Significance
Aluminum (Al3+) on acidic soils, which represent half of the world’s agricultural lands, damages plant roots. In Africa, where sorghum is a staple food, 20% of the agricultural soils are acidic, significantly reducing yields. SbMATE confers sorghum Al tolerance via root citrate exudation into the soil, where citrate binds and detoxifies Al3+, but shows reduced expression in some genetic backgrounds. This phenomenon results from the action of a variable tandem repeat flanking a transposon in the SbMATE promoter working in concert with WRKY and zinc finger-DHHC proteins, which bind to the SbMATE promoter and regulate expression in response to Al3+. We can now select for superior alleles of these transcription factors to maximize SbMATE expression, thereby contributing to global food security.
Keywords: transcriptional regulation, abiotic stress, transporters, expression QTL, MITE transposon
Abstract
Acidic soils, where aluminum (Al) toxicity is a major agricultural constraint, are globally widespread and are prevalent in developing countries. In sorghum, the root citrate transporter SbMATE confers Al tolerance by protecting root apices from toxic Al3+, but can exhibit reduced expression when introgressed into different lines. We show that allele-specific SbMATE transactivation occurs and is caused by factors located away from SbMATE. Using expression-QTL mapping and expression genome-wide association mapping, we establish that SbMATE transcription is controlled in a bipartite fashion, primarily in cis but also in trans. Multiallelic promoter transactivation and ChIP analyses demonstrated that intermolecular effects on SbMATE expression arise from a WRKY and a zinc finger-DHHC transcription factor (TF) that bind to and trans-activate the SbMATE promoter. A haplotype analysis in sorghum RILs indicates that the TFs influence SbMATE expression and Al tolerance. Variation in SbMATE expression likely results from changes in tandemly repeated cis sequences flanking a transposable element (a miniature inverted repeat transposable element) insertion in the SbMATE promoter, which are recognized by the Al3+-responsive TFs. According to our model, repeat expansion in Al-tolerant genotypes increases TF recruitment and, hence, SbMATE expression, which is, in turn, lower in Al-sensitive genetic backgrounds as a result of lower TF expression and fewer binding sites. We thus show that even dominant cis regulation of an agronomically important gene can be subjected to precise intermolecular fine-tuning. These concerted cis/trans interactions, which allow the plant to sense and respond to environmental cues, such as Al3+ toxicity, can now be used to increase yields and food security on acidic soils.
Decisions in plant breeding often reflect the complex interplay between noncoding DNA sequences, acting locally in chromatin, and trans-regulatory elements driving gene expression via intermolecular interactions. Studies in Drosophila have shown that cis elements are evolutionarily important (1), whereas trans factors play pivotal roles in regulating plant stress responses in Arabidopsis (2). However, the manner in which cis and trans factors interact to control phenotypic expression is less clear.
Half the world’s agricultural soils are highly acidic (3), which solubilizes Al3+ into the soil solution, damaging plant roots and reducing yields. The Al-activated root citrate transporter SbMATE, which underlies Al tolerance via formation of nontoxic Al–citrate complexes in the rhizosphere (4), increased grain yield by 0.6 ton⋅ha−1 for sorghum grown on acidic soil (5). SbMATE SNPs were associated to sorghum grain yield production in West Africa (6), where sorghum is a staple food. This makes SbMATE important for global food security.
SbMATE expression is up-regulated by Al3+ in a time-dependent fashion (4) and is highly correlated with Al tolerance (7). A Tourist-like miniature inverted repeat transposable element (MITE) (8) and its flanking sequences, which are repeated in tandem, were found 2 kb upstream of SbMATE. Variation in the number of these tandem repeats in different sorghum lines was positively correlated with Al tolerance. Nevertheless, introgression of the AltSB locus, where SbMATE resides (9), into Al-sensitive recurrent parents resulted in reduced SbMATE expression and Al tolerance; this suggested involvement of accessory loci acting in trans (7). The current study focuses on the elucidation of the role of the MITE repeats in SbMATE transcriptional regulation, and on the dissection of the genetic background effects that can reduce SbMATE expression.
Results
SbMATE Expression Is Influenced by the Genetic Background.
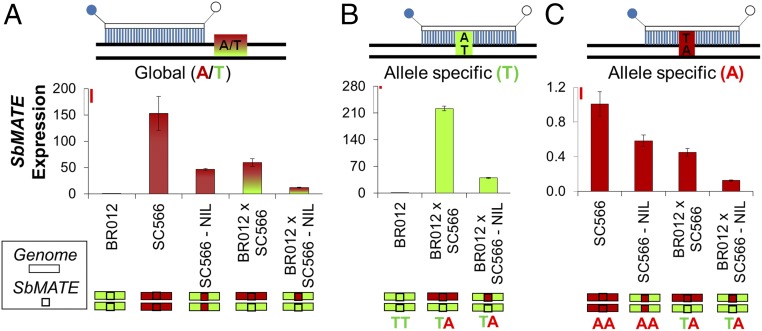
We studied global (i.e., joint expression of SbMATE alleles) and allele-specific expression of SbMATE (Fig. 1) to distinguish between cis and trans regulatory effects. This was done by using stocks derived from the low SbMATE-expressing Al-sensitive line BR012 crossed with SC566, a very Al-tolerant line with high SbMATE expression (7). We generated a homozygous stock in which the SbMATE allele from SC566 was introgressed into the BR012 genetic background [SC566-near isogenic line (NIL)]. BR012 × SC566 and BR012 × SC566-NIL both have SbMATE in heterozygosity, but they have different genetic backgrounds. Although BR012 × SC566 has a hybrid background, BR012 × SC566-NIL has the homozygous background of BR012. Compared with SC566, there was a consistent reduction of global SbMATE expression (Fig. 1A) in the SC566-NIL. SbMATE expression was higher in the hybrid background (BR012 × SC566) than in the BR012 background in the BR012 × SC566-NIL. Therefore, SbMATE expression is reduced in the Al-sensitive background.
Fig. 1.
Global and allele-specific expression of SbMATE assessed with TaqMan probes. The global assay assesses the joint expression of SbMATE alleles, and allele-specific expression was based on an SNP (T, present in Al-sensitive BR012; A, present in Al-tolerant SC566) within SbMATE. Colored schematics indicate the genetic backgrounds (genome, rectangles) and the SbMATE alleles (squares): SC566 (red) and BR012 (green). (A) Global expression, allele-specific expression of the (B) Al-sensitive (T, green) allele and (C) Al-tolerant (A, red) allele, with the probes depicted on Top. The red/green gradient in A shows the joint expression of both the A and T alleles in stocks heterozygous for SbMATE. Global relative expression values are fold changes relative to the Al-sensitive line. Expression of the T and A alleles are fold changes relative to expression in the parents, BR012 and SC566, which are homozygous for the T and A alleles, respectively. The sorghum genotypes were grown with {27} µM Al3+ in nutrient solution at pH 4.0 for 5 d, and the root apex (1 cm) was collected for RNA isolation. Values are mean ± SD; n = 3. Least significant difference (Fisher’s LSD, α = 0.05) bars (in red) are drawn to scale (Top of the y axis).
SbMATE allele-specific expression was quantified relative to expression in the Al-tolerant and Al-sensitive parents, SC566 and BR012 (Fig. 1 B and C). Here, allele-specific expression was based on a T/A single nucleotide polymorphism (SNP) in the first exon of SbMATE, with the A allele present in SC566 and the T allele present in BR012 (7). A marked, 210-fold up-regulation of the Al-sensitive allele (T) was observed in the BR012 × SC566 hybrid (Fig. 1B). Expression of the Al-sensitive allele of SbMATE was greatly reduced when present in the BR012 genome (BR012 × SC566-NIL) compared with the hybrid genome (BR012 × SC566; Fig. 1B). Expression changes for the Al-tolerant allele (A) were relatively subtle (Fig. 1C), but expression was reduced in all stocks harboring the BR012 genome compared with SC566.
These findings indicate that SbMATE expression is influenced by trans-acting factors whose favorable alleles are donated by the Al-tolerant line SC566. These factors are unlinked to AltSB, as allele-specific expression was higher in hybrids (BR012 × SC566) compared with the NIL hybrid (BR012 × SC566-NIL), where the AltSB locus is heterozygous but within a fully Al-sensitive background.
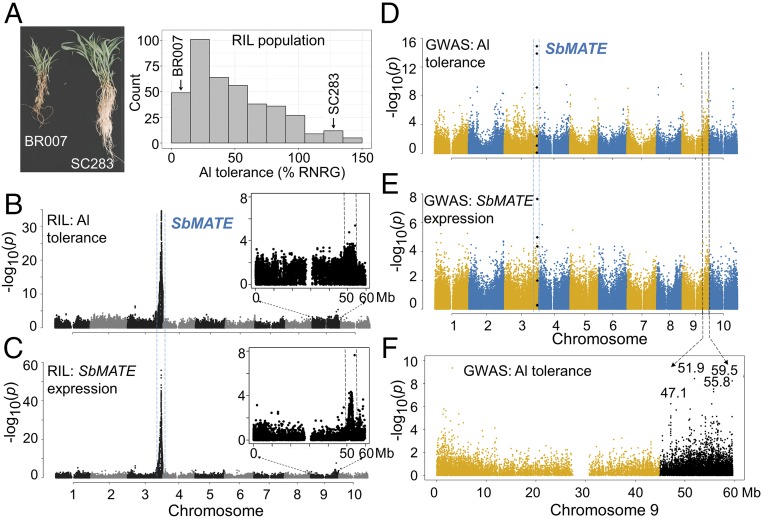
Trans-Acting Loci Influencing SbMATE Expression Are Present Within an SbMATE Expression/Al Tolerance QTL on Chromosome 9.
Next, we undertook QTL mapping in a BR007 × SC283 recombinant inbred line (RIL) population. SC283 is highly tolerant to Al toxicity, whereas BR007 is highly Al-sensitive (Fig. 2A), and this RIL population was previously used to positionally clone SbMATE (4). The availability of a large population size for this highly contrasting cross (9) justified its choice for QTL mapping. Major QTL for both Al tolerance (Fig. 2B) and SbMATE expression (Fig. 2C) were colocated with SbMATE on chromosome 3. This indicates that higher SbMATE expression and Al tolerance in SC283 (7) is achieved predominantly in cis. A comparatively smaller Al tolerance and SbMATE expression QTL (eQTL) was detected at ∼51 to ∼54 Mb [2 < −log10(p) < 8 for the eQTL] on chromosome 9 (zoomed in; Fig. 2 B and C). Although other loci with similar P values are found elsewhere, the chromosome 9 QTL was chosen because of its clear joint effect on both Al tolerance and SbMATE expression. Multilocus mapping was also undertaken and revealed a possible interaction between the QTL on chromosomes 3 and 9 (SI Appendix, Table S1).
Fig. 2.
Trans-factor positional cloning based on expression GWAS. (A) Contrasting phenotypes in BR007 (Al-sensitive) and SC283 (Al-tolerant) based on root damage and relative net root growth (%RNRG) assessed in nutrient solution with {27} µM Al3+, in the context of the BR007 × SC283 RILs. QTL mapping was carried out for (B) Al-tolerance and (C) SbMATE expression (eQTL) in the BR007 × SC283 RILs. The chromosome 9 region, where a colocated Al-tolerance/eQTL was detected, is expanded. GWAS with (D) Al tolerance and (E) SbMATE expression. SNPs near or within SbMATE previously associated with Al tolerance (20) are depicted as black diamonds. Blue and black dashed lines indicate the SbMATE region (chr 3) and the region harboring significant SNPs on chromosome 9, respectively, which overlap in the RIL QTL map (B and C) and in the GWAS plots (D and E). (F) Details of the chromosome 9 region showing physical positions for SNPs with the strongest association signals with Al tolerance (black dots).
Genome-wide association mapping (GWAS) identified SNPs associated both with Al tolerance and SbMATE expression in the Al tolerance/eQTL region on chromosome 9 (Fig. 2 D and E). In that region, SNP loci in linkage disequilibrium were detected across distances exceeding 2 Mb (SI Appendix, Fig. S1). Next, we identified SNPs associated with both Al tolerance and SbMATE expression, with −log(p) between 3 and 9. We overlapped the resulting physical interval with that of the QTL identified in the RIL population, and defined an extended region between ∼45 and ∼59 Mb (Fig. 2F) as a search region to identify candidate genes with regulatory signatures (https://phytozome.jgi.doe.gov/pz/portal.html, v1.4).
SbMATE Promoter.
The structure of the SbMATE promoter region that contains the MITE insertion is shown in SI Appendix, Fig. S2 A and B. The MITE element (unit “b”) is flanked by 100-bp (unit “a”) and 20-bp (unit “c”) sequences. This MITE-containing a-b-c triplet (designated hereafter simply as “MITE repeats”) is followed by a single terminal (unrepeated) 100-bp “a” unit with either an 8-bp deletion (present in SC283 and Tx430) or a 12-bp deletion (present in BR012; SI Appendix, Fig. S2B). Henceforth, the 100-bp “a” sequence within the MITE repeats will be designated as the 100-bp repeat and the terminal, unrepeated units, as the 88- or 92-bp terminal. Natural, allelic variation at the SbMATE promoter arises from tandem variations in the number of identical a-b-c units, which are present either as a singlet or as repeated units in different sorghum lines, with Al-tolerant lines showing in general more repeats compared with Al-sensitive lines (4). For example, the parents of the RIL population, BR007 (Al-sensitive) and SC283 (tolerant), have three and five repeats, respectively. For transactivation assays, we synthesized SbMATE promoters containing one MITE repeat (promoter from the Al-sensitive line Tx430, designated as Tx430p), four repeats (BR012p, from BR012, which is Al-sensitive), and five repeats (SC283p, from SC283, Al-tolerant), with the flanking, 1,749-bp and 2,010-bp sequences from the sorghum BAC where SbMATE resides (4). Promoter truncations were amplified from SC283.
Transcription Factors on the Chromosome 9 QTL Transactivate the SbMATE Promoter in Yeast.
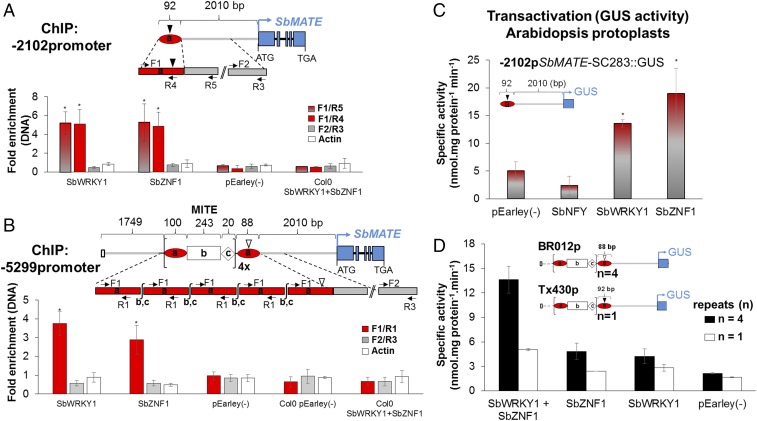
A qualitative analysis based on the yeast one-hybrid assay indicated that, in the Al tolerance/eQTL region, a WRKY-like transcription factor (TF), Sb09g023500 (SbWRKY1) at 53.14 Mb, and Sb09g021530, a gene encoding a zinc finger DHHC (zf-DHHC) domain-containing protein (SbZNF1) at 50.98 Mb (SI Appendix, Fig. S3 A and B), were both capable of trans-activating SbMATE promoter alleles harboring one, four, and five copies of the MITE repeats (SI Appendix, Fig. S2 C and D). In contrast, SbNFY1, a NFY-like TF (Sb09g022810) located within the same QTL, did not activate the SbMATE promoter, confirming the specificity of SbWRKY1 and SbZNF1 transcriptional activation of SbMATE.
Qualitative promoter deletion analysis (SI Appendix, Fig. S2 C and D) showed that a proximal SC283 promoter fragment, extending to position −2102 relative to the SbMATE start codon (−2102pSC283, where “p” stands for promoter), was sufficient for trans-activation by both TFs, but trans-activation was lost when the 92-bp terminal was deleted (−2010pSC283).
SbZNF1 and SbWRKY1 Bind Both to the 100-bp Repeat and to the 92-bp Terminal.
Leaf protoplasts from transgenic Arabidopsis transformed with constructs containing the first 2,010 bp of the SbMATE promoter including either the 92-bp terminal (−2102p, from the SC283 promoter) or the MITE repeats in BR012 (−5299p; Fig. 3 A and B) were transformed along with constructs encoding 35S-driven YFP::TF cDNA. The immunoprecipitated chromatin (ChIP) fragments obtained with anti-GFP antibody were analyzed by PCR and qPCR. For both SbWRKY1-expressing and SbZNF1-expressing protoplasts, qPCR with IP DNA showed that a fragment within the 92-bp terminal, amplified with primers F1/R4 and F1/R5, was significantly enriched over the input (control) DNA, confirming binding to the 92-bp terminal (Fig. 3A). In contrast, no enrichment was observed with primers annealing either to the 2,010-bp fragment (F2/R3), which lacks both the 100-bp repeat and the 92-bp terminal, or the actin gene (endogenous control).
Fig. 3.
SbWRKY1 and SbZNF1 bind to and transactivate the SbMATE promoter. For both (A and B) ChIP-qPCR and (C and D) transactivation assays, protoplasts from Arabidopsis stably transformed with promoter constructs were transfected with pEarleyGate104 containing the indicated TFs or with the empty vector (minus sign). Schematics of the SbMATE promoter constructs are shown: the general structure MITE region is depicted in B: the 243-bp MITE element (unit “b”) is flanked by 100-bp (unit “a”, 100-bp repeat) and 20-bp (unit “c”) sequences, and the number of identical a-b-c triplets varies in different promoter alleles. The MITE-containing a-b-c triplets (MITE repeats) is terminated by a single 100-bp “a” unit with either an 8-bp deletion (depicted by an inverted black triangle, present in SC283 and Tx430) or a 12-bp deletion (inverted white triangle, present in BR012), which results in the 88- or 92-bp terminal fragments. ChIP-qPCR with (A) a truncated SbMATE promoter from the SC283 line extending to position −2,102 bp [−2102promoter(p)] and (B) from BR012 extending to position −5,299 bp [−5299promoter(p)] and containing four copies of the MITE repeats (4×), followed by the 88-bp terminal. Arrows represent primer (SI Appendix, Table S2) positions. Data were normalized to the input (control) for each sample and are expressed as the fold-enrichment vs. preimmune IgG serum controls. Error bars indicate SEM (n = 3), and asterisks indicate significant differences (P < 0.05). (C and D) Transactivation in Arabidopsis protoplasts. Protoplasts stably transformed with (C) −2102pSC283::GUS and transfected with pEarleyGate104 with or without [pEarley(−)] the sorghum TFs. Results are the mean ± SD of three independent experiments. *P < 0.05; Scott–Knott test. (D) Transactivation with the promoter region of Tx430 (−4214pTx430::GUS) and BR012 (−5299pBR012::GUS), with one and four MITE repeats (n), respectively. The error bars represent the limits of the nonparametric bootstrap confidence interval of 95% with n = 4.
ChIP was also undertaken with the −5299 promoter from BR012, which has four MITE repeats followed by the terminal 88-bp fragment (Fig. 3B). Amplification of IP DNA with primers F1/R1, which are specific to the 100-bp repeat, was significantly enriched over the input DNA (Fig. 3B). Collectively, the results in Fig. 3 A and B show that SbWRKY1 and SbZNF1 bind both to the 100-bp “a” unit within the MITE repeats and to the 92-bp terminal. The amplification profiles of IP DNA (SI Appendix, Fig. S4) with primers flanking (F3 and F4) and within (R1) the 100-bp repeat (lanes 1 and 3 in −5299p) confirm such binding. Because the R1 primer in Fig. 3B does not anneal to the 88-bp terminal in −5299p, we cannot rule out that the TFs do not bind to that fragment because of its additional 4-bp deletion compared with the 92-bp terminal fragment in −2102p (see SI Appendix, Fig. S2B for “a” unit alignments).
The Number of MITE Repeats Correlates With Enhanced SbWRKY1 and SbZNF1 Transactivation Activity.
Because transactivation assays in yeast are qualitative, we quantified transactivation activity in Arabidopsis. Protoplasts were isolated from Arabidopsis transformed with the truncated −2102 promoter from Al-tolerant SC283 containing the 92-bp terminal (Fig. 3C). In addition, promoter alleles containing one (Tx430p) and four (BR012p) copies of the MITE repeats followed by the 92-bp and 88-bp terminal (Fig. 3D), respectively, were tested. Reporter gene-specific activity was higher with protoplasts isolated from Arabidopsis cotransformed with the 92-bp terminal and with SbWRKY1 and SbZNF1 compared with the negative controls (Fig. 3C). In addition, elimination of the 92-bp terminal abolished reporter gene activity (SI Appendix, Fig. S5 A–H), which is consistent with the transactivation results in yeast.
Reporter gene activity was significantly higher with promoter alleles containing four (BR012p) compared with one (Tx430p) copies of the MITE repeats (Fig. 3D). For both promoter constructs, a cotransactivation assay indicated a synergistic mode of action, with a greater effect on SbMATE promoter activity when both TFs are present (SbWRKY1 + SbZNF1) compared with their individual effects (Fig. 3D). These two promoter alleles vary both for the number of MITE repeats and for the presence of an additional 4-bp deletion specifically in the 88-bp unique terminal of the four-repeat promoter. However, we established previously that both TFs bind to the 100-bp “a” repeat and to the unrepeated 92-bp “a” terminal (Fig. 3 A and B). In view of that, even if both TFs did not bind to the terminal “a” 88-bp fragment of the four MITE-repeat promoter, this promoter would still harbor four binding sites for the TFs within its MITE region, in contrast to only two binding sites in the one MITE-repeat promoter (within its single 100-bp sequence and in the 92-bp terminal). It is thus unlikely that the additional 4-bp deletion was the cause of the higher transactivation in the 4 MITE-repeat (BR012p) promoter compared with the 1 MITE-repeat promoter (Tx430p). These results strongly suggest that increased binding site abundance in promoters where the number of MITE repeats has expanded leads to enhanced TF recruitment.
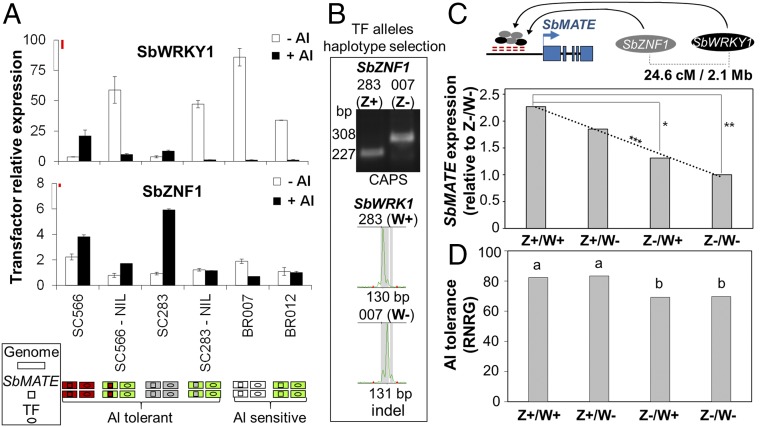
SbZNF1 and SbWRKY1 Alleles from Al-Tolerant and Al-Sensitive Lines Are Differentially Regulated by Al3+.
For clarity, alleles are designated in the text with the gene names (TF is used when referring to both SbWRKY1 and SbZNF1) subscribed with numbers indicating the allele donors. We looked at the expression profiles of SbWRKY1 and SbZNF1 alleles derived from unrelated Al-tolerant (SC566 and SC283) and Al-sensitive (BR007 and BR012) lines (10) (Fig. 4A). Genetic backgrounds are depicted by the colored rectangles beneath Fig. 4A. The SC566- and SC283-NILs carry the respective SbMATE allele (SbMATE566/283, depicted by squares) in the BR012 genetic background, and as such, their TF alleles (green ovals) are the same as the ones in BR012 (TF012).
Fig. 4.
Transcription factor expression profile and effect on SbMATE expression and Al tolerance. (A) SbWRKY1 and SbZNF1 expression in Al-tolerant (SC283 and SC566) and Al-sensitive (BR007 and BR012) lines, and in the SC566-NIL and SC283-NIL (SC566/SC283 SbMATE in the BR012 background). Colored schematics indicate the genetic backgrounds (genome, rectangles), the SbMATE alleles (squares), and TF alleles (ovals), along with the Al tolerance phenotype (7). Plants were grown on ±{27} µM Al3+ for 5 d in nutrient solution at pH 4.0 (brackets denote free Al3+ activity estimated with GEOCHEM; see SI Appendix, Supplementary Methods), and the root apices (1 cm) were collected. Values are mean ±SD (n = 2). Least significant difference (Fisher’s LSD, α = 0.10) bars (in red) are drawn to scale (Top of the y axis). This experiment was repeated with similar results (SI Appendix, Fig. S6, 5 d; n = 3). (B) SbWRKY1 and SbZNF1 polymorphisms in the RIL parents, BR007 (007, Al-sensitive) and SC283 (283, Al-tolerant), which were used to select RILs with all combinations of TF alleles (i.e., TF haplotypes). (C) SbWRKY1 and SbZNF1 effect on SbMATE expression estimated based on RILs homozygous for the SC283 (Al-tolerant) alleles at both TF loci (Z+/W+), for the BR007 (Al-sensitive) allele (Z−/W−), or showing alternate TF alleles (Z+/W− and Z−/W+). Significant differences based on 5% (**) and 12% (*) confidence intervals. A linear regression model fit to haplotype SbMATE expression was highly significant (***α = 0.01). Physical (Mb) and genetic (cM) distances between TFs are depicted at the Top. (D) Effect of SbWRKY1 and SbZNF1 on Al tolerance as measured by relative net root growth (%RNRG). Different letters indicate statistical differences (Fisher’s least significant difference, α = 0.08).
In the presence of Al3+, both SbWRKY1 and SbZNF1 were more highly expressed in Al-tolerant (SC283 and SC566) compared with Al-sensitive (BR012 and BR007; Fig. 4A) lines. In NILs in which tolerant SbMATE alleles (SbMATE283/566) were introgressed into the Al-sensitive BR012 background (SC283- and SC566-NILs), TF expression was reduced compared with their respective Al-tolerant donors (SC283 and SC566). These responses are similar to their transcriptional target SbMATE, which also showed reduced expression in the SC566- and SC283-NILs compared with the Al-tolerant parents (SI Appendix, Fig. S6, 5 d, and ref. 7). Strikingly, the SbWRKY1 allele derived from the Al-tolerant line SC566, SbWRKY1566, was markedly up-regulated by Al3+ (SbWRKY1283 also shows a consistent tendency for Al3+ up-regulation, but slighter). In contrast, the Al-sensitive alleles in BR007 (SbWRKY1007), BR012, and NILs (SbWRKY1012) were strongly down-regulated by Al3+ (Fig. 4A). SbWRK1 and SbZNF1 exhibited different transcriptional responses to Al3+ in different Al-tolerant lines, as Al-induced SbWRKY1 expression was greater in SC566 compared with SC283, whereas SbZNF1 Al3+ up-regulation and expression was higher in SC283.
In Sorghum, SbWRK1 and SbZNF1 Alleles Derived from SC283 (Al-Tolerant) Increase SbMATE Expression.
A genetic analysis in the BR007 × SC283 RIL population was conducted using SbWRKY1 (W) and SbZNF1 (Z) gene-specific markers, which were designed based on polymorphisms that differentiate the TF283 and TF007 alleles (Fig. 4B). This was done to select RILs with different combinations between parental alleles of SbZNF1 and SbWRKY1 (i.e., TF haplotypes). For this analysis, we compared SbMATE expression and Al tolerance of RILs selected to contain both TF alleles from the Al-tolerant parent (TF283, Z+/W+), from the Al-sensitive parent (TF007, Z−/W−), and one TF allele from each parent (Z+/W−, Z−/W+).
A linear regression model fit to all haplotype classes indicated that both TFs enhanced SbMATE expression (Fig. 4C and SI Appendix, Fig. S7). SbMATE expression for the double homozygous haplotypic class containing the Al-tolerant SC283 allele at both loci (Z+/W+) produced the largest increase in SbMATE expression compared with the other haplotype classes, and was more than 2.3-fold higher than the class containing Al-sensitive alleles from BR007 at both loci (Z−/W−; Fig. 4C).
SbZNF1 exerted a stronger effect on both SbMATE expression (Fig. 4C) and Al tolerance (Fig. 4D) compared with SbWRKY1, the individual effect of which on Al tolerance was below the statistical power of our haplotype-based approach. This result is likely population-specific, resulting from stronger Al3+ up-regulation of SbZNF1 expression compared with SbWKRY1 specifically in the Al-tolerant parent of the RIL population, SC283 (Fig. 4A).
Time-Dependent Expression in Root Apices Exposed to Al3+ for SbWRKY1 and SbZNF1 Is Similar to SbMATE.
Al-induced expression of both SbWRKY1 and SbZNF1 was higher in root apices of Al-tolerant lines compared with the rest of the root system and shoots (SI Appendix, Fig. S8). This response favoring preferential expression in root apices was larger for SbWRKY1 in SC566 compared with SbZNF1 in SC283, which are the genotypes that display the highest expression of each TF gene under Al3+ (Fig. 4A). A time-course analysis indicated a general trend for time-dependent increase in TF expression in Al-tolerant lines between 1 and 5 d of Al exposure, which was higher for SbWRKY1 (4.2–4.8-fold) compared with SbZNF1 (1.1–1.6-fold). In general, preferential, time-dependent expression in root apices exposed to Al3+ for SbZNF1 and SbWRKY1 parallels the SbMATE expression measured under the same period in Al. For the Al-sensitive lines, BR007 and BR012, SbWRKY1 and SbZNF1 expression in the presence of Al3+ decreased over the same 1-, 3-, and 5-d periods (SI Appendix, Fig. S6).
Discussion
We discovered that SbMATE expression is influenced by a cis-acting tandemly repeated sequence flanking a MITE insertion upstream of SbMATE, which provides sites in which SbWRKY1 and SbZNF1 bind and transcriptionally regulate SbMATE. Possible binding motifs in the binding fragment are the recognition core for Dof1/MNB1a zf-TFs (11, 12) and a motif similar to the WT-box, where a WRKY TF has been shown to bind (13) (see SI Appendix, Table S3 for cis elements identified in silico).
Our results indicate that SbMATE and SbWRKY1 are coregulated (r = 0.3; P = 0.08; SI Appendix, Fig. S6). This suggests that SbWRKY1 functionally evolved to regulate SbMATE expression in response to Al3+, which is consistent with the active and adaptable nature of Group III C2H-type zfs (14). SbZNF1 is a DHHC-like S-acyl transferase zf, and such proteins have been implicated in abiotic stress tolerance (15). SbZNF1 is preferentially expressed in roots of Al-tolerant lines, but its expression is localized to the root tip to a lesser extent than SbWRKY1. This pattern may reflect the more general physiological role of DHHC proteins, stemming from the DHHC cognate function in increasing protein hydrophobicity (16).
Our quantitative analysis of transactivation in Arabidopsis protoplasts positively associated SbWRKY1 and SbZNF1 transactivation activity and the number of MITE repeats in the SbMATE promoter, suggesting a dosage dependency. Hence, we propose that the singular (17), tandemly repeated structure of the MITE repeats has led to differential TF recruitment (SI Appendix, Fig. S9 A and B), resulting in the previously observed positive correlation between the size of the MITE insertion region and the Al tolerance phenotype (4).
Synergistic transactivation in Arabidopsis protoplasts, in conjunction with our haplotype analysis of SbWRKY1 and SbZNF1 in RILs derived from parents harboring different TF alleles, suggest that, in sorghum, these TFs cooperate to increase SbMATE expression. The cDNA sequences of SbWRKY1 and SbZNF1 alleles in Al-tolerant and Al-sensitive lines were found to be identical. Therefore, differential, time-dependent regulation by Al3+ of TF alleles appears to be a critical step in the cis/trans interactions that control SbMATE expression. Although the SC283 (Al-tolerant) allele of both TFs is up-regulated by Al3+, the alternative, BR007 (Al-sensitive allele), is down-regulated (Fig. 4A), which helps to explain why RILs fixed for the SC283 alleles of both SbWRKY1 and SbZNF1 show a 2.3-fold increase in SbMATE expression compared with RILs fixed for the BR007 alleles (Fig. 4C). The estimated TF effect on Al tolerance (18% increase; Fig. 4D: Z+/W+ vs. Z−/W−) is equal to the decrease in Al tolerance when the SC283 allele of SbMATE was introgressed into the background of the Al-sensitive line, BR012 (∼18% in SC283 vs. SC283-NIL; figure 2 in ref. 7), which we show here has low-expressing alleles for both SbWRKY1 and SbZNF1 (Fig. 4A). This suggests that allelic variation at the TF loci is responsible for our previously observed genetic background effects, which lead to reduced expression of Al-tolerant alleles of SbMATE when introgressed into Al-sensitive backgrounds (7).
Our cis/trans interaction model (SI Appendix, Fig. S9) depicts a possible compensatory mode of action for cis and trans effects in highly Al-tolerant lines. Accordingly, the loss of one MITE repeat in SC566 compared with SC283 would be expected to reduce TF occupancy and, hence, reduce SbMATE expression in SC566. However, the loss of one binding unit appears to be compensated for by higher Al3+ up-regulation of SbWRKY1 expression in SC566, resulting in higher SbMATE expression in the presence of Al3+. Conversely, in the absence of Al3+, high expression of SbWRKY1 and SbZNF1 in Al-sensitive lines (BR007 and BR012, with three and four MITE repeats, respectively; SI Appendix, Fig. S9) does not lead to enhanced SbMATE expression (Fig. 4A and SI Appendix, Fig. S6). This suggests the occurrence of independent cis-acting repressor components acting upstream of the MITE repeats in their role of providing TF binding sites. Although these components likely control extreme Al tolerance and sensitive phenotypes, SbWRKY1 and SbZNF1 appear to act to regulate SbMATE in Al-tolerant lines.
Compensatory cis/trans effects (2, 18), which appear to be a rather widespread mechanism that cells use to stabilize gene expression (19), are implicated in coevolution between cis and trans mutations. Although cis variants may provide a more stable control of gene expression under stress, trans regulation is important for environmental responses (2). In the absence of Al3+, both SbWRKY1 and SbZNF1 are down-regulated in Al-tolerant genotypes (SC283 and SC566), reducing SbMATE expression precisely when root citrate release, which can be costly to the plant, is not needed because of the lack of Al toxicity. Therefore, the interplay between cis-acting elements and TFs that are responsive to Al3+ stress may be advantageous, as a result of a balancing effect on SbMATE expression, which would otherwise be more inflexibly controlled in cis, resulting in genetic load related to the loss of carbon during unnecessary root citrate release.
In light of the molecular nature of cis and trans variants that modulate SbMATE expression, we can now both predict and circumvent genetic background effects that reduce SbMATE expression to increase grain yield production on acidic, Al toxic soils across the world.
Materials and Methods
Genetic Stocks.
Development of NILs, RILs, and hybrid stocks, and the association panel used for GWAS (20), are described in SI Appendix, Supplementary Methods.
Al Tolerance in Hydroponics.
Al tolerance was assessed based on root growth inhibition, relative net root growth (RNRG), in nutrient solution with and without {27} µM Al3+ at pH 4.0 (20) (brackets denote free Al3+ activity estimated with GEOCHEM; see SI Appendix, Supplementary Methods).
Gene Expression via Quantitative RT-PCR.
Sorghum plants were grown in nutrient solution ±{27} µM Al3+ for 1 and/or 3 and 5 d, depending on the experiment. Gene expression was assessed either with the TaqMan Gene Expression or SYBR Green assay (Applied Biosystems). Allele-specific expression was assessed (TaqMan) based on an A/T SNP in the first exon of SbMATE (7), with the A allele present in SC566 and the T allele present in all other lines. See SI Appendix, Supplementary Methods.
QTL Mapping in a RIL Population.
Al tolerance and SbMATE expression data were obtained in nutrient solution with {27} µM Al3+ at pH 4.0 for 5 d, and QTL mapping with SNP markers was undertaken with TASSEL (GLM) and by multiple regression.
Genomewide Association Mapping.
Genomewide association mapping was undertaken based on a mixed linear model (Q + K) with TASSEL. SNP markers were tested for associations with Al tolerance [RNRG; SI Appendix, Table S4 (20)] and SbMATE expression (ΔΔCt) at 5 d of Al exposure.
Transactivation Assays.
Full-length promoter fragments and trans-factor cDNA (v1.4 of the sorghum genome) sequences were commercially synthesized, and transactivation assays were conducted as described in SI Appendix, Supplementary Methods. The experiments were repeated four times with similar results.
Transcription Factor Effects on SbMATE Expression and Al Tolerance via Haplotype Analysis in an RIL Population.
SbWRKY1 and SbZNF1 genotyping was based on an indel and a SNP polymorphism, respectively, as described in the SI Appendix, Supplementary Methods.
Chromatin Immunoprecipitation Assay.
Leaf protoplasts were isolated from transgenic Arabidopsis thaliana plants transformed with different SbMATE promoter fragments and then transformed with the 35S::YFP::SbWRKY1 and 35S:YFP::SbZNF1 vectors. See SI Appendix, Supplementary Methods.
Supplementary Material
Acknowledgments
We thank Veridiana Cano for assistance with allele-specific expression assays and William Lucas (University of California, Davis) for critically reading the manuscript. We acknowledge grants from the CGIAR Generation Challenge Program, the Embrapa Macroprogram, the Fundação de Amparo a Pesquisa do Estado de Minas Gerais, and the National Council for Scientific and Technological Development.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Data associated with this paper are available to download from the Dryad Digital Repository (doi:10.5061/dryad.18p3h04). The uploaded data (December 10, 2018) include SNP physical positions and association P values with Al tolerance and SbMATE expression.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808400115/-/DCSupplemental.
References
- 1.Wittkopp PJ, Haerum BK, Clark AG. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet. 2008;40:346–350. doi: 10.1038/ng.77. [DOI] [PubMed] [Google Scholar]
- 2.Cubillos FA, et al. Extensive cis-regulatory variation robust to environmental perturbation in Arabidopsis. Plant Cell. 2014;26:4298–4310. doi: 10.1105/tpc.114.130310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Uexküll HR, Mutert E. Global extent, development and economic impact of acid soils. Plant Soil. 1995;171:1–15. [Google Scholar]
- 4.Magalhaes JV, et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet. 2007;39:1156–1161. doi: 10.1038/ng2074. [DOI] [PubMed] [Google Scholar]
- 5.Carvalho G, Jr, et al. Back to acid soil fields: The citrate transporter SbMATE is a major asset for sustainable grain yield for sorghum cultivated on acid soils. G3 Genes Genomes Genet. 2016;6:475–484. doi: 10.1534/g3.115.025791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leiser WL, et al. Two in one sweep: Aluminum tolerance and grain yield in P-limited soils are associated to the same genomic region in West African sorghum. BMC Plant Biol. 2014;14:206. doi: 10.1186/s12870-014-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melo JO, et al. Incomplete transfer of accessory loci influencing SbMATE expression underlies genetic background effects for aluminum tolerance in sorghum. Plant J. 2013;73:276–288. doi: 10.1111/tpj.12029. [DOI] [PubMed] [Google Scholar]
- 8.Wessler SR, Bureau TE, White SE. LTR-retrotransposons and MITEs: Important players in the evolution of plant genomes. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 9.Magalhaes JV, et al. Comparative mapping of a major aluminum tolerance gene in sorghum and other species in the poaceae. Genetics. 2004;167:1905–1914. doi: 10.1534/genetics.103.023580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caniato FF, et al. Genetic diversity for aluminum tolerance in sorghum. Theor Appl Genet. 2007;114:863–876. doi: 10.1007/s00122-006-0485-x. [DOI] [PubMed] [Google Scholar]
- 11.Yanagisawa S. A novel DNA-binding domain that may form a single zinc finger motif. Nucleic Acids Res. 1995;23:3403–3410. doi: 10.1093/nar/23.17.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanagisawa S, Schmidt RJ. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999;17:209–214. doi: 10.1046/j.1365-313x.1999.00363.x. [DOI] [PubMed] [Google Scholar]
- 13.Machens F, Becker M, Umrath F, Hehl R. Identification of a novel type of WRKY transcription factor binding site in elicitor-responsive cis-sequences from Arabidopsis thaliana. Plant Mol Biol. 2014;84:371–385. doi: 10.1007/s11103-013-0136-y. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y, et al. Members of WRKY group III transcription factors are important in TYLCV defense signaling pathway in tomato (Solanum lycopersicum) BMC Genomics. 2016;17:788. doi: 10.1186/s12864-016-3123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou L-Z, et al. Protein S-ACYL Transferase10 is critical for development and salt tolerance in Arabidopsis. Plant Cell. 2013;25:1093–1107. doi: 10.1105/tpc.112.108829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chamberlain LH, Shipston MJ. The physiology of protein S-acylation. Physiol Rev. 2015;95:341–376. doi: 10.1152/physrev.00032.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Zhang L, Meyer E, Matz MV. Characterization of a group of MITEs with unusual features from two coral genomes. PLoS One. 2010;5:e10700. doi: 10.1371/journal.pone.0010700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo D, et al. Coevolution within a transcriptional network by compensatory trans and cis mutations. Genome Res. 2010;20:1672–1678. doi: 10.1101/gr.111765.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goncalves A, et al. Extensive compensatory cis-trans regulation in the evolution of mouse gene expression. Genome Res. 2012;22:2376–2384. doi: 10.1101/gr.142281.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caniato FF, et al. Association mapping provides insights into the origin and the fine structure of the sorghum aluminum tolerance locus, AltSB. PLoS One. 2014;9:e87438. doi: 10.1371/journal.pone.0087438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.