Abstract
Background
MicroRNAs (miRNAs) have been widely recognized as essential regulators in human cancers, including colorectal cancer (CRC). Whether miR-769 is implicated in CRC progression remains elusive. The present study aimed to determine the function of miR-769 in CRC.
Material/Methods
MiR-769 expression in CRC tissues and adjacent normal tissues were measured by quantitative real-time polymerase chain reaction (qRT-PCR) and in situ hybridization. Kaplan-Meier curve analysis was utilized to determine the association between miR-769 expression and prognosis in CRC patients. The effects of miR-769 overexpression on CRC cell proliferation, cell cycle and invasion were analyzed using Cell Counting Kit-8 (CCK8), fluorescence activated cell sorting (FACS), and Transwell assays. Western blot was utilized to assess the effect of miR-769 on HEY1 expression.
Results
MiR-769 expression was decreased in CRC tissues. MiR-769 level was negatively correlated with the prognosis of CRC patients. Additionally, miR-769 overexpression remarkably inhibited cell proliferation, arrested CRC cells in G0 stage, and reduced cellular invasion. As to the mechanism, HEY1 was a direct target of miR-769; HEY1 level was inversely correlated with that of miR-769 in CRC tissues. Finally, overexpression of HEY1 reversed the effects of miR-769 on cell proliferation and invasion in CRC.
Conclusions
Our findings demonstrated that miR-769 suppressed the proliferation and invasion of CRC cells through targeting HEY1, which implied that miR-769 might be a novel therapeutic target for CRC treatment.
MeSH Keywords: Cell Proliferation, Colorectal Neoplasms, MicroRNAs, Neoplasm Invasiveness
Background
Colorectal cancer (CRC) is one of the most common cancers around the world and leads to a large number of cancer-related mortalities every year [1]. The main cause of CRC malignancy is cancer cell invasion and metastasis [2]. Despite some advances made in CRC therapy, the outcomes for CRC patients still remain very poor. And the incidence of CRC is gradually increasing year by year [3]. Therefore, elucidating the underlying molecular mechanism of CRC development and progression will greatly contribute to the development of therapeutic methods against CRC.
MicroRNA (miRNA) is a group of short noncoding RNAs with about 22 nucleotides; miRNA could control gene expression post-transcriptionally through targeting 3′-UTR regions of mRNAs [4]. Increasing evidence demonstrates that miRNAs exert very important roles in a diversity of physiological processes, such as survival, proliferation, and metastasis [5,6]. Dysregulated expression of miRNA is correlated with tumorigenesis and cancer progression [7]. For example, Zhou et al. reported that miR-143 suppresses breast cancer proliferation through regulating ERK5 and MAP3K7 expression [8]. Guo et al. reported that miR-302b-3p represses AKT signaling to prevent gastric cancer growth [9]. Therefore, looking for tumor-specific miRNAs and determining their functional mechanism are critical for the development of therapeutic targets against cancers.
MiR-769 has been shown to suppress non-small cell lung carcinoma growth [10]. Whether miR-769 is implicated in CRC development remains largely unclear. In this study, we found that miR-769 expression was decreased in CRC tissues compared to adjacent normal tissues. We also showed that miR-769 was a biomarker of CRC prognosis. Moreover, we showed that miR-769 upregulation significantly suppressed the growth and invasion of CRC cells. In terms of mechanisms, we found that miR-769 targeted HEY1 in CRC cells. Overexpression of HEY1 remarkably reversed the effects of miR-769 on CRC cells. Taken together, our results showed that miR-769 served as an oncogene in CRC by targeting HEY1, which suggested miR-769 might act as a therapeutic target for CRC intervention.
Material and Methods
Human samples
A total of 53 pairs of CRC (35stage I/II and 18 stage III/IV) and paired non-tumor tissues were collected at Peace Hospital Affiliated to Changzhi Medical College. All tissues were immediately frozen in liquid nitrogen after collection. We obtained written informed consents from all patients. Our study was approved by the Ethics Committee at Peace Hospital Affiliated to Changzhi Medical College.
Cell lines and cell culture
Five CRC cell lines (HCT-116, HCT-15, HT-29, SW480, and SW620) and a normal colon cell line (FHC) were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). Cells were cultured in DMEM medium (Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% FBS (Gibco) and maintained in a humidified incubator at 37°C and 5% CO2.
Cell transfection
MiR-769 mimics and negative control (NC) were synthesized by GenePharm (Shanghai, China). Cells transfection was performed using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer’s protocol.
Cell proliferation assays
We plated 2000 HCT-116 or SW480 cells into a 96-well plate and cultured them for 24 hours, 48 hours, or 72 hours. Then CCK8 solution (Beyotime, Shanghai, China) was added and cell viability was determined by measuring the absorbance at 450 nm.
In vitro transwell assay
Transwell cell culture chambers (Corning Incorporated, Corning, NY, USA) were used to measure cell invasion. We seeded 1×105 CRC cells in 200 μL serum-free medium in the upper chamber pre-coated with Matrigel. The lower chamber contained 600 μL DMEM supplemented with 10% FBS. Then, 24 hours later, cells in the lower chamber were fixed and then stained with 0.2% crystal violet. The cell number was determined using an inverted microscope (Olympus Corporation, Tokyo, Japan) at 200× magnification.
Real-time quantitative PCR
Total RNA was isolated using TRIzol extraction (Invitrogen) and reversely transcribed into cDNA utilizing a PrimeScript RT reagent kit (Takara, Dalian, China), followed by q-RTPCR analysis using SYBR Green Kit (Takara, Dalian China). Gene expression was normalized to U6 or GAPDH and calculated according to the 2−ΔΔCt method.
Luciferase reporter assay
The 3′-UTR region of HEY1 containing the wild-type (WT) or mutant (Mut) putative binding site of miR-769 was constructed into the pmirGLO dual-luciferase vector (Promega, Madison, WI, USA). Then CRC cells were co-transfected with pmirGLO-HEY1-WT or pmirGLO-HEY1-Mut and miR-769 mimics or NC using Lipofectamine 2000. Then 48 hours later, the luciferase intensity was determined using the Dual-luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA) according to the manufacturer’s instructions.
Statistical analysis
All data analyzed using SPSS 19.0 software (SPSS, Chicago, IL, USA) were displayed as mean ± standard deviation. Student’s t-test, one-way ANOVA analysis or Pearson’s correlation analysis was used to calculate the significant difference. The 5-year survival rate was analyzed using Kaplan-Meier curve and log-rank test was used for P-value. P<0.05 was considered as statistically significant.
Results
The expression of miR-769 was downregulated in CRC tissues
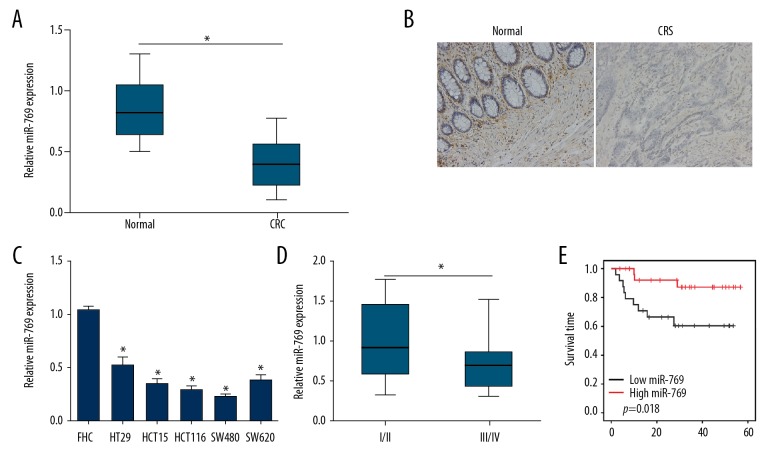
To investigate the role of miR-769 in CRC carcinogenesis, we analyzed miR-769 expression in 53 CRC tissues and matched normal tissues through qRT-PCR. The results showed that miR-769 was underexpressed in tumor tissues compared to normal tissues (Figure 1A). Consistent with this, in situ RNA hybridization results also suggested that miR-769 level was lower in CRC tissues (Figure 1B). Besides, qRT-PCR analysis showed that miR-769 expression was decreased in CRC cell lines compared to FHC cells (Figure 1C). And miR-769 expression was lower in samples with advanced stages (Figure 1D). Furthermore, we analyzed the survival rate by Kaplan-Meier analysis based on miR-769 expression (high expression group versus low expression group). As shown, lower expression of miR-769 in CRC patients was linked to poorer prognosis (Figure 1E). Taken together, these results indicated that miR-769 downregulation might be implicated in CRC progression.
Figure 1.
The expression of miR-769 was downregulated in colorectal cancer (CRC) tissues. (A) qRT-PCR analysis indicated that miR-769 was downregulated in CRC tissues (n=53) compared to adjacent normal tissues (n=53). (B) RNA in situ hybridization was used to measure the expression of miR-769 in paired of CRC tissues and normal tissues. (C) Relative expression of miR-769 in CRC cell lines and FHC cells. (D) Relative expression of miR-769 in CRC samples of stage I/II (n=35) and stage III/IV (n=18) by qRT-PCR. (E) Kaplan-Meier survival rate analysis based on miR-769 expression levels in CRC tissues. * P<0.05, ** P<0.01 and *** P<0.01. qRT-PCR – quantitative real-time polymerase chain reaction.
MiR-769 overexpression suppressed CRC cell proliferation and invasion
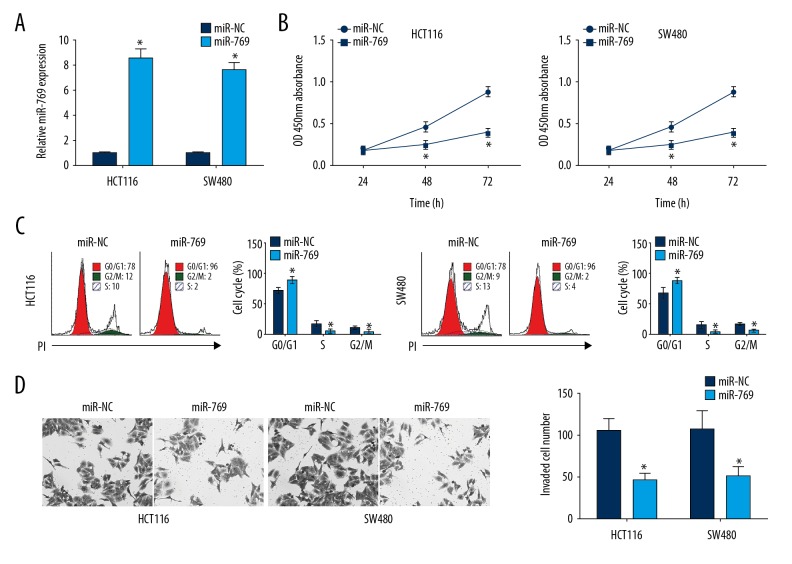
To investigate the role of miR-769, we measured the effects of miR-769 overexpression on CRC growth and metastasis by CCK8 and Transwell assay. Following transfection with miR-769 mimics in HCT116 and SW480 cells, miR-769 was significantly upregulated (Figure 2A). We then observed that miR-769 overexpression significantly suppressed cell proliferation (Figure 2B). Cell cycle arrest was a direct cause for reduced proliferation. We then assessed the effect of miR-769 on cell cycle by FACS and found that overexpression of miR-769 led to increased HCT116 and SW480 cells in G0/G1 phase, but decreased cells in S or G2/M phase (Figure 2C). Furthermore, Transwell assays revealed that HCT116 and SW480 cells transduced with miR-769 exhibited remarkably greater numbers of cells compared with NC group (Figure 2D).
Figure 2.
MiR-769 overexpression suppressed colorectal cancer (CRC) cell proliferation and invasion. (A) qRT-PCR analysis for evaluation of miR-769 expression in HCT116 and SW480 cells transduced with miR-769 mimics or negative control (miR-NC). (B) CCK8 assay was used to detect the proliferation of HCT116 and SW480 cells. (C) Cell-cycle distribution was determined by fluorescence activated cell sorting in HCT116 and SW480 cells. (D) Transwell assay was used for analysis of cell invasion. All data were obtained from three independent experiments and represented as mean ±SD. * P<0.05, ** P<0.01 and *** P<0.01. qRT-PCR, quantitative real-time polymerase chain reaction.
HEY1 was a target of miR-769
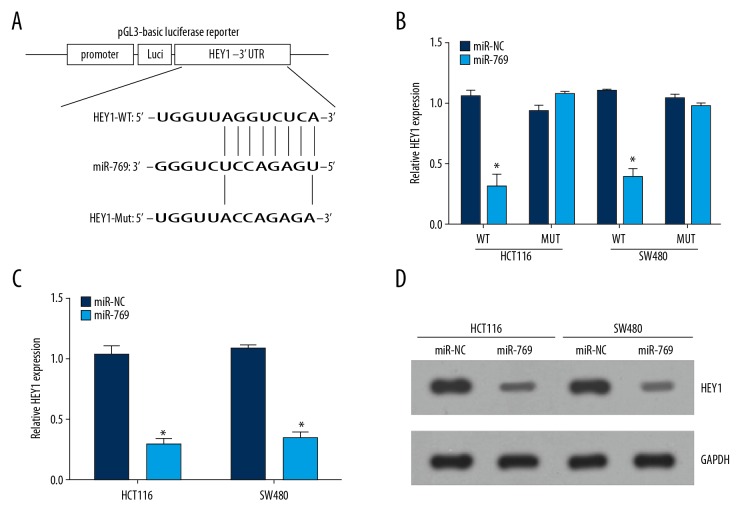
To determine the underlying molecular mechanism of miR-769 in CRC, 3 available databases (TargetScan, PicTar, and miRanda) were utilized to look for potential target genes of miR-769. The predicted targets were arranged based on the binding probability score. We then chose the target with the high score. Among all targets, HEY1 was chosen for further analysis. MiR-769 has conserved binding site in the 3′-UTR region of HEY1 mRNA (Figure 3A). To elucidate this predication, we constructed wild-type and mutant luciferase reporter plasmids and performed luciferase activity reporter assays. The results indicated that ectopic expression of miR-769 decreased the luciferase intensity in CRC cells transduced with wild-type luciferase reporter plasmid, whereas the activity of the mutant reporter plasmid was not altered by miR-769 overexpression (Figure 3B). Furthermore, we showed that miR-769 upregulation dramatically suppressed HEY1 expression in HCT116 and SW480 cells (Figure 3C, 3D).
Figure 3.
HEY1 was a target of miR-769. (A) Wild-type and mutant luciferase activity reporter plasmids were constructed. (B) Luciferase activity was detected in HCT116 and SW480 cells transduced with miR-769 or miR-NC. (C, D) Relative mRNA and protein levels of HEY1 were measured by qRT-PCR and western blot. * P<0.05 and ** P<0.01.
HEY1 was upregulated in CRC tissue
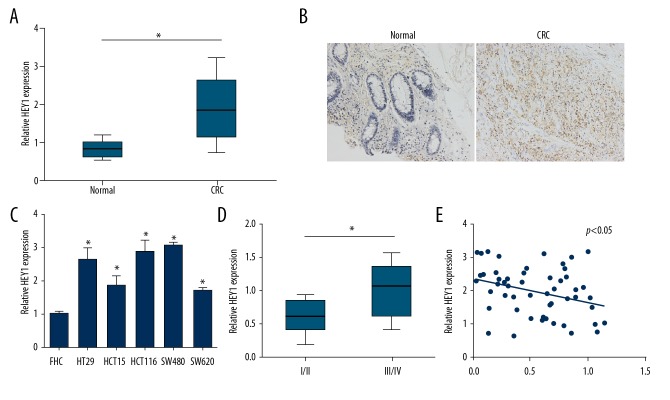
We analyzed the expression patterns of HEY1 in CRC by qRT-PCR and in situ RNA hybridization. We found that HEY1 was highly expressed in CRC tissues (Figure 4A, 4B). Similarly, the expression of HEY1 was elevated in CRC cell lines (Figure 4C). Moreover, HEY1 expression was higher in CRC tissues with advanced stage disease (Figure 4D). Furthermore, we observed an inverse correlation between the expression of miR-769 and HEY1 in CRC tissues (Figure 4E).
Figure 4.
HEY1 was upregulated in colorectal cancer (CRC) tissues. (A) Relative expression of HEY1 was measured in CRC tissues and adjacent normal tissues by qRT-PCR. (B) Expression level of HEY1 was determined by IHC. (C) qRT-PCR analysis of HEY1 expression in CRC cell lines. (D) qRT-PCR analysis for HEY1 expression in different stages of CRC samples. (E) Expression correlation between HEY1 and miR-769 in CRC tissues. * P<0.05 and ** P<0.01. qRT-PCR – quantitative real-time polymerase chain reaction.
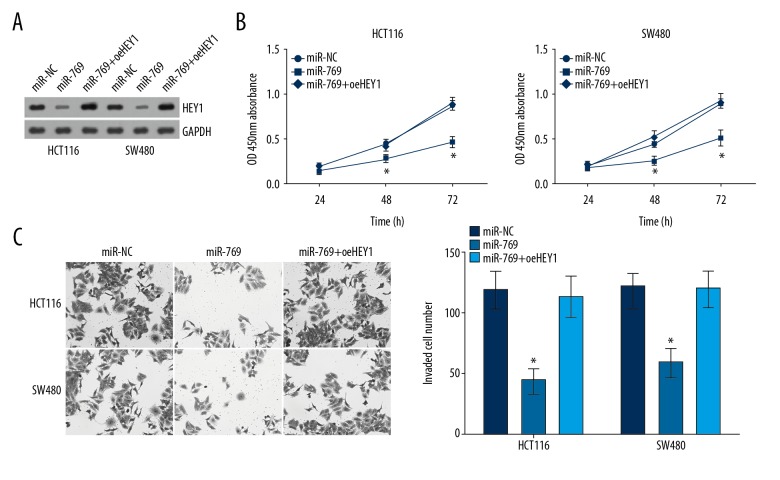
Restoration of HEY1 reversed the effects of miR-769
To further determine whether miR-769 regulates CRC through targeting HEY1, we restored HEY1 protein levels in miR-769-transduced HCT116 and SW480 cells. As shown in Figure 5A, HEY1 level was effectively increased in HCT116 and SW480 cells after ectopic expression. Then CCK8 and Transwell assays were conducted. The results indicated that HEY1 restoration significantly rescued the proliferation and invasion of miR-769-overexpressing HCT116 and SW480 cells (Figure 5B, 5C). In conclusion, these data indicated that miR-769 inhibited the proliferation and invasion of CRC cells through repressing HEY1 mRNA level.
Figure 5.
Restoration of HEY1 reversed the effects of microR-769. (A) Protein levels of HEY1 in indicated cell lines. (B) CCK8 assay was used for examination of cell proliferation. (C) Transwell assay was used for detection of cell migration. * P<0.05 and ** P<0.01. CCK8 – cell counting Kit-8.
Discussion
The initiation and development of CRC are a very complex process induced by various factors and multi-step changes. Until today, the mechanism underlying CRC carcinogenesis remains largely unknown. To develop effective CRC therapies, thoroughly investigating the molecular mechanism in CRC has been urgently required.
MiRNAs have been shown to regulate the development and progression of various cancers through modulating cell survival, proliferation, apoptosis, invasion or other aspects [11]. For instance, Li et al. reported that miR-223 enhances the growth and metastasis of gastric cancer through inhibiting EPB41L3 [12]. Ru et al. reported that miR-29b inhibits cell invasion in prostate cancer [13]. In addition, Akoa et al. showed that let-7 suppresses colon cancer progression [14]. Many studies have proven miR-769 is dysregulated in different human cancers. It is reported that miR-769 contributes to melanoma progression via targeting GSK3B [15]. Also, it has been reported that miR-769 promotes apoptosis of breast cancer cells [16]. In addition, observations have demonstrated that miR-769 suppressed non-small cell lung cancer growth and metastasis through inhibiting TGFBR1 [10]. However, whether miR-769 regulates CRC progression needs to be investigated. Our study found that miR-769 was underexpressed in CRC tissues and correlated with patients’ prognosis (5-year survival rates). Moreover, miR-769 represses CRC cell proliferation and invasion in vitro. Collectively, these findings indicate a tumor suppressive role of miR-769 in CRC. Notably, miR-769 could either exert roles of promotion or suppression in different cancers. The potential mechanism is that miR-769 specially targets different genes in respective cancers; this requires more investigation.
NPTCH signaling pathway is a particularly important signaling in human cancers. Aberrant activation often leads to the initiation or aggression of various cancers, including prostate cancer [17], breast cancer [18], glioblastoma [19], endometrial cancer [20], oral squamous cell carcinoma [21], and CRC [22]. Li et al. reported that increased activation of NOTCH signaling promotes non-small cell lung cancer progression [23]. Zhang et al. showed that Notch signaling pathway activated by SNHG1 enhance esophageal squamous cell cancer growth [24]. HEY1 is a member of the hairy and enhancer of split-related (HESR) family of basic helix-loop-helix (bHLH)-type transcriptional repressors and is a classic downstream effector of NOTCH signaling pathway. After NOTCH activation, HEY1 expression was initiated and consequently regulates pathological processes, such as proliferation, death, and invasion [25,26]. How HEY1 is post-transcriptionally regulated in CRC cells remains elusive. In our study, we found that HEY1 is a direct target of miR-769 in CRC cells. We showed that miR-769 overexpression significantly inhibited HEY1 levels in HCT116 and SW480 cells. Moreover, we demonstrated that HEY1 was upregulated in CRC tissues and its expression was negatively correlated with that of miR-769 in CRC tissues. What’s more, we found that restoration of HEY1 abrogated the effects of miR-769 on CRC cells, which suggested that miR-769 depended on HEY1 to exert biological functions in CRC cells.
Conclusions
Our study demonstrated that miR-769 serves as a tumor suppressor by targeting HEY1 in CRC cells. Our findings implied that miR-769 might be a promising prognostic biomarker and therapeutic target for CRC treatment.
Footnotes
Source of support: Departmental sources
Conflict of interests
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Ke J, Shao W, Jiang Y, et al. MicroRNA103 regulates tumorigenesis in colorectal cancer by targeting ZO1. Mol Med Rep. 2018;17:783–88. doi: 10.3892/mmr.2017.8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Lundberg IV, Wikberg ML, Ljuslinder I, et al. MicroRNA expression in KRAS- and BRAF-mutated colorectal cancers. Anticancer Res. 2018;38:677–83. doi: 10.21873/anticanres.12272. [DOI] [PubMed] [Google Scholar]
- 6.Shan Y, Liu Y, Zhao L, et al. MicroRNA-33a and let-7e inhibit human colorectal cancer progression by targeting ST8SIA1. Int J Biochem Cell Biol. 2017;90:48–58. doi: 10.1016/j.biocel.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Zhou LL, Dong JL, Huang G, et al. MicroRNA-143 inhibits cell growth by targeting ERK5 and MAP3K7 in breast cancer. Braz J Med Biol Res. 2017;50:e5891. doi: 10.1590/1414-431X20175891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo B, Zhao Z, Wang Z, et al. MicroRNA-302b-3p Suppresses cell proliferation through AKT pathway by targeting IGF-1R in human gastric cancer. Cell Physiol Biochem. 2017;42:1701–11. doi: 10.1159/000479419. [DOI] [PubMed] [Google Scholar]
- 10.Yang Z, He J, Gao P, et al. miR-769-5p suppressed cell proliferation, migration and invasion by targeting TGFBR1 in non-small cell lung carcinoma. Oncotarget. 2017;8:113558–70. doi: 10.18632/oncotarget.23060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15:38. doi: 10.1186/s12935-015-0185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li XH, Zhang Y, Zhang HW, et al. miRNA-223 Promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9:824–33. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- 13.Ru P, Steele R, Newhall P, et al. miRNA-29b Suppresses prostate cancer metastasis by regulating epithelial-mesenchymal transition signaling. Mol Cancer Ther. 2012;11:1166–73. doi: 10.1158/1535-7163.MCT-12-0100. [DOI] [PubMed] [Google Scholar]
- 14.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–6. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 15.Qiu HJ, Lu XH, Yang SS, et al. MiR-769 promoted cell proliferation in human melanoma by suppressing GSK3B expression. Biomed Pharmacother. 2016;82:117–23. doi: 10.1016/j.biopha.2016.04.052. [DOI] [PubMed] [Google Scholar]
- 16.Luo EC, Chang YC, Sher YP, et al. MicroRNA-769-3p down-regulates NDRG1 and enhances apoptosis in MCF-7 cells during reoxygenation. Sci Rep. 2014;4:5908. doi: 10.1038/srep05908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Li Y, Banerjee S, et al. Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-B signaling pathways (Retraction of vol 109, pg 726, 2010) J Cell Biochem. 2016;117:1960. doi: 10.1002/jcb.22451. [DOI] [PubMed] [Google Scholar]
- 18.Robinson DR, Kalyana-Sundaram S, Wu YM, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;17:1646–U163. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J, Yi L, Ouyang Q, et al. Neurotensin signaling regulates stem-like traits of glioblastoma stem cells through activation of IL-8/CXCR1/STAT3 pathway. Cell Signal. 2014;26:2896–902. doi: 10.1016/j.cellsig.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 20.Wei YA, Zhang ZB, Liao H, et al. Nuclear estrogen receptor-mediated Notch signaling and GPR30-mediated PI3K/AKT signaling in the regulation of endometrial cancer cell proliferation. Oncol Rep. 2012;27:504–10. doi: 10.3892/or.2011.1536. [DOI] [PubMed] [Google Scholar]
- 21.Lee SH, Hong HS, Liu ZX, et al. TNF alpha enhances cancer stem cell-like phenotype via Notch-Hes1 activation in oral squamous cell carcinoma cells. Biochem Biophys Res Commun. 2012;424:58–64. doi: 10.1016/j.bbrc.2012.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pannequin J, Bonnans C, Delaunay N, et al. The Wnt target Jagged-1 mediates the activation of notch signaling by progastrin in human colorectal cancer cells. Cancer Res. 2009;69:6065–73. doi: 10.1158/0008-5472.CAN-08-2409. [DOI] [PubMed] [Google Scholar]
- 23.Li SW, Zhao H, Li JQ, et al. Downregulation of long non-coding RNA LET predicts poor prognosis and increases Notch signaling in non-small cell lung cancer. Oncotarget. 2018;9:1156–68. doi: 10.18632/oncotarget.23452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang YJ, Jin XT, Wang Z, et al. Downregulation of SNHG1 suppresses cell proliferation and invasion by regulating Notch signaling pathway in esophageal squamous cell cancer. Cancer Biomark. 2018;21:89–96. doi: 10.3233/CBM-170286. [DOI] [PubMed] [Google Scholar]
- 25.Sethi N, Dai XD, Winter CG, Kang YB. Tumor-derived Jagged1 promotes osteolytic bone metastasis of breast cancer by engaging Notch signaling in bone cells. Cancer Cell. 2011;19:192–205. doi: 10.1016/j.ccr.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolos V, Mira E, Martinez-Poveda B, et al. Notch activation stimulates migration of breast cancer cells and promotes tumor growth. Breast Cancer Res. 2013;15:R54. doi: 10.1186/bcr3447. [DOI] [PMC free article] [PubMed] [Google Scholar]