Significance
The presence of heterogeneous subsets of cancer stem cells (CSCs) remains a clinical challenge. These subsets often occupy different regions in the primary tumor and have varied epithelial–mesenchymal phenotypes. Here, we devise a theoretical framework to investigate how the tumor microenvironment (TME) modulates this spatial patterning. We find that a spatial gradient of EMT-inducing signal, coupled with juxtacrine Notch-JAG1 signaling triggered by inflammatory cytokines in TME, explains this spatial heterogeneity. Finally, in vitro JAG1 knockdown in triple-negative breast cancer SUM149 cells severely restricts the growth of tumor organoid, hence validating the association between JAG1 and CSC fraction. Our results offer insights into principles of spatiotemporal patterning in TME and identify a relevant target to alleviate multiple CSC subsets: JAG1.
Keywords: cancer stem cells, epithelial–mesenchymal transition, Notch signaling, inflammation, breast tumor organoids
Abstract
The epithelial–mesenchymal transition (EMT) and cancer stem cell (CSC) formation are two paramount processes driving tumor progression, therapy resistance, and cancer metastasis. Recent experiments show that cells with varying EMT and CSC phenotypes are spatially segregated in the primary tumor. The underlying mechanisms generating such spatiotemporal dynamics in the tumor microenvironment, however, remain largely unexplored. Here, we show through a mechanism-based dynamical model that the diffusion of EMT-inducing signals such as TGF-β, together with noncell autonomous control of EMT and CSC decision making via the Notch signaling pathway, can explain experimentally observed disparate localization of subsets of CSCs with varying EMT phenotypes in the tumor. Our simulations show that the more mesenchymal CSCs lie at the invasive edge, while the hybrid epithelial/mesenchymal (E/M) CSCs reside in the tumor interior. Further, motivated by the role of Notch-Jagged signaling in mediating EMT and stemness, we investigated the microenvironmental factors that promote Notch-Jagged signaling. We show that many inflammatory cytokines such as IL-6 that can promote Notch-Jagged signaling can (i) stabilize a hybrid E/M phenotype, (ii) increase the likelihood of spatial proximity of hybrid E/M cells, and (iii) expand the fraction of CSCs. To validate the predicted connection between Notch-Jagged signaling and stemness, we knocked down JAG1 in hybrid E/M SUM149 human breast cancer cells in vitro. JAG1 knockdown significantly restricted tumor organoid formation, confirming the key role that Notch-Jagged signaling can play in tumor progression. Together, our integrated computational–experimental framework reveals the underlying principles of spatiotemporal dynamics of EMT and CSCs.
The tumor microenvironment (TME) offers intriguing questions about the spatiotemporal dynamics of pattern formation. Abundant phenotypic and functional heterogeneity (1–3), coupled with varying concentrations of nutrient availability (4) and bidirectional cross-talk among constituent cells (5), can lead to the formation of complex patterns correlated with aggressive pathological behaviors. Two interconnected hallmarks of cellular plasticity that contribute to this spatiotemporal heterogeneity are the epithelial–mesenchymal transition (EMT) and the generation of cancer stem cells (CSCs). These are a “dangerous duo” that can cooperatively promote tumor progression, metastasis, therapy resistance, and tumor relapse (6). Remarkably, the initial proposition of the involvement of EMT in metastasis was based on spatially varying levels of β-catenin (7), where cells at the invasive edge of the tumor and those in the tumor interior contained varying subcellular localization of β-catenin and E-cadherin, thus affecting cell adhesion and migration. Extensive characterization since then has identified that the activation of EMT and the localization of different subsets of CSCs is spatially quite heterogeneous within a tumor (8, 9). This heterogeneity can arise from bidirectional interplay among tumor cells and stromal cells, thus highlighting the contributions that the noncell autonomous effects in the activation of EMT and CSCs can have on spatiotemporal dynamics in the TME.
These noncell autonomous effects can be modulated by cell–cell communication pathways that cross-talk with intracellular signaling networks governing EMT and CSCs. One canonical case is the Notch-Jagged signaling pathway, which has been implicated in metastasis, drug resistance, and tumor relapse (10). Notch-Jagged signaling has been studied in multiple developmental contexts for its role in cellular decision making and tissue patterning (11–13). Recent in silico, in vitro, and in vivo observations have suggested that Notch-Jagged signaling can promote a hybrid epithelial/mesenchymal (E/M) phenotype and traits similar to CSCs (5, 14–17). Moreover, Notch-Jagged signaling among cancer cells can facilitate the formation of clusters of circulating tumor cells (CTCs)—the “bad actors” of cancer metastasis (15, 16, 18). Furthermore, Notch-Jagged signaling between cancer cells and stromal cells and/or cells in metastatic niche can aggravate tumor progression (19). Put together, hybrid E/M cells with CSC-like traits and activated Notch-Jagged signaling perhaps represents the “fittest” phenotype for metastasis (20–23).
Several factors in the TME can activate the Notch-Jagged pathway. For instance, many inflammatory cytokines such as IL-6, IL-1β, and TNF-α can promote Notch-Jagged signaling by increasing the intracellular production of Jagged. Some of them can also decrease the production of Delta, a competing ligand that binds to the Notch receptor (24–27). Inflammation is a hallmark of damaged tissues: acute inflammation plays a crucial role in wound healing; chronic inflammation predisposes the damaged tissue to the development of cancer. Also, cancer patients treated with chemotherapy and radiation have elevated inflammation (28, 29), often via the abovementioned inflammatory cytokines that can aggravate tumor progression by both promoting EMT and CSC formation (30, 31). The operating principles behind the complex spatiotemporal interplay between signaling cues diffusing in the TME, noncell autonomous signaling via Notch-Jagged, and the intracellular molecular machinery regulating EMT and CSC, however, remain largely elusive.
Here, we propose a mathematical modeling framework that captures the experimentally identified interconnections among inflammatory cytokines, EMT, CSCs, and Notch signaling and decodes the emergent spatial patterns of different subsets of CSCs with different EMT phenotypes. First, we show that the production and diffusion of an EMT-inducing signal (such as TGF-β), together with Notch signaling, can robustly recapitulate experimental observations that while mesenchymal CSCs are localized toward the tumor–stroma periphery, hybrid E/M CSCs are typically located in the tumor interior (9). Further, modeling the effect of inflammatory cytokines in the TME shows that these cytokines can stabilize a hybrid E/M phenotype and increase the frequency of CSCs by activating Notch-Jagged signaling. We experimentally validate the functional relation between Notch-Jagged signaling and stemness by knocking down JAG1 in hybrid E/M SUM149 inflammatory breast cancer (IBC) cells and showing a significant impairment of tumor formation in vitro. Put together, our model yields valuable insights into the interconnected spatiotemporal dynamics of EMT, CSCs, and Notch-Jagged signaling, identifies the mechanisms underlying experimental observations, and generates testable predictions that have been validated in SUM149 breast cancer cells.
Results
A Gradient of EMT-Inducing Signal Recapitulates the Experimentally Observed Spatial Segregation of Different EMT Phenotypes.
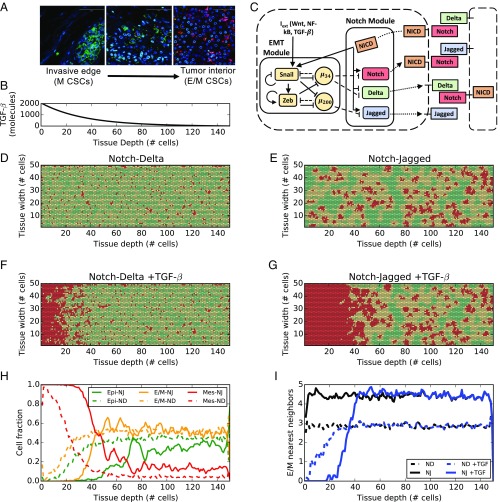
The interplay of different microenvironmental signals can give rise to intricate spatial arrangements of cells with different EMT phenotypes and/or different subsets of CSCs. Immunofluorescence analysis of breast carcinoma tissue revealed that different populations of breast CSCs with different EMT profiles can localize in anatomically distinct regions of a tumor (Fig. 1A) (9). A CD24−CD44+ mesenchymal-like breast CSC (BCSC) population was localized at the invasive edge of tumor, while an ALDH1+ epithelial-like BCSC population was localized in the interior close to the tumor stroma (Fig. 1A). Follow-up studies have characterized these ALDH1+ cells as showing a hybrid E/M, and not a purely epithelial, signature (32).
Fig. 1.
EMT phenotype patterning in the presence of EMT-induced Notch signaling. (A) Immunofluorescent staining of CD24 (magenta), CD44 (green), ALDH1 (red), and DAPI (blue) in human invasive breast carcinoma. Adapted from ref. 9. (Scale bars: 100 μm.) (B) Concentration of EMT-inducing signal as a function of tissue depth in the layer. Because the signal is secreted uniformly at one end of the layer, the profile is constant across the layer width. (C) Coupling between Notch pathway and core EMT regulatory circuit. Solid arrows/bars represent transcriptional activation/inhibition, while dashed lines represent posttranslational inhibition by microRNAs. Dotted lines represent translocation of Notch, Delta, and Jagged to the cell surface. (D) EMT phenotype distribution in the cell layer after 120 h of equilibration starting from randomized initial conditions and for a strong Notch-Delta signaling (gD = 90 molecules per hour, gJ = 20 molecules per hour). Green, yellow, and red colors denote epithelial (E), hybrid (E/M), and mesenchymal (M) cells, respectively. (E) Same as D, but for strong Notch-Jagged signaling (gD = 20 molecules per hour, gJ = 50 molecules per hour). (F) Same as D in the presence of TGF-β gradient in the tissue layer. (G) Same as E in the presence of TGF-β gradient in the tissue layer. (H) Fraction of E, hybrid E/M, and M cells as a function of tissue depth corresponding to F (dashed lines) and (G) (continuous lines). (I) Average number of E/M nearest neighbors of the hybrid E/M cells as a function of tissue depth for the four cases of D–G. H and I present an average over 10 simulations starting from random initial conditions.
To decipher the signaling mechanisms that may underlie such heterogeneous distribution of EMT phenotypes, we extended our previously developed mathematical model that couples a core EMT regulatory circuit with the juxtacrine Notch signaling pathway (5). Here, we consider the effect of a diffusing EMT-inducing signaling (such as TGF-β) on our multicell lattice setup consisting of (50 × 150) cells. A spatial gradient of TGF-β diffuses from one end of the layer (the invasive edge of tumor, as shown in Fig. 1A) toward the tumor interior, mimics the secretion of TGF-β by stromal cells (33) (Fig. 1B). The width/depth ratio of the lattice was chosen to evaluate the effect of gradients of signaling molecules in different tissue areas that represent anatomically distinct regions of a tumor.
The Notch signaling pathway is activated when one of its transmembrane ligands (Delta or Jagged) binds to the transmembrane receptor Notch belonging to a neighboring cell, leading to the release of NICD (Notch Intracellular Domain). NICD transcriptionally regulates several target genes resulting in the activation of Notch and Jagged and the inhibition of Delta (34) (Fig. 1C, Notch module). Therefore, Notch-Jagged signaling between two cells enables a similar cell fate (“lateral induction”) where both cells can send and receive signals, due to the presence of ligand (Jagged) and receptor (Notch), thus called a hybrid Sender/Receiver phenotype. However, Notch-Delta signaling promotes opposite cell fates (“lateral inhibition”) where one cell has high Notch (thus, acting as a Receptor) and the other cell has high Delta (thus, behaving as a Sender) (35, 36). In our model, Jagged refers to JAG1 that has been found to be particularly involved in lateral induction as well as tumor progression (15, 19, 37). Further, NICD activates EMT by upregulating the EMT-inducing transcription factor (EMT-TF) SNAIL (5) (Fig. 1C). A core regulatory circuit for EMT is composed of two families of EMT-TFs (SNAIL and ZEB) and two families of EMT-inhibiting microRNAs (miR-34 and miR-200) that mutually inhibit each other (38). Additional inputs from pathways such as TGF-β can activate EMT (38) (Fig. 1C, EMT module). This EMT circuit can enable three cell phenotypes: epithelial (high microRNAs, low EMT-TFs), hybrid E/M (intermediate microRNAs, intermediate EMT-TFs), and mesenchymal (low microRNAs, high EMT-TFs) (38). These microRNAs can inhibit the activation of Notch signaling (5); thus, the activation of EMT can also lead to activated Notch signaling.
In the absence of a TGF-β gradient, Notch-Jagged signaling generates spatial patterns with clusters of hybrid E/M cells, but Notch-Delta signaling leads to an alternating arrangement of epithelial and hybrid E/M cells, similar to our previous results (5, 39) (Fig. 1 D and E and Movies S1 and S2). Introducing the gradient of the EMT-inducing signal TGF-β through the tissue, however, generates spatial segregation of different EMT phenotypes. Cells close to the invasive edge, where TGF-β is secreted, undergo a complete EMT, while cells in the interior, at low TGF-β exposure, are mostly epithelial and hybrid E/M (Fig. 1 F and G and Movies S3 and S4). Specifically, the fraction of hybrid E/M cells is similar in the Notch-Delta and Notch-Jagged cases (Fig. 1H and SI Appendix, Fig. S1), but the patterning is remarkably different. Notch-Jagged signaling, but not Notch-Delta signaling, enables a pattern where the hybrid E/M cells are much more likely to be surrounded by other hybrid E/M cells, thus leading to the formation of clusters of hybrid E/M cells in the tissue interior (Fig. 1I). JAG1 has been observed to be among top differentially expressed genes in clusters of CTCs (16), thus supporting the prediction of our model.
Therefore, the secretion and diffusion of EMT-inducing signals such as TGF-β from the tumor–stroma boundary can generate the spatial segregation of cells with different EMT phenotypes experimentally observed in carcinomas (7, 9) in either scenarios when cells are communicating via Notch-Delta or Notch-Jagged. Furthermore, experimental results indicate that the generation of CSCs—a process intricately tied with EMT (40, 41)—can be produced both at the tumor edge (mesenchymal CSCs) and in the interior region at lower exposure to EMT inducer (hybrid E/M CSCs). Thus, a more complete characterization of the tumor tissue requires understanding the signaling mechanisms in the TME that can mediate CSC properties.
Inflammatory Cytokines Can Promote a Hybrid E/M Phenotype by Increasing Notch-Jagged Signaling.
Previous observations have suggested a key role of the Notch-Jagged signaling axis in mediating CSC properties (14, 42–44). Therefore, we next focused on what signals in the TME can promote Notch-Jagged signaling. Inflammatory cytokines such as IL-6, IL-1β, TNF-α, and the pathways they involve (such as NF-κB) can enhance the production of Jagged and, in some cases, also inhibit that of Delta (24–27). To investigate the effect of these cytokines, we introduced an external variable () that acts as an activator of Jagged and an inhibitor of Delta (Methods). We vary to characterize the effect of different inhibition/activation strengths arising in a concentration-specific or cytokine-specific way.
As a first step toward understanding the effect of inflammatory cytokines on Notch signaling and the plasticity of tumor cells, we analyzed the dynamics of an individual cell that is exposed to variable levels of inflammatory cytokines () and Jagged ligands () that can bind to the Notch receptor present at the cell surface and induce the EMT regulatory cascade.
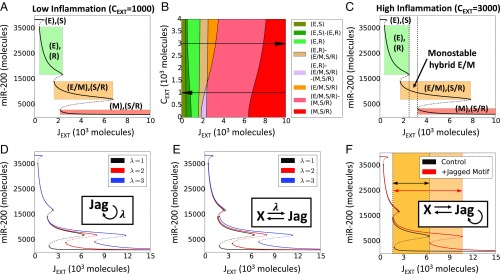
We first plotted the levels of miR-200 [gatekeepers of epithelial phenotype (38, 45)] upon full equilibration of the system, as a function of the external ligand available ), under conditions of low levels of inflammatory cytokines ( molecules). The cell is initially in an epithelial (E) phenotype (high levels of miR-200), and exhibits a “Sender” (S) Notch state characterized by a low expression of Notch receptor and a high expression of ligand Delta [Fig. 2A, (E), (S)]. For increasing values of , Notch signaling is activated, triggering a shift to a “Receiver” Notch (R) state characterized by a high expression of Notch receptor and a low expression of ligand Delta. EMT, however, is not yet triggered [green shaded region in Fig. 2A, (E), (R)]. A larger level of further activates Notch signaling and induces a partial EMT, or a transition to a hybrid E/M phenotype. Concomitantly, intracellular Jagged production is also elevated as the inhibition of Jagged by miR-200 is relieved. Thus, the cell attains a hybrid Sender/Receiver (S/R) Notch state [orange shaded region in Fig. 2A, (E/M), (S/R)]. A further increase in induces a stronger activation of the EMT circuitry, driving the cells toward a mesenchymal state [red shaded region in Fig. 2A, (M), (S/R)]. It is worth pointing out that our model operates in a parameter regime where the Notch signaling is functionally active in epithelial cells. It is possible, however, that the epithelial cells have “inactive” Notch signaling (SI Appendix, Fig. S2), as seen in some contexts.
Fig. 2.
Inflammation stabilizes a hybrid E/M phenotype. (A) Bifurcation curves of miR-200 as a function of JEXT for low inflammation (CEXT = 1,000 molecules). (B) Cell phenotype diagram as a function of external Jagged (JEXT) and external cytokines (CEXT). The different colors represent portions of parameter space characterized by monostability or multistability of the coupled Notch-EMT system. (C) Represents the same case as A, but for high inflammation (CEXT = 3,000 molecules). Solid lines represent stable steady states, and dotted lines represent unstable steady states. Vertical dotted lines in C depict the range of control parameter values that allows for monostability of the (E/M, S/R) state. The colored rectangles in A and C elucidate the interval of (JEXT) values for which different states of coupled EMT-Notch circuitry are stable, and the corresponding level of miR-200. For this simulation, the external concentrations of Notch and Delta are fixed at NEXT = 10,000 molecules, DEXT = 0 molecules (36). Bifurcation diagrams for all model’s variables are presented in SI Appendix, Fig. S3B. (D–F) Bifurcation diagram of miR-200 in presence of self-activation of Jagged (D), positive feedback loop between Jagged and another additional component (E), and a combination of both (F). Color shading in F shows the increased stability of the hybrid E/M phenotype in presence of the Jagged motif. Hill coefficient(s) is(are), unless stated otherwise, n = 2. In D, λ is the fold change in production rate of Jagged due to the activation by X, while in E it represents the fold change of both interactions. In F, all λ = 2.
Next, to better understand the role of inflammatory cytokines in mediating this bifurcation diagram, we plotted a 2D phenotype diagram, varying the levels of both and (Fig. 2B). This diagram displays multiple phases (i.e., sets of coexisting phenotypes under the same set of physiological conditions). The population distribution, when cells can attain more than one phenotype, depends on the local microenvironment and the varied genetic and epigenetic landscapes of individual cells. Remarkably, at low levels of inflammatory cytokines, the hybrid (S/R, E/M) state is found only in multistable regions, that is, only in combination with other possible cell states (Fig. 2B). Conversely, this cell state can exist by itself (monostable phase) when the cytokine levels are high. This remarkable difference can be visualized through a bifurcation diagram of miR-200 for a case of high levels of inflammatory cytokines ( molecules), where the region of stability of a hybrid E/M phenotype significantly increases (shown by dotted rectangle in Fig. 2C). A similar effect is observed in a single-cell model driven by the external ligand Delta (SI Appendix, Fig. S3 A–C).
The effect of inflammatory cytokines on Notch signaling can often be mediated by the inflammatory response transcription factor NF-κB. For instance, NF-κB governs the effect of TNF-α on Jagged levels (24). Further, Jagged can activate NF-κB (46, 47), thus indirectly leading to its self-activation. Also, IL-6/IL-6R signaling can be promoted via autocrine effects mediated through EMT-Notch circuitry (48). To assess the effect of such amplification and/or autocrine signaling, we tested various circuit motifs that result in Jagged overexpression, by computing the bifurcation diagram of miR-200. Our motifs include Jagged self-activation (Fig. 2D), mutual positive activation (Fig. 2E), and the combination of these two possibilities (Fig. 2F). All motifs contributed to further stabilizing a hybrid E/M phenotype (Fig. 2 D–F). These results suggest that autocrine and/or paracrine effects of inflammatory cytokines capable of up-regulating Notch-Jagged signaling, such as IL-6, can stabilize a hybrid E/M phenotype.
Generalizing the model to a multicell scenario, these cytokines increase both the fraction of E/M cells in the tissue (SI Appendix, Fig. S4 A and B) and the likelihood of physical contact between these hybrid E/M cells (SI Appendix, Fig. S4C) when the signaling is dominated by Notch-Jagged signaling, but not by Notch-Delta signaling (SI Appendix, Fig. S5), reminiscent of what is observed in Fig. 1 G–I.
Together, these results indicate that inflammation can expand the range of physiological conditions for which cells can adopt a hybrid state in terms of cellular plasticity (E/M) and intercellular signaling (S/R) by activating Notch-Jagged signaling. This behavior is reminiscent of previous observations that JAG1 can act as a “phenotypic stability factor” for a hybrid E/M phenotype (5).
Inflammatory Cytokines Increase the CSC Population by Enhancing Notch-Jagged Signaling.
We have discussed how the gradients of EMT-inducing signals, together with Notch-Jagged signaling promoted by inflammatory cytokines such as IL-6, can enable a subpopulation of cancer cells to exhibit a hybrid E/M phenotype. A hybrid E/M phenotype has been proposed to often possess the typical properties of CSCs (20–23, 49). Furthermore, recent experiments showed that various inflammatory cytokines increase the CSC population in a tumor (50). Thus, we explored the effect of cytokines on stemness. Within our framework, we define stemness based on activation of Notch-Jagged signaling (SI Appendix, Fig. S6A). Therefore, a cell is classified as CSC if it has high levels of NICD and Jagged. This classification is based on observations in multiple cancer types such as glioblastoma, pancreatic cancer, colon cancer, and basal-like breast cancer (BLBC) that Notch and Jagged are overexpressed in CSCs compared with non-CSCs (51–54). Further, our previous in silico results indicated that enhanced Notch-Jagged signaling is likely to correlate with the acquisition of stem-like traits (14). Thus, to elucidate the connection between inflammation, EMT, and CSCs, we determined the frequency of CSCs in a population of a (100 × 100) multicell lattice. We chose a square lattice because no spatial gradient was applied in this simulation.
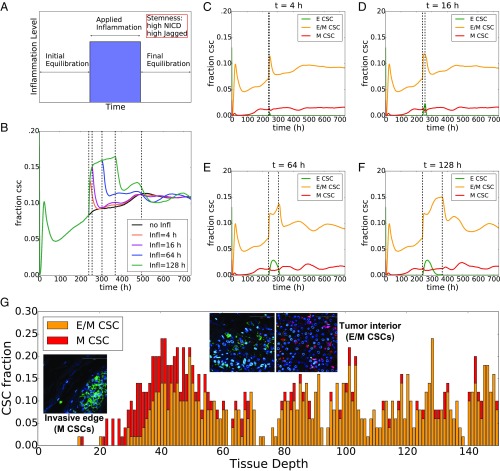
To probe the variation in CSC population due to inflammatory cytokines, we set up a simulation where the multicell layer is exposed to these cytokines for a variable amount of time (ti) and then allowed to equilibrate (Fig. 3A). The fraction of CSC initially increases sharply as soon as inflammation is applied. Afterward, if the inflammation is maintained for long enough, the conversion rate of non-CSC to CSC diminishes. Once the inflammatory stimulus is removed, the CSC fraction decreases (Fig. 3B). Interestingly, this rate of return to initial state depends on the length of the inflammation period. If inflammation is applied for a short period, the fraction of CSCs in the tissue quickly returns to the initial value (i.e., corresponding fraction of CSC with no applied inflammation). Conversely, compared with the control value, chronic inflammation maintains a significantly higher CSC population for a longer period after the inflammation is removed (compare the 4-h and 16-h curves with the 64-h and 128-h curves in Fig. 3B and SI Appendix, Fig. S6B). This observation suggests that the effects of inflammation on a tissue can be a long-lasting phenomenon and prevail for a significant time after the stimulus is removed.
Fig. 3.
Inflammation increases the CSC population. (A) Schematic of simulation setup: the 2D layer of cells undergoes initial equilibration for 240 h (initial conditions for proteins and microRNAs are extracted randomly); an inflammatory signal constant through the layer (CEXT = 3,000 molecules) is applied for a variable time interval (blue region); after the inflammation is removed, the system equilibrates. (B) Temporal dynamics of the fraction of CSC for different durations of applied inflammation and comparison with control (no applied inflammation, black curve). The first vertical dotted line from the left indicates the time when the inflammation is applied (same for all curves); the four successive dotted lines depict the end of the applied inflammation for the different curves. (C–F) Temporal dynamics of the fraction of epithelial, hybrid E/M, and mesenchymal CSC. In C, for a short inflammation period (t = 4 h) the spike in CSC population is due to hybrid E/M cells. In this simulation, the production rates of Jagged and Delta are gJ = 50 molecules per hour, gD = 25 molecules per hour, respectively (as in Fig. 2). (G) Fraction of CSCs with different EMT phenotypes as a function of tissue depth in the cell layer for the (Notch-Jagged, TGF-β gradient) of panel Fig. 1G. Insets show the spatial distribution of M-CSC by the invasive edge of the tumor and E/M-CSC in the tumor interior. Adapted from ref. 9. (Scale bars: 100 μm.)
Specifically, the initial spike in CSC fraction in response to inflammation is essentially a conversion of hybrid E/M non-CSCs into hybrid E/M CSCs (Fig. 3 C–F, orange curves and SI Appendix, Fig. S6C). In other words, these are hybrid E/M cells that had relatively low levels of either NICD or Jagged and hence were not classified as CSCs before the inflammatory stimulus was applied. After the initial spike, the fraction of hybrid E/M CSC remains nearly constant, while the fraction of epithelial CSCs increases (Fig. 3 D and E, green curves and SI Appendix, Fig. S6C). If the inflammation is maintained for longer time, epithelial CSCs undergo partial EMT and become hybrid E/M CSCs (see the reduction of epithelial CSCs and the corresponding increase in hybrid E/M CSCs in Fig. 3F). Interestingly, inflammation does not alter the fraction of mesenchymal CSCs (Fig. 3 C–F, red curves and SI Appendix, Fig. S6C). Overall, these results suggest a strong correlation between the cytokine-induced hybrid E/M phenotype and the acquisition of stem-like traits.
Finally, the quantification of stemness based on Notch-Jagged signaling can explain the segregation of CSCs with different EMT phenotypes as observed experimentally (9) in the case of a tissue layer exposed to TGF-β gradient and strong Notch-Jagged signaling. Specifically, our model predicts a front of mesenchymal CSC toward the tumor–stroma interface, where the exposure to TGF-β is high, and a high fraction of hybrid E/M CSC in tumor interior (Fig. 3G and Movies S1–S4, Bottom), in excellent agreement with experimental data (Fig. 3G). This prediction is in good agreement with our previous observation that an E/M-CSC phenotype can switch to an M-CSC phenotype upon activation of TGF-β downstream targets (14). These results offer a unifying framework for the concepts of intratumoral heterogeneity, EMT, and CSCs and indicate the crucial role of JAG1 in simultaneously promoting a hybrid E/M phenotype and the acquisition of stem properties. In this framework, stemness is envisioned as a feature that can be reversibly gained or lost by cancer cells, as opposed to a more classical model where stemness can be only lost via cell differentiation (55, 56). Importantly, Notch signaling need not to be the only factor that dictates the spatial segregation of different CSC subsets, and the “hunt” for a complete set of variables that characterize the EMT and CSC status remains open.
JAG1 Knockdown Restricts Tumor Emboli Formation in IBC Cells SUM149.
To validate our prediction that enhanced Notch-Jagged signaling can enhance the acquisition of a stem-like, more aggressive phenotype, we investigated the proliferation in both 2D and 3D cultures of a triple-negative, basal-type, IBC patient primary tumor-derived cell line SUM149 in response to knockdown of JAG1. SUM149 cells constitutively express epidermal growth factor receptor and coexpress E-cadherin and N-cadherin and have been previously characterized as exhibiting a hybrid E/M phenotype (57, 58). Further, flow cytometry analysis of SUM149 identifies that a large percentage of cells are CD24hi CD44hi (57) and exhibit increased expression of ALDH+ cancer stem-like marker (59, 60), a proposed signature of the hybrid E/M phenotype that has the potential to influence clinical outcomes (20, 59).
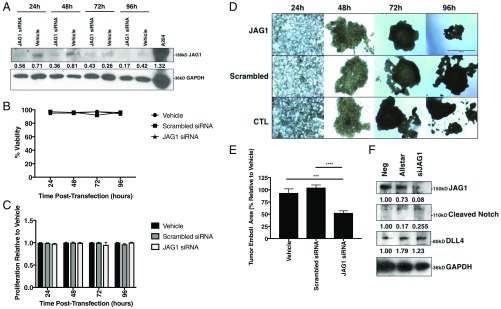
JAG1 knockdown was effective in cell culture for up to 96 h (Fig. 4A) but had no effect on viability or 2D proliferation in SUM149 cells (Fig. 4 B and C). Interestingly, the IBC patient-derived SUM149 cells, like most cancer cells, are not only capable of spontaneously forming tumor 3D tumor spheroids/mammospheres (61) when seeded in ultralow attachment plates in vitro but can also be cultured as tumor organoids under conditions simulating the viscosity and shear force of the lymphatic system (62, 63). Thus, we utilized the tumor emboli/organoid model allowing for the physiologically relevant 3D growth that is similar to the clinicopathological hallmark of collective tumor cell aggregates/lymphatic emboli seen in patients with IBC (62). Once the tumor emboli were formed at 48 h, JAG1 siRNA-treated emboli were significantly smaller and less dispersed than vehicle (P < 0.001) or scrambled siRNA-treated emboli (P < 0.0001) (Fig. 4 D and E). JAG1 siRNA-treated emboli were harvested and lysed following 96 h of growth and found by immunoblot analysis to contain depleted levels of JAG1 and cleaved Notch proteins, as well as increased levels of DLL-4 protein, compared with vehicle-treated cells (Fig. 4F). Overall, these findings indicate that the Notch-Jagged is a crucial axis that regulates the acquisition of proliferative traits.
Fig. 4.
JAG1 knockdown reduces organoid size and decreases Notch-JAG1 signaling in SUM149 cells. (A) Western blot of SUM149 cells for JAG1 and GAPDH proteins following treatment with JAG1 siRNA or transfection reagent alone for 24, 48, 72, or 96 h. JAG1 protein levels normalized to GAPDH. (B) Percent viability and (C) 2D proliferation relative to vehicle of SUM149 cells at 24, 48, 72, and 96 h time points following JAG1 siRNA, scrambled siRNA, or vehicle treatment. (D) Representative 40× magnification images and (E) area quantification of SUM149 tumor emboli treated with JAG1 siRNA, scrambled siRNA, or vehicle alone at days 1, 2, 3, and 4 posttransfection. (Scale bar: 500 μm.) (F) Western immunoblot analysis for JAG1, cleaved Notch, DLL4, and IL-6 proteins at t = 96 h from SUM149 tumor emboli lysates, normalized to GAPDH and to negative untreated cells. A204, human muscle rhabdomyosarcoma cell lysate used for antibody control. ***P < 0.001, ****P < 0.0001 by one-way ANOVA and Fisher's least significant difference post hoc test, n = 6.
Discussion
Targeting the ubiquitous intratumoral phenotypic and functional heterogeneity remains an unsolved clinical challenge. Such heterogeneity is exacerbated by noncell autonomous effects of phenotypic plasticity. Phenotypic plasticity mediated by reversible activation of EMT or CSC pathways can fuel the acquisition of metastatically competent, drug-refractory, and adaptive phenotypes (23, 64). A canonical example of this heterogeneity is the presence of different subsets of CSCs with varying EMT phenotypes in spatially distinct regions of the tumor (9).
Here, using a mathematical modeling framework, we demonstrate that the spatial gradients of EMT-inducing signals such as TGF-β, coupled with cell–cell communication via Notch signaling, can generate distinct subsets of CSC with varying EMT phenotypes in spatially segregated regions of the tumor. Our model predicts that mesenchymal-like CSCs localize at the invasive edge of the tumor and hybrid E/M CSCs localize in the tumor interior. This prediction agrees well, at least qualitatively, with recent observations in human breast carcinoma tissue, where CD44+/CD24− cells (mesenchymal CSCs) were present at the tumor invasive edge, while ALDH1+ cells [hybrid E/M CSCs (21, 32)] were present in the interior of the tumor (9). However, we acknowledge that our model has multiple limitations. First, we assumed a spatial profile of TGF-β based on its secretion from stromal cells; however, there may be local variations in TGF-β signal intensity due to a variety of mechanical and/or chemical stimuli, including its secretion by cancer cells. Second, the model does not account for migration of hybrid E/M and mesenchymal cancer cells, which may modify the spatial patterning, particularly by overestimating the effects of mesenchymal cancer cells in inducing EMT in neighboring cells. Third, cell proliferation might play a relevant role, especially given the observed correlation between a hybrid E/M phenotype and stemness. The proliferation of hybrid E/M cells could give rise to regions with locally high density of E/M cells, and therefore facilitating cluster formation. Fourth, it considers EMT as a discrete three-step process. While some recent studies point to a set of discrete hybrid E/M states (six in the case of ref. 22), other investigations have also suggested a more continuous spectrum of phenotypic plasticity (65, 66). Despite these limitations, our framework offers promising insights into the spatial juxtaposition of different phenotypes within a given tumor tissue; such patterns are only beginning to be uncovered by advances in imaging techniques. Further progress in multiplexed imaging over large tumor sections can not only unravel the new layers of underlying biological complexity, but also present an opportunity to develop more representative and predictive computational models.
Our results highlight how inflammatory cytokines such as IL-6 can amplify Notch-Jagged signaling, thus facilitating cells to maintain a hybrid E/M stem-like phenotype, as well as form clusters of hybrid E/M cells that may dislodge from primary tumors as clusters of CTCs—the primary “villains” of metastasis (16, 67). The presence of both the transmembrane receptor Notch and the transmembrane ligand Jagged can facilitate bidirectional signaling not only among cancer cells but also in tumor–stroma cross-talk. For instance, in BLBC, where Notch, Jagged, and IL-6 receptor are overexpressed relative to other breast cancer subtypes (10, 27), IL-6 up-regulates JAG1 and boosts communication among cells through Notch3 and JAG1 (27). Notch3, a member of the Notch family, can facilitate the autocrine production of IL-6, hence ensuring sustained high levels of inflammatory cytokines (27). The IL-6/Notch3/JAG1 axis can sustain mammosphere growth; an autocrine activation of this axis seems to be required for higher invasive potential (27). These observations are reinforced by our results that JAG1 knockdown significantly impairs tumor organoid formation potential in triple-negative breast cancer (TNBC) SUM149 cells. Furthermore, the association between Notch-Jagged signaling and hybrid E/M state is strengthened by observations such as (i) loss of glycosyltransferase Fringe—a suppressor of Notch-Jagged signaling (68)—in BLBC (69), (ii) enrichment of hybrid E/M cells in primary tumor biopsies of TNBC patients (70), and (iii) association of enhanced JAG1 levels with poor outcome in BLBC (51). Beyond breast cancer, similar results for JAG1 knockdown are also seen in ovarian cancer, where silencing JAG1 in stromal cells as well as in tumor cells impaired tumor growth, and silencing it both in the tumor and stroma had synergistic effects (71). This synergistic effect can be explained by the observation that deleting JAG1 in stromal cells can limit the possibility of activation of Notch-Jagged signaling among tumor cells by limiting the strength of lateral induction, as noted experimentally (19).
Our results proposing that IL-6–driven Notch-Jagged signaling can mediate stemness are also consistent with multiple previous studies in IBC and other subtypes. First, IBC emboli tend to exhibit activated Notch 3 intracellular domain; depletion of Notch 3 can lead to a loss of stem cell markers and induce apoptosis (72). Second, IBC patients have significantly higher levels of IL-6 levels in serum and carcinoma tissues compared with non-IBC ones (73). Third, CD24hi CD44hi and ALDH+ cells that represent a hybrid E/M phenotype (20) and form aggressive tumors in vivo (74, 75) are enriched in IBC upon drug treatment (15) and exhibit up-regulated Notch-Jagged signaling (5). Fourth, Notch-JAG1 signaling mediates resistance to tamoxifen (44) and is enriched upon inhibition of HER2 (42), suggesting how adaptive resistance mediated by JAG1 can contribute to tumor relapse. Put together, these studies strongly emphasize the role of inflammatory cytokines and Notch-Jagged signaling in cancer cell survival and aggressive potential.
Methods
Modeling the Interplay Between Notch Signaling, EMT and Inflammatory Cytokines.
The levels of every miR, TF, and protein in the circuit is described by an ordinary differential equation that takes the form
| [1] |
where is the production rate and is the degradation rate constant, assuming first-order degradation kinetics (see SI Appendix, section 1 for the complete set of equations). Additionally, the presence of an activator/inhibitor () increases/decreases the basal production rate constant by a factor . is a Hill function for transcriptional regulation, while it assumes a more complex form to describe protein degradation due to binding of noncoding RNAs to their target mRNAs (SI Appendix, section 2). Additionally, Notch receptors and ligands irreversibly bind via a term , where is a binding rate constant and is the receptor/ligand binding to . This term is present only in equations where is a Notch receptor/ligand. Binding affinity of Notch receptor is different for Delta and Jagged ligands. Additionally, ligands can inhibit the signaling by binding to Notch receptors of the same cell, which leads to the degradation of the complex (cis inhibition). In this scenario, both and belong to the same cell.
In the model, inflammatory cytokines such as TNF-α, IL-6, and IL-1β are described by a variable , which represent the number of inflammatory molecules that interact with the cell, inhibiting the production of Delta and activating Jagged according to
| [2] |
| [3] |
where are negative and positive shifted Hill functions, respectively (38).
This framework is extended to model the dynamics of a 2D hexagonal lattice of cells. In the multicell model, Delta and Jagged ligands of a cell can bind to the Notch receptors of a neighbor cell. Therefore, the number of receptors and ligands available to bind to one cell’s receptors and ligands are
| [4] |
| [5] |
| [6] |
where , , and are the levels of Notch, Delta, and Jagged in the neighbor cells n, and the index n represents the set of nearest neighbors in the lattice.
The cell tissue can be exposed to an EMT-inducing signal such as TGF-β that is secreted at one end of the lattice and removed at the opposite end (a source-sink dynamics). This profile mimics the situation that this diffusible molecule can be secreted by many stromal cells that are present next to the tumor invasive edge. Therefore, this signal obeys a diffusion equation:
| [7] |
with boundary conditions such that at the tissue end that secrets the signal () and at , where is the length of the tissue.
Cell Culture and Reagents.
SUM149 (Asterland, Inc.) IBC cells were cultured as per manufacturer’s instructions and previous studies (76). Cells were cultured with 1% penicillin/streptomycin (Invitrogen) supplemented in their respective media. Cells were cultured in growth medium at 37 °C under an atmosphere of 5% CO2.
Cell Transfection.
Cells were plated at 20,000 cells per well in a 24-well plate and transfected 24 h later with 37.5 ng siJAG1 (SI02780134; Qiagen), Allstar scrambled siRNA (SI03650318; Qiagen), or no siRNA with HiPerfect (301704; Qiagen) transfection reagent added alone. siRNA was incubated with 3 μL of transfection reagent at room temperature for 10 min and added dropwise to each respective well on the plate. Cells were incubated at 37 °C.
Trypan Blue Exclusion Viability Assay Analysis.
Cells were plated in a 12-well plate at 3 × 104 cells per well for 24 h, then treated with PAH mixture, chlorothalonil, or left untreated. Cells were trypsinized after 24 h and resuspended in media then centrifuged at 501 × g for 4 min. The supernatant was aspirated and 70 μL cold DPBS was added to the pellet. An aliquot of cell suspension was mixed with an equal volume of 0.4% trypan blue solution, and cell numbers were recorded in 10 μL of the resultant mixture using a hemocytometer. Viability is reported as the ratio of live cells to total cells.
MTT Proliferation Assay Analysis.
Cell toxicity and proliferation were assessed by MTT assay (3-[4,5- dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide), based on the conversion of MTT into formazan crystals by living cells, which determines metabolic activity and is an acceptable measure of viable, proliferating cells. Cells were seeded at 4,000 cells per well into 96-well flat-bottom plates and transfected. Cells were grown for 24, 48, 72, or 96 h. Proliferation was assessed using 3(4,5- dimethylthlthiazol-2-yl)-2,5-diphenyltetrazolium, after which cells were incubated at 37 °C for 2 h, DMSO was added to each well, and absorbance was read at 570 nm on a Fluostar Optima plate reader.
Tumor Organoid Formation Analysis.
SUM149 cells were plated at 10,000 cells per well in an ultralow-attachment 24-well plate with filtered media supplemented with 2.25% polyethylene glycol as described previously (62). The resultant tumor cell emboli in culture were left to form for 48 h, verified formed and uniform by microscopy, then assessed for growth and size. Tumor emboli in culture were then allowed to grow for 72–96 h, after which they were imaged at 4× magnification and their gross particle area was analyzed using NIH ImageJ.
Western Immunoblot Analysis.
Tumor emboli in culture were harvested after 96 h growth and lysed using Poly + Halt Lysis Buffer (Promega) and 1 M DTT (Ameresco) by gently pipetting to dislodge the emboli, placed in a heat block at 50 °C for 30 min with intermittent vortexing for 10 s every 5 min. The disrupted emboli cells were spun at 501 × g for 4 min and the pellet was resuspended in phosphate buffer saline. Protein concentration measured by Nanodrop 2000 and 20 μg of the lysates were loaded for immunoblot analysis as described previously (77). Two-dimensional monolayer cells were also treated and harvested following transfection for similar immunoblot analysis. Membranes were incubated overnight at 4 °C with primary antibodies JAG1, cleaved Notch, DLL4, IL-6 (1:1,000 dilution; Cell Signaling Technologies), or GAPDH (1:2,000 dilution). Membranes were washed and incubated with anti-mouse or anti-rabbit HRP-conjugated antibodies (Cell Signaling Technologies) for 1 h at room temperature. Chemiluminescent substrate was applied for 5 min, and then membranes were exposed to radiographic film. Densitometric analysis was performed using NIH ImageJ software with GAPDH as a loading control, with all values normalized to the untreated lanes.
Supplementary Material
Acknowledgments
This work was sponsored by NSF Grants PHY-1427654 (Center for Theoretical Biological Physics), PHY-1605817, CHE- 1614101, and MCB-1241332 and by Department of Defense Breast Cancer Research Program Breakthrough Award W81XWH-17-1-0297 (to G.R.D.) and Duke School of Medicine Bridge Research Funds (to G.R.D.). L.G.-S. was supported by a Duke University predoctoral training program in environmental health (NIEHS T32-ES021432). F.B. was partially supported by the Hasselman Fellowship for academic excellence in Chemistry. M.K.J. was also supported by a training fellowship from the Gulf Coast Consortia, on the Computational Cancer Biology Training Program (CPRIT Grant RP170593). M.B. was supported by São Paulo Research Foundation Grant 2013/14438-8.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815345116/-/DCSupplemental.
References
- 1.Carmona-Fontaine C, et al. Emergence of spatial structure in the tumor microenvironment due to the Warburg effect. Proc Natl Acad Sci USA. 2013;110:19402–19407. doi: 10.1073/pnas.1311939110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dai Z, Locasale JW. Metabolic pattern formation in the tumor microenvironment. Mol Syst Biol. 2017;13:915. doi: 10.15252/msb.20167518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan Y. Spatial heterogeneity in the tumor microenvironment. Cold Spring Harb Perspect Med. 2016;6:a026583. doi: 10.1101/cshperspect.a026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmona-Fontaine C, et al. Metabolic origins of spatial organization in the tumor microenvironment. Proc Natl Acad Sci USA. 2017;114:2934–2939. doi: 10.1073/pnas.1700600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boareto M, et al. Notch-Jagged signalling can give rise to clusters of cells exhibiting a hybrid epithelial/mesenchymal phenotype. J R Soc Interface. 2016;13:20151106. doi: 10.1098/rsif.2015.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.May CD, et al. Epithelial-mesenchymal transition and cancer stem cells: A dangerously dynamic duo in breast cancer progression. Breast Cancer Res. 2011;13:202. doi: 10.1186/bcr2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brabletz T, et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci USA. 2001;98:10356–10361. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 9.Liu S, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Reports. 2013;2:78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li D, Masiero M, Banham AH, Harris AL. The notch ligand JAGGED1 as a target for anti-tumor therapy. Front Oncol. 2014;4:254. doi: 10.3389/fonc.2014.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartman BH, Reh TA, Bermingham-McDonogh O. Notch signaling specifies prosensory domains via lateral induction in the developing mammalian inner ear. Proc Natl Acad Sci USA. 2010;107:15792–15797. doi: 10.1073/pnas.1002827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saravanamuthu SS, Gao CY, Zelenka PS. Notch signaling is required for lateral induction of Jagged1 during FGF-induced lens fiber differentiation. Dev Biol. 2009;332:166–176. doi: 10.1016/j.ydbio.2009.05.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loerakker S, et al. Mechanosensitivity of Jagged-Notch signaling can induce a switch-type behavior in vascular homeostasis. Proc Natl Acad Sci USA. 2018;115:E3682–E3691. doi: 10.1073/pnas.1715277115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bocci F, Jolly MK, George JT, Levine H, Onuchic JN. A mechanism-based computational model to capture the interconnections among epithelial-mesenchymal transition, cancer stem cells and Notch-Jagged signaling. Oncotarget. 2018;9:29906–29920. doi: 10.18632/oncotarget.25692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jolly MK, et al. Inflammatory breast cancer: A model for investigating cluster-based dissemination. NPJ Breast Cancer. 2017;3:21. doi: 10.1038/s41523-017-0023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung KJ, et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc Natl Acad Sci USA. 2016;113:E854–E863. doi: 10.1073/pnas.1508541113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakrabarti R, et al. Notch ligand Dll1 mediates cross-talk between mammary stem cells and the macrophageal niche. Science. 2018;360:eaan4153. doi: 10.1126/science.aan4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jolly MK, et al. Implications of the hybrid epithelial/mesenchymal phenotype in metastasis. Front Oncol. 2015;5:155. doi: 10.3389/fonc.2015.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, et al. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell. 2013;23:171–185. doi: 10.1016/j.ccr.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grosse-Wilde A, et al. Stemness of the hybrid epithelial/mesenchymal state in breast cancer and its association with poor survival. PLoS One. 2015;10:e0126522. doi: 10.1371/journal.pone.0126522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jolly MK, et al. Towards elucidating the connection between epithelial-mesenchymal transitions and stemness. J R Soc Interface. 2014;11:20140962. doi: 10.1098/rsif.2014.0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pastushenko I, et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 23.Jolly MK, Mani SA, Levine H. Hybrid epithelial/mesenchymal phenotype(s): The “fittest” for metastasis? Biochim Biophys Acta Rev Cancer. 2018;1870:151–157. doi: 10.1016/j.bbcan.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 24.Johnston DA, Dong B, Hughes CCW. TNF induction of jagged-1 in endothelial cells is NFkappaB-dependent. Gene. 2009;435:36–44. doi: 10.1016/j.gene.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foldi J, et al. Autoamplification of Notch signaling in macrophages by TLR-induced and RBP-J-dependent induction of Jagged1. J Immunol. 2010;185:5023–5031. doi: 10.4049/jimmunol.1001544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benedito R, et al. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137:1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 27.Sansone P, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest. 2007;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith AK, et al. Epigenetic changes associated with inflammation in breast cancer patients treated with chemotherapy. Brain Behav Immun. 2014;38:227–236. doi: 10.1016/j.bbi.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Multhoff G, Radons J. Radiation, inflammation, and immune responses in cancer. Front Oncol. 2012;2:58. doi: 10.3389/fonc.2012.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riehl A, Németh J, Angel P, Hess J. The receptor RAGE: Bridging inflammation and cancer. Cell Commun Signal. 2009;7:12. doi: 10.1186/1478-811X-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korkaya H, Liu S, Wicha MS. Regulation of cancer stem cells by cytokine networks: Attacking cancer’s inflammatory roots. Clin Cancer Res. 2011;17:6125–6129. doi: 10.1158/1078-0432.CCR-10-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colacino JA, et al. Heterogeneity of human breast stem and progenitor cells as revealed by transcriptional profiling. Stem Cell Reports. 2018;10:1596–1609. doi: 10.1016/j.stemcr.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costanza B, Umelo IA, Bellier J, Castronovo V, Turtoi A. Stromal modulators of TGF-β in cancer. J Clin Med. 2017;6:E7. doi: 10.3390/jcm6010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersson ER, Sandberg R, Lendahl U. Notch signaling: Simplicity in design, versatility in function. Development. 2011;138:3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- 35.Shaya O, Sprinzak D. From Notch signaling to fine-grained patterning: Modeling meets experiments. Curr Opin Genet Dev. 2011;21:732–739. doi: 10.1016/j.gde.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Boareto M, et al. Jagged-Delta asymmetry in Notch signaling can give rise to a Sender/Receiver hybrid phenotype. Proc Natl Acad Sci USA. 2015;112:E402–E409. doi: 10.1073/pnas.1416287112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meurette O, Mehlen P. Notch signaling in the tumor microenvironment. Cancer Cell. 2018;34:536–548. doi: 10.1016/j.ccell.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 38.Lu M, Jolly MK, Levine H, Onuchic JN, Ben-Jacob E. MicroRNA-based regulation of epithelial-hybrid-mesenchymal fate determination. Proc Natl Acad Sci USA. 2013;110:18144–18149. doi: 10.1073/pnas.1318192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bocci F, et al. Numb prevents a complete epithelial-mesenchymal transition by modulating Notch signalling. J R Soc Interface. 2017;14:20170512. doi: 10.1098/rsif.2017.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jolly MK, et al. Coupling the modules of EMT and stemness: A tunable ‘stemness window’ model. Oncotarget. 2015;6:25161–25174. doi: 10.18632/oncotarget.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shah D, et al. Inhibition of HER2 increases JAGGED1-dependent breast cancer stem cells: Role for membrane JAGGED1. Clin Cancer Res. 2018;24:4566–4578. doi: 10.1158/1078-0432.CCR-17-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarkar S, et al. Activation of NOTCH signaling by Tenascin-C promotes growth of human brain tumor-initiating cells. Cancer Res. 2017;77:3231–3243. doi: 10.1158/0008-5472.CAN-16-2171. [DOI] [PubMed] [Google Scholar]
- 44.Simões BM, et al. Anti-estrogen resistance in human breast tumors is driven by JAG1-NOTCH4-dependent cancer stem cell activity. Cell Rep. 2015;12:1968–1977. doi: 10.1016/j.celrep.2015.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Somarelli JA, et al. Mesenchymal-epithelial transition in sarcomas is controlled by the combinatorial expression of MicroRNA 200s and GRHL2. Mol Cell Biol. 2016;36:2503–2513. doi: 10.1128/MCB.00373-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morga E, et al. Jagged1 regulates the activation of astrocytes via modulation of NFkappaB and JAK/STAT/SOCS pathways. Glia. 2009;57:1741–1753. doi: 10.1002/glia.20887. [DOI] [PubMed] [Google Scholar]
- 47.Wang Z, et al. Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-kappaB signaling pathways. J Cell Biochem. 2010;109:726–736. doi: 10.1002/jcb.22451. [DOI] [PubMed] [Google Scholar]
- 48.Rokavec M, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bierie B, et al. Integrin-β4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc Natl Acad Sci USA. 2017;114:E2337–E2346. doi: 10.1073/pnas.1618298114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pietilä M, Ivaska J, Mani SA. Whom to blame for metastasis, the epithelial-mesenchymal transition or the tumor microenvironment? Cancer Lett. 2016;380:359–368. doi: 10.1016/j.canlet.2015.12.033. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto M, et al. NF-κB non-cell-autonomously regulates cancer stem cell populations in the basal-like breast cancer subtype. Nat Commun. 2013;4:2299. doi: 10.1038/ncomms3299. [DOI] [PubMed] [Google Scholar]
- 52.Sikandar SS, et al. NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer. Cancer Res. 2010;70:1469–1478. doi: 10.1158/0008-5472.CAN-09-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu TS, et al. Endothelial cells create a stem cell niche in glioblastoma by providing NOTCH ligands that nurture self-renewal of cancer stem-like cells. Cancer Res. 2011;71:6061–6072. doi: 10.1158/0008-5472.CAN-10-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z, et al. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 2009;69:2400–2407. doi: 10.1158/0008-5472.CAN-08-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bocci F, Levine H, Onuchic JN, Jolly MK. August 28, 2018. Deciphering the dynamics of epithelial-mesenchymal transition and cancer stem cells in tumor progression. arXiv:1808.09113.
- 56.Sellerio AL, et al. Overshoot during phenotypic switching of cancer cell populations. Sci Rep. 2015;5:15464. doi: 10.1038/srep15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robertson FM, et al. Genomic profiling of pre-clinical models of inflammatory breast cancer identifies a signature of epithelial plasticity and suppression of TGFβ signaling. J Clin Exp Pathol. 2012;2:119. [Google Scholar]
- 58.Kai K, et al. CSF-1/CSF-1R axis is associated with epithelial/mesenchymal hybrid phenotype in epithelial-like inflammatory breast cancer. Sci Rep. 2018;8:9427. doi: 10.1038/s41598-018-27409-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Charafe-Jauffret E, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Allensworth JL, et al. Disulfiram (DSF) acts as a copper ionophore to induce copper-dependent oxidative stress and mediate anti-tumor efficacy in inflammatory breast cancer. Mol Oncol. 2015;9:1155–1168. doi: 10.1016/j.molonc.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nath S, Devi GR. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol Ther. 2016;163:94–108. doi: 10.1016/j.pharmthera.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arora J, et al. Inflammatory breast cancer tumor emboli express high levels of anti-apoptotic proteins: Use of a quantitative high content and high-throughput 3D IBC spheroid assay to identify targeting strategies. Oncotarget. 2017;8:25848–25863. doi: 10.18632/oncotarget.15667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lehman HL, et al. Modeling and characterization of inflammatory breast cancer emboli grown in vitro. Int J Cancer. 2013;132:2283–2294. doi: 10.1002/ijc.27928. [DOI] [PubMed] [Google Scholar]
- 64.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Font-Clos F, Zapperi S, La Porta CAM. Topography of epithelial-mesenchymal plasticity. Proc Natl Acad Sci USA. 2018;115:5902–5907. doi: 10.1073/pnas.1722609115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bhatia S, Monkman J, Toh AKL, Nagaraj SH, Thompson EW. Targeting epithelial-mesenchymal plasticity in cancer: Clinical and preclinical advances in therapy and monitoring. Biochem J. 2017;474:3269–3306. doi: 10.1042/BCJ20160782. [DOI] [PubMed] [Google Scholar]
- 67.Aceto N, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jolly MK, et al. Operating principles of Notch-Delta-Jagged module of cell-cell communication. New J Phys. 2015;17:55021. [Google Scholar]
- 69.Xu K, et al. Lunatic fringe deficiency cooperates with the Met/Caveolin gene amplicon to induce basal-like breast cancer. Cancer Cell. 2012;21:626–641. doi: 10.1016/j.ccr.2012.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu M, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Steg AD, et al. Targeting the notch ligand JAGGED1 in both tumor cells and stroma in ovarian cancer. Clin Cancer Res. 2011;17:5674–5685. doi: 10.1158/1078-0432.CCR-11-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiao Y, Ye Y, Yearsley K, Jones S, Barsky SH. The lymphovascular embolus of inflammatory breast cancer expresses a stem cell-like phenotype. Oncogene. 2011;30:287–300. doi: 10.1038/onc.2010.405. [DOI] [PubMed] [Google Scholar]
- 73.Mohamed MM, Al-Raawi D, Sabet SF, El-Shinawi M. Inflammatory breast cancer: New factors contribute to disease etiology: A review. J Adv Res. 2014;5:525–536. doi: 10.1016/j.jare.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goldman A, et al. Temporally sequenced anticancer drugs overcome adaptive resistance by targeting a vulnerable chemotherapy-induced phenotypic transition. Nat Commun. 2015;6:6139. doi: 10.1038/ncomms7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Evans MK, et al. XIAP regulation by MNK links MAPK and NFκB signaling to determine an aggressive breast cancer phenotype. Cancer Res. 2018;78:1726–1738. doi: 10.1158/0008-5472.CAN-17-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Allensworth JL, Aird KM, Aldrich AJ, Batinic-Haberle I, Devi GR. XIAP inhibition and generation of reactive oxygen species enhances TRAIL sensitivity in inflammatory breast cancer cells. Mol Cancer Ther. 2012;11:1518–1527. doi: 10.1158/1535-7163.MCT-11-0787. [DOI] [PubMed] [Google Scholar]
- 77.Evans MK, et al. X-linked inhibitor of apoptosis protein mediates tumor cell resistance to antibody-dependent cellular cytotoxicity. Cell Death Dis. 2016;7:e2073. doi: 10.1038/cddis.2015.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.