spatial and temporal differences between the two migration types facilitate use of heterogeneous habitats, buffer populations against environmental variability, and provide variation upon which future evolution can act (2, 26, 28).
Many rivers historically hosted large numbers of both phenotypes (29, 30). However, because they rely on clean, cold water throughout hot summer months, spring-run Chinook are more vulnerable than fall-run Chinook to anthropogenic activities that affect river conditions such as logging, mining, dam construction, and water diversion (11, 13, 26, 29, 31). Consequently, in locations where both phenotypes existed historically, the spring-run phenotype has either dramatically declined in relative frequency or disappeared completely since the arrival of Europeans (24, 32). Despite their broad and well-recognized value [e.g., spring-run Chinook play important roles in the indigenous cultures of the Pacific Northwest (33–35), are widely considered to be the most desirable of any salmon for consumption due to their high fat content (36), and transport marine-derived nutrients higher into watersheds than fall-run Chinook (26, 37)], the widespread declines and extirpations of spring-run Chinook have been met with limited conservation concern. Previous research found that coastal (i.e., noninterior) spring-run and fall-run Chinook within a river usually exhibit little overall genetic differentiation and are more closely related to each other than to populations of the same phenotype from other watersheds (38, 39). This was interpreted to suggest the spring-run phenotype could rapidly reemerge from fall-run populations if favored by future conditions (e.g., habitat was restored) (13). Here we investigate the mechanism underlying the dramatic decline of the spring-run phenotype and its future recovery potential.
Results
Rapid Genetic Change from Strong Selection at a Single Locus Explains Phenotypic Shift in Rogue Chinook.
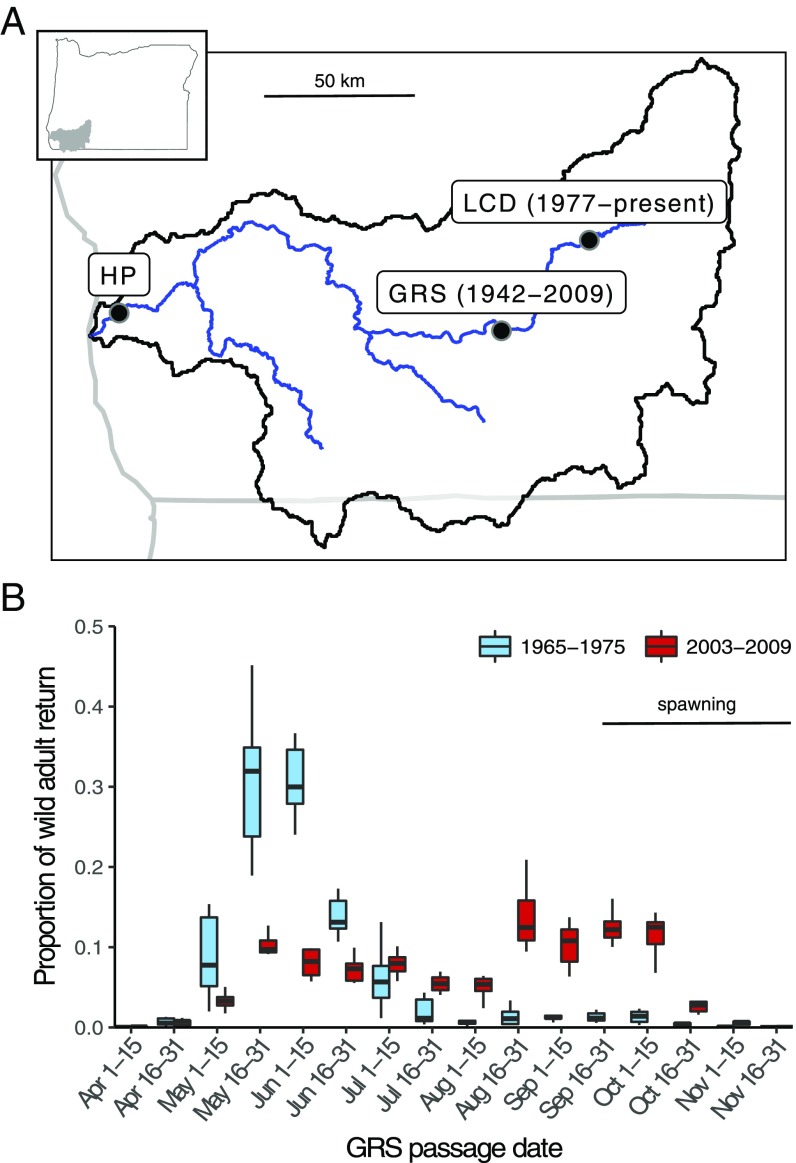
As one of the few remaining locations with a significant number of wild spring-run Chinook (40), the Rogue River in Oregon (Fig. 1A) presents a prime opportunity to examine the mechanism behind anthropogenically induced changes in Chinook migration characteristics. Before construction of Lost Creek Dam (LCD) in 1977, Chinook entered the upper basin (i.e., crossed the Gold Ray Fish Counting Station [GRS]) almost exclusively in the spring. After dam construction, the Chinook population experienced a phenotypic shift that, by the 2000s, had resulted in a striking increase in the number of individuals entering the upper basin in summer and fall, and a corresponding decrease in the number entering in the spring (Fig. 1B and Dataset S1, Table S1) (25). This shift occurred despite the majority of Chinook spawning habitat existing below the dam site (25). Because the dam altered downstream temperature and flow regimes (e.g., SI Appendix, Fig. S1) (25), this shift may have resulted from phenotypic plasticity, where postdam environmental conditions cue fish to migrate later. Alternatively, or in addition, the phenotypic shift may have resulted from rapid genetic evolution due to selection caused by postdam conditions.
Fig. 1.
Phenotypic change in Rogue River Chinook. (A) Map of Rogue River; dates indicate presence of features. (B) Bimonthly proportion of annual wild adult Chinook return across GRS before (1965–1975; 1968 was excluded due to incomplete data) and after (2003–2009; counts before 2003 included hatchery fish and GRS was removed in 2010) LCD construction; horizontal bar depicts Chinook spawn timing.
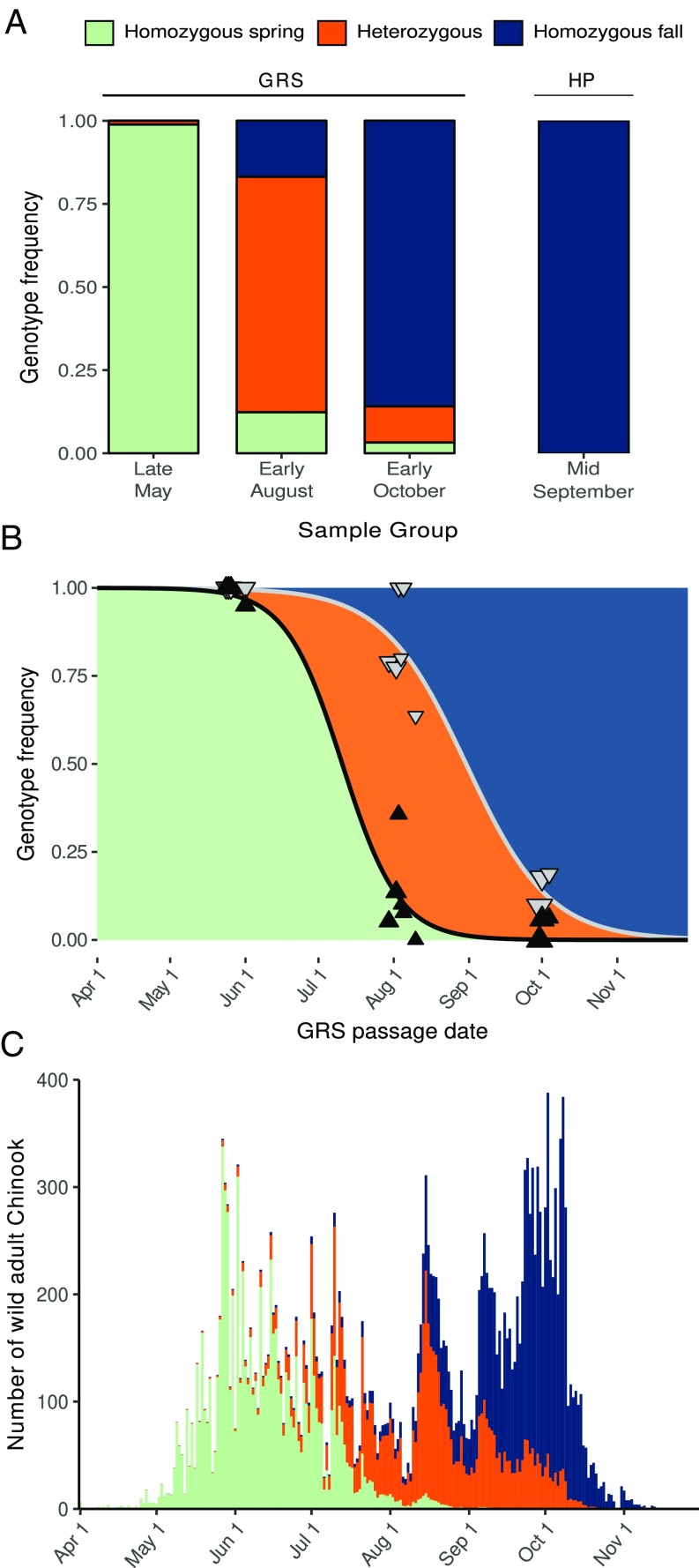
To begin investigating the shift in Rogue Chinook migration characteristics, we analyzed 269 fish that crossed GRS during three approximately week-long intervals in late May (n = 88), early August (n = 89), and early October (n = 92). Each fish was genotyped at a locus (the GREB1L region) previously found to be associated with migration type (i.e., spring-run or fall-run) across a wide array of Chinook populations (41, 42), using a newly developed marker (Materials and Methods and Dataset S1, Tables S2 and S5). Strikingly, the three groups had dramatically different genotype frequencies (Fig. 2A and Dataset S1, Table S3). All but one late May fish were homozygous for the allele associated with the spring-run phenotype (hereafter referred to as the spring-run allele), with the single heterozygote passing GRS on the last day of that collection period. The majority of early August fish were heterozygous. The early October group was overwhelming homozygous for the allele associated with the fall-run phenotype (hereafter referred to as the fall-run allele). However, a few early October individuals were heterozygous or homozygous for the spring-run allele. GRS is located ∼200 km from the river mouth (Fig. 1A) and thus the heterozygous and homozygous spring-run fish that passed GRS in early October may have entered freshwater earlier but held below GRS for an extended period before passage. We conclude that there is a strong association between GREB1L genotype and GRS passage date in Rogue Chinook and that heterozygotes have an intermediate migration phenotype.
Fig. 2.
Genetic basis of adult migration phenotype in Rogue River Chinook. (A) Stacked bar graph representing observed GREB1L genotype frequencies in GRS and HP sample groups. (B) Scatter plot representing observed GREB1L genotype frequencies in GRS samples across 13 collection days; triangles represent homozygous spring-run (black) and homozygous spring-run plus heterozygous (gray) genotype frequencies; triangle size is proportional to the number of fish analyzed each day (minimum 10, maximum 42). For fish that pass GRS during a specific time interval (e.g., a single day), the area below the black line represents the expected frequency of the homozygous spring-run genotype, the area between the lines represents heterozygotes, and the area above the gray line represents the homozygous fall-run genotype. (C) Stacked bar graph representing number of wild adult Chinook passing GRS in 2004; colors represent estimated proportion of each GREB1L genotype.
To further investigate the association between GREB1L and the migration characteristics of Rogue Chinook, we genotyped 38 fish collected in mid-September at Huntley Park (HP; Fig. 1A). HP is located on the mainstem Rogue ∼13 km from the river mouth so, unlike GRS samples, HP fish are unlikely to have been in freshwater for an extended period before collection. Strikingly, all HP samples were homozygous for the fall-run allele (Fig. 2A), a significantly lower homozygous spring-run/heterozygous frequency than GRS early October samples (P = 0.003; binomial distribution). This suggests that heterozygous and homozygous spring-run fish from GRS in early October likely entered freshwater earlier in the year but held for an extended period below GRS before crossing. We conclude that genotype at the GREB1L locus is a better predictor of general migration type (spring-run, fall-run, or intermediate) than passage date at GRS.
We next estimated the total number of fish of each genotype that passed GRS during the year our samples were collected by extrapolating the genotype frequencies across the entire run year. Briefly, we fit the genotype frequencies with sigmoidal curves to estimate the probability that a fish ascending GRS on any specific day would be each of the three possible genotypes (Fig. 2B). We then multiplied the observed number of individuals passing on each day by the genotype probabilities for the same day (Fig. 2C and Dataset S1, Table S1). Finally, we performed bootstrap resampling of the daily genotype data to determine 95% confidence intervals for this and subsequent analyses. The analysis suggested that, of the 24,332 individuals that passed GRS in 2004 (Dataset S1, Table S1), 8,561 (7,825–9,527) were homozygous for the spring-run allele, 6,636 (5,077–7,798) were heterozygous, and 9,135 (8,124–10,253) were homozygous fall-run. These abundance estimates correspond to homozygous spring-run, heterozygous, and homozygous fall-run genotype frequencies of 0.352 (0.322–0.392), 0.273 (0.209–0.320), and 0.375 (0.334–0.421), respectively, as well as a spring-run allele frequency of 0.488 (0.457–0.518) and a fall-run allele frequency of 0.512 (0.482–0.543). Notably, the estimated homozygous spring-run migration date distribution was strikingly similar to the empirical migration date distribution before LCD construction (Figs. 1B and 2C), suggesting the predam population was predominantly homozygous spring-run and the migration time of this genotype has not changed since dam construction. This was further supported by an analysis of 36 predam samples collected near the historical late-May/early-June GRS migration peak (Fig. 1B), all of which were homozygous for the spring-run allele (Materials and Methods and Dataset S1, Table S3). We conclude that the phenotypic shift after dam construction is explained by rapid allele and genotype frequency shifts at the GREB1L locus.
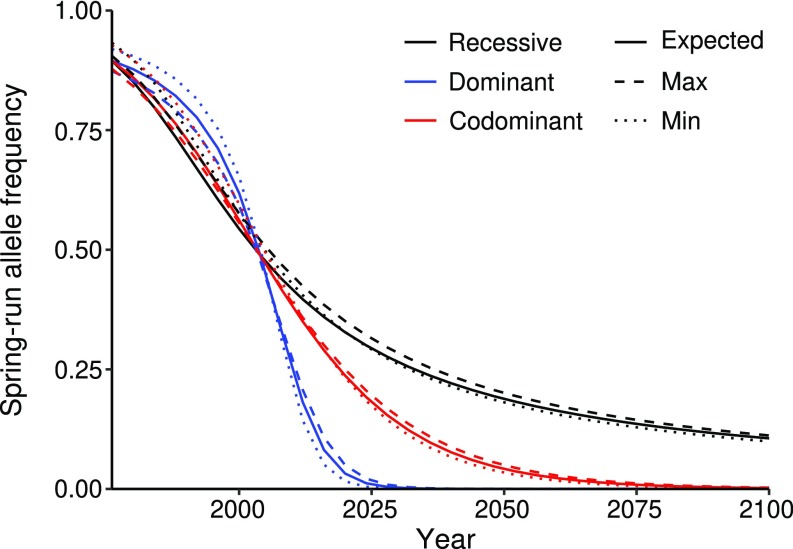
To explore selection regimes that could produce this genetic change in such a short time frame (approximately seven generations), we estimated the spring-run allele frequency before LCD and the selection coefficients required to reach the observed 2004 allele frequency under a simple model assuming the spring-run allele was either recessive, dominant, or codominant with respect to fitness (Materials and Methods) (21). Under the recessive scenario, heterozygous and homozygous fall-run genotypes have equal fitness (selection coefficients: sFF = sSF = 0, 0 ≤ sSS ≤ 1). Under the dominant scenario, heterozygous and homozygous spring-run genotypes have equal fitness (sFF = 0, 0 ≤ sSF = sSS ≤ 1). Under the codominant scenario, heterozygotes have an intermediate fitness (sFF = 0, sSF = 1/2sSS, 0 ≤ sSS ≤ 1). Applying the genotype probability distribution (Fig. 2B) to the predam fish counts (Fig. 1B) suggested a predam spring-run allele frequency of 0.895 (0.873–0.919), which the predam sample analysis (discussed above) supports as a reasonable estimate (Materials and Methods and Dataset S1, Table S3). Next, the modeling estimated selection coefficients for the homozygous spring-run genotype (sSS) of 0.367 (0.348–0.391), 0.646 (0.594–0.712), and 0.447 (0.424–0.480) under the recessive, dominant, and codominant scenarios, respectively. Furthermore, we explored the potential consequences of continued selection against the spring-run phenotype by extrapolating our modeling into the future. This predicted a spring-run allele frequency in 2100 of 0.106 (0.099–0.112), 3.24 × 10–11 (2.44 × 10–13 to 7.96 × 10–10), and 0.002 (0.001–0.003) under the recessive, dominant, and codominant scenarios, respectively (Fig. 3). Thus, our modeling demonstrates that selection strong enough to explain the rapid phenotypic and genotypic shifts could lead to loss of the spring-run allele in a relatively short time. We conclude that, under continual selection against the spring-run phenotype, the spring-run allele cannot be expected to persist unless it is recessive with respect to fitness.
Fig. 3.
Selection modeling in Rogue Chinook. Line graph representing the spring-run allele frequency over time under recessive, dominant, and codominant scenarios. Estimated spring-run allele frequencies in 1976 (1 y before LCD construction) and 2004 were used to determine selection coefficients for each scenario [recessive: sFF = sSF = 0, sSS = 0.367; dominant: sFF = 0, sSF = sSS = 0.646; codominant: sFF = 0, sSF = 1/2(sSS), sSS = 0.447]. The modeling assumes random mating and no genetic drift.
Ancient and Contemporary Klamath Chinook Reveal Hindered Spring-Run Restoration Potential.
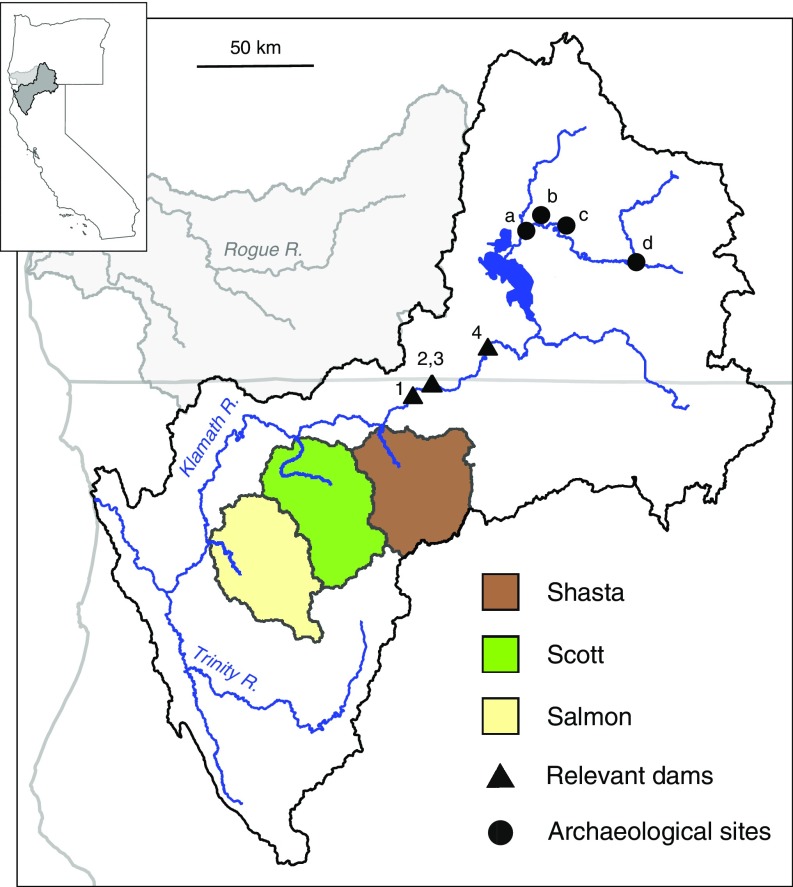
The Klamath River in northern California and southern Oregon (Fig. 4) presents an opportunity to empirically examine the consequences of longer-term selection against the spring-run phenotype. The Klamath historically hosted hundreds of thousands of adult spring-run Chinook annually, with the spring-run phenotype possibly exceeding the fall-run phenotype in frequency (30). While the fall-run phenotype remains relatively abundant (i.e., tens to hundreds of thousands of adults per year) (43), dam construction and habitat degradation beginning in the late 1800s led to severe declines in the spring-run phenotype, with virtually complete loss of wild spring-run Chinook in the mainstem and tributaries except the Salmon River (Fig. 4) (24, 44). In the last decade, Salmon River spring-run Chinook have ranged from ∼200–1,600 individuals (45) and are expected to be extirpated within 50 y (24). In 2021, the largest-scale dam removal project in history is scheduled to remove four dams in the upper basin (46) and reopen hundreds of miles of historical Chinook habitat inaccessible since 1912 (47) (Fig. 4). This dam removal provides an opportunity unprecedented in scale to restore extirpated populations, including spring-run Chinook (48). However, while historical documentation supports the presence of early-migrating Chinook in the upper Klamath (47), the extent to which above-dam populations relied on the same spring-run allele as the Rogue (discussed above) and other contemporary Chinook populations (41) (Materials and Methods and Dataset S1, Table S5) is unknown. Furthermore, since most contemporary Klamath populations have lost the spring-run phenotype, it is unclear which, if any, are acting as reservoirs of the spring-run allele and therefore could serve as a source population for restoration of spring-run Chinook in the upper basin.
Fig. 4.
Map of the Klamath Basin. Klamath dams scheduled for removal in 2021: 1, Iron Gate; 2, Copco 1; 3, Copco 2; and 4, J. C. Boyle. Archaeological site locations of ancient samples: a, Williamson River Bridge; b, Bezuksewas Village; c, Kawumkan Springs Midden; and d, Beatty Curve. R., River.
To investigate the genetic composition of historical upper Klamath Chinook, we genotyped nine Chinook samples collected from four archaeological sites in the upper basin known to be historically important fishing places for Klamath peoples (49) (Fig. 4). The samples ranged in age from post-European contact to ∼5,000 y old and, based on the presence of all body parts in the archaeological sites, were likely caught locally as opposed to being acquired through trade (49–51) (Table 1). Strikingly, three of the locations had only homozygous spring-run samples, while the remaining location had only homozygous fall-run samples (Table 1). The spring-run sample locations are known to have been occupied by humans in the spring or throughout the year and are also near major cold-water input sources [suitable oversummering habitat for spring-run Chinook (52)], whereas the fall-run samples came from a location with a documented historical fall fishery (53). We conclude that the upper basin harbored the same allelic variants as contemporary populations, and these spring-run alleles are expected to be necessary for restoration of the spring-run phenotype in the upper basin (discussed above) (41).
Table 1.
Ancient upper Klamath Chinook sample information and genotyping results, listing Simon Fraser University (SFU) sample identification number and Oregon state site numbers
| SFU sample ID | Site name (no.) | Age* | Genotype |
| SBC01 | Beatty Curve (35KL95) | AD 1860–20th century | Homozygous fall-run |
| SBC13 | Beatty Curve (35KL95) | AD 1860–20th century | Homozygous fall-run |
| SBC14 | Beatty Curve (35KL95) | AD 1860–20th century | Homozygous fall-run |
| SBC26 | Bezuksewas Village (35KL778) | AD 1390–1860 | Homozygous spring-run |
| SBC53 | Bezuksewas Village (35KL778) | AD 1390–1860 | Homozygous spring-run |
| SBC36 | Kawumkan Springs Midden (35KL9-12) | Unknown (likely before AD 1860) | Homozygous spring-run |
| SBC33 | Kawumkan Springs Midden (35KL9-12) | 3160–3110 BC | Homozygous spring-run |
| SBC42 | Williamson River Bridge (35KL677) | 450 BC–20th century | Homozygous spring-run |
| SBC43 | Williamson River Bridge (35KL677) | 450 BC–20th century | Homozygous spring-run |
To test if lower (i.e., below-dam) Klamath populations that have lost the spring-run phenotype are serving as reservoirs of the spring-run allele, we genotyped juvenile Chinook collected from the Shasta River (Fig. 4) throughout the juvenile out-migration season in 2008–2012 (Dataset S1, Table S4) (54). The Shasta, where spring-run Chinook were last observed in the 1930s (30), is a major Klamath tributary that shares many environmental characteristics with the habitat above the dams (e.g., large spring water input sources, dry climate, etc.) (55). Thus, Shasta Chinook may contain additional adaptive variation suitable for the upper Klamath, which makes them an attractive restoration stock candidate (56). Strikingly, out of the 437 successfully genotyped individuals, only 2 were heterozygous and all others were homozygous for the fall-run allele, corresponding to a spring-run allele frequency of 0.002 (binomial distribution 95% CI: 3 × 10−4 to 0.008; Table 2). This is at least an order of magnitude below the expected frequency if the spring-run allele was recessive with respect to fitness (Discussion; e.g., Fig. 3) (21) and, interestingly, very similar to the codominant scenario in our Rogue Chinook modeling (0.002 vs. 0.002; Fig. 3) after a similar period of selection against the spring-run phenotype (late 1800s-early 2000s vs. 1977–2100). Given the recent annual adult returns to the Shasta River (mean during the years our samples were spawned: 5486) (57) and Ne/N ratios in Chinook (58), such frequencies suggest the spring-run allele is highly vulnerable to complete loss through continued selection and/or genetic drift (Discussion). We conclude the contemporary Shasta Chinook population cannot be considered a sustainable reservoir of the spring-run allele.
Table 2.
Klamath Chinook smolt information and genotyping results
| River | Date last spring-run Chinook observed | No. | Year(s) | Homozygous spring-run | Heterozygous | Homozygous fall-run | Spring-run allele frequency |
| Shasta | 1930s* | 437 | 2008–2012 | 0 | 2 | 435 | 0.002 (3 × 10−4 to 0.008)† |
| Scott | 1970s | 425 | 2007–2013 | 0 | 2 | 423 | 0.002 (3 × 10−4 to 0.008)† |
| Salmon | Present | 116 | 2017 | 14 | 19 | 83 | 0.20 |
Spring-run Chinook were still observed just upstream of the Shasta River mouth at Iron Gate Dam into the 1970s.
Ninety-five percent CI calculated using binomial probability distribution.
To test if locations with disparate environmental conditions are acting as reservoirs of the spring-run allele, we genotyped Chinook juveniles collected over a similar time range in the Scott River (Fig. 4 and Dataset S1, Table S4), a Klamath tributary that exhibits a hydrologic regime driven by surface water, which is typical of the lower Klamath basin but very different from the Shasta River (55). The spring-run phenotype was last observed in the Scott River in the 1970s (30). We also genotyped 116 juveniles from the Salmon River (see above; Fig. 4 and Dataset S1, Table S4) as a positive control. Out of 425 successfully genotyped Scott samples, we found only two heterozygotes (spring-run allele frequency: 0.002; binomial distribution 95% CI: 3 × 10−4 to 0.008), whereas the Salmon River samples had an overall spring-run allele frequency of 0.20 (Table 2), corresponding well with spring-run phenotype frequency estimates based on annual dive and carcass surveys in the Salmon River (45, 59). We conclude the Scott River is also not acting as a sustainable reservoir of the spring-run allele, and diverse environments are susceptible to rapid loss of the spring-run allele upon extirpation of the spring-run phenotype.
Discussion
Phenotypic variation in natural populations facilitates resilience in heterogeneous or variable environments (2, 5). The genetic architecture of natural phenotypic variation, though usually unknown, is typically assumed to be complex (i.e., polygenic and influenced by the environment) (60). A recent study identified a single locus (the GREB1L region) associated with migration type in Chinook as well as the closely related species steelhead (Oncorhynchus mykiss) (41). However, the relatively low marker resolution and poor phenotypic information in the Chinook analysis obscured the strength of association and phenotype of heterozygotes (41). Our analysis of samples with more detailed phenotypic information [i.e., specific migration dates at GRS and HP (Results and Dataset S1, Table S3) as well as the lower South Fork Trinity (Materials and Methods and Dataset S1, Table S5)] using a new marker identified through a high-resolution, multipopulation analysis of GREB1L (Materials and Methods and Dataset S1, Tables S2 and S5) suggests that (i) the association of migration characteristics with variation at GREB1L is extremely robust and (ii) heterozygotes have an intermediate migration phenotype (Fig. 2A). Therefore, while phenotypic variation within each genotype (e.g., precise freshwater entry and spawning dates) is yet to be explained, general migration type (i.e., premature/spring-run or mature/fall-run) appears to have a relatively simple genetic architecture (i.e., a locus of very large effect). Furthermore, the association of a single haplotype with the spring-run phenotype in diverse locations (Materials and Methods and Dataset S1, Table S5) supports previous evidence that spring-run alleles arose from a single evolutionary event and cannot be expected to readily reevolve (41, 61). Thus, important natural phenotypic variation can be underpinned by relatively simple modes of inheritance and rare allelic evolutionary events.
Selection results from the balance between benefits and costs of specific phenotypes (62), and anthropogenic habitat alteration can potentially disrupt this balance (9, 12, 63, 64). The large and rapid decline in the Rogue spring-run phenotype and allele frequency suggests strong selection against spring-run Chinook after LCD construction. Furthermore, our modeling demonstrates that such selection, if sustained, could rapidly result in complete loss of the spring-run allele. A main benefit of the spring-run phenotype is thought to be access to exclusive temporal and/or spatial habitat, while a major cost is reduced gametic investment (e.g., smaller egg size) because energy must be dedicated to maintenance and maturation while fasting in freshwater (26, 65). River flow regimes can be a major driver of life history evolution in aquatic systems (12, 64), and LCD altered downstream temperature and flow in a way that may allow fall-run Chinook access to spawning habitat that was previously exclusive to spring-run Chinook (25). An analysis of carcass samples from the Rogue revealed substantial spatial and temporal overlap in spawning distributions of all three genotypes (SI Appendix, Fig. S2 and Dataset S1, Table S3), supporting the hypothesis that anthropogenically induced habitat alterations have reduced the historical benefit of the spring-run phenotype, contributing to its decline. Regardless of exact mechanisms, our results provide a clear example where anthropogenic factors induced rapid phenotypic change through genetic evolution as opposed to phenotypic plasticity.
Population genetics theory and our selection modeling predicts that, for loci with a large phenotypic effect, alleles promoting negatively selected phenotypes will be eliminated from a population unless they are masked in the heterozygous state (i.e., recessive with respect to fitness) (21). The intermediate migration phenotype of heterozygotes, in combination with typical lower river conditions at intermediate times (i.e., conditions inhospitable to salmonids), suggests their fitness will be at least somewhat different, and likely lower, than that of fall-run Chinook in most locations (66). Therefore, where the spring-run phenotype is lost, spring-run alleles cannot be expected to be maintained in the heterozygous state. This prediction is empirically supported by our results from the Shasta and Scott Rivers where, based on adult run size estimates during the years our samples were spawned (∼5,000 per year in each river) (57, 67), the observed spring-run allele frequency (0.002) would correspond to an average of ∼20 heterozygous adults per year in each river. Given that adult Chinook have highly variable reproductive success (58), such a low frequency makes the spring-run allele extremely vulnerable to complete loss through genetic drift regardless of selection (21) (something that may conceivably have already occurred, given our samples were collected several years ago). Notably, while habitat alterations extirpated the spring-run phenotype from the Shasta and Scott, the total Chinook census sizes (i.e., adults of any migration type) of both rivers are considered robust (57, 67). Thus, both theory and empirical evidence suggest heterozygotes cannot be expected to act as a sustainable reservoir for spring-run alleles, and important adaptive variation can be vulnerable to loss from human impacts regardless of total population size.
Adaptive variation is likely important to the success of species restoration efforts (56, 68). The planned removal of Klamath dams provides an opportunity to restore Chinook to historical habitat that is unprecedented in scale and provides a lens through which to evaluate the challenges of recovering the spring-run phenotype. Historical documentation (47) and our analysis of ancient samples suggest both migration types existed above the dams. Furthermore, an evaluation of the upper basin environment suggests habitat suitable for both phenotypes will be available after dam removal (48, 52, 69), with some locations likely favoring the earlier migration and spawning times of the spring-run phenotype (52). While abundant Klamath fall-run Chinook are likely to naturally recolonize the upper basin, the current scarcity of the spring-run phenotype and allele in the Klamath will likely hinder natural recolonization of spring-run Chinook. Similarly, natural recolonization via straying from out-of-basin populations is improbable on short timescales and tenuous on longer timescales given the ongoing declines and extirpations of spring-run Chinook throughout their range. Human-facilitated restoration may also be challenged by limited options for appropriate source populations. The Shasta River’s environmental similarities with the upper basin (55, 69) would have made it an attractive candidate if spring-run alleles were more abundant (52, 56, 70). Salmon River spring-run Chinook are severely depressed in number (24, 45, 52) and may lack other adaptive variation important for the upper basin due to the major environmental differences between the locations (55, 70). Spring-run alleles are present in a within-basin hatchery population (i.e., Trinity River Hatchery), but hatchery salmonids are partially domesticated, have reduced reproductive success in the wild, and can negatively impact wild populations (71–74). Introducing an out-of-basin wild stock [e.g., Rogue spring-run Chinook, the most proximate spring-run population to the Klamath (Figs. 1A and 4 and refs. 41 and 52)] could be an option but may also be challenged by incompatibilities stemming from local adaptation (52, 70). Given that wild spring-run Chinook are expected to disappear from the lower Klamath within 50 y and are declining across their range (24), the current challenges of restoring spring-run Chinook upon Klamath dam removal are a preview of even greater challenges that will be faced in future spring-run Chinook restoration projects if the spring-run phenotype continues to decline. Thus, the decline and loss of adaptive variation due to anthropogenic habitat alterations can hinder the ability to recover previous characteristics and restore wild populations.
Humans impact phenotypic variation across taxa and traits (9, 10) through diverse means (e.g., hunting and fishing, habitat modification, climate change, etc.; refs. 20, 64, and 75–78). While a substantial body of work has discussed the theoretical consequences of human-driven selection (5, 10, 12, 15, 18, 20, 75), empirical explorations have been challenged by the historical difficulty of uncovering the genetic basis of natural phenotypic variation. Although recent work has begun to characterize the genetic basis of phenotypic variation and identified large-effect loci in species of conservation concern (79–84), empirical work evaluating the consequences of anthropogenic selection for the long-term persistence and/or recovery potential of adaptive variation is still rare. The results presented here demonstrate that human-induced phenotypic change can have severe consequences with respect to the ability of previous variation to reemerge. Given the broad impacts of anthropogenic activities on phenotypic diversity, future research examining the consequences for the persistence and recovery of variation in other species will be important for informing conservation and management actions.
Although this study provides important insights into the genetics and conservation of spring-run Chinook, additional information would be useful to further inform conservation and restoration actions. In the Klamath, more extensive evaluation of the adaptive suitabilities of potential restoration source stocks (e.g., Salmon, Trinity, and Rogue River spring-run Chinook) would be valuable. On a broader scale, work characterizing the distribution of spring-run alleles, especially in populations that appear to lack the spring-run phenotype, is needed to identify if and where the genetic potential for the phenotype still exists (e.g., in heterozygotes) (85). Ongoing monitoring of allele frequencies will likely also be essential, as spring-run alleles may be present but in decline. Importantly, a better understanding of the ecology (i.e., spawning and rearing locations), phenotype (i.e., range of river entry and spawning dates, fecundity, etc.), and fitness (i.e., relative reproductive success) of each genotype would be useful for understanding selection mechanisms and targeting conservation strategies, as would a thorough exploration of the roles hatchery fish may play in the decline or persistence of spring-run alleles in wild populations. Given that spring-run Chinook have historically been prominent on the southernmost edge of the species range (26), the phenotype may carry substantial adaptive importance for more northern locations under climate change (86). A more extensive evaluation of this would be valuable. Finally, although the genetic marker used here is currently the best available to distinguish between migration types (see Dataset S1, Table S5 for marker comparison), continued marker development [e.g., identification of the causative polymorphism(s)] would reduce the potential for misclassification of migration type due to factors such as rare recombination events.
The combination of results from this study provides important insights into the mechanisms and consequences of phenotypic change induced by anthropogenic habitat alteration. First, our results demonstrate that natural phenotypic variation can have a relatively simple genetic architecture and that anthropogenically induced phenotypic change can be caused by rapid genetic evolution from strong selection at individual loci. Furthermore, our results (both modeled and empirical) demonstrate such a situation can lead to the rapid loss of important adaptive alleles, including from populations that are healthy from a total population size perspective. In cases where adaptive alleles are the product of mutational events that are very rare from an evolutionary perspective [such as the spring-run allele in Chinook (41, 42)], their loss will create a major challenge for future restoration as well as limit resilience and evolutionary potential. Taken together, our results highlight the need to conserve and restore critical adaptive variation before the potential for recovery is lost.
Materials and Methods
GREB1L Marker Discovery.
Previous research identified a significant association between variation in the GREB1L region and adult migration type (i.e., premature or mature) in both Chinook and steelhead (O. mykiss) (41, 42, 87). Although the strongest associated SNP in Chinook [position 569200 on scaffold79929e (41)] had a large allele frequency difference between premature and mature migrating populations in several locations (41), this association was notably weaker than observed in steelhead. We reasoned the weaker association could have resulted from technical reasons (e.g., lower SNP resolution of the Chinook analysis) as opposed to biological reasons (e.g., smaller influence of the GREB1L locus in Chinook compared with steelhead).
We therefore used capture baits to isolate and sequence the GREB1L region in 64 Chinook samples (across eight locations in California, Oregon, and Washington; Dataset S1, Table S5) from the previous association study (41) for additional SNP identification and association testing. The two most strongly associated SNPs identified by this process (positions 640165 and 670329 on scaffold79929e) were ∼30 kb apart just upstream of GREB1L and revealed much stronger associations than the most strongly associated SNPs from the previous study (41) (Dataset S1, Table S5). These results confirm that the relatively weak association between GREB1L and migration type previously observed in Chinook (compared with steelhead) (41) was due to lower SNP resolution as opposed to a smaller influence on phenotype.
SNP Assay Design and Validation.
We designed TaqMan-based genotyping assays for the two newly discovered SNPs to facilitate rapid and inexpensive genotyping of the GREB1L locus across large numbers of samples. Approximately 300 bp of Chinook sequence surrounding each SNP (Dataset S1, Table S2) was submitted to the Custom TaqMan Assay Design Tool (Applied Biosystems) to generate primer and probe sequences for each SNP. Additional polymorphic sites in the surrounding sequence identified in the capture sequencing were masked to avoid primer or probe design across these sites. Assays were run using 5 µL 2× TaqMan Genotyping Master Mix, 0.5 µL 20× genotyping assay [final concentrations of 900 nM (primers) and 200 nM (MGB probes)], 2.5 µL DNA-grade water, and 2 µL sample DNA for each reaction. Reporter dyes were Vic and Fam. Each 96-well SNP assay plate also contained one positive control for each genotype (taken from samples used in capture sequencing) and two negatives controls substituting water or low TE (0.1 mM EDTA and 10 mM Tris, pH 8.0) for DNA. No negative controls ever amplified. Each SNP assay was run separately (not multiplexed) for each sample. The assays were run on either a Chromo4 or QuantStudio-3 Real Time PCR machine for 10 min at 95 °C followed by 40 cycles of 15 s at 95 °C and 1 min at 58–59 °C (snp640165) or 62–64 °C (snp670329).
SNP assays were validated with the samples used for capture sequencing. All results were consistent with sequencing-based genotype calls (Dataset S1, Table S5). Our genotyping results from GRS and HP (Fig. 2A and Dataset S1, Table S3) serve as further validation of the assays in the Rogue River. For additional validation in the Klamath, we genotyped 62 samples from Chinook with known migration dates through a weir on the lower South Fork Trinity River (Dataset S1, Table S5). All South Fork Trinity samples phenotyped as spring-run (i.e., weir passages dates between mid-May and end of July) were homozygous for the spring-run allele except for a single heterozygote collected on July 31. All samples phenotyped as fall-run (i.e., weir passages dates between mid-October and mid-November) were homozygous for the fall-run allele (Dataset S1, Table S5).
Contemporary Sample Collection and DNA Extraction.
Rogue GRS samples were obtained from wild Chinook salmon, defined as lacking an adipose fin clip, that returned to spawn in the Rogue River during 2004. Fish were trapped by Oregon Department of Fish and Wildlife (ODFW) personnel at a fish-count station (GRS) located at Gold Ray Dam (erected in 1941). Tissue was sampled from the operculum of each fish and placed in 100% ethanol for storage and subsequent DNA extraction using Qiagen DNeasy kits following the manufacturer’s protocols. Following sampling, fish were released unharmed upstream of the dam barrier. Approximately 300 samples were evenly obtained across three temporal sampling windows (May 24 to June 1; July 30 to August 10; and September 30 to October 4) that targeted spring, intermediate, and fall runs.
Rogue HP samples were collected from wild Chinook caught in beach seines near HP in September 2014 (Dataset S1, Table S3). Rogue pre-LCD samples were collected in the lower river during May of 1975 and 1976 (Dataset S1, Table S3) and stored in the ODFW scale archive. Rogue carcass samples were collected during ODFW spawning surveys of the upper Rogue in 2014 (Dataset S1, Table S3). Juvenile Chinook from the Salmon, Shasta, and Scott Rivers in the Klamath Basin were caught in screwtraps during smolt outmigration across several years (Dataset S1, Table S4) (54). South Fork Trinity samples were collected from live adult Chinook during passage through Sandy Bar weir, except for three samples that were collected at Forest Glen (Dataset S1, Table S5). Fin clip (HP, Rogue carcass, and Salmon) or scale (Rogue pre-LCD, Shasta, and Scott) samples were collected, dried on filter paper, and stored at room temperature. DNA was extracted using a magnetic bead-based protocol (88) and stored at −20 °C.
Archaeological Sample Collection and DNA Extraction.
The archaeological samples were recovered from archaeological excavation projects led by research teams from the University of Oregon Museum of Natural and Cultural History between the late 1940s and the late 2000s (49, 89). The four sites represent fishing camps or year-round villages occupied by ancestral people to the Klamath Tribes of Oregon (Table 1 and Dataset S1, Table S4). Three sites are located on the Sprague River: Kawumkan Springs Midden (90), Beatty Curve (89), and Bezuksewas Village (91). A fourth, Williamson River Bridge (92), is located near the confluence of the Williamson and Sprague Rivers (Fig. 4). The sites range in age from 7,500 y ago to the early 20th century (49). Because of severe stratigraphic disturbance by burrowing rodents, the materials can typically only be assigned to very broad time periods (Table 1 and Dataset S1, Table S4). Deposits were assigned to AD 1860 or later based on presence of artifacts of Euro-American origin, as AD 1860 marks the establishment of Fort Klamath and time of sustained Euro-American contact in the upper Klamath Basin. Klamath people continued to fish and occupy the Beatty Curve and Williamson River Bridge site locations into the 20th century, so the end date is uncertain. All other ages were based on multiple radiocarbon samples (49), calibrated using OxCal v4.2 (93).
Previous projects (49) assigned the fish remains to the finest taxon possible using modern reference skeletons from known species. To obtain species-level identification, a sample of salmonid remains was sent to the dedicated Ancient DNA Laboratory in the Department of Archaeology at Simon Fraser University, Burnaby, BC, Canada. Twelve vertebra samples (nine Chinook and three steelhead as controls) were included in this study (Dataset S1, Table S4). Samples were chemically decontaminated through submersion in commercial bleach (4–6% sodium hypochlorite) for 10 min, rinsed twice with ultrapure water, and UV-irradiated for 30 min each on two sides. Bones were crushed into powder and incubated overnight in a lysis buffer (0.5 M EDTA, pH 8.0, 0.25% SDS, and 0.5 mg/mL proteinase K) in a rotating hybridization oven at 50 °C. Samples were then centrifuged and 2.5–3.0 mL of supernatant from each sample was concentrated to <100 μL using Amicon Ultra-4 centrifugal filter devices (10 kDa, 4 mL; Millipore). Concentrated extracts were purified using QIAquick spin columns based on previously developed methods (94, 95); 100 μL of DNA from each sample was eluted from QIAquick columns for PCR amplifications.
Species identification was accomplished by targeting salmonid mitochondrial d-loop (249 bp) and cytochrome b (cytb) (168 bp) fragments as previously described (96). Successfully amplified products were sequenced at Eurofins MWG Operon Ltd. using forward and/or reverse primers. The resulting sequences were compared with GenBank reference sequences through the BLAST application to determine their closest match, and species identifications were confirmed through multiple alignments of the ancient sequences and published salmonid reference sequences conducted using ClustalW (97) through BioEdit (98), as well as the construction of neighbor-joining phylogenetic trees using Kimura’s 2-parameter model in the Mega 6.0 software program (99). Nine of the 12 samples were identified as Chinook (Dataset S1, Table S4) and the remaining three as steelhead.
Rogue and Contemporary Klamath Genotyping.
After DNA extraction, samples were genotyped using the assays (snp640165 and snp670329; Dataset S1, Table S2) and qPCR protocol described above. All samples were tested at both SNPs, and a genotype call (homozygous spring-run, heterozygous, or homozygous fall-run; Dataset S1, Tables S3 and S4) was made only if both SNPs were successfully genotyped and consistent with each other. The causative polymorphism(s) in the GREB1L region are currently unknown, so requiring successful and consistent calls at both associated SNPs provides greater confidence that the genotype (homozygous spring-run, heterozygous, or homozygous fall-run) was not miscalled due to biological factors such as rare recombination events and is more conservative than using a single SNP. Of the 1,390 samples tested from live-caught fish, 1,333 (95.9%) successfully genotyped at both SNPs, 31 (2.2%) failed at one SNP, and 26 (1.9%) failed at both SNPs. Of the 96 Rogue River carcass samples tested, 86 (89.6%) successfully genotyped at both SNPs, 2 (2.1%) failed at one SNP, and 8 (8.3%) failed at both SNPs. Of the successful live and carcass samples (1,419 total), 1,406 (99%) had the same genotype call at both SNPs, indicating near perfect linkage disequilibrium (LD) between the SNPs. The remaining 13 samples [all from the Rogue (2.9% of successfully genotyped Rogue samples) and mostly from the GRS August group] had a homozygous genotype at one SNP and a heterozygous genotype at the other (Dataset S1, Table S3). Because we do not know which, if either, SNP is in stronger LD with the causative polymorphism(s), these samples were called as ambiguous (Dataset S1, Table S3) and excluded from further analyses.
Ancient Klamath Genotyping.
Multiple sealed aliquots of extracted ancient DNA from 12 archaeological samples were shipped from Simon Fraser University to the University of California, Davis on dry ice. Nine samples were from Chinook and the remaining three were from steelhead, which are known to have the same alleles as fall-run Chinook at the two SNPs based on the O. mykiss reference genome (100). Genotyping was conducted under blinded conditions with respect to species, location, and age. SNP assays were run using 10 µL 2× TaqMan Genotyping Master Mix, 1 µL 20× genotyping assay [final concentrations of 900 nM (primers) and 200 nM (MGB probes)], 5 µL DNA-grade water, and 4 µL of sample DNA diluted in low TE (either 1:10 or 1:50) for each reaction. The assays were run on a QuantStudio-3 Real Time PCR machine for 10 min at 95 °C followed by 80 cycles of 15 s at 95 °C and 1 min at 58 °C (snp640165) or 64 °C (snp670329). Fluorescence after each amplification cycle was measured and checked to prevent erroneous calls due to high cycle number. All plates contained positive controls for each genotype diluted at ratios similar to the unknown samples and at least 12 negative controls substituting the low TE used in sample dilutions in place of DNA. No amplification was ever observed in a negative control in either the ancient sample plates or any plates containing contemporary samples. All results were replicated using separately sealed aliquots on different days. Due to the extremely high LD in contemporary samples and the precious nature of the ancient samples, genotypes were called even if only one SNP was successfully genotyped (Dataset S1, Table S4). Requiring both SNPs to be successfully genotyped would have reduced the number of ancient Chinook samples with a migration type call from nine to five (two fall-run and three spring-run; Dataset S1, Table S4) but would not have altered our conclusions.
Curve Fitting and Selection Modeling.
Sigmoidal curves were fit to the genotype frequencies measured for each collection day at GRS (Fig. 2B and Dataset S1, Table S3). The curves were fit using the nonlinear least squares (nls) function in R (101) for a sigmoidal model, optimizing for b and m values: S = 1/(1 + e−b(x − m)). The R command used was nls(gf∼1/(1 + exp(−b * (x − m))), weights = w, start = list(b = (−0.01), m = 90)) where gf was either a list of the homozygous spring-run or homozygous spring-run plus heterozygous frequencies (a.k.a. 1 - homozygous fall-run frequency) with each frequency corresponding to a specific sample collection day, x was a list of numeric dates (April 1 was set to day 1) corresponding to each collection day, and w was the number of samples from each day. The resulting equations represent the estimated probability of each genotype on any given day (Fig. 2B), and were applied to daily empirical GRS fish counts from 2004 to estimate allele frequencies in 2004.
Pre-LCD allele frequencies were estimated by applying the genotype probability distribution calculated from the 2004 GRS samples (Fig. 2B) to the average biweekly fish counts (using mean probability across the biweekly bin) in the decade before LCD construction (Fig. 1B, see ref. 25) and resulted in a pre-LCD spring-run allele frequency estimate of ∼90% (Results). This approach was used because a pre-LCD sample set adequate to perform a direct estimate of the pre-LCD allele frequencies (e.g., pre-LCD samples collected at GRS throughout the migration season) was not available. However, this approach assumes that the relationship between GREB1L genotype and GRS passage date was not substantially different pre- and post-LCD. If this assumption is inaccurate (e.g., the association of GREB1L with GRS passage date was weaker in the pre-LCD environment), the pre-LCD population may have had a spring-run allele frequency significantly lower than 90%.
We investigated this possibility by genotyping 36 pre-LCD adult Chinook sampled in May (mean date May 20) from the lower Rogue (mean river mile 17) at the GREB1L locus (Dataset S1, Table S3). Based on measured migration rates of Rogue Chinook (25), these fish would likely have passed GRS near or somewhat after the pre-LCD migration peak in late May/early June (Fig. 1B). Strikingly, all 36 samples were homozygous for the spring-run allele (Dataset S1, Table S3). This demonstrates that pre-LCD individuals that passed GRS around the spring migration peak overwhelmingly contained the spring-run allele and, since very few pre-LCD individuals passed GRS later in the year, suggests our pre-LCD spring-run allele frequency is unlikely to be an overestimate. Furthermore, because the curves are fit to genotype frequencies from post-LCD conditions where heterozygotes are likely more frequent, the pre-LCD allele frequency results likely underestimate the true spring-run allele frequency before LCD. Thus, the true change in allele frequency after LCD is probably somewhat greater than what is estimated here, and therefore our estimated allele frequencies and selection coefficients are likely conservative.
The strength of selection against the spring-run phenotype [i.e., the homozygous spring-run selection coefficient (sSS)] was estimated by calculating values of sSS that explain the estimated change in spring-run allele frequencies between pre-LCD and 2004 using the equation p′ = (sSS p2 + sSF p(1 − p)/(sSS p2 + sSF 2p(1 − p) + sFF (1 − p)2) (21), where sxx is the selection coefficient of each genotype, p is the spring-run allele frequency in the current generation, and p′ is the spring-run allele frequency in the next generation. The estimated pre-LCD spring-run allele frequency was used as the starting value of p, and the equation was run recursively using the p′ value from the current run as the next value of p to find values of sSS that resulted in the estimated 2004 spring-run allele frequency after seven generations (assuming 4-y generations). Calculations were conducted under three relative fitness scenarios: recessive (sSF = sFF), dominant (sSS = sSF), and codominant (sSS = 2sSF). The homozygous fall-run genotype was always assumed to have the lowest selection coefficient (sFF = 0). This approach assumes Hardy–Weinberg Equilibrium (HWE), which is probably violated because the slightly earlier mean spawning date of spring-run Chinook likely creates some level of assortative mating (e.g., Fig. 2C and SI Appendix, Fig. S2). Under assortative mating, the overrepresentation of homozygous spring-run individuals could lead to an even more rapid decrease in the spring-run allele frequency because homozygous spring-run experiences the strongest selection in our modeling. Thus, assuming HWE likely produces conservative selection coefficient and future allele frequency estimates.
Supplementary Material
Acknowledgments
We thank C. Adams, C. Bean, R. Bowden, J. Bull, B. Chesney, A. Corum, J. Crawford, M. Johnson, D. Jacobson, L. Ketchum, J. Minch, K. O’Malley, M. Pepping, B. Quinter, S. Richardson, T. Satterthwaite, T. Soto, and P. Tronquet for help with sample acquisition; T. Satterthwaite for GRS sampling design and fish count data; D. Van Dyke and S. Clements for valuable feedback on initial Rogue results; J. Hamilton for advice on references; R. Peak for assistance with map visualization; and M. Hereford and R. Waples for valuable comments on an earlier version of the manuscript. We also acknowledge the help and support of the Klamath Tribes, particularly P. Chocktoot, Jr. and L. Dunsmoor, as well as P. Endzweig, T. Connolly, D. Jenkins, and J. Erlandson (Museum of Natural and Cultural History, Eugene, Oregon) for facilitating access to archaeological fish remains. Partial funding for this work was provided by the Gordon and Betty Moore Foundation through a Science Capacity Grant to the Wild Salmon Center. Part of the ancient DNA work was funded by National Oceanic and Atmospheric Administration Contract AB133F09CQ0039, through the efforts of J. Simondet and I. Lagomarsino with help from J. Hamilton and B. Tinniswood. Additional support for the ancient DNA work was provided by Social Sciences and Humanities Research Council of Canada grants (to D.Y.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811559115/-/DCSupplemental.
References
- 1.Braun DC, Moore JW, Candy J, Bailey RE. Population diversity in salmon: Linkages among response, genetic and life history diversity. Ecography. 2016;39:317–328. [Google Scholar]
- 2.Carlson SM, Satterthwaite WH. Weakened portfolio effect in a collapsed salmon population complex. Can J Fish Aquat Sci. 2011;68:1579–1589. [Google Scholar]
- 3.Greene CM, Hall JE, Guilbault KR, Quinn TP. Improved viability of populations with diverse life-history portfolios. Biol Lett. 2010;6:382–386. doi: 10.1098/rsbl.2009.0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilborn R, Quinn TP, Schindler DE, Rogers DE. Biocomplexity and fisheries sustainability. Proc Natl Acad Sci USA. 2003;100:6564–6568. doi: 10.1073/pnas.1037274100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mimura M, et al. Understanding and monitoring the consequences of human impacts on intraspecific variation. Evol Appl. 2016;10:121–139. doi: 10.1111/eva.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore JW, McClure M, Rogers LA, Schindler DE. Synchronization and portfolio performance of threatened salmon. Conserv Lett. 2010;3:340–348. [Google Scholar]
- 7.Schindler DE, et al. Population diversity and the portfolio effect in an exploited species. Nature. 2010;465:609–612. doi: 10.1038/nature09060. [DOI] [PubMed] [Google Scholar]
- 8.Moore JW, Yeakel JD, Peard D, Lough J, Beere M. Life-history diversity and its importance to population stability and persistence of a migratory fish: Steelhead in two large North American watersheds. J Anim Ecol. 2014;83:1035–1046. doi: 10.1111/1365-2656.12212. [DOI] [PubMed] [Google Scholar]
- 9.Alberti M, et al. Global urban signatures of phenotypic change in animal and plant populations. Proc Natl Acad Sci USA. 2017;114:8951–8956. doi: 10.1073/pnas.1606034114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendry AP, Farrugia TJ, Kinnison MT. Human influences on rates of phenotypic change in wild animal populations. Mol Ecol. 2008;17:20–29. doi: 10.1111/j.1365-294X.2007.03428.x. [DOI] [PubMed] [Google Scholar]
- 11.McClure MM, et al. Evolutionary consequences of habitat loss for Pacific anadromous salmonids. Evol Appl. 2008;1:300–318. doi: 10.1111/j.1752-4571.2008.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waples RS, Zabel RW, Scheuerell MD, Sanderson BL. Evolutionary responses by native species to major anthropogenic changes to their ecosystems: Pacific salmon in the Columbia River hydropower system. Mol Ecol. 2008;17:84–96. doi: 10.1111/j.1365-294x.2007.03510.x. [DOI] [PubMed] [Google Scholar]
- 13.Williams TH, et al. 2013. Upper Klamath and Trinity River Chinook salmon biological review team report (US Department of Commerce, Washington, DC)
- 14.Traill LW, Schindler S, Coulson T. Demography, not inheritance, drives phenotypic change in hunted bighorn sheep. Proc Natl Acad Sci USA. 2014;111:13223–13228. doi: 10.1073/pnas.1407508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stockwell CA, Hendry AP, Kinnison MT. Contemporary evolution meets conservation biology. Trends Ecol Evol. 2003;18:94–101. [Google Scholar]
- 16.Franklin IR, Frankham R. How large must populations be to retain evolutionary potential? Anim Conserv. 1998;1:69–70. [Google Scholar]
- 17.Williams SE, Shoo LP, Isaac JL, Hoffmann AA, Langham G. Towards an integrated framework for assessing the vulnerability of species to climate change. PLoS Biol. 2008;6:2621–2626. doi: 10.1371/journal.pbio.0060325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chevin L-M, Lande R, Mace GM. Adaptation, plasticity, and extinction in a changing environment: Towards a predictive theory. PLoS Biol. 2010;8:e1000357. doi: 10.1371/journal.pbio.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forsman A. Rethinking phenotypic plasticity and its consequences for individuals, populations and species. Heredity (Edinb) 2015;115:276–284. doi: 10.1038/hdy.2014.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allendorf FW, Hard JJ. Human-induced evolution caused by unnatural selection through harvest of wild animals. Proc Natl Acad Sci USA. 2009;106:9987–9994. doi: 10.1073/pnas.0901069106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charlesworth B, Charlesworth D. Elements of Evolutionary Genetics. 1st Ed Freeman; Greenwood Village, CO: 2010. [Google Scholar]
- 22.Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 2007;17:240–248. doi: 10.1101/gr.5681207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shafer ABA, et al. Genomics and the challenging translation into conservation practice. Trends Ecol Evol. 2015;30:78–87. doi: 10.1016/j.tree.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Moyle PB, Lusardi RA, Samuel PJ, Katz JVE. 2017. State of the salmonids: Status of California’s emblematic fishes 2017 (UC Davis Center for Watershed Sciences, Davis, CA and California Trout, San Francisco, CA)
- 25.ODFW 2000. Effects of Lost Creek Dam on spring Chinook salmon in the Rogue River. Phase II completion report (Oregon Dept of Fish and Wildlife, Salem, OR)
- 26.Quinn TP, McGinnity P, Reed TE. The paradox of ‘premature migration’ by adult anadromous salmonid fishes: Patterns and hypotheses. Can J Fish Aquat Sci. 2015;73:1015–1030. [Google Scholar]
- 27.Hearsey JW, Kinziger AP. Diversity in sympatric chinook salmon runs: Timing, relative fat content and maturation. Environ Biol Fishes. 2014;98:413–423. [Google Scholar]
- 28.Belchik M, Hillemeier D, Pierce RM. 2004. The Klamath River fish kill of 2002; Analysis of contributing factors (Yurok Tribal Fisheries Program, Klamath, CA)
- 29.Meyers JM, et al. 1998. Status review of Chinook salmon from Washington, Idaho, Oregon, and California (National Oceanic and Atmospheric Administration, Washington, DC)
- 30.Moyle PB. Inland Fishes of California: Revised and Expanded. 1st Ed Univ of California Press; Berkeley, CA: 2002. [Google Scholar]
- 31.Committee on Endangered and Threatened Fishes in the Klamath River Basin, Board on Environmental Studies and Toxicology, Division on Earth and Life Studies, National Research Council . Endangered and Threatened Fishes in the Klamath River Basin: Causes of Decline and Strategies for Recovery. National Academies Press; Washington, DC: 2004. [Google Scholar]
- 32.Gustafson RG, et al. Pacific salmon extinctions: Quantifying lost and remaining diversity. Conserv Biol. 2007;21:1009–1020. doi: 10.1111/j.1523-1739.2007.00693.x. [DOI] [PubMed] [Google Scholar]
- 33.Swezey SL, Heizer RF. Ritual management of salmonid fish resources in California. J Calif Anthropol. 1977;4:6–29. [Google Scholar]
- 34.Campbell SK, Butler VL. Archaeological evidence for resilience of Pacific Northwest salmon populations and the socioecological system over the last ∼7,500 years. Ecol Soc. 2010;15:1–17. [Google Scholar]
- 35.Trosper RL. Resilience in pre-contact Pacific Northwest social ecological systems. Conserv Ecol. 2003;7:6–17. [Google Scholar]
- 36.Cook L. Upstream: Searching for Wild Salmon, from River to Table. Random House; New York: 2017. [Google Scholar]
- 37.Quinn TP. The Behavior and Ecology of Pacific Salmon and Trout. Univ of Washington Press; Seattle: 2005. [Google Scholar]
- 38.Waples RS, Teel DJ, Myers JM, Marshall AR. Life-history divergence in Chinook salmon: Historic contingency and parallel evolution. Evolution. 2004;58:386–403. [PubMed] [Google Scholar]
- 39.Narum SR, Hess JE, Matala AP. Examining genetic lineages of Chinook salmon in the Columbia river basin. Trans Am Fish Soc. 2010;139:1465–1477. [Google Scholar]
- 40.ODFW 2005. Oregon native fish status report (Oregon Dept of Fish and Wildlife, Fish Division, Salem, OR), Vol II.
- 41.Prince DJ, et al. The evolutionary basis of premature migration in Pacific salmon highlights the utility of genomics for informing conservation. Sci Adv. 2017;3:e1603198. doi: 10.1126/sciadv.1603198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narum SR, Di Genova A, Micheletti SJ, Maass A. Genomic variation underlying complex life-history traits revealed by genome sequencing in Chinook salmon. Proc R Soc B. 2018;285:20180935. doi: 10.1098/rspb.2018.0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.CDFW 2017. Klamath River basin fall Chinook salmon spawner escapement, in-river harvest and run-size estimates, 1978-2016 a/ (California Dept of Fish and Wildlife, Fisheries Branch, Sacramento, CA)
- 44.Klamath River Basin Fisheries Task Force William M. Kier Associates 1991. Long range plan for the Klamath River basin conservation area fishery restoration program (US Fish Wildlife Service, Yreka, CA)
- 45.USFS 2017. 2016 Spring Chinook salmon spawning ground survey (Salmon-Scott Rivers Ranger District, Fort Jones, CA)
- 46.KBRA 2010. Klamath basin restoration agreement for the sustainability of public and trust resources and affected communities (US Dept of Interior, Washington, DC)
- 47.Hamilton JB, et al. The persistence and characteristics of Chinook salmon migrations to the upper Klamath river prior to exclusion by dams. Oreg Hist Q. 2016;117:326–377. [Google Scholar]
- 48.Hamilton J, et al. 2011. Synthesis of the effects to fish species of two management scenarios for the secretarial determination on removal of the lower four dams on the Klamath River (US Fish and Wildlife Service, Yreka, CA)
- 49.Stevenson AE, Butler VL. The holocene history of fish and fisheries of the upper Klamath basin, Oregon. J Calif Gt Basin Anthropol. 2015;35:169–188. [Google Scholar]
- 50.Butler VL, et al. 2010. The use of archaeological fish remains to establish predevelopment salmonid biogeography in the upper Klamath Basin (National Marine Fisheries Service, Yreka, CA)
- 51.Lubinski PM, Partlow MA. Evidence for local fish catch in zooarchaeology. J Ethnobiol. 2012;32:228–245. [Google Scholar]
- 52.Huntington CW, Claire EW, Al Espinosa,F, Jr, House R. 2006. Reintroduction of anadromous fish to the upper Klamath basin: An evaluation and conceptual plan (Oregon Dept of Fish and Wildlife, Salem, OR)
- 53.Lane and Lane Associates 1981. The Copco dams and the fisheries of the Klamath Tribe (US Department of Interior, Bureau of Indian Affairs, Portland, OR)
- 54.CDFG 2010. Final report Shasta and Scott River juvenile salmonid outmigrant study, 2010 (Dept of Fish and Game, Sacramento, CA)
- 55.National Research Council . Hydrology, Ecology, and Fishes of the Klamath River Basin. National Academies Press; Washington, DC: 2008. [Google Scholar]
- 56.Anderson JH, et al. Planning Pacific salmon and steelhead reintroductions aimed at long-term viability and recovery. N Am J Fish Manage. 2014;34:72–93. [Google Scholar]
- 57.CDFW 2017. Shasta River Chinook and Coho salmon observations in 2016, final report (California Dept of Fish and Wildlife, Sacramento, CA)
- 58.Waples RS. Evolution Illuminated: Salmon and Their Relatives. Oxford Univ Press; Oxford: 2004. Salmonid insights into effective population size; pp. 295–314. [Google Scholar]
- 59.USFS 2017. 2016 Fall Chinook salmon spawning ground survey (Salmon-Scott Rivers Ranger District, Fort Jones, CA)
- 60.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sinauer; Sunderland, MA: 1998. [Google Scholar]
- 61.Miller MR, et al. A conserved haplotype controls parallel adaptation in geographically distant salmonid populations. Mol Ecol. 2012;21:237–249. doi: 10.1111/j.1365-294X.2011.05305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Darwin C. On the Origin of Species, 1859. Routledge; Abingdon, UK: 1859. [Google Scholar]
- 63.Sullivan AP, Bird DW, Perry GH. Human behaviour as a long-term ecological driver of non-human evolution. Nature Ecol Evol. 2017;1:0065. doi: 10.1038/s41559-016-0065. [DOI] [PubMed] [Google Scholar]
- 64.Bunn SE, Arthington AH. Basic principles and ecological consequences of altered flow regimes for aquatic biodiversity. Environ Manage. 2002;30:492–507. doi: 10.1007/s00267-002-2737-0. [DOI] [PubMed] [Google Scholar]
- 65.Healey MC. Patterns of gametic investment by female stream-and ocean-type Chinook salmon. J Fish Biol. 2001;58:1545–1556. [Google Scholar]
- 66.Spencer L. A Temporary Refuge: Fourteen Seasons with Wild Summer Steelhead. Patagonia; Ventura, CA: 2017. [Google Scholar]
- 67.CDFW 2017. 2016 Scott River salmon studies, final report (California Dept of Fish and Wildlife, Yreka, CA)
- 68.Lo Cascio Sætre C, et al. Rapid adaptive phenotypic change following colonization of a newly restored habitat. Nat Commun. 2017;8:14159. doi: 10.1038/ncomms14159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huntington CW. 2012. Summer holding habitat for adult spring Chinook in the Klamath-Trinity River system of California and Oregon (US Fish and Wildlife Service, Yreka, CA)
- 70.Peterson DA, Hilborn R, Hauser L. Local adaptation limits lifetime reproductive success of dispersers in a wild salmon metapopulation. Nat Commun. 2014;5:3696. doi: 10.1038/ncomms4696. [DOI] [PubMed] [Google Scholar]
- 71.Araki H, Cooper B, Blouin MS. Genetic effects of captive breeding cause a rapid, cumulative fitness decline in the wild. Science. 2007;318:100–103. doi: 10.1126/science.1145621. [DOI] [PubMed] [Google Scholar]
- 72.Chilcote MW, Goodson KW, Falcy MR. Reduced recruitment performance in natural populations of anadromous salmonids associated with hatchery-reared fish. Can J Fish Aquat Sci. 2011;68:511–522. [Google Scholar]
- 73.Christie MR, Marine ML, French RA, Blouin MS. Genetic adaptation to captivity can occur in a single generation. Proc Natl Acad Sci USA. 2012;109:238–242. doi: 10.1073/pnas.1111073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Willoughby JR, Christie MR. Long-term demographic and genetic effects of releasing captive-born individuals into the wild. Conserv Biol. August 31, 2018 doi: 10.1111/cobi.13217. [DOI] [PubMed] [Google Scholar]
- 75.Hard JJ, et al. Evolutionary consequences of fishing and their implications for salmon. Evol Appl. 2008;1:388–408. doi: 10.1111/j.1752-4571.2008.00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Darimont CT, et al. Human predators outpace other agents of trait change in the wild. Proc Natl Acad Sci USA. 2009;106:952–954. doi: 10.1073/pnas.0809235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mainwaring MC, et al. Climate change and nesting behaviour in vertebrates: A review of the ecological threats and potential for adaptive responses. Biol Rev Camb Philos Soc. 2017;92:1991–2002. doi: 10.1111/brv.12317. [DOI] [PubMed] [Google Scholar]
- 78.Réale D, McAdam AG, Boutin S, Berteaux D. Genetic and plastic responses of a northern mammal to climate change. Proc Biol Sci. 2003;270:591–596. doi: 10.1098/rspb.2002.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barson NJ, et al. Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature. 2015;528:405–408. doi: 10.1038/nature16062. [DOI] [PubMed] [Google Scholar]
- 80.Veale AJ, Russello MA. An ancient selective sweep linked to reproductive life history evolution in sockeye salmon. Sci Rep. 2017;7:1747. doi: 10.1038/s41598-017-01890-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pearse DE, Miller MR, Abadía-Cardoso A, Garza JC. Rapid parallel evolution of standing variation in a single, complex, genomic region is associated with life history in steelhead/rainbow trout. Proc Biol Sci. 2014;281:20140012. doi: 10.1098/rspb.2014.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ayllon F, et al. The vgll3 locus controls age at maturity in wild and domesticated Atlantic salmon (Salmo salar L.) males. PLoS Genet. 2015;11:e1005628. doi: 10.1371/journal.pgen.1005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Margres MJ, et al. Large-effect loci affect survival in Tasmanian devils (Sarcophilus harrisii) infected with a transmissible cancer. Mol Ecol. 2018;27:4189–4199. doi: 10.1111/mec.14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Toews DP, et al. Plumage genes and little else distinguish the genomes of hybridizing warblers. Curr Biol. 2016;26:2313–2318. doi: 10.1016/j.cub.2016.06.034. [DOI] [PubMed] [Google Scholar]
- 85.Waples RS, Lindley ST. Genomics and conservation units: The genetic basis of adult migration timing in Pacific salmonids. Evol Appl. 2018;11:1518–1526. doi: 10.1111/eva.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Visser ME. Keeping up with a warming world; assessing the rate of adaptation to climate change. Proc Biol Sci. 2008;275:649–659. doi: 10.1098/rspb.2007.0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hess JE, Zendt JS, Matala AR, Narum SR. Genetic basis of adult migration timing in anadromous steelhead discovered through multivariate association testing. Proc Biol Sci. 2016;283:20153064. doi: 10.1098/rspb.2015.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ali OA, et al. RAD Capture (Rapture): Flexible and efficient sequence-based genotyping. Genetics. 2016;202:389–400. doi: 10.1534/genetics.115.183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Connolly TJ, Ruiz DL, Jenkins DL, Deur D. 2015. This place is home: Exploring heritage and community of the Klamath Tribes at the Beatty Curve Site (35KL95) (Museum of Natural and Cultural History & State Museum of Anthropology, Univ of Oregon, Eugene, OR)
- 90.Cressman L. Klamath prehistory: The prehistory of the culture of the Klamath Lake area, Oregon. Trans Am Philos Soc New Ser. 1956;46:375–513. [Google Scholar]
- 91.Cheatham RD, et al. 1995. Archaeological investigations at the Bezuksewas Village Site (35KL778), Klamath County, Oregon (Eugene, OR)
- 92.Cheatham RD. 1991. Archaeological investigations at the Williamson River Bridge site (35KL677): A riverside fishing camp in Klamath County, Oregon (Eugene, OR)
- 93.Bronk Ramsey C. 2014 OxCal. Available at https://c14.arch.ox.ac.uk/oxcal/OxCal.html. Accessed March 22, 2014.
- 94.Yang DY, Cannon A, Saunders SR. DNA species identification of archaeological salmon bone from the Pacific Northwest Coast of North America. J Archaeol Sci. 2004;31:619–631. [Google Scholar]
- 95.Yang DY, Eng B, Waye JS, Dudar JC, Saunders SR. Technical note: Improved DNA extraction from ancient bones using silica-based spin columns. Am J Phys Anthropol. 1998;105:539–543. doi: 10.1002/(SICI)1096-8644(199804)105:4<539::AID-AJPA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 96.Yang DY, Speller CF. Co-amplification of cytochrome b and D-loop mtDNA fragments for the identification of degraded DNA samples. Mol Ecol Resour. 2006;6:605–608. [Google Scholar]
- 97.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hall TA. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, (Information Retrieval Ltd., London), pp 95–98, c1979–c2000.
- 99.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Berthelot C, et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 2014;5:3657. doi: 10.1038/ncomms4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Team RC. 2017. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.