Abstract
Background
Itaconic acid (IA), a C5-dicarboxylic acid, has previously been identified as one of the top twelve biochemicals that can be produced by biotechnological means. IA is naturally produced by Aspergillus terreus, however, heterologous production in the related species Aspergillus niger has been proposed earlier. Remarkably, we observed that during high producing conditions and elevated titers A. niger detoxifies the extracellular medium of IA. In order to determine the genes responsible for this decline in IA titers a transcriptome analysis was performed.
Results
Transcriptome analysis has led to the identification of two novel and previously unknown IA bioconversion pathways in A. niger. One pathway is proposed to convert IA into pyruvate and acetyl-CoA through the action of itaconyl-CoA transferase (IctA), itaconyl-CoA hydratase (IchA) and citramalyl-CoA lyase, similar to the pathway identified in A. terreus. Another pathway putatively converts IA into 1-methyl itaconate through the action of trans-aconitate methyltransferase (TmtA). Upon deleting the key genes ictA and ichA we have observed increased IA production and titers and cessation of IA bioconversion. Surprisingly, deletion of tmtA lead to strong reduction of heterologous IA production.
Conclusion
Heterologous IA production in A. niger induces the expression of IA bioconversion pathways. These pathways can be inhibited by deleting the key genes ictA, ichA and tmtA. Deletion of ictA and ichA resulted in increased IA production. Deletion of tmtA, however, resulted in almost complete cessation of IA production.
Electronic supplementary material
The online version of this article (10.1186/s40694-018-0062-5) contains supplementary material, which is available to authorized users.
Background
Rising carbon emissions due to increased industrialization and its effect on the climate are raising awareness to organize our economy in more sustainable ways. However, to transition from our current fossil resource-based economy to a bio-based economy is not easily achieved given the huge dependency on fossil fuels for energy and commodity needs. Biotechnologically produced organic acids have great potential as sustainable alternative for petrochemicals and its use as commodities [1]. The main bottleneck for industrial application of biochemicals, however, is the high price compared with petrochemicals. Many yeasts and bacteria have been exploited in the production of industrially relevant biochemicals, e.g. succinic acid [2]. However, filamentous fungi are due to their natural lifestyle as saprophytic organisms well equipped to break down complex carbohydrate structures e.g. lignocellulose and produce industrially relevant biochemicals. Apart from this fact filamentous fungi are also known as efficient organic acid producers, in particular members of the genus Aspergillus [3].
Itaconic acid (IA), a C5-dicarboxylic acid, has previously been identified as one of the top twelve biochemicals that can be produced by biotechnological means [4]. The potential applications of IA in green chemistry are numerous and an exciting overview of novel applications is provided by Robert and Friebel [5]. Industrial production of IA is performed using the natural producer A. terreus. Currently IA and its chemical derivatives are seen as niche chemical with low industrial relevance. The main applications of IA and its chemical derivatives are as superabsorbent polymers, synthetic latex, detergent builders, polymers and polyester resins [6, 7]. Lowering production costs and selling price of IA could result in the promotion of IA into a platform chemical that would lead to an tenfold increase in its market size and open market applications such as thermoplastics [8, 9]. In order to achieve this feat the selling price of IA should be competitive with fossil-based end use chemicals such as maleic anhydride. The high production cost and selling price of IA can be attributed to the sensitivity of A. terreus to impurities in industrial cultivation medium and tight control of fermentation processes [10–12].
Heterologous IA production in the related species Aspergillus niger has been proposed earlier based on the superior organic acid production capabilities of A. niger as exemplified by industrial citric acid (CA) production that mainly employs A. niger [13–15]. Additionally A. niger is a more robust production organism able to withstand impurities in industrial cultivation medium; a feat that is evident by the low selling prices of biotechnologically produced CA [16, 17]. In our previous report we have communicated the rewiring of A. niger secondary metabolism citrate synthase (CitB) leading to an increased IA yield, titer and productivity [18]. Metabolic engineering of ATP-citrate lyase in our rewired pathway has resulted in further improvement of IA production (Hossain AH et al. manuscript in preparation). However, we also observed in cultivations with these engineered strains that IA titer reaches a plateau after which titers start to decline rapidly (Hossain AH et al. manuscript in preparation). This decline in IA is supposedly brought about by putative IA bioconversion. In order to determine the genes responsible for IA bioconversion a transcriptome analysis was performed. In combination with a previous transcriptome analysis of primary metabolism genes related to glycolysis, TCA cycle and organic acid transport (de Vries et al. [3]), this analysis led to the identification of previously unknown genes of which the expression is highly upregulated in IA producing conditions.
Materials and methods
Strains and media components
Aspergillus niger strain CitB#99 (CBS141659) [18] was used in this study in which deletion of ictA, ichA and tmtA was performed. The strains used for transcriptome analysis are listed in Table 1. All strains were stored in 30% glycerol at − 80 °C and maintained on agar containing minimal medium (MM) plates (16 g/L agar, 6 g/L NaNO3, 0.52 g/L KCl, 1.52 g/L KH2PO4, 10 g/L glucose, 0.0022 g/L ZnSO4.7H2O, 0.0011 g/L H3BO3, 0.0005 g/L MnCl2.4H2O, 0.0005 g/L FeSO4.7H2O, 0.00017 g/L CoCl2.6H2O, 0.00016 g/L CuSO4.5H2O, 0.00015 g/L NaMoO4.2H2O, 0.005 g/L Na2EDTA and 0.5 g/L Mg2SO4). Spore suspensions were prepared using physiological salt solution (0.9% NaCl) and stored at 4 °C for up to 1 year. Fresh spore suspensions were prepared for inoculation during each shake flask and batch fermentation experiment.
Table 1.
List of strains used for transcriptome analysis
| Strain | Abbreviation | Strain description |
|---|---|---|
| AB1.13 pyrG+ | AB1.13 | Uridine prototroph of AB1.13 pyrG—[19] |
| AB1.13 CAD 4.1 | AB1.13 CAD | Selected pyrG + transformant of cadA expressing transformant (CAD10.1) of AB1.13 [20] |
| AB1.13 CAD + MTT + MFS_3 | AB1.13 CAD + MTT + MFS | Selected mfsA expressing transformant of MTT 1.4 [19] |
| AB1.13 CAD + MFS + MTT #49B; | AB1.13 #49B | Selected mttA expressing transformants of AB1.13 CAD + MFS 3.9 [18] |
| AB1.13 CAD + MFS + MTT + CitB #99; #113 | CitB#99; CitB#113 | Selected citB overexpressing transformants of AB1.13 CAD + MFS + MTT #49B [18] |
Split marker deletion construct and transformation
Auxotrophic pyrE-strains were generated by cultivating CitB#99 on 5-fluoroorotic acid selective plates [21]. Plates were incubated in a 33 °C stove for 3–5 days until colony formation was visible. Organic acid production of CitB#99 pyrE-colonies was tested by cultivation in microtiter plates (see section screening). Knock-out of ictA, ichA and tmtA was performed using the split-marker method [22]. An schematic overview of the split-marker approach is shown in Additional file 1. Split-marker flanks for an ictA and ichA knock-out were generated with fusion PCR and split-marker flanks for a tmtA knock-out were in vitro synthesized by GeneArt Gene Synthesis (Thermo Scientific) and individually introduced into pJET1.2/blunt via blunt-end ligation using the CloneJET PCR Cloning Kit (Thermo Scientific). The primers used to create the split-marker fragments for ictA and ichA are listed in Additional file 2. The pyrE gene of Aspergillus oryzae RIB40 (AO090026000521) was employed to complement the pyrE deficient phenotype. The split-marker fragments were co-transformed in an ratio of 1:1 (5 µg flank 5′: 5 µg flank 3′). All transformations were carried out according to the protocol as reported by Punt et al. [23]. Transformed protoplasts were plated on MM agar plates containing sorbitol and grown at 33 °C for 3–5 days until colonies were visible. Successful targeted integration of bi-partite fragments were determined with diagnostic colony PCR [24] using various primer combinations. Colonies that showed the expected PCR fragments were selected for further screening in microtiter plate and shake flask experiments.
Microtiter plate transformant screening
Plates carrying transformed cells were allowed to grow and sporulate for 1–2 weeks after which individual colonies were transferred to a selective MM plate. Individual colonies from this plate were each streaked on a separate selective MM plate to isolate single colonies that in turn were used to inoculate 1 mL liquid cultures in a 96-wells deepwell plate (Axygen; Corning, NY) containing M12 ++ medium (1.43 g/L NH4NO3, 0.11 g/L KH2PO4, 0.5 g/L MgSO4 × 7 H2O, 0.005 g/L CuSO4 × 5 H2O, 0.0006 g/L FeIIICl3 × 6 H2O, 0.0006 g/L ZnSO4 × 7 H2O, 0.074 g/L NaCl, 0.13 g/L CaCl2 × 2 H2O and 100 g/L glucose) [20]. This 96-wells plate was incubated for 72 h at 33 °C and 850 RPM. Supernatant was filtered over a 0.22 µM filter (Corning; Corning, NY) and analyzed on an HPLC for IA production (see below).
Flask cultivations
MM agar plates were streaked with conidia from glycerol stocks or from isolated single colonies that were determined by colony PCR. These plates were incubated at 33 °C for several days till plates were fully grown. Fresh conidia suspensions were prepared by harvesting conidia from these plates with sterile 0.9% NaCl solution. The harvested conidia were counted on the LUNA II cell counter (Logos Biosystems). Non-baffled shake flasks (500 mL) were filled with 100 mL M12 ++ medium and inoculated with 1.0 × 106/mL conidia and incubated at 35 °C and 0 RPM. Flasks were weighed when empty, after inoculation and each day before sampling. Evaporation is calculated from the measured weight of the flasks and used to correct measured concentrations of organic acids and glucose by HPLC (see below). Error bars in graphs of flask cultivations indicate the standard error of the mean. All flask cultivations were performed in duplicate.
Controlled-batch cultivations
Controlled-batch cultivations were performed on 5L scale benchtop New Brunswick Scientific fermenters (BioFlo 3000) at 33 °C. Starting pH was 3.5 after inoculation and medium was allowed to naturally acidify till pH 2.3 and then kept at pH 2.3 by addition of 4 M KOH. Dissolved oxygen (DO) tension was 25% at the moment of inoculation and when DO dropped till 20% it was kept at 20%. The system was calibrated with 100% sterile air as 100% DO and 100% N2 as 0% DO. The fermenter was inoculated by 72 h old 100 mL non-baffled shake flask cultures containing 1.0 × 108 spores. Medium composition for fermentation and pre-culture (M12 ++) is described above.
HPLC
Metabolite analysis was performed using a WATERS e2695 Separations Module equipped with an Aminex HPX-87H column (Bio-Rad) and 5 mM H2SO4 as eluent. Detection of peaks occurred simultaneously by a refractive index detector (WATERS 2414) and a dual-wavelength detector (WATERS UV/Vis 2489). Data processing was done with Empower Pro software (Empower 2 Software, copyright 2005–2008, Waters Corporation, Milford, MA, USA).
RNA isolation and transcriptome analyses
Biomass samples for RNA isolation were taken at several time points during fermentation and washed with distilled water and frozen in liquid N2. The mycelium was disrupted by bead-beating with 0.1 mm acid-washed Zirconium-Silica beads and RNA extraction proceeded using the ChargeSwitch RNA extraction protocol from Invitrogen (Carlsbad, CA, USA). Quality control was checked on 1 × 3-(N-morpholino)propanesulfonic acid/6% Formaldehyde agarose gels and stained with ethidium bromide.
BaseClear in Leiden, NL performed digital gene expression profiling experiments based on RNA-Seq with an Illumina HiSeq 2000 System. Approximately 8–32 M unfiltered paired-end (PE) reads (99 bp/read on ~ 320 bp cDNA inserts) were obtained. Reads were trimmed of the first 2 bases of the 5′ end because these bases showed an aberrantly low GC content. The reads were then further filtered, such that all quality Phred scores after filtering are at least 22, with a read-length of at least 40 bases. Around 70–80% of the bases passed these criteria (including a 2% loss because of clipping). After filtering the # PE-reads/samples were between 7.6 and 19.8 M for all the samples respectively.
Reads were aligned to the 20 contigs in a FastA file of the Aspergillus niger CBS 513.88 reference genome (from http://www.ebi.ac.uk/ena). Source EMBL annotations were converted to GFF format. The embl data appeared to be derived from multiple sources with different feature tags. These were converted to one uniform GFF format that could be accepted by our third-party software (consistent gene_ids across all contigs). Missing gene definitions (e.g. inserted genes for IA production) were inserted. The reads were aligned to the reference genome using software based on a Burrows–Wheeler Transform (BWT) algorithm. A mismatch rate of 4% was allowed for the alignment. The maximum insertion length was 3. The maximum deletion length was 3. All samples had more than 85% of the reads aligned, resulting in SAM alignment files. Gene expression was measured as the number of aligned reads to reference genes and was normalized to RPKM values (Reads Per Kb per Million reads; Mortazavi et al. [25]). Hierarchical clustering was performed with TIGR MEV 4.0. A stringent cut-off at 2logR value of 4.0 for upregulated genes and − 4.0 for downregulated genes was held for data analysis. A more relaxed cutoff of > 2.0 or smaller − 2.0 was used to explore the data for identifying novel differentially expressed gene clusters.
Results
Transcriptome analysis of high IA producing A. niger strains
Previously we have reported the IA production of A. niger strain CitB#99 that reaches a final titer of 26.2 g/L with max productivity of 0.35 g/L/h and yield of 0.37 g/g [18]. IA production was further improved by metabolic engineering of ATP-citrate lyase (Hossain et al. [18]). Remarkably, we have also observed IA bioconversion in A. niger during IA producing cultivations. This observation manifested in strongly reduced IA titers after achieving a peak IA titer. In addition, IA bioconversion was also observed in cultures were exogenous IA was added to shake flask cultures, showing a reduction of IA levels (Additional file 3). In order to identify the genes involved in IA bioconversion we have analyzed a transcriptome dataset of biomass isolated from batch fermentations with low, medium and high IA producing A. niger strains. In Table 2 transcriptome results of differentially regulated genes between the high IA producing strain CitB#99 and AB1.13 WT, that does not produce IA, is given. As expected, the four genes that constitute the engineered part of the heterologous IA biosynthesis pathway i.e. cadA, mfsA, mttA and citB are all highly expressed in CitB#99 (Table 2A).
Table 2.
Differential expressed genes in high IA producing strains
| A | 2logR values | RPKM values | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus tag | Old locus tag | Annotation | TargetP | AB1.13 | AB1.13 CAD | AB1.13 #49B | CitB#99 | AB1.13 | AB1.13 CAD | AB1.13 #49B | CitB#99 |
| ANI_1_1474074 | An08g10920 | Citrate synthase (CitB) | Cytosol | 0.02 | 0.04 | − 0.05 | 11.40 | 3.05 | 3.10 | 2.87 | 10,838.65 |
| cadA (A. terreus) | Cis-aconitate decarboxylase | Cytosol | 0.05 | 10.62 | 9.47 | 10.72 | 3.54 | 6887.96 | 3116.06 | 7417.64 | |
| ANI_1_1906104 | An12g05750 | MFS phosphate transporter | PM | 0.13 | 1.02 | 10.28 | 10.55 | 1.09 | 2.87 | 2375.80 | 2852.93 |
| hph (S. hindustanus) | Hygromycin B 4-O-kinase | Cytosol | 0.23 | 0.17 | 8.84 | 10.04 | 0.94 | 0.86 | 759.61 | 1743.36 | |
| mttA (A. terreus) | Putative mitochondrial tricarboxylite transporter | Mito | − 0.18 | 0.18 | 8.46 | 9.86 | 0.19 | 0.54 | 477.02 | 1257.78 | |
| mfsA (A. terreus) | Putative major facilitator superfamily transporter A | PM | 0.07 | 1.44 | 7.49 | 9.19 | 0.30 | 2.36 | 222.56 | 721.88 | |
| amdS (A. nidulans) | Acetamidase A | Cytosol | 0.00 | 8.57 | 7.57 | 7.90 | 0.37 | 517.76 | 259.47 | 325.00 | |
| ANI_1_2022014 | An01g14940 | Phospholipase C PLC-C | Secreted | 0.06 | 0.08 | 4.99 | 7.18 | 8.40 | 8.53 | 286.39 | 1310.11 |
| ANI_1_1486074 | An08g11030 | 3-Phytase B | Secreted | − 0.03 | 0.21 | 5.94 | 6.99 | 27.36 | 32.28 | 1771.34 | 3663.34 |
| ANI_1_2368024 | An02g02840 | Sialidase | Secreted | − 0.05 | 0.06 | 7.22 | 6.81 | 0.21 | 0.30 | 184.78 | 139.52 |
| ANI_1_236084 | An09g02180 | Lipase | Secreted/Mito | − 0.34 | − 0.30 | 3.96 | 6.78 | 2.43 | 2.55 | 67.01 | 479.51 |
| ANI_1_1848144 | An16g06190 | Metabolite transport protein GIT1 | PM | 0.04 | 0.73 | 6.80 | 6.46 | 6.81 | 11.62 | 846.83 | 669.87 |
| ANI_1_166144 | An16g01340 | Glyoxalase domain-containing protein 5 | Mito/Cytosol | 0.24 | 4.70 | 5.70 | 6.20 | 16.28 | 379.47 | 761.69 | 1080.98 |
| ANI_1_248124 | An14g01550 | Acid phosphatase | Secreted | − 0.16 | − 0.27 | 4.91 | 5.89 | 4.20 | 3.82 | 174.16 | 345.29 |
| ANI_1_1500104 | An12g02320 | Short-chain dehydrogenase/reductase | Cytosol | 0.01 | 3.07 | 4.70 | 5.65 | 2.33 | 26.63 | 84.56 | 164.88 |
| ANI_1_1330084 | An09g03700 | Hypothetical protein | Secreted | − 0.14 | 0.10 | 4.05 | 5.64 | 0.46 | 0.72 | 25.72 | 79.39 |
| ANI_1_460094 | An11g03340 | Alpha-amylase A type-1/2 | Secreted | − 0.12 | 0.33 | 1.56 | 5.52 | 12.80 | 17.81 | 43.27 | 683.63 |
| ANI_1_542164 | An18g04140 | Acid phosphatase | Secreted | 0.04 | 0.07 | 4.04 | 5.50 | 1.42 | 1.45 | 37.62 | 104.94 |
| ANI_1_1576144 | An16g02440 | Phytase-like | Secreted | − 0.03 | 0.04 | 4.27 | 5.48 | 3.98 | 4.23 | 97.10 | 226.06 |
| ANI_1_260144 | An16g01850 | BYS1 domain protein | Secreted | − 0.64 | − 0.16 | 2.35 | 5.30 | 9.22 | 13.19 | 79.91 | 623.78 |
| ANI_1_1250124 | An14g02660 | NPP1 domain protein | Secreted | 0.12 | 0.24 | 4.08 | 5.29 | 3.11 | 3.48 | 63.27 | 147.19 |
| ANI_1_2836024 | An02g08830 | Histone transcription regulator 3 | Nucleus | 0.00 | 0.09 | − 0.03 | 5.15 | 9.48 | 10.11 | 9.28 | 371.63 |
| ANI_1_652114 | An13g01750 | Acid phosphatase | Secreted | 0.06 | 0.10 | 4.67 | 5.13 | 70.73 | 72.83 | 1754.06 | 2422.06 |
| ANI_1_1440124 | An14g04660 | Aromatic ring-opening dioxygenase, catalytic LigB subunit | Cytosol/Mito/Per | 0.13 | 0.32 | 0.32 | 5.13 | 2.59 | 3.10 | 3.09 | 113.67 |
| ANI_1_1432064 | An07g00760 | Putative itaconyl-CoA transferase (ictA) | Mito | 0.32 | 3.27 | 3.92 | 5.06 | 13.07 | 108.35 | 170.34 | 375.89 |
| ANI_1_1812184 | An04g04240 | Phosphate transporter | PM | 0.27 | 1.05 | 4.83 | 4.99 | 2.94 | 5.74 | 91.74 | 102.55 |
| ANI_1_126034 | An03g01120 | Aromatic-amino-acid aminotransferase | Nucleus | − 0.34 | 3.62 | 4.24 | 4.98 | 29.79 | 478.24 | 735.44 | 1226.92 |
| ANI_1_246114 | An13g01760 | Oligopeptide transporter | PM | 0.33 | 0.46 | 2.43 | 4.97 | 1.30 | 1.51 | 8.88 | 56.23 |
| ANI_1_1908104 | An12g05810 | Multicopper oxidase | Secreted | 0.04 | 0.20 | 4.33 | 4.76 | 0.18 | 0.31 | 21.98 | 30.01 |
| ANI_1_2118064 | An07g09220 | Putative itaconyl-CoA hydratase (ichA) | Mito | − 0.11 | 2.78 | 4.04 | 4.71 | 7.32 | 60.28 | 146.61 | 232.65 |
| ANI_1_1324074 | An08g09850 | Acid phosphatase | Secreted | − 0.18 | − 0.36 | 3.70 | 4.58 | 128.06 | 112.96 | 1896.57 | 3490.63 |
| ANI_1_1520134 | An15g04760 | Glycosyl hydrolase family 71 protein | Secreted | − 0.29 | − 0.82 | 2.21 | 4.57 | 1.58 | 0.79 | 13.68 | 74.04 |
| ANI_1_2122184 | An04g08320 | MFS monocarboxylate transporter | PM | 0.17 | 0.49 | 1.25 | 4.46 | 7.11 | 9.16 | 16.14 | 158.52 |
| ANI_1_3198024 | An02g13080 | Phenylacetate-coenzyme A ligase | Cytosol | − 0.28 | − 0.51 | 0.35 | 4.28 | 0.27 | 0.08 | 0.96 | 28.87 |
| ANI_1_1430064 | An07g00750 | Fungal transcription factor | Nucleus/Mito | 0.02 | 3.04 | 3.73 | 4.28 | 0.52 | 11.37 | 19.01 | 28.19 |
| ANI_1_820034 | An03g06550 | Glucoamylase | Secreted | 0.13 | − 0.02 | 1.76 | 4.18 | 243.81 | 218.86 | 756.84 | 4054.75 |
| ANI_1_1020134 | An15g07520 | Phosphoglycerate mutase family protein | Cytosol | − 0.20 | − 0.60 | 4.75 | 4.18 | 12.67 | 9.31 | 420.93 | 282.80 |
| ANI_1_1810184 | An04g04230 | Phosphate transporter (reannotated) | Nucleus/Cytosol | − 0.18 | 0.33 | 4.60 | 4.17 | 4.67 | 7.06 | 154.52 | 114.01 |
| ANI_1_294044 | An05g02340 | Extracellular dihydrogeodin oxidase/laccase | Secreted | − 0.08 | − 0.07 | 4.48 | 4.11 | 0.44 | 0.44 | 32.96 | 25.14 |
| ANI_1_864144 | An16g06510 | Trans-aconitate 2-methyltransferase (tmtA) | Cytosol | 0.08 | 0.73 | 2.40 | 4.08 | 12.71 | 20.51 | 67.34 | 218.46 |
| ANI_1_342114 | An13g02590 | Sugar transporter | PM | − 0.01 | 0.28 | 2.16 | 4.06 | 3.21 | 4.16 | 18.01 | 69.60 |
| ANI_1_1098184 | An04g06920 | Alpha-glucosidase | Secreted | 0.00 | − 0.33 | 0.57 | 4.04 | 174.34 | 138.54 | 258.87 | 2884.10 |
| ANI_1_1820094 | An11g03450 | Small secreted protein | Secreted | 0.33 | 1.68 | 2.25 | 4.02 | 58.07 | 149.69 | 222.30 | 763.49 |
| ANI_1_330024 | An02g02480 | MFS phosphate transporter | PM | − 0.20 | 1.65 | 3.18 | 4.02 | 3.12 | 13.82 | 41.86 | 75.60 |
| ANI_1_762014 | An01g05900 | Cytochrome P450 | Secreted | − 0.13 | − 0.10 | 3.86 | 4.00 | 0.63 | 0.66 | 24.97 | 27.63 |
| ANI_1_1778074 | An08g02590 | GPI anchored protein | Secreted | − 0.06 | 0.21 | 3.26 | 4.00 | 10.62 | 13.07 | 115.12 | 192.81 |
| B | 2logR values | RPKM values | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus tag | Old locus tag | Annotation | TargetP | AB1.13 | AB1.13 CAD | AB1.13 #49B | CitB99 | AB1.13 | AB1.13 CAD | AB1.13 #49B | CitB99 |
| ANI_1_542134 | An15g03800 | Hydrophobin | Secreted | 0.06 | − 0.55 | − 2.13 | − 8.78 | 981.57 | 641.93 | 213.11 | 1.14 |
| ANI_1_1416134 | An15g04010 | Hypothetical protein | Nucleus | 0.27 | − 0.20 | − 5.87 | − 7.79 | 288.73 | 208.03 | 3.11 | 0.09 |
| ANI_1_92174 | An10g00820 | Oxaloacetate acetylhydrolase | Cytosol | − 0.13 | − 0.81 | − 2.61 | − 7.51 | 1803.48 | 1122.69 | 323.44 | 9.81 |
| ANI_1_220174 | An10g00800 | Purine nucleoside permease | Secreted | − 0.13 | − 0.33 | − 2.72 | − 6.47 | 375.32 | 326.25 | 61.40 | 3.64 |
| ANI_1_1046034 | An03g01450 | Major Facilitator Superfamily | PM | − 0.20 | − 0.83 | − 2.49 | − 6.29 | 73.83 | 47.18 | 14.31 | 0.10 |
| ANI_1_1062034 | An03g01590 | Arginine permease | PM | 0.13 | 0.41 | − 2.09 | − 6.08 | 243.24 | 295.39 | 51.56 | 2.30 |
| ANI_1_964034 | An03g00580 | Cytochrome P450 monooxygenase | Mito | − 0.29 | − 1.86 | − 3.52 | − 6.00 | 228.74 | 76.05 | 23.45 | 3.39 |
| ANI_1_966034 | An03g00590 | Trichodiene synthase Tri5 | Cytosol | − 0.31 | − 2.27 | − 4.02 | − 5.87 | 94.11 | 23.35 | 6.27 | 1.01 |
| ANI_1_170034 | An03g01460 | 1-Aminocyclopropane-1-carboxylate oxidase | Cytosol | − 0.10 | − 0.86 | − 2.52 | − 5.43 | 114.81 | 67.35 | 20.61 | 1.88 |
| ANI_1_1914064 | An07g06460 | C-7 hydroxycephem methyltransferase | Cytosol | − 0.26 | − 0.32 | − 4.38 | − 5.28 | 36.47 | 35.03 | 1.16 | 0.16 |
| ANI_1_182034 | An03g01540 | FAD dependent oxidoreductase | Secreted/Cytosol | 0.05 | − 0.42 | − 1.58 | − 5.26 | 78.94 | 56.69 | 24.80 | 1.02 |
| ANI_1_1770144 | An16g05290 | Purine-cytosine permease | PM | − 0.05 | 0.00 | − 1.13 | − 5.22 | 838.03 | 867.97 | 395.45 | 22.38 |
| ANI_1_990084 | An09g00660 | OPT oligopeptide transporter protein | PM | 0.01 | 0.10 | − 0.27 | − 5.09 | 279.04 | 298.75 | 230.29 | 7.20 |
| ANI_1_2008094 | An11g05190 | Type 1 glutamine amidotransferase | Cytosol | − 0.02 | 0.24 | − 1.25 | − 5.06 | 70.06 | 84.12 | 29.32 | 1.16 |
| ANI_1_1366064 | An07g00170 | Hypothetical protein | Secreted | 0.37 | 0.34 | − 1.87 | − 5.04 | 56.14 | 54.96 | 11.10 | 0.35 |
| ANI_1_1572014 | An01g11640 | Ammonium transporter | PM | 0.22 | 0.03 | − 1.36 | − 5.02 | 222.82 | 195.30 | 74.16 | 4.91 |
| ANI_1_1862104 | An12g05440 | Urea active transporter 1 | PM | − 0.15 | − 0.26 | − 2.63 | − 5.01 | 68.04 | 63.09 | 11.38 | 1.38 |
| ANI_1_2238104 | An12g09660 | Phosphoribosyl transferase | Cytosol | 0.64 | 0.85 | − 1.49 | − 4.94 | 126.25 | 145.43 | 27.94 | 1.66 |
| ANI_1_1456124 | An14g04760 | Male sterility domain containing protein | Cytosol | 0.26 | − 0.49 | − 2.51 | − 4.91 | 258.07 | 152.77 | 36.93 | 6.18 |
| ANI_1_708024 | An02g05060 | Hypothetical protein | Secreted | − 0.03 | − 1.15 | − 1.98 | − 4.81 | 810.00 | 371.87 | 208.67 | 28.41 |
| ANI_1_2764024 | An02g08200 | Short-chain dehydrogenase/reductase | Cytosol | 1.05 | 2.54 | − 0.64 | − 4.80 | 308.27 | 867.28 | 94.68 | 4.34 |
| ANI_1_2236104 | An12g09640 | Uracil-regulated protein 1 | Cytosol | 0.44 | 0.67 | − 0.72 | − 4.80 | 107.20 | 125.82 | 47.45 | 1.86 |
| ANI_1_46014 | An01g00370 | Aspartic endopeptidase (AP1) | Nucleus | 0.10 | − 0.18 | − 1.58 | − 4.79 | 1351.68 | 1115.18 | 422.54 | 44.77 |
| ANI_1_1054034 | An03g01520 | 12-oxophytodienoate reductase | Mito | − 0.03 | − 1.01 | − 1.20 | − 4.71 | 42.16 | 20.82 | 18.17 | 0.68 |
| ANI_1_1850094 | An11g03640 | OPT oligopeptide transporter family | PM | − 0.11 | − 0.52 | − 0.49 | − 4.71 | 104.23 | 78.34 | 79.93 | 3.35 |
| ANI_1_1990074 | An08g05320 | Short-chain dehydrogenase/reductase | Cytosol | − 1.43 | − 1.61 | − 3.15 | − 4.70 | 16.50 | 14.51 | 4.33 | 0.82 |
| ANI_1_474154 | An17g01540 | GABA permease | PM | 0.07 | 0.26 | − 2.34 | − 4.67 | 85.82 | 98.28 | 15.40 | 2.26 |
| ANI_1_1748104 | An12g04260 | NAD dependent epimerase/dehydratase | Mito | 1.23 | 2.79 | − 2.00 | − 4.66 | 69.28 | 206.65 | 6.50 | 0.19 |
| ANI_1_726064 | An07g05830 | Formamidase | Cytosol | − 0.03 | 0.02 | − 0.57 | − 4.58 | 267.27 | 276.94 | 184.40 | 10.45 |
| ANI_1_3344014 | An01g14400 | Cercosporin toxin resistance protein CRG1 | Cytosol | 0.04 | 0.02 | − 2.87 | − 4.56 | 63.05 | 62.21 | 7.56 | 1.65 |
| ANI_1_176044 | An05g01410 | Acyl-CoA oxidase | Mito | − 0.30 | − 0.96 | − 2.25 | − 4.55 | 178.03 | 111.60 | 45.07 | 8.38 |
| ANI_1_568054 | An06g02150 | DUF3328 protein | Secreted | 0.11 | 0.34 | − 4.28 | − 4.55 | 48.49 | 57.10 | 1.36 | 0.96 |
| ANI_1_1554124 | An14g05980 | MFS transporter | PM | 0.01 | 0.48 | − 3.43 | − 4.54 | 29.12 | 40.79 | 1.78 | 0.29 |
| ANI_1_1048034 | An03g01470 | d-galactonate transporter | PM | − 0.07 | − 1.24 | − 2.35 | − 4.52 | 37.53 | 16.20 | 6.96 | 0.76 |
| ANI_1_806074 | An08g05670 | Nitrate transporter | PM | 0.13 | 1.08 | − 1.66 | − 4.44 | 44.15 | 86.17 | 12.00 | 0.90 |
| ANI_1_3376014 | Fe(II)/2-oxoglutarate (2OG) oxygenase | Cytosol | − 0.17 | − 0.07 | − 1.01 | − 4.38 | 97.28 | 104.78 | 54.14 | 4.31 | |
| ANI_1_1340084 | An09g03750 | MAC/Perforin domain | Cytosol | 0.10 | 0.11 | − 2.61 | − 4.37 | 22.58 | 22.73 | 2.58 | 0.06 |
| ANI_1_1514034 | An03g06020 | 4-Carboxymuconolactone decarboxylase | Cytosol | 0.19 | − 0.43 | − 1.29 | − 4.31 | 5568.62 | 3603.74 | 1987.79 | 245.12 |
| ANI_1_2954014 | An01g09970 | Surface layer protein | Cytosol | 0.37 | − 0.03 | − 1.62 | − 4.29 | 66.37 | 50.05 | 16.01 | 1.67 |
| ANI_1_2450014 | An01g03790 | Urea active transporter 1 | PM | − 0.05 | − 0.17 | − 0.10 | − 4.29 | 116.82 | 107.59 | 113.28 | 5.27 |
| ANI_1_616114 | An13g01470 | Laminin gamma 3 | Secreted | − 0.15 | − 1.01 | − 2.79 | − 4.27 | 110.93 | 60.59 | 16.95 | 5.42 |
| ANI_1_1456104 | An12g01910 | Phytase | Secreted | − 0.04 | − 0.95 | − 4.39 | − 4.26 | 30.69 | 15.78 | 0.55 | 0.70 |
| ANI_1_142174 | An10g00180 | Cupin_like | Cytosol/Mito | 0.34 | − 0.10 | − 1.24 | − 4.24 | 424.35 | 310.92 | 140.90 | 16.72 |
| ANI_1_1362014 | An01g09980 | Asp-hemolysin | Secreted | 0.32 | − 0.14 | − 1.07 | − 4.21 | 347.94 | 252.51 | 132.21 | 14.06 |
| ANI_1_878114 | An13g03990 | Nucleobase-cation-symport-1 (NCS1) transporter | PM | − 0.08 | 0.15 | − 2.27 | − 4.21 | 30.66 | 36.14 | 5.91 | 0.81 |
| ANI_1_2456094 | S-adenosylmethionine-dependent methyltransferase | 0.08 | − 0.27 | − 3.01 | − 4.20 | 69.27 | 54.39 | 7.29 | 2.63 | ||
| ANI_1_2086074 | An08g07010 | DUF3445 protein | Mito | 0.00 | − 0.25 | − 2.44 | − 4.17 | 37.07 | 31.01 | 6.01 | 1.11 |
| ANI_1_212174 | Nitrilase | − 0.38 | − 0.59 | − 2.64 | − 4.12 | 18.92 | 16.23 | 3.15 | 0.49 | ||
| ANI_1_1828184 | An04g04370 | Phenylalanine ammonia-lyase | PM | 0.17 | − 0.10 | − 2.11 | − 4.11 | 277.24 | 229.84 | 56.44 | 13.37 |
| ANI_1_76154 | An17g00640 | Zinc-binding alcohol dehydrogenase domain-containing protein cipB | Cytosol | 1.04 | 2.86 | − 2.13 | − 4.11 | 119.10 | 425.44 | 12.36 | 2.40 |
| ANI_1_1094134 | An15g00280 | Polysaccharide deacetylase | Cytosol | 0.00 | − 0.07 | − 0.98 | − 4.08 | 204.72 | 194.38 | 102.72 | 11.10 |
| ANI_1_1144084 | An09g01920 | FAD binding domain protein | Secreted | 0.75 | 2.11 | − 1.49 | − 4.04 | 39.96 | 103.97 | 7.67 | 0.48 |
| ANI_1_294064 | An07g02310 | Glutathione S-transferase Ure2-like protein | Cytosol | 0.01 | − 0.86 | − 2.83 | − 4.02 | 187.81 | 102.94 | 25.42 | 10.62 |
(A) Genes upregulated in high IA producing conditions. Cutoff of 2logR values of 4.00 or higher was held. (B) Genes downregulated in high IA producing conditions. Cutoff of 2logR values of − 4.00 was held
Interestingly, among the highly upregulated genes are a large number of genes that encode gene products with domains that have functions in phosphate liberation and translocation. This result is in line with the fact that in the IA production medium phosphate is limiting [26]. Remarkably, also the expression of major secreted enzymes such as amylase and glucoamylase, but also a number of other secreted proteins is upregulated in high IA producing conditions. The mechanism behind this result is not entirely clear, but may also be related to the fact that in high IA secretion conditions stress responses are induced which may lead to improved protein secretion (Table 2A) [27, 28]. Upon closer inspection of the results and in particular when expanding the results using a more relaxed cut-off, many of the upregulated and down regulated genes are clustered together in the genome in putative metabolic pathway clusters. This becomes even more apparent when we look at all genes for which the cut-off for differential expression is 2log = 2. Several phosphate and iron related gene clusters were identified showing co-induction in high IA producing strains (Additional file 4).
Heterologous IA production in A. niger also results in the downregulation of many genes compared with the AB1.13 strain (Table 2B). In relation to organic acid production it is interesting to note that the expression of the oahA gene coding for oxaloacetate acetylhydrolase, the enzyme that hydrolyses oxaloacetate into oxalate and acetate, is downregulated in CitB#99. This observation is in line with our previous report were oxalic acid could not be detected in batch fermentations of CitB#99 [18]. Upon closer inspection also genes encoding malate synthase and isopropylmalate synthase are downregulated, assuming further rewiring of the organic acid pathway in our IA production hosts (Additional file 5). Remarkably among the downregulated genes are many that are involved with N transport and utilization. This could be caused by the use of NH4SO4, as sole N-source in IA production media, which could mediate nitrogen metabolite repression under high IA production conditions. In closer inspection of the selection of genes showing at least fourfold repression in IA overproducing strains besides the many N-source utilisation related functions (permeases etc.) also several secondary metabolite gene clusters were identified (Additional file 5).
Interestingly, in high IA producing conditions CitB#99 upregulates the expression of An07g00760 and An07g09220, when compared to AB1.13. This upregulation can already be observed in AB1.13 CAD strain that produces low titers of IA (Table 3) [20]. Genes An07g00760 and An07g09220 share sequence similarity with ATEG_06299 and ATEG_03709 that are identified in A. terreus as genes responsible for the biological degradation of IA [29] (Additional file 6). Sasikaran et al. [30] have also elucidated a similar pathway in pathogenic bacteria, suggesting a role in pathogenicity. As Chen et al. show the product of genes ATEG_06299 and ATEG_03709 are itaconyl-CoA transferase (IctA) and itaconyl-CoA hydratase (IchA) that together with citramalyl-CoA lyase (CclA) (ATEG_03186) degrade IA into the cellular building block chemicals pyruvate and acetyl-CoA. In A. terreus all three genes ictA, ichA and cclA are induced whereas in A. niger only ictA and ichA are induced. In contrast to what may have been expected for a catabolic pathway of a secondary metabolite as IA is, these IA bioconversion genes are not clustered in the genome of A. niger. More dedicated sequence analysis of the encoded proteins reveal that all three carry predicted mitochondrial targeting sequences, suggesting that IA conversion occurs in this compartment.
Table 3.
Transcriptome data of genes involved in IA biosynthesis and bioconversion
| Locus tag | Enzyme | Old locus tag | TargetP | AB 1.13 WT | AB 1.13 CAD | AB1.13 #49B | CitB#99 |
|---|---|---|---|---|---|---|---|
| RPKM | RPKM | RPKM | RPKM | ||||
| IA biosynthesis cluster | |||||||
| Major facilitator superfamily transporter (mfsA) | PM | 0.30 | 2.36 | 222.56 | 721.88 | ||
| Cis-aconitate decarboxylase (cadA) | Cytosol | 3.54 | 6887.96 | 3116.06 | 7417.64 | ||
| Mitochondrial tricarboxylite transporter (mttA) | Mito | 0.19 | 0.54 | 477.02 | 1257.78 | ||
| Citrate synthase | |||||||
| ANI_1_876084 | Citrate synthase (citA) | An09g06680 | Mito | 284.82 | 269.02 | 255.71 | 238.09 |
| ANI_1_1226134 | Citrate synthase (mcsA) | An15g01920 | Mito | 76.60 | 61.89 | 82.55 | 85.78 |
| ANI_1_1474074 | Citrate synthase (citB) | An08g10920 | Cytosol | 3.05 | 3.10 | 2.87 | 10,838.65 |
| ANI_1_2950014 | Citrate synthase (citC) | An01g09940 | Cytosol | 463.10 | 370.88 | 438.08 | 96.44 |
| PrpD family (CadA like) | |||||||
| ANI_1_1536084 | Immune-responsive protein | An09g06220 | Cytosol | 0.30 | 0.23 | 0.36 | 0.43 |
| ANI_1_2952014 | Immune-responsive protein | An01g09950 | Cytosol | 552.06 | 456.98 | 383.21 | 165.41 |
| ANI_1_2948014 | 2-Methylcitrate dehydratase | An01g09930 | Cytosol | 308.68 | 261.08 | 288.92 | 145.66 |
| ANI_1_3352024 | MmgE_PrpD superfamily protein | An02g14730 | Cytosol | 5.97 | 7.29 | 5.81 | 10.08 |
| OahA class family | |||||||
| ANI_1_92174 | Oxaloacetate acetylhydrolase (oahA) | An10g00820 | Cytosol | 1803.48 | 1122.69 | 323.44 | 9.81 |
| ANI_1_2054064 | Oxaloacetate acetylhydrolase | An07g08390 | Cytosol | 10.41 | 7.31 | 7.55 | 11.70 |
| ANI_1_1800134 | Oxaloacetate hydrolase class protein | An15g07720 | Cytosol | 49.63 | 70.43 | 95.09 | 67.85 |
| IA bioconversion | |||||||
| ANI_1_1432064 | Itaconyl-CoA transferase A (ictA) | An07g00760 | Mito | 13.07 | 108.35 | 170.34 | 375.89 |
| ANI_1_676164 | CoA transferase superfamily enzyme | An18g05120 | Mito | 11.10 | 9.36 | 11.79 | 9.99 |
| ANI_1_2118064 | Itaconyl-CoA hydratase A (ichA) | An07g09220 | Mito | 7.32 | 60.28 | 146.61 | 232.65 |
| ANI_1_316154 | HTD2 protein | An17g02190 | Mito | 23.69 | 22.26 | 22.39 | 26.75 |
| ANI_1_1156014 | Citramalate-CoA lyase (cclA) | An01g08610 | Mito | 10.17 | 12.59 | 21.43 | 19.54 |
| ANI_1_864144 | Trans-aconitate 2-methyltransferase (tmtA) | An16g06510 | Mito | 12.71 | 20.51 | 67.34 | 218.46 |
| ANI_1_830134 | UMTA methyltransferase family protein | An15g06160 | Per/Cytosol | 15.52 | 11.45 | 52.11 | 23.02 |
Interestingly, also another previously uncharacterized gene showed similar induction in expression as ictA and ichA in high producing IA strains. Upon closer inspection this gene product (An16g06510) shares 43% homology with Escherichia coli trans-aconitate 2-methyltransferase Tam, which is identified by Zhao et al. as potential gene product that esterifies itaconate into 1-methyl itaconate in the yeast Saccharomyces cerevisiae [31]. Tam reportedly shares the same molecular function as the yeast trans-aconitate 3-methyltransferase TMT1 i.e. methylation of spontaneously formed trans-aconitate in order to relieve cytosolic toxicity by trans-aconitate mediated inhibition of aconitase [32, 33].
Deletion of ictA and ichA
In our transcriptome analysis we observed that the expression of ictA and ichA is upregulated in high IA producing strain CitB99. In A. terreus homologues of both gene products IctA and IchA have been shown to participate in a pathway that intracellularly convert IA into pyruvate and acetyl-CoA [29]. Based on these observations it was considered that knocking-out ictA and ichA would improve IA production. Bi-partite fragments were generated and transformed into CitB99. After transformation 89 colonies were visible on transformation plates. 24 colonies of each transformation were cultivated in microtiter plates for colony PCR. 4 colonies of CitB99 ΔICT were shown to be positive for deletion of the ictA gene and 6 colonies CitB99 ΔICH were positive for deletion of ichA (data not shown). To test for the effect of ΔictA and ΔichA on IA production in-time one CitB99 ΔICT and one CitB99 ΔICH strain were analyzed in flask experiments.
Flask cultivation of CitB99 ΔICT and CitB99 ΔICH
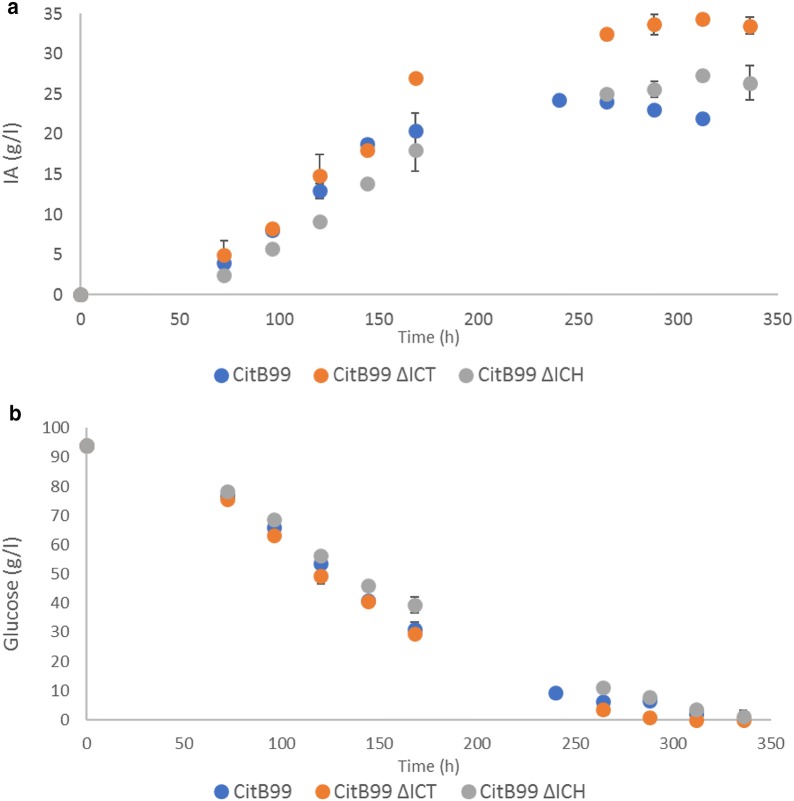
The IA production of CitB99 ΔICT and CitB99 ΔICH were analyzed in flask experiments in order to test for the effect of ΔictA and ΔichA on IA production. In line with results obtained for CA production in A. niger [17] we have observed that under non-shaken conditions more reproducible CA and IA production levels could be obtained (Hossain AH et al. unpublished). Therefore Erlenmeyer flasks were inoculated with 1.0 × 106 conidia/mL and incubated at 33 °C without shaking. From the results depicted in Fig. 1a it can be seen that IA production starts and proceeds very similar between CitB99 and CitB99 ΔICT up until 144 h of incubation, after which CitB99 ΔICT continues producing IA at a higher rate than CitB99. IA production in CitB99 reaches a plateau at 24.2 g/L after 240 h incubation before IA levels start to decline, due to degradation or bioconversion. Interestingly IA production in CitB99 ΔICT continues and reaches a final titer of 33.52 g/L IA after 336 h of incubation even after glucose is depleted in CitB99 ΔICT cultivation after 288 h (Fig. 1b). IA production in CitB99 ΔICH however, proceeds at a slower rate compared to CitB99 and CitB99 ΔICT. The final titer that is reached with this strain is much lower than CitB99 ΔICT at 26.39 g/L after 336 h but higher than the final titer of CitB99 due to the lack of degradation most likely. No side-product formation e.g. citrate or oxalate has been observed using HPLC analysis. These results show that IA detoxification can be inhibited by deleting ictA or ichA in A. niger. CitB99 ΔICT and CitB99 ΔICH were further tested in controlled-batch cultivations at 5-L scale.
Fig. 1.
Non-shaking flask cultivation of CitB99 ΔICT, CitB99 ΔICH and CitB99. Non-baffled 500 mL shake flasks were inoculated with 1.0 × 106 conidia/mL medium and incubated at 33 °C 0 RPM. The experiment was performed in duplicate. a Itaconic acid production, b glucose consumption during cultivation
Controlled-batch cultivation
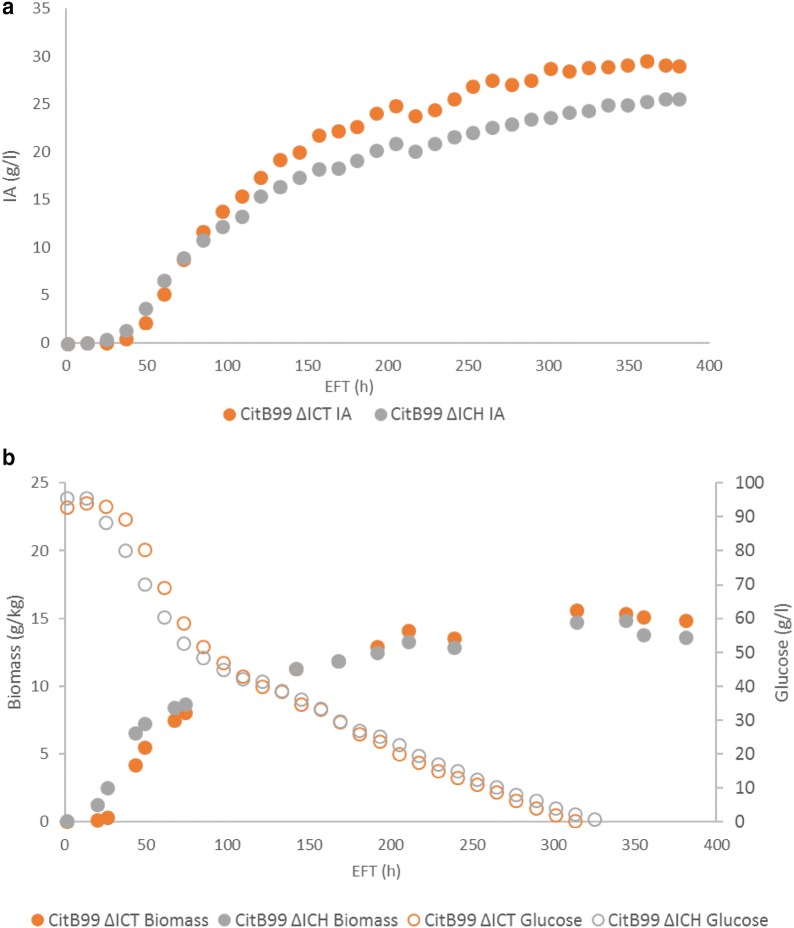
Previously, we have observed a slight decrease in IA titers during the end of controlled-batch fermentations [13, 18]. In order to further test the IA production of strains CitB99 ΔICT and CitB99 ΔICH we performed controlled-batch cultivations at 5L scale. The fermenters were inoculated with 100 mL 3 days old pre-cultures and DO was set at 20% saturation after inoculation. Glucose consumption, biomass formation and IA production started after 24 h of incubation. Glucose was depleted after 312 h for CitB99 ΔICT and after 324 h for CitB99 ΔICH at which point IA titer reached 28.7 and 24.3 g/L IA respectively (Fig. 2a). The fermentation was allowed to run for two more days to see if IA would drop in the glucose depleted cultures (Fig. 2b). The experiment was terminated after 380 h incubation in which no drop in IA levels were measured even after glucose had been depleted. The final titers reached were 29.2 g/L for CitB99 ΔICT and 25.7 g/L for CitB99 ΔICH.
Fig. 2.
Controlled-batch cultivation of CitB99 ΔICT and CitB99 ΔICH. Three day grown pre-cultures were used to inoculate 5L controlled-batch cultivations. a IA production of CitB99 ΔICT and CitB99 ΔICH. b Glucose consumption and biomass formation of CitB99 ΔICT and CitB99 ΔICH
Deletion of tmtA
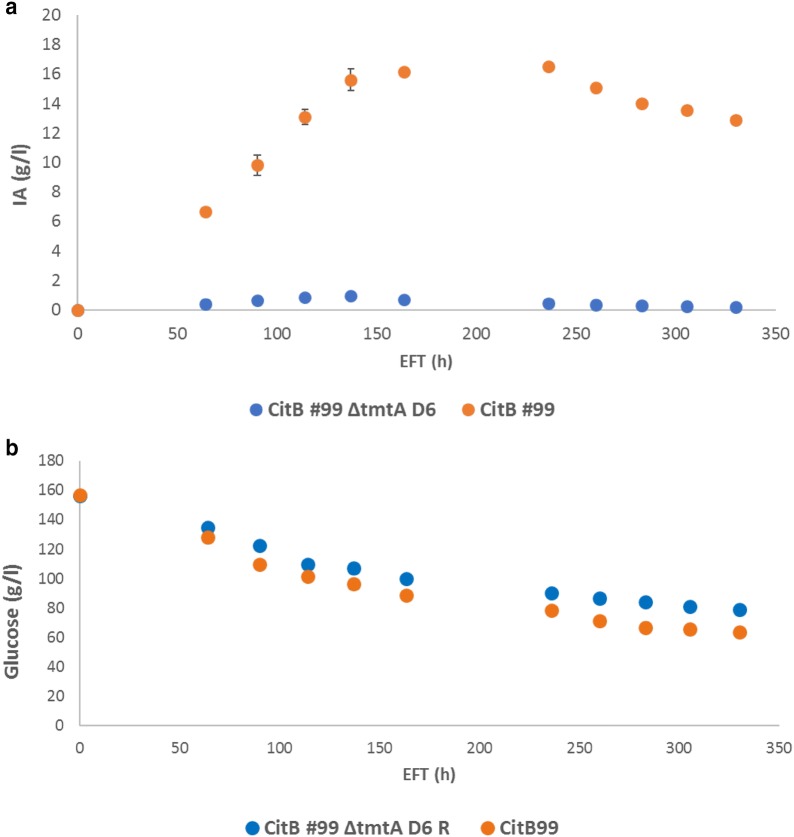
In order to investigate the role of TmtA in itaconate bioconversion, tmtA was disrupted in CitB99. After transformation 96 colonies were screened for knock-out using colony PCR and strain CitB99 ΔtmtA D6 was found to be a clean knock-out. IA production of CitB99 ΔtmtA D6 was further investigated by cultivation in shake flasks. However, HPLC analysis of shake flask cultivations with CitB99 ΔtmtA D6 and CitB99 (parental strain) in M12 ++ medium showed that IA production of CitB99 ΔtmtA D6 was negatively affected, achieving a max. titre of only 0.9 g/L IA compared to a max titre of 15.6 g/L IA achieved by the parental strain (Fig. 3a). Interestingly, glucose consumption was comparable between the two strains, suggesting that primary metabolism of glucose might not be affected in the ΔtmtA strain despite the low production of IA (Fig. 3b). Elevated levels of other organic compounds, including trans-aconitate and citrate, were not detected in the supernatant (data not shown). These results indicate that knock-out of tmtA does not result in the same desired phenotype in IA production as ΔictA and ΔichA has and as a result we did not proceed with 5L controlled-batch cultivations with this strain.
Fig. 3.
IA production of CitB99 ΔtmtA D6 and the parental CitB99 strain. Non-baffled 500 mL shake flasks were inoculated with 1.0 × 106 conidia/mL medium and incubated at 33 °C and 250 RPM. a Itaconic acid production, b glucose consumption during cultivation
Discussion
In our attempt at identifying unknown IA bioconversion pathways in A. niger we have performed a transcriptome analysis of high, medium and low IA producing strains. Interestingly, this analysis led to the identification of at least two novel organic acid bioconversion pathways that were not observed before (Fig. 4). IA is putatively converted into pyruvate and acetyl-CoA through the combined activity of IctA, IchA and CclA. Also, IA is putatively converted into 1-methyl itaconate through the activity of TmtA in yet another bioconversion pathway.
Fig. 4.
Hypothetical model of IA bioconversion in A. niger. Itaconate bioconversion presumably takes place in the mitochondrion where itaconate is converted to itaconyl-CoA by action of IctA and further hydrated to citramalyl-CoA by IchA. CclA facilitates the final conversion of citramalyl-CoA into pyruvate and acetyl-CoA. Concomitantly an parallel pathway can convert itaconate into 1-methyl itaconate
Our observation that knock-out strains of ictA and ichA show increased production of IA and are not able to degrade IA corroborate with the results of Chen et al. [29], that this pathway indeed converts IA intracellularly. Although CitB99 ΔICT strain is able to achieve higher titers than the CitB99 ΔICH strain, we observed that deletion of ictA appears to have the same effect as ichA i.e. both knock-out strains are unable to degrade IA. One remarkable feature of the parental strain CitB99 is that the strain performs variably in fermentations where high and low IA production can be seen (data not shown), whereas IA production appears to be more stable in CitB99 ΔICT (data not shown). Our results also suggest that no other enzyme is able to convert itaconate into itaconyl-CoA in the absence of IctA and also that itaconate bioconversion cannot proceed without the activity of IchA. Interestingly, CitB99 ΔICH achieves lower IA end titers than CitB99 ΔICT, possibly due to intracellular accumulation of itaconyl-CoA. However, the fate of IctA-mediated itaconyl-CoA accumulation in the CitB99 ΔICH strain remains unclear. Interestingly, the intracellular accumulation of itaconyl-CoA has been linked to decreased vitamin B12 levels in human brown adipocytes by Shen H et al. These researchers found that itaconyl-CoA can have a toxic influence by competitive inhibition of the mitochondrial vitamin B12-dependent methylmalonyl-CoA mutase (mut). This inhibition is mediated by converting vitamin B12 into the chemically inactive cob(II)alamin, thereby decreasing intracellular vitamin B12 levels [34]. Although fungi are not reported in literature to be able to synthesize nor use vitamin B12 as cofactor in biochemical reactions it cannot be excluded that a similar itaconyl-CoA mediated toxicity might be elicited [35]. Intracellular itaconyl-CoA accumulation might also exert a similar toxicity response as propionyl-CoA in Aspergillus nidulans. Brock M and Buckel W found that intracellular accumulation of propionyl-CoA mainly affects enzymes involved in glucose metabolism, thereby severely retarding growth [36]. However, as no apparent toxic effects of itaconyl-CoA accumulation were detected on growth and biomass formation in the CitB99 ΔICH strain, further research is required to investigate any potential toxic side-effects of itaconyl-CoA accumulation on fungal physiology.
Interestingly, the identified IA bioconversion pathway appears to share common features with bacterial C5-dicarboxylic acid metabolism, which can use C5 dicarboxylates such as itaconate, mesaconate and citramalate as growth substrates [37]. Pathogenic bacteria have been shown to use this pathway as a means to evade the hosts cellular defense mechanism during infection [30, 38]. Similarly A. niger may use this IA bioconversion pathway as defense mechanism during biological warfare. In nature A. niger and A. terreus share many common growth habitats and are constantly in conflict over scarce resources. However, the link between Aspergillus niger IA bioconversion and central metabolism is not known and is subject to further investigation.
Remarkably, in the high IA producing strain CitB#99 ictA and ichA show similar high levels of induction in expression compared to expression in the AB1.13 strain, however cclA does not show the same induction as ictA and ichA, suggesting that another protein has its function. An alternative possibility might be that IA is converted into an unknown compound in A. niger. We have detected unknown peaks in the HPLC samples of cultivations with high IA producing strains. The identification of these peaks and unraveling the link to IA detoxification however, is topic of ongoing research.
Interestingly, the second identified putative IA bioconversion pathway involves TmtA that supposedly converts IA into 1-methyl itaconate [31]. tmtA shows similar induction in expression as ictA and ichA from AB1.13 to CitB#99, possibly suggesting a role in IA bioconversion. A. niger TmtA shows sequence similarity of 28% with S. cerevisiae Tmt1 at a query coverage of 26% and 44% sequence similarity with E. coli Tam at 91% query coverage. Functional characterization of Tmt1 and Tam has led to the identification of trans-aconitate methyltransferase activity of both enzymes [32]. Remarkably, Tmt1 was also found to have low-level affinity for IA [33]. Katz et al. further elucidated that the major endogenous substrate for Tmt1 is an intermediate in the leucine biosynthesis pathway, 3-isopropylmalate, and that Tmt1 functions as a so called ‘moonlighting’ enzyme i.e. an enzyme that can perform multiple functions in different pathways [39]. The proposed role that Tmt1 plays in the citric acid cycle is detoxification of spontaneously formed trans-aconitate, an potent inhibitor of aconitase. However the physiologic function of methylating 3-isopropylmalate still requires elucidation [33, 39]. TmtA could perform a similar moonlighting function in A. niger where it methylates trans-aconitate and itaconate. Interestingly, deleting tmtA results in almost complete shutdown of IA production in A. niger. This may be caused by an accumulation of trans-aconitate in the mitochondrion and concomitantly the inhibition of aconitase, decreasing the flux to cis-aconitate. Interestingly, no other side-product such as CA was observed in the ΔtmtA strain. How TmtA exactly fits in heterologous IA production and overall central metabolism in A. niger is not entirely clear and is subject of further research. However from the data presented it is apparent that deletion of tmtA does not play any role in reducing IA bioconversion.
Remarkably, all of the enzymes involved in IA bioconversion IctA, IchA, CclA and TmtA carry predicted mitochondrial targeting sequences (Table 3). However, for IA bioconversion to take place in the mitochondrion, IA has to be first transported into the cytosol from the extracellular medium and concomitantly transported into the mitochondrion. It is currently unknown which transporters are involved in the transport across the plasma and mitochondrial membrane. However, in our transcriptome data multiple solute transporters have been identified making this a topic for further investigation.
In our research, apart from IA bioconversion pathways, we also looked at other potentially competing biosynthetic pathways that could hamper IA production. One possible way in which IA production might be hampered is when competing organic acid biosynthesis pathways are pulling precursor molecules, e.g. pyruvic acid or oxaloacetate, towards them. In A. niger one such competing pathway might lead to the formation of l-lactate [40]. Although a l-lactate dehydrogenase gene is annotated by sequence homology in Aspergillus niger (An04g08220) a functional l-lactate dehydrogenase enzyme has not been characterized yet [41]. Although A. niger strains are known to be potent oxalic acid producers, we have seen in our transcriptome data that the expression of the key gene for oxalate production i.e. oahA is significantly downregulated in CitB#99, indicating less competition from this pathway for precursor molecules.
In our research (Table 3), we have also found a non-canonical cytosolic citrate synthase similar to the previously identified citB [18]. This citC gene (An01g09940) is actually downregulated under citB-mediated improvement of IA production. Interestingly, also upon overexpression of citC we have observed similar positive effects on IA production (Additional file 7). Interestingly, similar as citB also citC appears to be clustered in a secondary metabolite gene cluster carrying two genes encoding CadA-like enzymes (An01g09930 and An01g09950). These results suggest that rewiring of secondary metabolism of A. niger towards IA production is much more intricate than we previously suspected.
Interestingly, our transcriptome data also indicates upregulation of phosphorus scavenging enzymes like acid phosphatase and phytase. Organic acid production in A. niger has been linked with phosphate depletion in the past [42, 43]. Recently, Upton et al. have shown that in phosphate limited citric acid production medium phosphate is quickly taken up by A. niger and stored as polyphosphate. The researchers further suggest that polyphosphate hydrolysis poses a constraint that limits growth and enables flux of carbon to organic acid production [44]. Whether a similar response occurs and if polyphosphate putatively fulfills a similar role during heterologous IA production in A. niger is not clear and warrants further research. Furthermore we have observed strong reduction in expression of genes encoding products that are involved in N transport and utilization, whose functional relation with heterologous IA production is not clear and also topic for further research.
Conclusion
In our attempt to identify genes involved in putative IA bioconversion in A. niger we have analyzed a transcriptome dataset from batch fermentations of low, medium and high IA producing A. niger strains.
Transcriptome analysis has led us to two novel IA conversion pathways in A. niger. These pathways are not induced in non-IA producing conditions, strongly suggesting that they are specific for IA. One pathway shares homology with a recently identified IA degradation pathway identified in A. terreus through the combined action of IctA, IchA, and CclA. Upon deleting ictA or ichA we observed cessation of IA bioconversion and an increase in IA production titers.
Furthermore we identified a second putative IA bioconversion pathway in A. niger that supposedly converts IA into 1-methyl itaconate through the methylating activity of TmtA. Upon deleting tmtA we observed almost complete cessation of IA production, whereas overall growth and glucose consumption did not appear to be affected. Based on these observations we postulate that TmtA is an enzyme that esterifies spontaneously formed trans-aconitate and thereby relieves the cell of potential toxic effects of trans-aconitate accumulation. The results presented in this research contribute in further elucidation of heterologous IA production in A. niger.
Additional files
Additional file 1. Schematic overview of the split-marker method.
Additional file 2. List of primers that were used to generate the split-markers for ictA and ichA deletion.
Additional file 3. Bioconversion of externally added IA to the cultivation medium. Strains AB1.13 CAD and CitB#99 were cultivated in IA supplemented medium (20 g/L) containing 0.1 and 0.5% glucose respectively. IA bioconversion was compared with medium containing 0.1% glucose.
Additional file 4. Transcriptome data of upregulated genes in CitB#99 vs AB1.13 with a 2logR cut off at 2.0.
Additional file 5. Transcriptome data of downregulated genes in CitB#99 vs AB1.13 with a 2logR cut off at -2.0.
Additional file 6. Protein BLAST alignments of IctA, IchA and CclA.
Additional file 7. IA production of citC overexpressing A. niger strain AB1.13 #49B-citC A12 Z.
Authors’ contributions
AH, ATB and PJP designed the experiments and analysed the results; AH performed the experiments; AH and PJP wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors are very grateful to Prof. Dr Stanley Brul and Dr. Gertien Smits from University of Amsterdam for discussions and providing ideas and insights. Also are the authors grateful for the contribution from Roy van Gerven in obtaining tmtA knockout strains.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The transcriptome data will be uploaded on GEO.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This research was fully funded by Dutch DNA Biotech BV.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sauer M, Porro D, Mattanovich D, Branduardi P. Microbial production of organic acids: expanding the markets. Trends Biotechnol (Internet). 2008;26:100–8. http://linkinghub.elsevier.com/retrieve/pii/S0167779907003228. [DOI] [PubMed]
- 2.Jung Ho A, Yu-Sin J, Yup Lee S. Production of succinic acid by metabolically engineered microorganisms. Curr Opin Biotechnol. 2016;42:54–66. doi: 10.1016/j.copbio.2016.02.034. [DOI] [PubMed] [Google Scholar]
- 3.de Vries RP, Riley R, Wiebenga A, Aguilar-Osorio G, Amillis S, Uchima CA, et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus (Internet). Genome Biol. 2017. Available from: http://genomebiology.biomedcentral.com/articles/10.1186/s13059-017-1151-0. [DOI] [PMC free article] [PubMed]
- 4.Werpy T, Petersen G. Top value added chemicals from biomass. Program. 2004; 1–76.
- 5.Robert T, Friebel S. Itaconic acid—a versatile building block for renewable polyesters with enhanced functionality. Green Chem (Internet). 2016;18:2922–34. Available from: http://xlink.rsc.org/?DOI=C6GC00605A.
- 6.Okabe M, Lies D, Kanamasa S, Park EY. Biotechnological production of itaconic acid and its biosynthesis in Aspergillus terreus. Appl Microbiol Biotechnol. 2009;84:597–606. doi: 10.1007/s00253-009-2132-3. [DOI] [PubMed] [Google Scholar]
- 7.Weastra SRO. Determination of market potential for selected platform chemicals: itaconic acid, succinic adis, 2,5-furandicarboxylic acid. 2011; 1–173
- 8.Nieder-Heitmann M, Haigh KF, Görgens JF. Process design and economic analysis of a biorefinery co-producing itaconic acid and electricity from sugarcane bagasse and trash lignocelluloses. Bioresour Technol (Internet). 2018;262:159–68. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0960852418305984. [DOI] [PubMed]
- 9.Cruz JC, Castro AM, Sérvulo EFC. World market and biotechnological production of itaconic acid Juliana. 3 Biotech (Internet). Springer, Berlin; 2017;1–27. Available from: 10.1007/s13205-018-1151-0. [DOI] [PMC free article] [PubMed]
- 10.Gyamerah MH. Oxygen requirement and energy relations of itaconic acid fermentation by Aspergillus terreus NRRL 1960. Appl Microbiol Biotechnol. 1995;44:20–26. doi: 10.1007/BF00164475. [DOI] [Google Scholar]
- 11.Karaffa L, Díaz R, Papp B, Fekete E, Sándor E, Kubicek C. A deficiency of manganese ions in the presence of high sugar concentrations is the critical parameter for achieving high yields of itaconic acid by Aspergillus terreus. Appl Microbiol Biotechnol (Internet). 2015;1–8. Available from: 10.1007/s00253-015-6735-6. [DOI] [PubMed]
- 12.Kuenz A, Krull S. Biotechnological production of itaconic acid—things you have to know. Appl Microbiol Biotechnol. 2018;102:3901–3914. doi: 10.1007/s00253-018-8895-7. [DOI] [PubMed] [Google Scholar]
- 13.Li A, van Luijk N, ter Beek M, Caspers M, Punt P, van der Werf M. A clone-based transcriptomics approach for the identification of genes relevant for itaconic acid production in Aspergillus. Fungal Genet Biol. 2011;48:602–611. doi: 10.1016/j.fgb.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 14.van der Straat L, Vernooij M, Lammers M, van den Berg W, Schonewille T, Cordewener J, et al. Expression of the Aspergillus terreus itaconic acid biosynthesis cluster in Aspergillus niger. Microb Cell Fact (Internet). 2014;13:11. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3898256&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed]
- 15.Steiger MG, Blumhoff ML, Mattanovich D, Sauer M. Biochemistry of microbial itaconic acid production. Front Microbiol. 2013;4:1–5. doi: 10.3389/fmicb.2013.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vandenberghe LPS, Soccol CR, Pandey A, Lebeault JM. Review: microbial production of citric acid. Braz Arch Biol Technol. 1999;42:263–276. doi: 10.1590/S1516-89131999000300001. [DOI] [Google Scholar]
- 17.Soccol CR, Vandenberghe LPS, Rodrigues C. New perspectives for citric acid production and application. Food Technol Biotechnol. 2006;44:141–149. [Google Scholar]
- 18.Hossain AH, Li A, Brickwedde A, Wilms L, Caspers M, Overkamp K, et al. Rewiring a secondary metabolite pathway towards itaconic acid production in Aspergillus niger. Microb Cell Fact. 2016;15:130. doi: 10.1186/s12934-016-0527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li A, Pfelzer N, Zuijderwijk R, Brickwedde A, van Zeijl C, Punt P. Reduced by-product formation and modified oxygen availability improve itaconic acid production in Aspergillus niger. Appl Microbiol Biotechnol (Internet). 2013;97:3901–11. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23397482. [DOI] [PubMed]
- 20.Li A, Pfelzer N, Zuijderwijk R, Punt P. Enhanced itaconic acid production in Aspergillus niger using genetic modification and medium optimization. BMC Biotechnol (Internet). 2012;12:57. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3472327&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed]
- 21.Boeke JD, Lacroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 22.Arentshorst M, Niu J, Ram AFJ. Genetic transformation systems in Fungi, Volume 1. 2015;1:263–72. Available from: http://link.springer.com/10.1007/978-3-319-10142-2.
- 23.Punt PJ, Van Den Hondel C a MJJ. Transformation of filamentous fungi based on Hygromycin B and Phleomycin Resistance Markers. Methods Enzymol (Internet). 1992. p. 447–57. Available from: http://www.sciencedirect.com/science/article/pii/007668799216041H#. [DOI] [PubMed]
- 24.Van Zeijl CMJ, Van De Kamp EHM, Punt PJ, Selten GCM, Hauer B, Van Gorcom RFM, et al. An improved colony-PCR method for filamentous fungi for amplification of pcr-fragments of several kilobases. J Biotechnol. 1998;59:221–224. doi: 10.1016/S0168-1656(97)00170-3. [DOI] [PubMed] [Google Scholar]
- 25.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 26.Klement T, Büchs J. Itaconic acid—a biotechnological process in change. Bioresour Technol (Internet). 2013;135:422–31. Available from: 10.1016/j.biortech.2012.11.141. [DOI] [PubMed]
- 27.Guillemette T, Ram AFJ, Carvalho NDSP, Joubert A, Simoneau P, Archer DB. Methods for investigating the UPR in Filamentous Fungi. Unfolded Protein Response Cell. Stress Part B (Internet). 2011 [cited 2018 Sep 14];490:1–29. Available from: https://ac.els-cdn.com/B9780123851147000015/1-s2.0-B9780123851147000015-main.pdf?_tid=d0522153-ae40-4041-93dc-32453a9abbda&acdnat=1536913645_a8a8387cca7001d4a2e6173cd3636227. [DOI] [PubMed]
- 28.S P Carvalho ND, Arentshorst M, Kooistra R, Stam H, Sagt CM, M J J van den Hondel CA, et al. Effects of a defective ERAD pathway on growth and heterologous protein production in Aspergillus niger. Appl Genet Mol Biotechnol (Internet) (cited 2018 Sep 14); Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3016150/pdf/253_2010_Article_2916.pdf. [DOI] [PMC free article] [PubMed]
- 29.Chen M, Huang X, Zhong C, Li J, Lu X. Identification of an itaconic acid degrading pathway in itaconic acid producing Aspergillus terreus. Appl Microbiol Biotechnol (Internet) 2016 doi: 10.1007/s00253-016-7554-0. [DOI] [PubMed] [Google Scholar]
- 30.Sasikaran J, Ziemski M, Zadora PK, Fleig A, Berg IA. Bacterial itaconate degradation promotes pathogenicity. Nat Chem Biol (Internet). 2014;10:371–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24657929. [DOI] [PubMed]
- 31.Zhao Z, Meijrink B, Van der Hoeven R, Roubos A, van Gelder M. Cells for itaconic acid production. United States; 2015.
- 32.Cai H, Strouse J, Dumlao D, Jung ME, Clarke S. Distinct reactions catalyzed by bacterial and yeast trans-aconitate methyltransferases. Biochemistry. 2001;40:2210–2219. doi: 10.1021/bi0022902. [DOI] [PubMed] [Google Scholar]
- 33.Cai H, Dumlao D, Katz JE, Clarke S. Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from saccharomyces cerevisiae. Biochemistry. 2001;40:13699–13709. doi: 10.1021/bi011380j. [DOI] [PubMed] [Google Scholar]
- 34.Shen H, Campanello GC, Flicker D, Luo C, Banerjee R, Mootha VK, et al. The Human Knockout Gene CLYBL Connects The Human Knockout Gene CLYBL Connects Itaconate to Vitamin B 12:771–82. [DOI] [PMC free article] [PubMed]
- 35.Roth JR, Lawrence JG, Bobik TA. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 36.Brock M, Buckel W. On the mechanism of action of the antifungal agent propionate. Propionyl-CoA inhibits glucose metabolism in Aspergillus nidulans. Eur J Biochem. 2004;271:3227–3241. doi: 10.1111/j.1432-1033.2004.04255.x. [DOI] [PubMed] [Google Scholar]
- 37.Fuchs G, Berg IA. Unfamiliar metabolic links in the central carbon metabolism. J Biotechnol. 2014;192:314–322. doi: 10.1016/j.jbiotec.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Cordes T, Michelucci A, Hiller K. Itaconic acid: the surprising role of an industrial compound as a mammalian antimicrobial metabolite. Annu Rev Nutr (Internet). 2014;35:150514143029003. Available from: http://www.annualreviews.org/doi/abs/10.1146/annurev-nutr-071714-034243. [DOI] [PubMed]
- 39.Katz JE, Dumlao DS, Wasserman JI, Lansdown MG, Jung ME, Faull KF, et al. 3-Isopropylmalate is the major endogenous substrate of the Saccharomyces cerevisiae trans-aconitate methyltransferase. Biochemistry. 2004;43:5976–5986. doi: 10.1021/bi049784+. [DOI] [PubMed] [Google Scholar]
- 40.Li A, Punt P. Industrial production of organic acids by Fungi. Appl Microb Eng (Internet). 2013:52–74. Available from: 10.1201/b15250-3. [DOI]
- 41.Dave KK, Punekar NS. Expression of lactate dehydrogenase in aspergillus Niger for l-lactic acid production. PLoS ONE. 2015;10:1–16. doi: 10.1371/journal.pone.0145459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider KD, Van Straaten P, De Orduña RM, Glasauer S, Trevors J, Fallow D, et al. Comparing phosphorus mobilization strategies using Aspergillus niger for the mineral dissolution of three phosphate rocks. J Appl Microbiol. 2010;108:366–374. doi: 10.1111/j.1365-2672.2009.04489.x. [DOI] [PubMed] [Google Scholar]
- 43.Chuang C-C, Kuo Y-L, Chao C-C, Chao W-L. Solubilization of inorganic phosphates and plant growth promotion by Aspergillus niger. Biol Fertil Soils (Internet). 2007 (cited 2018 Apr 14);44:415–6. Available from: http://link.springer.com/10.1007/s00374-007-0236-4.
- 44.Upton DJ, McQueen-Mason SJ, Wood AJ. An accurate description of Aspergillus niger organic acid batch fermentation through dynamic metabolic modelling. Biotechnol Biofuels (Internet). 2017;10:258. Available from: https://biotechnologyforbiofuels.biomedcentral.com/articles/10.1186/s13068-017-0950-6. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Schematic overview of the split-marker method.
Additional file 2. List of primers that were used to generate the split-markers for ictA and ichA deletion.
Additional file 3. Bioconversion of externally added IA to the cultivation medium. Strains AB1.13 CAD and CitB#99 were cultivated in IA supplemented medium (20 g/L) containing 0.1 and 0.5% glucose respectively. IA bioconversion was compared with medium containing 0.1% glucose.
Additional file 4. Transcriptome data of upregulated genes in CitB#99 vs AB1.13 with a 2logR cut off at 2.0.
Additional file 5. Transcriptome data of downregulated genes in CitB#99 vs AB1.13 with a 2logR cut off at -2.0.
Additional file 6. Protein BLAST alignments of IctA, IchA and CclA.
Additional file 7. IA production of citC overexpressing A. niger strain AB1.13 #49B-citC A12 Z.
Data Availability Statement
The transcriptome data will be uploaded on GEO.