Abstract
Background
Long noncoding RNAs (lncRNA) contained in exosomes have been recognized as promising stable biomarkers in cancers. H19 is a well-known oncogenic lncRNA, but whether extracellular H19 is contained in exosomes and the clinical significance of exosomal H19 are poorly understood. The aim of this study was to assess serum-derived exosomal H19 lncRNA as a cancer predictive marker.
Material/Methods
Exosomes from serum of bladder cancer (BC) patients were isolated using the ExoQuick purification method and identified using transmission electron microscopy and nanoparticle tracking analysis. RT-qPCR was used for the measurement of H19 expression in serum or tissue samples.
Results
Serum H19 was little influenced by the treatment of RNase A alone but was dramatically downregulated when treated with RNase A and Triton ×100 simultaneously. The concentration of H19 was significantly higher in serum exosomes than in exosome-depleted supernatants in serum. Then, we validated that exosomal H19 is stable in serum by exposing serum samples to prolonged conditions of room temperature, 4°C, multiple freeze-thaw cycles, and low/high pH. Serum exosomal H19 of BC patients was positively correlated with total H19 level in paired BC tissues, and exosomal H19 was significantly downregulated in postoperative samples when compared to the paired preoperative samples. In addition, exosomal H19 level was significantly increased in serum of BC patients when compared to healthy people and benign disease patients. More importantly, Kaplan-Meier survival curve analysis showed that higher serum exosomal H19 level in BC patients was correlated with poorer survival.
Conclusions
Detection of serum exosomal H19 shed light on using exosomal lncRNAs as a noninvasive diagnostic and prognostic biomarker for BC patients.
MeSH Keywords: Diagnosis; Exosomes; Prognosis; RNA, Long Noncoding
Background
Bladder cancer (BC) is one of the most common urological malignancies worldwide, with an estimated 79 030 new cases and 16 870 BC-related deaths in 2017 in the United States [1]. Early screening and monitoring are essential for early and improved treatment of BC. Currently, voided urine cytology lacks the diagnostic sensitivity necessary to rule out cancer, especially for low-grade cancer. Cystoscopy is the criterion standard for BC surveillance and has high diagnostic accuracy, but is invasive and expensive and has low patient acceptance. Therefore, there is a need for new markers to help in noninvasive detection and surveillance of BC [2,3].
lncRNAs are a class of long noncoding RNAs of greater than 200 nucleotides in length, which play a crucial role in regulating gene expression [4,5]. It has been observed that lncRNAs are promising diagnostic and predictive marker for BC patients [6]. Exosomes are membrane-derived vesicles and have a size range of 20–200 nm when released into body fluids such as blood, urine, and malignant ascites. These vesicles contain DNA, protein fragments, and coding or noncoding RNAs secreted by their parental cell cytoplasm. Moreover, cancerous cells secrete exosomes into the peripheral circulation, and exosomal RNAs can more accurately reflect changes in cancer cells during tumor progression [7].
Recently, accumulating studies have revealed that exosomal lncRNAs are enriched and more stable in the circulatory system and protected from ribonuclease (RNase) degradation [8,9]. The identification of exosomal lncRNAs in bodily fluids suggested their predictive value in clinical diagnosis or prognosis for different types of cancer [10–12]. The imprinted H19 gene, which encodes an untranslated RNA, lies at the end of a cluster of imprinted genes, and is strongly expressed in embryonic cells [13]. Recent studies showed that H19 is overexpressed in several malignancies and may serve as a useful biomarker [14–16]. However, the clinical significance of exosomal H19 and whether serum H19 is contained in exosomes are poorly understood.
In this study, we hypothesized that tumor-released serum H19 in BC patients is contained in exosomes, and serum exosomal H19 serves as a more reliable circulating biomarker for diagnosis and prognosis of BC. To test this hypothesis, we extracted and purified exosomes from serum of BC patients and determined the expression level of H19 in exosomes. By establishing receiver operating characteristic (ROC) curves and performing Kaplan-Meier analysis, we further explored the diagnostic and prognostic potential of serum exosomal H19 for patients with BC.
Material and Methods
Clinical samples and reagents
In total, 52 preoperative serum samples and primary tissue specimens were collected from 52 BC patients. At the same time, 52 serum samples from patients with benign disease – 19 benign prostatic hyperplasia (BPH), 15 urolithiasis and 18 cystitis – and another 52 serum samples from healthy subjects without BC history were obtained from Hunan Provincial People’s Hospital between June 2009 and May 2010. In addition, 32 postoperative serum samples were also obtained from 32 of the 52 BC patients. For serum samples, 5 mL of venous blood from each participant was collected by venipuncture before any treatment. Serum was separated via centrifugation at 1600 g for 10 min at 4°C within 2 h after collection, followed by a second round of centrifugation at 16 000 g for 10 min at 4°C to remove the residual cell debris. Each serum supernatant was put into RNase-free tubes until use. Tissue specimens obtained from surgery resection were immediately stored in liquid nitrogen until further use. Written informed consent was obtained from each participant prior to blood and tumor sample collection. The study protocol was approved by the Clinical Research Ethics Committee of Hunan Provincial People’s Hospital (approval number WHYX0013). RNase A was purchased from Sigma (Cambridge, MA) and was used at 1 μg/mL alone or combined with 0.1% Triton ×100 (Beyotime, Shanghai, China) for 30 min at room temperature.
Serum exosomes isolation and RNA extraction
Serum was centrifuged at 3000 rpm for 20 min to remove precipitation and impurities. The extraction of exosomes from serum was done using the ExoQuick™ Exosome Precipitation Solution (System Biosciences, mountainite, CA). First, 250 μL of the supernatant was mixed with 63 μL of ExoQuick precipitation kit and incubated at 4°C for 30 min after brief mixing. Then, the master mixture was centrifuged at 1500 rpm for 25 min. Finally, the supernatant was removed by careful aspiration, followed by another 5 min of centrifugation to remove the residual liquid. The exosome-containing pellet was subsequently resuspended in 250 μL phosphate-buffered saline (PBS). Extraction of RNA from exosomes was performed using the commercial miRNeasy Serum/Plasma kit (QIAGEN, #217184). RNA extraction from tissue samples was performed using Trizol (Invitrogen, Carlsbad, CA) after tissues were homogenized manually with homogenizing medium (0.01 mol/L Tris-Hcl, 1 mmol/L EDTA-2Na, 0.01 mol/L sucrose, and 0.8% NaCl). Non-tissue RNA elution steps were carried out at 12 000 g for 15 s, and the RNA was finally eluted in 15 μl RNase-free ultra-pure water.
Reverse transcription (RT) and real-time quantitative PCR (RT-qPCR)
RT and qPCR kits were used to evaluate the expression of H19 in serum exosomes samples. The 20 μL RT reactions were performed using a PrimeScript® RT reagent kit (Takara, Dalian, China) and incubated for 30 min at 37°C, 5 s at 85°C, and then maintained at 4°C. For real-time quantitative PCR, 2 μL of diluted RT product was mixed with 12.5 μL of 2×SYBR®Premix Ex Taq™, 0.5 μL of 50×ROX reference Dye II (Takara, Dalian, China), 2 μL forward and reverse primers (10 μM), and 8 μL nuclease-free water to a final volume of 25 μL. All reactions were carried out using an Eppendorf Master cycler EP Gradient S (Eppendorf, Germany) under the following conditions: 95°C for 30 s, followed by 45 cycles of 95°C for 5 s and 60°C for 30 s. The RT-qPCR was run on the CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, USA). All reactions were carried out in triplicate. The expression levels of H19 were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the comparative 2−ΔΔCq method [17,18]. The primers used in this study were: H19: Forward 5′-ATCGGTGCCTCAGCGTTCGG-3′, Reverse 5′-CTGTCCTCGCCGTCACACCG-3′; GAPDH: Forward 5′-GCACCGTCAAGGCTGAGAAC-3′, Reverse 5′-ATGGTGG TGAAGACGCCAGT-3′.
Transmission electron microscopy (TEM)
We used 50 μL PBS to suspend the exosomes pellets and then put 1 drop of this suspension on the parafilm. A copper mesh coated with carbon was then used to drift on the drop for 5 min at 25°C. Then, the grid was removed and excess liquid was drained by touching the grid edge against a piece of clean filter paper. The grid was then placed onto a drop of 2% phosphotungstic acid with pH 7.0 for approximately 5 s, and excess liquid was drained off. The grid was allowed to dry for several minutes and then examined using a JEM-1200 EX microscope (JEOL, Akishima, Japan) at 80 kilo-electron volts.
Size distribution of exosomes
As a previous study described [19], exosome pellets were resuspended in PBS. Size distribution of exosomes was analyzed by using nanoparticle tracking analysis using a Nanosight LM10 instrument (Malvern Instruments, Ltd, Worcestershire, UK).
Urine cytology determination
Midstream urine sediments were separated via centrifugation at 1500 rpm for 15 min at room temperature. Three cytopathologists performed the cytological examination. The conclusion was categorized as positive only if all 3 cytopathologists identified cancer cells, and was classified as negative under other conditions.
Statistical analysis
The Kolmogorov-Smirnov test was applied for data analysis with the distribution of each group of samples. Data are presented as median (interquartile range). Comparisons between 2 groups were performed by nonparametric Mann-Whitney U tests. ROC curves were established to discriminate BC from controls. MedCalc 9.3.9.0 (MedCalc, Mariakerke, Belgium) was used for ROC analysis, MATLAB software (MATLAB, R2013a) was employed for logistic regression analysis, and others were calculated using SPSS version 19.0 software (SPSS, Chicago, IL). Cox proportional hazards analysis was used to calculate the hazard ratio (HR) and 95% confidence interval (CI). In addition, Kaplan-Meier survival analysis was applied to test the association between expression level and patients’ survival. A two-sided P<0.05 was considered statistically significant.
Results
Identification of exosomes in serum
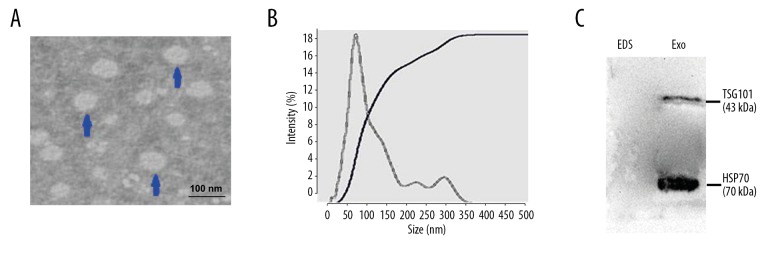
We used the ExoQuick exosome purification kit to purify and enrich exosomes contained in serum samples from BC patients, according to the manufacturer’s protocol. To confirm whether exosomes were extracted successfully by the method, exosomes were characterized by transmission electron microscopy (TEM). The representative micrographs showed vesicles with round or oval membranes, and a diameter of 30–150 nm under TEM (Figure 1A). Exosomes with size ranging from 30 nm to 150 nm in diameter accounted for 76.4%, with the mode value of 67.52 nm using Nanoparticle tracking analysis (Figure 1B). The exosome protein markers TSG101 and HSP70 were analyzed by Western blot analysis, which showed specific bands in isolated exosomes pellets, but not in exosome-depleted supernatant (Figure 1C). Taken together, these observations suggest that we successfully isolated exosomes from serum samples of BC patients and further verified the efficacy of our protocol for extracting exosomes from human serum samples.
Figure 1.
Characterization of exosomes isolated from serum samples. (A) Transmission electron microscopy (TEM) images of exosomes in serum of BC patients. HV=100.0 kv; Direct Mag: 100 000×. (B) Size distribution of exosomes were analyzed by performing Nanoparticle tracking analysis. (C) Western blot analysis of exosomal protein TSG101 and HSP70 in exosomes (Exo) and exosome-depleted serum samples (EDS).
Serum H19 is contained in exosomes of BC patients
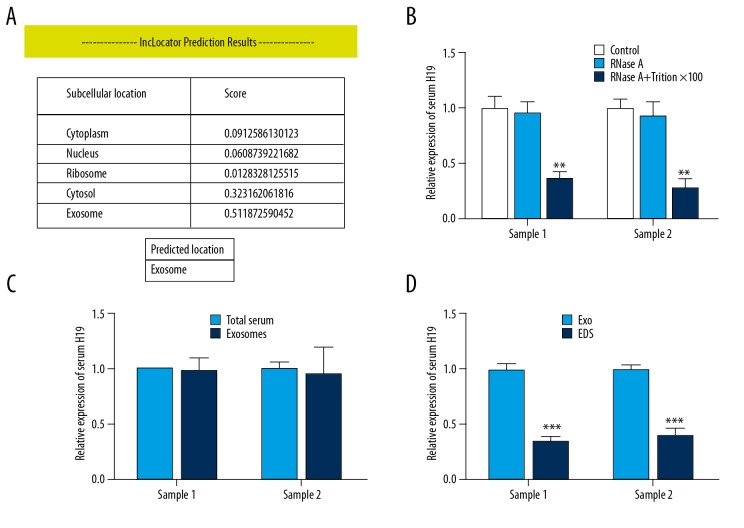
To investigate whether H19 in serum is contained in exosomes, we first localized its subcellular location by using the online lncRNA location prediction software lncLocator (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/). As shown in Figure 2A, the intracellular H19 was predicted to be located in exosomes, suggesting that the extracellular H19 may exist in exosomes. To confirm this hypothesis, we mixed the 52 serum samples (0.1 ml each) and divided them into 2 parts (Sample 1 and Sample 2). We detected the expression level change of serum H19 after treatment with RNase A and found that serum H19 was little influenced by treatment with RNase A alone but was dramatically downregulated when treated with RNase A and Triton ×100 simultaneously (Figure 2B), suggesting that serum H19 was protected by the membrane instead of being directly secreted. We then compared the expression level of H19 in total serum and purified exosomes from BC patients by RT-qPCR and found that there was no statistically significant difference in H19 levels between total serum and exosomes (Figure 2C). In addition, we examined the expression of H19 in exosomes and exosome-depleted serum supernatants from BC patients. The concentration of H19 was significantly higher in exosomes than in exosome-depleted supernatants (Figure 2D). Taken together, our results show that serum H19 is enriched in exosomes.
Figure 2.
Serum H19 is incorporated in exosomes of BC patients. (A) Presentation of score of H19 at different subcellular locations by lncLocator (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/). (B) RT-qPCR was used to determine the serum exosomal H19 level after dealing with 1 μg/mL RNase A alone or combined with 0.1% Triton ×100 for 30 min, ** P<0.01 compared to control by repeated-measures test. (C) RT-qPCR analysis of H19 in purified exosomes and total serum samples, and repeated-measures test was applied. (D) RT-qPCR analysis of H19 in purified exosomes (Exo) and exosome-depleted serum (EDS) samples, *** P<0.001 compared to Exo group by repeated-measures test.
Serum exosomal H19 is released from BC cells
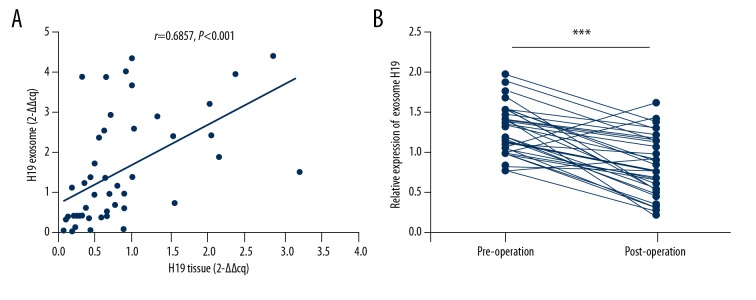
Subsequently, we performed a series of experiments to verify that the serum exosomal H19 in BC patients is released from cancer cells. First, we detected the expression level of H19 in serum exosomes of BC patients and the paired primary cancer tissues. The Spearman correlation testing showed that the expression level of serum exosomal H19 was positively correlated with total H19 of paired BC tissues (Figure 3A). Second, we detected the expression level of serum exosomal H19 in BC patients before and after surgery. As shown in Figure 3B, the expression level of exosomal H19 was significantly downregulated in postoperative samples when compared to the paired preoperative samples, indicating that exosomal H19 in serum is released from BC cells.
Figure 3.
Serum exosomal H19 was released from BC cells. (A) Spearman correlation analysis was used to determine the expression relationship of H19 in serum exosomes and paired BC tissues. (B) The changes of serum exosomal H19 expression was analyzed via RT-qPCR in 32 preoperative patients and paired postoperative (14 days) patients, *** P<0.001 by paired t test.
The stability of H19 in serum exosomes
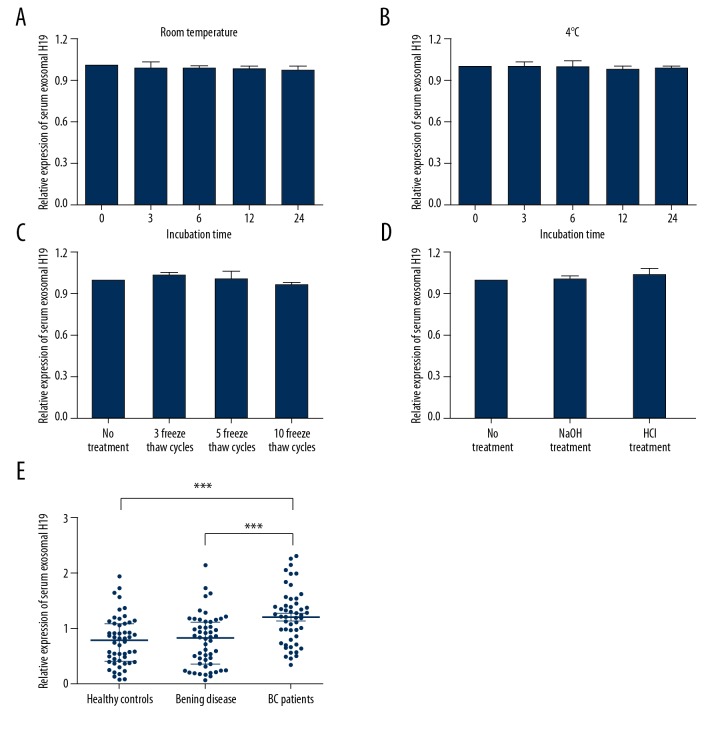
Since better stability is a critical prerequisite for tumor markers, we next tested the stability of H19 in serum exosomes by exposing serum samples to different conditions, including incubation at room temperature or 4°C for 0, 3, 6, 12, and 24 h, repeated freeze-thaw cycles, and low (pH=1) or high (pH=13) pH solution for 3 h. The expression level of H19 in serum exosomes was not significantly influenced in any of these experimental conditions (Figure 4A–4D), indicating that H19 was stable in serum exosomes.
Figure 4.
Exosomal H19 is stable in serum. (A–D) The expression levels of exosomal H19 remained stable when treated with prolonged exposure to room temperature (A), 4°C (B), multiple freeze-thaw cycles (C), and low/high pH (D). (E) RT-qPCR analysis of exosomal H19 for serum samples of BC patients, patients with benign disease, and healthy individuals. *** P<0.001.
Expression of exosomal H19 and clinicopathological characteristics in BC patients
After the validation of the existence, origin, and stability of exosomal H19 in serum, we determined the expression level of serum exosomal H19 in BC patients and healthy individuals. As expected, exosomal H19 level was significantly increased in serum of BC patients when compared to healthy controls or the benign disease group (Figure 4E). To further explore the potential of circulating exosomal H19 as a predictor for BC, we evaluated the association between H19 expression and clinical characteristics in BC patients. As shown in Table 1, exosomal H19 was significantly correlated with tumor stage, clinical TNM stage, and lymph node metastasis (P<0.05 for all). However, there was no significant correlation between the expression of exosomal H19 and the patients’ sex, age, or tumor grade (P>0.05 for all).
Table 1.
Correlation between serum exosomal H19 concentration and clinicopathological characteristics of patients with BC [median (interquartile range)].
| Factors | Case number | H19 level | P-value |
|---|---|---|---|
| Age | 0.48 | ||
| ≤65 | 26 | 1.20 (0.35–2.23) | |
| >65 | 26 | 1.44 (1.22–2.31) | |
| Gender | 0.67 | ||
| Male | 39 | 1.22 (0.35–2.30) | |
| Female | 13 | 1.31 (0.45–2.31) | |
| Tumor grade | 0.22 | ||
| High | 28 | 1.17 (0.73–2.21) | |
| Low | 24 | 1.36 (0.35–2.31) | |
| Tumor stage | |||
| T1–2 | 29 | 1.13 (0.35–2.31) | 0.04 |
| T3–4 | 23 | 1.44 (0.48–2.10) | |
| Lymphnode metastasis | 0.003 | ||
| Positive | 12 | 1.58 (0.76–2.31) | |
| Negative | 40 | 1.04 (0.35–1.90) | |
| Clinical Stage | <0.001 | ||
| I–II | 28 | 0.98 (0.35–2.03) | |
| III–IV | 24 | 1.73 (0.56–2.31) | |
BC – bladder cancer.
The diagnostic performance of serum exosomal H19 for BC patients
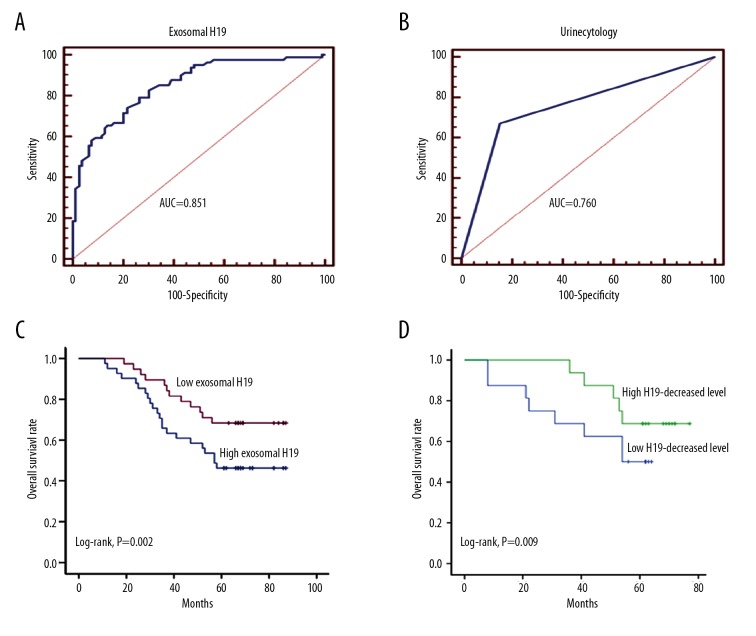
We then explored the diagnostic value of exosomal H19 in distinguishing BC patients from benign and healthy populations. ROC curve analysis showed an AUC (area under the curve) of 0.851 (95% CI: 0.787–0.903), with the diagnostic sensitivity and specificity reaching 74.07% and 78.08%, respectively (Figure 5A). We also investigated the diagnostic value of urine cytology in differentiating BC patients and individuals without cancer, and the ROC curve analysis showed that the AUC, diagnostic sensitivity, and specificity were 0.760 (95% CI: 0.666–0.838), 67.31%, and 84.62%, respectively (Figure 5B). Thus, serum exosomal H19 exhibited a higher AUC value and diagnostic sensitivity, whereas urine cytology showed a higher diagnostic specificity.
Figure 5.
The identification of serum exosomal H19 level as a noninvasive biomarker for BC patients. (A, B) ROC curves were established by using serum exosomal H19 level (A) and urine cytology (B) to explore their diagnostic potentials for BC patients. (C) Kaplan-Meier curve was established to investigate the prognostic value of exosomal H19 level for BC patient survival. (D) Kaplan-Meier analysis of the prognostic influence of decreased exosomal H19 level on overall survival.
Prognostic significance of exosomal lncRNA expression
Kaplan-Meier survival analysis was used to investigate the association between serum exosomal H19 expression (pre-operation) and prognosis of the 52 BC patients. As shown Figure 5C, patients with high H19 expression had a significantly poorer prognosis than those with low expression. Univariate Cox proportional hazards regression model analysis showed that the relative level of exosomal H19 expression, as well as tumor stage, lymph node metastasis, and clinical TNM stage, were correlated with the overall survival (OS) rate of BC patients (Table 2). Parameters significantly associated with OS in the univariate analysis were put into multivariate analysis to identify the independent factors for prognosis. The multivariate analysis revealed that exosomal H19 (HR: 2.193; 95% CI: 1.284–3.698, P=0.006) and clinical TNM stage (HR, 2.145; 95% CI: 1.266–4.890, P=0.009) were independent prognosis factors that affected OS of BC patients (Table 2).
Table 2.
Univariate and multivariate Cox proportional hazards regression model analysis of OS in BC patients.
| Factors | Categories | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Age | ≤65 vs. >65 | 0.922 (0.570–1.379) | 0.598 | ||
| Gender | Male vs. Female | 0.802 (0.543–1.321) | 0.330 | ||
| Tumor grade | High vs. low | 1.276 (0.966–1.902) | 0.070 | ||
| Tumor stage | T1–2 vs.T3–4 | 2.243 (1.235–2.998) | 0.015 | 1.308 (0.682–2.051) | 0.106 |
| Lymph node metastasis | Positive vs. negative | 3.060 (1.537–3.824) | 0.003 | 1.428 (0.456–2.754) | 0.063 |
| Clinical TNM Stage | I–II vs. III–IV | 3.374 (2.112–5.246) | <0.001 | 2.145 (1.266–4.890) | 0.009 |
| Exosomal H19 level | Low vs. high | 2.701 (1.715–4.458) | <0.001 | 2.193 (1.284–3.698) | 0.006 |
OS – overall survival; BC – bladder cancer; HR – hazard ratio; CI – confidence interval.
We also analyzed the decreased exosomal H19 level in the 32 patients after surgery compared to before surgery. A median value (0.465) was used to divide the patients into a high H19-decreased group (16 patients) and a low H19-decreased group (16 patients). We found that patients in the high H19-decreased group showed a longer survival time compared to the low H19-decreased group (Figure 5D), suggesting that patients who have decreased H19 after surgery will have a better prognosis.
Discussion
It is urgent to find an effective indicator to diagnose BC in earlier stage, thus allowing timely treatment [20]. Researchers have found that circulating exosomes in serum contain a group of genetic signatures in various diseases, especially cancer, thus presenting an enormous opportunity in terms of cancer diagnosis [21]. In this study, we investigated whether lncRNA serum H19 is contained in exosomes, and further explored the potential of serum exosomal H19 as a predictive biomarker of BC. Our results showed that serum H19 was contained in exosomes, and tumor-released exosomal H19 in serum is stable and significantly upregulated in BC patients compared to healthy individuals. Moreover, ROC analysis and Kaplan-Meier analysis indicated that serum exosomal H19 is a promising diagnostic and prognostic indicator.
Several attempts have been made to use lncRNAs in serum or plasma as diagnostic or prognostic predictors in BC [22,23]. Nevertheless, these potential tumor biomarkers are often in relatively low abundance and are impeded by the complexity of bodily fluids. Exosomes have drawn significant attention in the field of biomarker discovery. The release of exosomes into extracellular space affords an opportunity to examine exosomes in body fluids such as blood and urine. Most importantly, exosomes contain miRNAs, mRNAs, lncRNAs, and proteins, reflecting the features of cancer cells, which provides the ability to develop highly sensitive diagnostic strategies for monitoring the pathological conditions of cancer in a rapid and noninvasive manner [24]. On the other hand, determination of serum exosomal RNAs needs more time and effort compared to the detection of RNAs in serum directly. Recently, emerging evidence has uncovered the unique properties of exosomes, including their ability to embed specific lncRNAs, as well their stability and easy detection in circulatory system [25–27]. In this study, the exosomes morphology and size distribution were characterized by transmission electron microscopy and nanoparticle tracking analysis, respectively. These results demonstrated that we have successfully extracted exosomes from serum.
The roles of lncRNAs in cancer progression have long been researched, and H19 is widely accepted as an oncogene in cancers, including BC [28]. In addition, the potential of H19 as a predictive biomarker for BC has been investigated. Hua et al. demonstrated that genetic variants in lncRNA H19 are associated with the risk of bladder cancer in a Chinese population [29]. Ariel et al. revealed that H19 could serve as a prognostic tumor marker for the early recurrence of BC [30]; Gielchinsky et al. demonstrated that urinary cell H19 is a highly sensitive test for urothelial carcinoma [31]. For the role of H19 in exosomes, a report by Conigliaro et al. suggested that release of exosome-contained lncRNA H19 can modulate the endothelial cell phenotype of liver cancer cells [32]. However, the existence form of H19 in serum of bladder cancer patients is still unknown, and whether circulating exosomal H19 can serve as an ideal predictive biomarker warrants further investigations.
By using bioinformatics analysis followed by experimental verification, we found that serum H19 was contained in exosomes. Moreover, we conducted Spearman correlation testing followed by RT-qPCR analysis of expression change of serum exosomal H19 in preoperative and postoperative BC patients. Not surprisingly, we found that serum exosomal H19 was positively correlated with H19 expression in paired BC tissues, and the expression level of serum exosomal H19 was dramatically increased in BC patients when compared to healthy subjects or benign disease patients. Furthermore, we confirmed that exosomal H19 was stable in the serum samples. This is consistent with previous studies showing that exosomes in bodily fluids are a highly stable source of disease biomarkers [33]. We successfully obtained exosomes from serum and found that serum H19 were enriched and stable in exosomes, indicating that exosomal H19 in serum may be more suitable as diagnostic or prognostic biomarkers of BC.
We also investigated the diagnostic and prognostic value of serum exosomal H19 in BC patients. ROC curve analysis showed that serum exosomal H19 has a relatively high diagnostic potential when compared to urine cytology, and the noninvasive detection of exosomal H19 seems more promising. The diagnostic specificity of urine cytology was 84.6%, which could have been higher. However, the diagnostic value of serum exosomal H19 (sensitivity with 74.07%) was validated when compared to urine cytology. Kaplan-Meier survival curve showed that higher exosomal H19 level in serum of BC patients was correlated with poorer survival rate, which further suggests the clinical predictive value of H19 in exosomes. Moreover, patients experienced more decreased H19 level by surgical resection showed a longer survival time compared to the low H19-decreased patients. This result may be related to the influence of surgical resection on prognostic survival, as we believe that patients who have a large decrease in H19 might have had a more thorough resection. However, this needs further investigation.
Conclusions
Our study demonstrates that circulating exosomal H19 released from BC cells has considerable value for the diagnosis and prognosis of BC patients in clinical applications. This discovery of the lncRNA H19 in circulating exosomes could open new avenues for investigating BC progression, recovery, and therapy response.
Footnotes
Source of support: Departmental sources
Conflicts of interest
None.
References
- 1.Structure and function of poly(ADP-ribose) Hoppe Seylers Z Physiol Chem. 1972;353:843–51. [PubMed] [Google Scholar]
- 2.Ecke TH, Otto T. Illumination of a vision-how arthur rimbaud will give us motivation to find new input into bladder cancer biomarker research. Int J Mol Sci. 2017;18(11) doi: 10.3390/ijms18112463. pii: E2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim WT, Kim YH, Jeong P, et al. Urinary cell-free nucleic acid IQGAP3: A new non-invasive diagnostic marker for bladder cancer. Oncotarget. 2018;9:14354–65. doi: 10.18632/oncotarget.24436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Xing Z, Lin A, Li C, et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110–25. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du L, Duan W, Jiang X, et al. Cell-free lncRNA expression signatures in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med. 2018;22:2838–45. doi: 10.1111/jcmm.13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tkach M, Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164:1226–32. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 8.Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin Appl. 2015;9:358–67. doi: 10.1002/prca.201400114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah S, Wittmann S, Kilchert C, Vasiljeva L. lncRNA recruits RNAi and the exosome to dynamically regulate pho1 expression in response to phosphate levels in fission yeast. Genes Dev. 2014;28:231–44. doi: 10.1101/gad.230177.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Cai X, Yu J, et al. Exosome-mediated transfer of lncRNA RP11838N2.4 promotes erlotinib resistance in non-small cell lung cancer. Int J Oncol. 2018;53(2):527–38. doi: 10.3892/ijo.2018.4412. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Gao T, Liu X, He B, et al. Exosomal lncRNA 91H is associated with poor development in colorectal cancer by modifying HNRNPK expression. Cancer Cell Int. 2018;18:11. doi: 10.1186/s12935-018-0506-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isin M, Uysaler E, Ozgur E, et al. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front Genet. 2015;6:168. doi: 10.3389/fgene.2015.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hark AT, Schoenherr CJ, Katz DJ, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–89. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 14.Ding D, Li C, Zhao T, et al. LncRNA H19/miR-29b-3p/PGRN axis promoted epithelial-mesenchymal transition of colorectal cancer cells by acting on Wnt signaling. Mol Cells. 2018;41(5):423–35. doi: 10.14348/molcells.2018.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun H, Wang G, Peng Y, et al. H19 lncRNA mediates 17beta-estradiol-induced cell proliferation in MCF-7 breast cancer cells. Oncol Rep. 2015;33:3045–52. doi: 10.3892/or.2015.3899. [DOI] [PubMed] [Google Scholar]
- 16.Sidi AA, Ohana P, Benjamin S, et al. Phase I/II marker lesion study of intravesical BC-819 DNA plasmid in H19 over expressing superficial bladder cancer refractory to bacillus Calmette-Guerin. J Urol. 2008;180:2379–83. doi: 10.1016/j.juro.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Xue M, Chen W, Xiang A, et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer. 2017;16:143. doi: 10.1186/s12943-017-0714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, Chen Y, Dong X, Wang X. Serum exosomal long noncoding RNAs ENSG00000258332.1 and LINC00635 for the diagnosis and prognosis of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2018;27:710–16. doi: 10.1158/1055-9965.EPI-17-0770. [DOI] [PubMed] [Google Scholar]
- 19.Alegre E, Zubiri L, Perez-Gracia JL, et al. Circulating melanoma exosomes as diagnostic and prognosis biomarkers. Clin Chim Acta. 2016;454:28–32. doi: 10.1016/j.cca.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Fang L, Zang Y, Xu Z. Identification of core genes and key pathways via integrated analysis of gene expression and DNA methylation profiles in bladder cancer. Med Sci Monit. 2018;24:3024–33. doi: 10.12659/MSM.909514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizutani K, Terazawa R, Kameyama K, et al. Isolation of prostate cancer-related exosomes. Anticancer Res. 2014;34:3419–23. [PubMed] [Google Scholar]
- 22.Zhang S, Zhong G, He W, et al. lncRNA up-regulated in nonmuscle invasive bladder cancer facilitates tumor growth and acts as a negative prognostic factor of recurrence. J Urol. 2016;196:1270–78. doi: 10.1016/j.juro.2016.05.107. [DOI] [PubMed] [Google Scholar]
- 23.Chen T, Xie W, Xie L, et al. Expression of long noncoding RNA lncRNA-n336928 is correlated with tumor stage and grade and overall survival in bladder cancer. Biochem Biophys Res Commun. 2015;468:666–70. doi: 10.1016/j.bbrc.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 24.Abels ER, Breakefield XO. Introduction to extracellular vesicles: Biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol. 2016;36:301–12. doi: 10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enderle D, Spiel A, Coticchia CM, et al. Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLoS One. 2015;10:e0136133. doi: 10.1371/journal.pone.0136133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang L, Vader P, Schiffelers RM. Extracellular vesicles for nucleic acid delivery: Progress and prospects for safe RNA-based gene therapy. Gene Ther. 2017;24:157–66. doi: 10.1038/gt.2017.8. [DOI] [PubMed] [Google Scholar]
- 27.Navakanitworakul R, Hung WT, Gunewardena S, et al. Characterization and small RNA content of extracellular vesicles in follicular fluid of developing bovine antral follicles. Sci Rep. 2016;6:25486. doi: 10.1038/srep25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo M, Li Z, Wang W, et al. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213–21. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 29.Takai D, Gonzales FA, Tsai YC, et al. Large scale mapping of methylcytosines in CTCF-binding sites in the human H19 promoter and aberrant hypomethylation in human bladder cancer. Hum Mol Genet. 2001;10:2619–26. doi: 10.1093/hmg/10.23.2619. [DOI] [PubMed] [Google Scholar]
- 30.Ariel I, Sughayer M, Fellig Y, et al. The imprinted H19 gene is a marker of early recurrence in human bladder carcinoma. Mol Pathol. 2000;53:320–23. doi: 10.1136/mp.53.6.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gielchinsky I, Gilon M, Abu-Lail R, et al. H19 non-coding RNA in urine cells detects urothelial carcinoma: A pilot study. Biomarkers. 2017;22:661–66. doi: 10.1080/1354750X.2016.1276625. [DOI] [PubMed] [Google Scholar]
- 32.Conigliaro A, Costa V, Lo Dico A, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155. doi: 10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell P, Petfalski E, Houalla R, et al. Rrp47p is an exosome-associated protein required for the 3’ processing of stable RNAs. Mol Cell Biol. 23:6982–92. doi: 10.1128/MCB.23.19.6982-6992.2003. 20003. [DOI] [PMC free article] [PubMed] [Google Scholar]