Significance
β-amyloid is regarded by some scientists to be the cause of Alzheimer’s disease (AD). One of the strongest arguments against this hypothesis is the presence of hundreds of AD-causing mutations in presenilin, but none in the other three components of γ-secretase. This observation implies a γ-secretase–independent function of presenilin. To understand such a putative function, discovery of presenilin-binding proteins represents an important first step. In this study, we report the identification of Bax-inhibitor 1 (BI1) as a stable interacting partner of presenilin 1 (PS1), but not the intact γ-secretase. Our results link PS1 to BI1, a protein thought to play a role in apoptosis and calcium channel regulation. This finding opens a range of possibilities for the investigation of PS1 function and AD genesis.
Keywords: presenilin, γ-secretase, Alzheimer’s disease, Bax inhibitor 1, β-amyloid
Abstract
Presenilin is the catalytic subunit of γ-secretase, a four-component intramembrane protease responsible for the generation of β-amyloid (Aβ) peptides. Over 200 Alzheimer’s disease-related mutations have been identified in presenilin 1 (PS1) and PS2. Here, we report that Bax-inhibitor 1 (BI1), an evolutionarily conserved transmembrane protein, stably associates with PS1. BI1 specifically interacts with PS1 in isolation, but not with PS1 in the context of an assembled γ-secretase. The PS1–BI1 complex exhibits no apparent proteolytic activity, as judged by the inability to produce Aβ40 and Aβ42 from the substrate APP-C99. At an equimolar concentration, BI1 has no impact on the proteolytic activity of γ-secretase; at a 200-fold molar excess, BI1 reduces γ-secretase activity nearly by half. Our biochemical study identified BI1 as a PS1-interacting protein, suggesting additional functions of PS1 beyond its involvement in γ-secretase.
Presenilin 1 (PS1) and, to a lesser extent, PS2 are frequently mutated in early-onset familial Alzheimer’s disease (FAD) (1). Presenilin interacts with nicastrin, Aph-1, and Pen-2 to form an intramembrane protease complex known as γ-secretase, in which presenilin serves as the catalytic subunit. γ-Secretase is responsible for the generation of β-amyloid (Aβ) peptides from the amyloid precursor protein (APP) (2). The longer forms of Aβ peptides, exemplified by Aβ42 and Aβ43, are thought to be more toxic than the shorter ones (3–5). Due to its suggested role in Alzheimer’s disease (AD) pathogenesis, γ-secretase has been a major target for therapeutic intervention (6, 7). However, none of the compounds that alter Aβ peptide production through modulation of γ-secretase has produced clear clinical efficacy (8). In some cases, such compounds even worsened the symptoms of patients with AD and inflicted unexpected side effects, presumably through alteration of the cleavage of other γ-secretase substrates, such as Notch (9, 10).
Since the discovery of presenilin as a component of γ-secretase, functional investigation of presenilin has been focused on its role in the context of γ-secretase, namely, the enzymatic generation of Aβ peptides. Over the years, the amyloid hypothesis has been proposed and refined to explain the pathogenesis of AD (11, 12). According to the hypothesis, aberrant cleavage of APP by γ-secretase, manifested by the increased ratios of Aβ42/Aβ40, results in accumulation of amyloid plaques in the brain that causes AD. However, clinical observations are not fully consistent with this hypothesis (13, 14). Individuals with substantial amyloid plaques may have no clinical symptoms of dementia (15, 16). In contrast, although the familial form of frontotemporal dementia is mainly linked to mutations in C9orf72, Progranulin, and Tau, a handful of presenilin mutations have been found to be associated with some disease families without amyloid deposition (17–19). A systematic analysis of 138 AD-derived PS1 mutations revealed no statistically significant correlation between the Aβ42/Aβ40 ratio produced by a specific mutant γ-secretase and the average age at onset for patients with AD who have the mutation (20).
The presenilin hypothesis postulates that loss of certain functions of presenilin may be more important to the development of AD (21). Indeed, PS1 was reported to form a calcium leak channel independent of γ-secretase, and the channel activity was affected by mutations in PS1 (22–24), although this conclusion has been challenged by subsequent studies (25). Overexpression of PS1 induces apoptosis (26–28), and PS1 ablation or mutation causes calcium dysregulation (29, 30) and impairment of long-term potentiation in neuronal cells (31). Deletion of PS1 in the plant Physcomitrella patens, which lacks APP and Notch, leads to abnormity in early development and sterility (32). Notably, the archaeal homolog of presenilin (PSH) assembles into a homotetramer and produces Aβ peptides similar to the intact γ-secretase in an in vitro assay (33, 34).
Despite all these studies, presenilin as a component of γ-secretase has dominated its functional investigation. All binding proteins of presenilin or γ-secretase have been scrutinized with respect to their impact on Aβ peptide generation. To date, no protein has been found to only interact with presenilin in isolation, but not with presenilin in the context of γ-secretase. In this study, we report such a case and identify Bax inhibitor 1 (BI1) (35, 36) as a γ-secretase–independent presenilin-binding protein. This finding opens a range of possibilities for functional investigation.
Results
Identification of BI1 as a PS1-Binding Protein.
To identify potential PS1-binding proteins, we transiently expressed FLAG-tagged full-length human PS1 in HEK293 cells and used an anti-FLAG affinity column for its purification (Fig. 1A). The eluted sample was subjected to mass spectrometry (MS) analysis. Despite its relatively low expression level, PS1 eluted from the affinity column is clearly visible on the denaturing SDS/PAGE gel stained by Coomassie blue (Fig. 1B). In addition to PS1, a number of other proteins are present, and their presence was made more apparent by silver staining (Fig. 1C). Comparison with the control, where empty vector was used for transfection, reveals several candidates that are uniquely present in the sample derived from PS1 transfection. Some of these proteins were identified by MS to be the usual contaminants (e.g., tubulin, calnexin) or chaperones for proper protein folding (e.g., hsp70). To systematically identify all potential binding partners of presenilin, we applied all eluted proteins in the sample to MS analysis. This analysis led to the identification of over 1,000 proteins above the normalized spectral abundance factor (NSAF) value of 2 × 10−4, whereas the NSAF value of PS1 is 4 × 10−3.
Fig. 1.
Identification of BI1 as a potential PS1-binding protein. (A) Procedure for the identification of potential PS1-binding proteins. The FLAG-tagged human PS1 was overexpressed in HEK293F cells and purified by the M2 affinity resin. The eluted proteins were precipitated by trichloroacetic acid (TCA) and analyzed by MS. (B) Affinity-purified proteins were examined on an SDS/PAGE gel and stained by Coomassie blue. The full-length human PS1 is clearly present in the sample derived from PS1 transfection, but not in the sample derived from the empty vector control. (C) Affinity-purified proteins were examined by silver staining. A number of proteins are uniquely present in the sample derived from PS1 transfection. (D) Amino acid sequence of BI1. Blue cylinders denote predicted transmembrane helices (TMs) based on the structure of the bacterial BI1 homolog BsYetJ (60).
Next, based on the results of three independent affinity purification experiments, we selected a small subset of the MS-identified common candidates using two criteria. First, each of the candidate proteins must be at least 10-fold more abundant in the PS1-expressing sample than in the control. Second, the NSAF value for each of the candidate proteins must be 1% or more of that for PS1. This analysis led to the identification of about 100 proteins as potential binding partners of PS1. As anticipated, some of the known γ-secretase components, such as nicastrin, were among these proteins. To select γ-secretase–independent PS1-binding proteins, we removed those candidates that are also present in the sample derived from the γ-secretase–transfected cells. The remaining 24 candidates were individually scrutinized. Notably, BI1, an evolutionarily conserved membrane protein (36, 37), is abundantly present in the MS results of the sample derived from PS1 transfection, but not in the sample derived from γ-secretase overexpression (Fig. 1D and SI Appendix, Table S1). BI1 is reported to play a role in apoptosis (35) and may function as a calcium leak channel (38–40).
BI1 Interacts with PS1 both in Vitro and in Cells.
To examine the potential interactions between PS1 and BI1, we prepared the postnuclear supernatant (PNS) of HEK293F cells that had been doubly transfected by PS1 and BI1. The PNS was applied to Percoll gradient centrifugation. The similar patterns of cellular membrane distribution between PS1 and BI1 suggest their association in cells (Fig. 2A). Next, we analyzed the subcellular localization of PS1 and BI1, which were fused to GFP and RFP, respectively (Fig. 2B). To address the potential problem of membrane insertion abnormality caused by fusion, GFP or RFP was individually tethered to the N terminus as well as the C terminus of the target protein. For all four possible combinations of the fusion, PS1 and BI1 appeared to colocalize, as judged by confocal microscopy (Fig. 2B).
Fig. 2.
BI1 and PS1 form a stable complex. (A) Subcellular fractionation shows colocalization of FLAG-tagged PS1 and His6-tagged BI1. The PNS of HEK293F cells was fractionated on 30% Percoll and subjected to Western blot analysis using a monoclonal antibody against the His6 or FLAG tag. (B) PS1 and BI1 colocalize in HEK293 cells. PS1 and BI1 were fused with GFP and RFP, respectively, at their N or C termini. In all four combinations, PS1 and BI1 colocalize in cells by confocal microscopy. (Scale bar, 10 μm.) (C) PS1 interacts with BI1 in the IP-Western analysis. The cellular extract was immunoprecipitated using an anti-FLAG monoclonal antibody, and the pellet was examined by Western blot (WB) using anti-FLAG and anti-His6 antibodies. IP, immunoprecipitation. (D) PS1 and BI1 form a stable complex. The recombinant FLAG-tagged PS1 and His6-tagged BI1 were individually or together applied to gel filtration. The fractions were examined by Western blot.
Using an immunoprecipitation-Western blot (IP-Western) assay, we investigated the interactions between PS1 and BI1 in HEK293 cells. His6-tagged PS1 was readily detectable in the anti-FLAG–immunoprecipitated pellet in the presence of, but not in the absence of, the FLAG-tagged BI1 (Fig. 2C, Left). Conversely, His6-tagged BI1 appeared in the anti-FLAG–immunoprecipitated pellet only in the presence of the FLAG-tagged PS1 (Fig. 2C, Right). These results corroborate the observed cellular colocalization between PS1 and BI1. Finally, we individually purified FLAG-tagged PS1, His6-tagged BI1, and the complex between the two proteins; we examined their solution behavior using gel filtration (Fig. 2D). FLAG-tagged PS1 and His6-tagged BI1, both in isolation, displayed elution volumes of ∼8.75 mL and ∼10.5 mL, respectively (Fig. 2D, Top, red- and green-outlined panels). The PS1–BI1 binary complex, however, exhibited an elution volume of about 9.5 mL, different from either protein in isolation (Fig. 2D, Bottom, blue-outlined panels). These observations indicate a stable nature of the PS1–BI1 interaction in vitro.
Proteolytic Activity of the PS1–BI1 Complex.
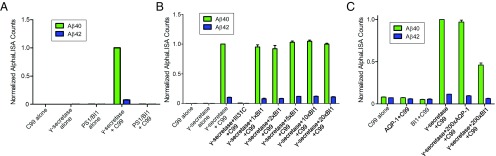
A variant of PS1, which has its exon 9 deleted (ΔE9), was reported to be proteolytically active by itself upon incorporation into the liposome (41). An archaeal PS1 homolog (PSH) faithfully recapitulates γ-secretase activity (33, 34). These observations suggest the possibility that the PS1–BI1 complex may represent another enzymatically active form of PS1. To examine this possibility, we conducted an in vitro bulk cleavage assay that was originally designed for γ-secretase using a 99-residue peptide derived from the C terminus of APP (APP C99) as the substrate (20, 33). In contrast to γ-secretase, the PS1–BI1 complex failed to produce any detectable amount of Aβ42 or Aβ40 using the sensitive, spectroscopic AlphaLISA detection method (Fig. 3A).
Fig. 3.
PS1–BI1 complex exhibits no proteolytic activity toward the APP-C99 substrate. (A) PS1–BI1 complex failed to generate Aβ42 or Aβ40 using the APP-C99 substrate. The spectroscopic reading of Aβ40 by γ-secretase is normalized as 1.0. The PS1–BI1 complex has no detectable activity. (B) Recombinant BI1 protein exhibits no apparent impact on the production of Aβ42 or Aβ40 by γ-secretase in vitro. Compared with γ-secretase, the amount of BI1 varied from equimolar to a 20-fold molar excess. (C) BI1 at a 200-fold molar excess inhibited the production of Aβ42 or Aβ40 by γ-secretase. The effect is specific because the control protein, AQP-1, at a 200-fold molar excess exhibited no impact on the proteolytic activity of γ-secretase.
Next, we examined the impact of the recombinant BI1 protein on the proteolytic activity of γ-secretase. The production of Aβ42 or Aβ40 by γ-secretase was unaffected by BI1 over a wide range of concentrations: from an equimolar amount to a 20-fold molar excess over γ-secretase (Fig. 3B). In contrast, the γ-secretase–specific inhibitor III-31C completely abrogated the generation of Aβ42 or Aβ40 by γ-secretase. Interestingly, however, when the concentration of BI1 reached a 200-fold molar excess over γ-secretase, the production of Aβ42 or Aβ40 by γ-secretase was markedly reduced (Fig. 3C). The presence of a control membrane protein, AQP-1, at a similar concentration failed to influence the proteolytic activity of γ-secretase, suggesting specific inhibition by BI1. We speculate that, at a sufficiently high concentration, BI1 dissociates PS1 from the γ-secretase complex, and hence cripples its enzymatic activity. Nonetheless, we cannot rule out the scenario, albeit highly unlikely, that the inhibition of γ-secretase by BI1 at the very high concentration is actually due to a trace amount of an unidentified factor that copurifies with the recombinant BI1 protein.
The absence of proteolytic activity by the PS1–BI1 complex toward the APP-C99 substrate, together with the lack of meaningful inhibition by BI1 toward γ-secretase, strongly argues that the PS1–BI1 complex might be involved in a cellular function that is independent of γ-secretase.
Characterization of the PS1–BI1 Interaction.
The way BI1 was identified and the in vitro cleavage studies both suggest BI1 to be a γ-secretase–independent binding protein of PS1. To corroborate the mutual exclusivity of PS1 in the PS1–BI1 complex versus γ-secretase, we performed an IP-Western assay using the HEK293F cells where PS1, Aph-1A, Pen-2, nicastrin, and BI1 were coexpressed. The expression of each protein was confirmed by a Western blot of the cell lysate (Fig. 4A, Left). The cell lysate was applied to an anti-FLAG column for the purification of FLAG-tagged PS1 and its associated proteins. As anticipated, the elution contains the other three components of γ-secretase (nicastrin, Aph-1A, and Pen-2), but BI1 was undetectable even with prolonged exposure of the Western blot (Fig. 4A, Right).
Fig. 4.
BI1 binds the isolated PS1, but not PS1 in the context of an assembled γ-secretase. (A) BI1 is absent in the purified γ-secretase. (Left) All four components of γ-secretase and BI1 are readily detectable upon cellular overexpression. (Right) Following affinity purification of the FLAG-tagged PS1, the other three components of γ-secretase, but not BI1, are detectable by Western blot. (B) Colocalization of PS1 and BI1 is altered by expression of the other three components of. γ-secretase. (Top) Coexpression of GFP-PS1 and RFP-BI1 reveals excellent colocalization. (Bottom) Extent of colocalization is markedly reduced by coexpression of the other three components of γ-secretase (Scale bar, 10 μm.) (C) Quantification of colocalization between PS1 and BI1 by Pearson’s coefficient in colocalized volume. (Left) Coefficient in the presence of PS1 overexpression alone is considerably higher than that in the presence of overexpression by all four components of γ-secretase. (Right) Typical 2D correlation diagrams. (D) Purification of the PS1–BI1 complex for structural studies. (Left) Representative gel filtration chromatograph of the two-step purified human PS1–BI1 complex. (Right) Aliquots of the peak fractions are visualized in an SDS/PAGE gel. (E) Analysis of the purified PS1–BI1 complex by negative-staining EM. Shown here is a representative micrograph of the PS1–BI1 complex.
We then examined the cellular colocalization of PS1 and BI1 in the presence of overexpression of the other three components of γ-secretase. Because the C terminus of PS1 is buried in a hydrophobic pocket within Aph-1 (42), we fused GFP to the N terminus of PS1. Compared with that in the absence of overexpression of the other three components of γ-secretase, the colocalization between the GFP-tagged PS1 and RFP-tagged BI1 is markedly reduced in the presence of such overexpression (Fig. 4B). Quantification of the results reveals that the colocalization ratio between GFP-tagged PS1 and RFP-tagged BI1 is significantly higher in the absence of overexpression of the other three components of γ-secretase than in the presence of their overexpression (Fig. 4C). The microscopy data, together with results of the IP-Western experiments, demonstrate that the interaction of PS1 with BI1 is mutually exclusive with the involvement of PS1 in γ-secretase.
More than 200 mutations in PS1 have been identified from patients with early-onset FAD (1). How these mutations cause FAD in a dominant negative fashion remains to be mechanistically characterized (34, 43). Although a large portion of the FAD mutations impair the proteolytic activity of γ-secretase and increase the Aβ42/Aβ40 ratio (44–47), some mutations exhibit the opposite effect on γ-secretase: with enhanced proteolytic activity and a lower Aβ42/Aβ40 ratio (20). These latter PS1 mutations run contrary to the Aβ hypothesis and may have an increased likelihood to have an impact on an aspect of cellular function that is unrelated to γ-secretase. Based on this rationale, we generated nine PS1 variants, each containing one such FAD mutation (SI Appendix, Fig. S1A). Compared with the WT PS1, each of the nine PS1 variants retained a qualitatively similar ability to interact with BI1 in an in vitro pulldown assay (SI Appendix, Fig. S1B).
To prepare the PS1–BI1 complex for structural studies, we coexpressed FLAG-tagged PS1 and His-tagged BI1 in HEK293 cells and isolated the binary complex through a two-step affinity purification procedure. The recombinant PS1–BI1 complex was eluted from gel filtration as a single but broad peak, suggesting mild aggregation and/or equilibrium among distinct states under the experimental condition (Fig. 4D). The elution volume corresponds to an apparent molecular mass of over 440 kDa, which exceeds the anticipated molecular mass of a PS1–BI1 heterodimer. The purified PS1–BI1 complex was examined by electron microscopy (EM) under negative staining (Fig. 4E). The particles are clearly identifiable without large aggregates; the varying sizes of the particles are consistent with the observed solution behavior on gel filtration and may pose a serious challenge for EM-based structural studies. The gel filtration analysis and the EM image are consistent with the notion that the PS1–BI1 complex may represent an equilibrium between higher order and lower order oligomers.
Conservation of the PS1–BI1 Interaction.
Phylogenetic analysis reveals a similar pattern of evolutionary conservation for both PS1 and BI1 in eukaryotes, suggesting functional importance (Fig. 5A). To examine this scenario, we investigated the potential interactions between PS1 and BI1 orthologs in four representative model organisms: plant (Arabidopsis thaliana), fruit fly (Drosophila melanogaster), zebrafish (Danio rerio), and frog (Xenopus laevis). Remarkably, in every case, the PS1 and BI1 orthologs formed a stable complex, as demonstrated by results of the IP-Western assay (Fig. 5B). Conservation of the PS1–BI1 interaction implies that this complex might be involved in an ancestral cellular function.
Fig. 5.
PS1–BI1 interaction is evolutionarily conserved. (A) Phylogenetic trees of PS1 and BI1. The evolutionary history was inferred using the maximum parsimony method. PS1 and BI1 exhibit a similar pattern of evolutionary history in eukaryotic species, suggesting coevolution of the two proteins. Evolutionary analyses were conducted in MEGA7 (61). (B) PS1 and BI1 homologs interact with each other in four representative model organisms. Shown here are results of the IP-Western assays. (C) BI1 also binds to PS2, which exhibits strong sequence homology to PS1. Shown here are results of the pulldown experiments.
Discussion
Despite an intimate link between presenilin and a small percentage of AD cases, how presenilin actually causes the disease remains unresolved (11, 14, 21, 44). The amyloid hypothesis restricts the function of presenilin to the context of γ-secretase and suffers from a number of clinical, biochemical, and genetic observations (13, 15–19). The presenilin hypothesis is technically appealing, but its exact meaning remains to be defined (21). Identification of a γ-secretase–independent function of PS1 and the discovery of PS1-binding proteins represent two key approaches in addressing the presenilin hypothesis.
Relying on affinity purification and MS, we have identified BI1 as a previously unknown PS1-binding protein (Fig. 1). BI1 stably interacts with PS1 in a γ-secretase–independent manner both in vitro and in cells (Figs. 2 and 4). Importantly, BI1 only binds free PS1, but not PS1 in the context of γ-secretase (Fig. 4). At an equimolar concentration, free BI1 is unable to snatch PS1 from γ-secretase, and consequently fails to inhibit the proteolytic activity of γ-secretase (Fig. 3). At a 200-fold molar excess over γ-secretase, however, BI1 markedly diminished its proteolytic activity. This is likely due to the dissociation of PS1 from, and subsequent disassembly of, the γ-secretase complex. Although PS1 contains two catalytic Asp residues, the PS1–BI1 complex exhibits no detectable proteolytic activity in terms of Aβ40 and Aβ42 production from the APP-C99 substrate. This result does not rule out the possibility that the PS1–BI1 complex may display proteolytic activity under other circumstances.
Intriguingly, the amount of FLAG-tagged BI1 brought down by the anti-FLAG monoclonal antibody was greatly diminished in the presence of His6-tagged PS1 (Fig. 2C, Left). This observation hints at a potential role of PS1 in the regulation of BI1 expression. In contrast, the amount of FLAG-tagged PS1 brought down by the anti-FLAG antibody was unaffected by the presence of BI1 (Fig. 2C, Right). FLAG-tagged PS1 alone was eluted from gel filtration in earlier fractions compared with the complex between FLAG-tagged PS1 and His6-tagged BI1 (Fig. 2D). This observation strongly suggests that free PS1 may exist in an oligomeric state, consistent with published results (43).
Both PS1 and BI1 are evolutionarily conserved membrane proteins (Fig. 5A) and appear to have similar cellular functions. Expression of PS1 is known to trigger apoptosis (26–28) and affects calcium signaling in cells (29, 30); expression of BI1 causes similar cellular phenotypes (35, 38). These similar functions are underscored by stable interactions between PS1 and BI1. PS2, which shares considerable sequence identity with PS1, is also known to induce apoptosis (48) and calcium dysregulation (49). In a preliminary IP-Western analysis, the human BI1 protein stably interacted with human PS2 (Fig. 5C).
Due to the low expression levels of PS1 and BI1 in cells, detection of the endogenous PS1–BI1 complex has proven to be technically challenging. This challenge is made tougher by the fact that PS1 shows a considerably stronger tendency to be part of γ-secretase compared with the PS1–BI1 complex. Nonetheless, we have tried to determine the interaction and localization patterns of endogenous PS1 and BI1 in cells. Endogenous PS1–BI1 interactions in HEK293 cells could be detected, and a small portion of PS1 was found to colocalize with BI1 in SH-SY5Y neuronal cells (data not shown). We speculate that the PS1–BI1 interaction might be greatly strengthened under certain conditions, such as excessive innate synthesis of PS1, PS1 mutations that impede its assembly into γ-secretase, cellular stress that disassembles γ-secretase, and conditions that entail the function of free PS1. Notably, for both PS1 and BI1, the transcription and protein expression levels are relatively high in a number of human organs (SI Appendix, Fig. S2). Investigation of the PS1–BI1 interaction in these organs may reveal unanticipated clues. In addition, it is possible that other unidentified protein(s) are involved in, or modulate, the PS1–BI1 interaction. Detection methods of a more sensitive nature may be required to further examine the endogenous PS1–BI1 interaction.
What is the cellular function of the PS1–BI1 complex? Is there a link between the function of the PS1–BI1 complex and AD genesis? Although conclusive answers are yet to be provided, there are tantalizing clues. Dysregulation of Ca2+ homeostasis and neuronal loss due to excessive cell death are hallmarks of AD (28, 50, 51). BI1 is thought to be a Ca2+ leak channel on the endoplasmic reticulum (38, 40, 52) (SI Appendix, Fig. S3). PS1 was also initially reported to function as a Ca2+ leak channel (22–24), although this claim has been rigorously contested by independent groups (25, 53, 54). Similar experiments using mouse embryonic fibroblasts generated contrasting results (24, 25). In more physiologically relevant experimental systems (e.g., presenilin-null hippocampal slices, primary neuronal cultures), reduced ryanodine receptor (RyR) activity or expression was seen to be correlated with presenilin inactivation (53, 54). In addition, these electrophysiology findings are consistent with calcium imaging data (53, 54). It is thus highly unlikely that PS1 conducts ion permeation by itself. Rather, the role of PS1 in calcium regulation may involve its functional interactions with other channels, such as RyR (53–55).
Our gel filtration analysis suggests that the PS1–BI1 complex may exist as a higher order oligomer (Fig. 4D). Examination of potential ion conductance of the PS1–BI1 complex and comparison of such data with those of BI1 alone may reveal unanticipated clues. On the other hand, overexpression of PS1 exhibits a proapoptotic effect (26–28), whereas BI1 was originally identified in a functional screening assay to suppress Bax-induced cell death in yeast (35). Can the PS1–BI1 complex function as a regulated apoptosis trigger in neuronal cells? Obviously, these speculations await experimental scrutiny.
Before our study, several presenilin-binding proteins have been identified. TMP21 binds and regulates the proteolytic activity of γ-secretase, altering Aβ peptide generation (56). Hif-1α, a protein expressed under hypoxic conditions, was found to interact with γ-secretase to modulate its activity (57). Recently, annexin A2 was reported to bind the Ser367-phosphorylated PS1, modulating Aβ peptide levels through autophagy (58, 59). All these studies impinge upon the Aβ peptide level through regulation of the γ-secretase. Unlike HiF-1α or TMP21, BI1 only binds free PS1, but not PS1, in the context of γ-secretase. PS1, on the other hand, has a much higher tendency to assemble into γ-secretase compared with formation of the PS1–BI1 complex. Therefore, the difficulty in detecting the PS1–BI1 interaction could be a major reason why such interaction has long been overlooked. Although functionally unresolved, BI1 represents a PS1 binding protein that does not interact with γ-secretase.
Materials and Methods
Clones and Plasmids.
The cDNAs were individually cloned into the pCAG vector. All plasmids used for transfection of mammalian cells were prepared using the EndoFree Plasmid Maxi Kit (CWBiotech).
Purification of PS1-Binding Proteins.
The FLAG-tagged PS1 or γ-secretase was individually transfected into HEK293F cells as described (42). After 48 h, the total cell lysate was prepared in lysis buffer [25 mM Hepes (pH 7.4), 150 mM NaCl] supplemented with 1% (wt/vol) digitonin before centrifugation. The supernatant was incubated with ANTI-FLAG M2 Affinity Gel (Sigma–Aldrich). The proteins were eluted by 200 μg/mL FLAG peptide and analyzed by MS.
IP-Western Assays.
Transfection and IP-Western assays were performed as described (20, 42).
Subcellular Fractionation.
FLAG-PS1– and His6-BI1–cotransfected HEK293 cells were suspended in the lysis buffer before homogenization and centrifugation. The PNS was centrifuged in 30% Percoll. PS1 and BI1 were detected by Western blots.
Protein Purification and Gel Filtration Analysis.
The human PS1, BI1, and γ-secretase were transfected or cotransfected individually into HEK293F cells as described (42). The target proteins were similarly affinity-purified as described above for PS1-binding proteins. The eluted proteins were analyzed by gel filtration and Western blots.
Purification of the PS1–BI1 Complex.
The complex between FLAG-tagged PS1 and His6-tagged BI1 was coexpressed and affinity-purified as described above. The eluted sample was further purified through a Ni2+-nitrilotriacetic acid column, followed by gel filtration. The peak fractions were visualized by negative-staining EM.
Cleavage Activity Assays.
The proteolytic activity, as indicated by the production of Aβ42 and Aβ40 for various samples, was measured using the AlphaLISA assay (PerkinElmer) as described (20).
Detection of Protein Localization in Cells.
Plasmids for various proteins were transfected into HEK293F cells using Lipofectamine LTX & PLUS Reagent (Invitrogen). After 24 h, cells were imaged by confocal microscopy, with wavelengths of 488 nm for GFP and 558 nm for RFP. The images were analyzed by Imaris software to obtain the Pearson’s coefficient in colocalized volume.
Supplementary Material
Acknowledgments
We thank the National Center for Protein Science (Beijing) Tsinghua for use of the EM Facility and the assistance of the Imaging Core Facility, Technology Center for Protein Sciences (Tsinghua University). This work was supported by funds from the National Natural Science Foundation of China (Grants 31621092 and 31430020).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810870116/-/DCSupplemental.
References
- 1.Cruts M, Theuns J, Van Broeckhoven C. Locus-specific mutation databases for neurodegenerative brain diseases. Hum Mutat. 2012;33:1340–1344. doi: 10.1002/humu.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Strooper B, Iwatsubo T, Wolfe MS. Presenilins and γ-secretase: Structure, function, and role in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2:a006304. doi: 10.1101/cshperspect.a006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burdick D, et al. Assembly and aggregation properties of synthetic Alzheimer’s A4/beta amyloid peptide analogs. J Biol Chem. 1992;267:546–554. [PubMed] [Google Scholar]
- 4.Jarrett JT, Berger EP, Lansbury PT., Jr The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer’s disease. Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki N, et al. An increased percentage of long amyloid beta protein secreted by familial amyloid beta protein precursor (beta APP717) mutants. Science. 1994;264:1336–1340. doi: 10.1126/science.8191290. [DOI] [PubMed] [Google Scholar]
- 6.Tomita T, Iwatsubo T. The inhibition of gamma-secretase as a therapeutic approach to Alzheimer’s disease. Drug News Perspect. 2004;17:321–325. doi: 10.1358/dnp.2004.17.5.829036. [DOI] [PubMed] [Google Scholar]
- 7.Crump CJ, Johnson DS, Li YM. Development and mechanism of γ-secretase modulators for Alzheimer’s disease. Biochemistry. 2013;52:3197–3216. doi: 10.1021/bi400377p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummings JL, Morstorf T, Zhong K. Alzheimer’s disease drug-development pipeline: Few candidates, frequent failures. Alzheimers Res Ther. 2014;6:37. doi: 10.1186/alzrt269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doody RS, et al. Alzheimer’s Disease Cooperative Study Steering Committee; Semagacestat Study Group A phase 3 trial of semagacestat for treatment of Alzheimer’s disease. N Engl J Med. 2013;369:341–350. doi: 10.1056/NEJMoa1210951. [DOI] [PubMed] [Google Scholar]
- 10.De Strooper B. Lessons from a failed γ-secretase Alzheimer trial. Cell. 2014;159:721–726. doi: 10.1016/j.cell.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 11.Hardy JA, Higgins GA. Alzheimer’s disease: The amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 12.Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 2016;8:595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrup K. The case for rejecting the amyloid cascade hypothesis. Nat Neurosci. 2015;18:794–799. doi: 10.1038/nn.4017. [DOI] [PubMed] [Google Scholar]
- 14.Pimplikar SW, Nixon RA, Robakis NK, Shen J, Tsai LH. Amyloid-independent mechanisms in Alzheimer’s disease pathogenesis. J Neurosci. 2010;30:14946–14954. doi: 10.1523/JNEUROSCI.4305-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aizenstein HJ, et al. Frequent amyloid deposition without significant cognitive impairment among the elderly. Arch Neurol. 2008;65:1509–1517. doi: 10.1001/archneur.65.11.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villemagne VL, et al. The ART of loss: Abeta imaging in the evaluation of Alzheimer’s disease and other dementias. Mol Neurobiol. 2008;38:1–15. doi: 10.1007/s12035-008-8019-y. [DOI] [PubMed] [Google Scholar]
- 17.Raux G, et al. Dementia with prominent frontotemporal features associated with L113P presenilin 1 mutation. Neurology. 2000;55:1577–1578. doi: 10.1212/wnl.55.10.1577. [DOI] [PubMed] [Google Scholar]
- 18.Tang-Wai D, et al. Familial frontotemporal dementia associated with a novel presenilin-1 mutation. Dement Geriatr Cogn Disord. 2002;14:13–21. doi: 10.1159/000058328. [DOI] [PubMed] [Google Scholar]
- 19.Dermaut B, et al. A novel presenilin 1 mutation associated with Pick’s disease but not beta-amyloid plaques. Ann Neurol. 2004;55:617–626. doi: 10.1002/ana.20083. [DOI] [PubMed] [Google Scholar]
- 20.Sun L, Zhou R, Yang G, Shi Y. Analysis of 138 pathogenic mutations in presenilin-1 on the in vitro production of Aβ42 and Aβ40 peptides by γ-secretase. Proc Natl Acad Sci USA. 2017;114:E476–E485. doi: 10.1073/pnas.1618657114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen J, Kelleher RJ., 3rd The presenilin hypothesis of Alzheimer’s disease: Evidence for a loss-of-function pathogenic mechanism. Proc Natl Acad Sci USA. 2007;104:403–409. doi: 10.1073/pnas.0608332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson O, et al. Mutagenesis mapping of the presenilin 1 calcium leak conductance pore. J Biol Chem. 2011;286:22339–22347. doi: 10.1074/jbc.M111.243063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson O, et al. Familial Alzheimer disease-linked mutations specifically disrupt Ca2+ leak function of presenilin 1. J Clin Invest. 2007;117:1230–1239. doi: 10.1172/JCI30447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu H, et al. Presenilins form ER Ca2+ leak channels, a function disrupted by familial Alzheimer’s disease-linked mutations. Cell. 2006;126:981–993. doi: 10.1016/j.cell.2006.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shilling D, Mak DO, Kang DE, Foskett JK. Lack of evidence for presenilins as endoplasmic reticulum Ca2+ leak channels. J Biol Chem. 2012;287:10933–10944. doi: 10.1074/jbc.M111.300491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terro F, et al. Neurons overexpressing mutant presenilin-1 are more sensitive to apoptosis induced by endoplasmic reticulum-Golgi stress. J Neurosci Res. 2002;69:530–539. doi: 10.1002/jnr.10312. [DOI] [PubMed] [Google Scholar]
- 27.Weihl CC, et al. Mutant presenilin-1 induces apoptosis and downregulates Akt/PKB. J Neurosci. 1999;19:5360–5369. doi: 10.1523/JNEUROSCI.19-13-05360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimohama S. Apoptosis in Alzheimer’s disease–An update. Apoptosis. 2000;5:9–16. doi: 10.1023/a:1009625323388. [DOI] [PubMed] [Google Scholar]
- 29.Kasri NN, et al. Up-regulation of inositol 1,4,5-trisphosphate receptor type 1 is responsible for a decreased endoplasmic-reticulum Ca2+ content in presenilin double knock-out cells. Cell Calcium. 2006;40:41–51. doi: 10.1016/j.ceca.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Yoo AS, et al. Presenilin-mediated modulation of capacitative calcium entry. Neuron. 2000;27:561–572. doi: 10.1016/s0896-6273(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 31.Auffret A, Gautheron V, Mattson MP, Mariani J, Rovira C. Progressive age-related impairment of the late long-term potentiation in Alzheimer’s disease presenilin-1 mutant knock-in mice. J Alzheimers Dis. 2010;19:1021–1033. doi: 10.3233/JAD-2010-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khandelwal A, Chandu D, Roe CM, Kopan R, Quatrano RS. Moonlighting activity of presenilin in plants is independent of gamma-secretase and evolutionarily conserved. Proc Natl Acad Sci USA. 2007;104:13337–13342. doi: 10.1073/pnas.0702038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang S, et al. Cleavage of amyloid precursor protein by an archaeal presenilin homologue PSH. Proc Natl Acad Sci USA. 2015;112:3344–3349. doi: 10.1073/pnas.1502150112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heilig EA, Gutti U, Tai T, Shen J, Kelleher RJ., 3rd Trans-dominant negative effects of pathogenic PSEN1 mutations on γ-secretase activity and Aβ production. J Neurosci. 2013;33:11606–11617. doi: 10.1523/JNEUROSCI.0954-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Q, Reed JC. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell. 1998;1:337–346. doi: 10.1016/s1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- 36.Hückelhoven R. BAX inhibitor-1, an ancient cell death suppressor in animals and plants with prokaryotic relatives. Apoptosis. 2004;9:299–307. doi: 10.1023/b:appt.0000025806.71000.1c. [DOI] [PubMed] [Google Scholar]
- 37.Chae HJ, et al. Evolutionarily conserved cytoprotection provided by Bax inhibitor-1 homologs from animals, plants, and yeast. Gene. 2003;323:101–113. doi: 10.1016/j.gene.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 38.Bultynck G, et al. The C terminus of Bax inhibitor-1 forms a Ca2+-permeable channel pore. J Biol Chem. 2012;287:2544–2557. doi: 10.1074/jbc.M111.275354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee GH, Lee HY, Li B, Kim HR, Chae HJ. Bax inhibitor-1-mediated inhibition of mitochondrial Ca2+ intake regulates mitochondrial permeability transition pore opening and cell death. Sci Rep. 2014;4:5194. doi: 10.1038/srep05194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bultynck G, Kiviluoto S, Methner A. Bax inhibitor-1 is likely a pH-sensitive calcium leak channel, not a H+/Ca2+ exchanger. Sci Signal. 2014;7:pe22. doi: 10.1126/scisignal.2005764. [DOI] [PubMed] [Google Scholar]
- 41.Ahn K, et al. Activation and intrinsic gamma-secretase activity of presenilin 1. Proc Natl Acad Sci USA. 2010;107:21435–21440. doi: 10.1073/pnas.1013246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai XC, et al. An atomic structure of human γ-secretase. Nature. 2015;525:212–217. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou R, Yang G, Shi Y. Dominant negative effect of the loss-of-function γ-secretase mutants on the wild-type enzyme through heterooligomerization. Proc Natl Acad Sci USA. 2017;114:12731–12736. doi: 10.1073/pnas.1713605114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chávez-Gutiérrez L, et al. The mechanism of γ-secretase dysfunction in familial Alzheimer disease. EMBO J. 2012;31:2261–2274. doi: 10.1038/emboj.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimojo M, Sahara N, Murayama M, Ichinose H, Takashima A. Decreased Abeta secretion by cells expressing familial Alzheimer’s disease-linked mutant presenilin 1. Neurosci Res. 2007;57:446–453. doi: 10.1016/j.neures.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Cacquevel M, Aeschbach L, Houacine J, Fraering PC. Alzheimer’s disease-linked mutations in presenilin-1 result in a drastic loss of activity in purified γ-secretase complexes. PLoS One. 2012;7:e35133. doi: 10.1371/journal.pone.0035133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heilig EA, Xia W, Shen J, Kelleher RJ., 3rd A presenilin-1 mutation identified in familial Alzheimer disease with cotton wool plaques causes a nearly complete loss of gamma-secretase activity. J Biol Chem. 2010;285:22350–22359. doi: 10.1074/jbc.M110.116962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolozin B, et al. Participation of presenilin 2 in apoptosis: Enhanced basal activity conferred by an Alzheimer mutation. Science. 1996;274:1710–1713. doi: 10.1126/science.274.5293.1710. [DOI] [PubMed] [Google Scholar]
- 49.Leissring MA, Parker I, LaFerla FM. Presenilin-2 mutations modulate amplitude and kinetics of inositol 1, 4,5-trisphosphate-mediated calcium signals. J Biol Chem. 1999;274:32535–32538. doi: 10.1074/jbc.274.46.32535. [DOI] [PubMed] [Google Scholar]
- 50.Supnet C, Bezprozvanny I. The dysregulation of intracellular calcium in Alzheimer disease. Cell Calcium. 2010;47:183–189. doi: 10.1016/j.ceca.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alzheimer’s Association Calcium Hypothesis W Calcium hypothesis of Alzheimer’s disease and brain aging: A framework for integrating new evidence into a comprehensive theory of pathogenesis. Alzheimers Dement. 2017;13:178–182.e17. doi: 10.1016/j.jalz.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 52.Robinson KS, Clements A, Williams AC, Berger CN, Frankel G. Bax inhibitor 1 in apoptosis and disease. Oncogene. 2011;30:2391–2400. doi: 10.1038/onc.2010.636. [DOI] [PubMed] [Google Scholar]
- 53.Zhang C, et al. Presenilins are essential for regulating neurotransmitter release. Nature. 2009;460:632–636. doi: 10.1038/nature08177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu B, Yamaguchi H, Lai FA, Shen J. Presenilins regulate calcium homeostasis and presynaptic function via ryanodine receptors in hippocampal neurons. Proc Natl Acad Sci USA. 2013;110:15091–15096. doi: 10.1073/pnas.1304171110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ho A, Shen J. Presenilins in synaptic function and disease. Trends Mol Med. 2011;17:617–624. doi: 10.1016/j.molmed.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen F, et al. TMP21 is a presenilin complex component that modulates gamma-secretase but not epsilon-secretase activity. Nature. 2006;440:1208–1212. doi: 10.1038/nature04667. [DOI] [PubMed] [Google Scholar]
- 57.Villa JC, et al. Nontranscriptional role of Hif-1α in activation of γ-secretase and notch signaling in breast cancer. Cell Reports. 2014;8:1077–1092. doi: 10.1016/j.celrep.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bustos V, et al. Bidirectional regulation of Aβ levels by presenilin 1. Proc Natl Acad Sci USA. 2017;114:7142–7147. doi: 10.1073/pnas.1705235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bustos V, et al. Phosphorylated presenilin 1 decreases β-amyloid by facilitating autophagosome-lysosome fusion. Proc Natl Acad Sci USA. 2017;114:7148–7153. doi: 10.1073/pnas.1705240114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang Y, et al. Structural basis for a pH-sensitive calcium leak across membranes. Science. 2014;344:1131–1135. doi: 10.1126/science.1252043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.