Abstract
Hematopoietic stem cells are Defined by their ability to self-renew and differentiate through progenitor cell stages into all types of mature blood cells. Gene-targeting studies in mice have demonstrated that many genes are essential for the generation and function of hematopoietic stem and progenitor cells. For Definitively analyzing the function of these cells, transplantation studies have to be performed. In this chapter, we describe methods to isolate and transplant fetal liver cells as well as how to analyze donor cell reconstitution. This protocol is tailored toward mouse models where embryonic lethality precludes analysis of adult hematopoiesis or where it is suspected that the function offetal liver hematopoietic stem and progenitor cells is compromised.
Keywords: Transplantation, Fetal liver, Hematopoietic stem cells, Hematopoietic progenitor cells, Knock out, Embryonic lethal, Donor reconstitution
1. Introduction
Hematopoietic cells are derived from a specific mesodermal precursor, the hemangioblast, between day 7 and 8 of mouse embryonic development (E7.0–8.0) (1, 2). The hemangioblast is bipotential and has the ability to differentiate into cells of the endothelial and hematopoietic lineages. The first hematopoietic cells are generated in yolk sac and are shown to differentiate into primitive nucleated erythroid cells. This is Defined as the first wave of hematopoiesis, which supplies the rapidly developing embryo oxygen before the onset of circulation. Definitive hematopoietic stem cells (HSCs), with repopulation activity in myeloablated recipients, do not appear until after E10.5 in the mouse yolk sac, aorta-gonad-mesonephros (AGM) region, and the placenta (1, 2). Around E11.5, the HSCs seed the fetal liver where a massive expansion of the cells takes place before they migrate to the bone marrow, which is the main site of hematopoiesis in the adult animal (3). The main characteristics of HSCs are their ability to self-renew and differentiate into all types of mature blood cells. In the adult bone marrow, the self-renewal capacity is almost exclusively found in the long-term hematopoietic stem cells (LT-HSCs), which are Defined as lineage negative (do not express mature hematopoietic lineage markers) c-kit+, Sca1+, CD34−, Flk2−, CD48−, CD150+ (4, 5). The LT-HSCs give rise to short-term HSCs, which in turn give rise to multipotent progenitors (MPPs), which have very limited or no self-renewal capacity.
Hematopoietic development during embryogenesis and in the adult is regulated by vast array of genes, e.g., genes encoding growth factors, growth factor receptors, intracellular signaling molecules, and transcription factors. The gold standard for studying the function of these genes in HSCs and progenitors has been by using gene targeting in mice. The most productive way to knock out genes is to use conditional targeting (Frt-flp and Cre-lox systems) since many genes are essential for normal embryonic development. This could lead to embryonic lethality, which would preclude studying the function of the genes at later stages of development. A conditional knockout of a gene with a hematopoietic phenotype (e.g., at the HSC level) allows researchers to study both fetal liver and adult hematopoiesis using different Cre strains (e.g., Vav-Cre and Mx1-Cre mice) (6, 7). Some models have shown a specific gene to be essential for fetal liver hematopoiesis but dispensable for adult hematopoiesis (e.g., Sox17) (8) as well as demonstrating that some genes are important for both fetal and adult hematopoiesis (e.g., MLL) (9).
In order to demonstrate that knocking out a specific gene leads to defects in HSCs and/or hematopoietic progenitors, the cells have to be transplanted into a lethally irradiated recipient and analyzed for short-term (30–60 days) and long-term engraftment (4 months). In simple terms, successful short-term engraftment indicates that the progenitor cell pool is intact, whereas a successful long-term engraftment indicates that the HSC are functional. However, even though a successful long-term engraftment in a primary transplant recipient indicates that the HSCs are intact, they might still have defects in proliferation potential or self-renewal capacity. Those issues can be resolved by serially transplanting the HSCs into secondary and tertiary recipients and by transplanting the targeted HSCs in competition with normal cells.
In this chapter, we will describe ways to isolate and transplant fetal liver cells in order to analyze the function of HSCs and progenitor cells. In addition, we will give special emphasis on mouse models with embryonic lethal phenotypes, where it is suspected that the function offetal liver HSCs is compromised. We will also describe how to genotype them and transplant within a Defined period of time. Finally, we will discuss how to track the donor cells in order to measure engraftment.
2. Materials
2.1. Preparation of C57BL/6J (CD45.1) Recipient Mice
C57BL/6J CD45.1 recipient mice (e.g., purchased from Jackson Laboratories).
Acidified water.
Amoxicillin (TEVA Pharmaceuticals).
2.2. Harvesting of Embryos and Fetal Livers
Micro-dissecting scissors and forceps (e.g., from Roboz Surgical Instruments).
Dulbecco’s phosphate-buffered saline; DPBS (Lonza).
Bovine albumin, fraction V (Sigma).
6-Well tissue culture plates (Costar).
2.3. Isolation of Genomic DNA and Genotyping
Micro-dissecting scissors for cutting embryonic tissues (Roboz).
DPBS (Lonza).
24-well tissue culture plates (Costar).
Disposable transfer pipettes (Fisherbrand).
NaOH.
EDTA.
Tris–HCl.
PCR reagents and thermal cycler (e.g., Applied Biosystems ABI 7500).
Agarose LE (Roche).
TAE buffer (10×) (Lonza).
GelRed, 10,000× in H2O (Biotium).
2.4. Preparation of Fetal Liver Cells and Counting
6-Well tissue culture plates (Costar).
40-μ m sterile cell strainer (Fisherbrand).
10-mL syringe (BD Bioscience).
DPBS/0.02% deionized BSA.
Refrigerated centrifuge (e.g., Eppendorf 5810R).
Trypan blue (0.4%).
Neubauer hemocytometer (Fisherbrand) or automatic cell counters (e.g., Cellometer from Nexcelcom).
2.5. Fetal Liver Cell Transplantation
Irradiator (Cesium source).
Box, cylinder, or Broome style mouse restrainer (e.g., from Stoelting or Harvard Apparatus).
1-mL syringes with 25G × 5/8 needles (BD Bioscience).
DPBS/0.02% deionized BSA.
Warming lamp (infrared bulb 250 W, 115–125 V).
2.6. Staining of Fetal Liver Cells for Flow Cytometric Analysis
Capillary tubes for blood collection.
ACK red cell lysis buffer.
Equipment for performing general anesthesia (iso flurane vaporizer).
Refrigerated centrifuge (e.g., Eppendorf 5810R).
DPBS/0.02% deionized BSA.
Fc block (anti-mouse CD16/32) (eBioscience).
Antibodies for flow cytometry: IgG2a-FITC (BD Pharmingen), IgG2a-PE (BD Pharmingen), CD45.2-FITC (BD Pharmingen), CD45.1-PE (BD Pharmingen).
1% paraformaldehyde solution (in DPBS/0.02% deionized BSA) for fixing cells if samples are to be stored before analysis.
3. Methods
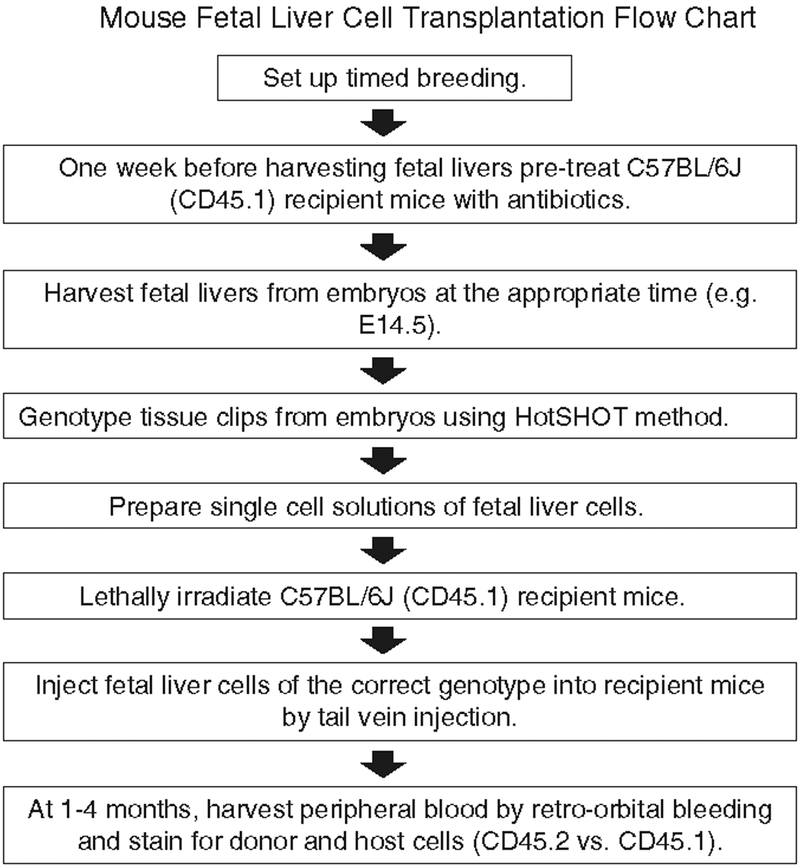
All experimental protocols should be reviewed and approved by your Institutional Animal Care and Use Committee. A flow diagram showing the transplantation procedure and detection of donor cells is presented in Fig. 1.
Fig. 1.
A flow diagram showing the mouse fetal liver cell transplantation procedure.
3.1. Setting Up Timed Breeding
In order to obtain sufficient numbers of embryos of a specific gestational age to work with for a particular experiment, at least 5–10 heterozygous females should be set up with heterozygous males (see Note 1).
The morning after, sexual activity can be monitored by checking for vaginal plugs which are composed of coagulated secretions from the male. The plugs generally fill the female vagina and persist for 8–24 h after breeding. Keep in mind that the presence of a plug does not guarantee pregnancy. That is highly strain speci fic and can vary considerably (see Note 2).
As a rule of thumb, you might expect 50% of plugged females to become pregnant. The morning a vaginal plug is found is often referred to as the first day of pregnancy (E1.0), but we considered it to be embryonic day 0.5 (E0.5) in concordance with Kaufman (10).
3.2. Preparation of C57BL/6J (CD45.1) Recipient Mice
One week before harvesting the embryonic liver cells, start pretreating the appropriate number of C57BL/6J recipient mice with acid water and antibiotics (amoxicillin 125 mg/250 mL acid water).
3.3. Harvesting of Embryos and Fetal Livers
Between E12.5 and E18.5 of development, fetal livers contain sufficient numbers of HSCs and progenitors to successfully rescue a lethally irradiated recipient without the use of any support cells such as adult bone marrow cells. Before E12.5, it is difficult to rescue the recipients without transplanting fetal liver cells in combination with support cells. Therefore, gene knockouts demonstrating lethality between E12.5 and E18.5 and with potential HSC and/or hematopoietic progenitor cell defects are optimal in such transplantation studies.
Euthanize pregnant mice using a CO2 chamber or cervical dislocation according to institutional guidelines (see Note 3).
Pin the mouse down on its back on a piece of styrofoam or cork board and sterilize the abdominal area by spraying it with a 70% EtOH solution.
Using forceps, pull up skin in the mid-abdominal area and cut laterally with scissors. Pull skin apart manually to expose the underlying abdomen. Cut open abdominal wall to expose the uterus. The embryos will be readily detectable within.
Cut each uterine horn at the distal end and by the cervix and transfer the uterus to the lid of a 6-well tissue culture plate as those allow easy maneuvering during the dissection steps.
Separate each embryo using scissors in order to make it easier to extract them from the yolk sac.
Using forceps, open the yolk sac and extract embryos. Cut the umbilical cord and remove the placenta.
Place each embryo in ice-cold DPBS/0.02% BSA in 6-well plates and keep on ice for 20 min to anesthetize the embryos (see Note 3).
Dissect fetal livers from each fetus using sterile forceps. This is best done under a stereomicroscope, but with increasing experience, it can be performed with the naked eye.
3.4. Isolation of Genomic DNA and Genotyping
In order to successfully genotype the embryos before trans-planting fetal liver cells, great care should be taken in ensuring that there is no contamination of maternal blood on the fetal tissues in question.
After obtaining the embryos, cut a very small piece of tail or toe tissue (approximately 0.2 cm) and place in a 24-well tissue culture plate containing 1 mL of ice-cold DPBS.
Serially rinse each piece of tissue three times in DPBS by transferring into new wells using a pipette (e.g., Rainin L200) or disposable transfer pipettes.
For isolation of genomic DNA, we use the HotSHOT method (11). Brie fly, to disintegrate the tissue pieces, incubate them in a thermal cycler at 95°C for 25 min in 50–75 μ L sodium hydroxide solution (pH 12). The pH is then adjusted by adding equal volume of 40 mM Tris–HCl solution (pH 5).
At this point, the DNA is ready for ampli fication using the PCR protocol of choice. As a reference, we use 10 μL DNA solution per 20 μL PCR reaction.
Separate the PCR products using 1% agarose gel in 1× TAE buffer. For staining the DNA, we use GelRed which is safer and more sensitive and gives less background than ethidium bromide.
3.5. Fetal Liver Cell Transplantation
In a tissue culture hood, place 40-μM cell strainers as needed into 6-well plates as needed and dispense 3–4 mL of ice-cold DPBS/0.02% BSA into each.
Place the fetal livers in the strainers and mash using the flat end of a plunger from a 5-mL syringe.
Rinse the strainer with ice-cold DPBS/0.02% BSA and collect the filtered cell suspensions into 15-μL conical tubes and put on ice. Allow clumps that might form during the collection to settle to the bottom of the tubes and transfer the supernatant to new tubes.
Collect cells by centrifugation in a chilled centrifuge (1,200 rpm, 5 min) and resuspend in 3–5 mL DPBS/0.02% BSA.
Count cells using trypan blue (0.4%) and a hemocytometer or an automatic cell counter. Resuspend cells to make a stock solution of 0.25–1 × 107 cells/mL, thereby allowing the injection of 0.5–2 × 106 cells/recipients in 0.2 mL DPBS/0.02% BSA (see Notes 4 and 5).
Before the recipient mice can be injected, they are irradiated with a lethal dose (see Note 6). Allow 2–3 h of resting after irradiation and before injection.
For injection, place the recipient mice into a restrainer, put the tail under a warming lamp, and inject 0.2 mL of the fetal liver cell suspension into the lateral tail vein using 1-mL syringes with 25G × 5/8 needle (see Note 7). In addition, inject 2–3 mice with saline solution as a control for the irradiation.
3.6. Analysis of Donor Fetal Liver Cell Repopulation
For analyzing short-term and long-term donor fetal liver cell repopulation, peripheral blood is collected by retro-orbital bleeding (see Note 8) at different time points (e.g., at 1, 2, and 4 months) and subject to CD45.1 and CD45.2 staining followed by flow cytometric analysis.
Anesthetize recipient mice using iso flurane (see Note 9).
Hold mice down by scruff on a stable surface covered with a paper towel and hold tail securely between ring finger and little finger in order to minimize any sudden movement should the mice start to come out of anesthesia during blood collection (generally 2–3 min).
While holding mouse, pull skin above and below eye using thumb and index finger until globe protrudes and insert blood collection capillary into the retro-orbital sinus at a 45° angle. Push steadily into the sinus rolling the capillary until blood starts to flow. By pulling the capillary slowly down and changing the angle, blood flow can be increased. Collect no more than approximately 200 μ L (see Note 8).
Remove capillary, immediately close eye, and apply pressure using gauze until bleeding has stopped (approximately 1 min).
Transfer blood to a 15-mL conical tube and add 2 mL ACK red cell lysis buffer.
Incubate on ice for 2 min and fill tube with ice-cold DPBS/0.02% BSA.
Collect cells by centrifugation in a chilled centrifuge (1,200 rpm, 5 min) and resuspend in 1–2 mL DPBS/0.02% BSA.
Count cells using trypan blue (0.4%) and a hemocytometer or an automatic cell counter.
Prepare single cell solution (2 × 106 cells/mL) in DPBS/0.02% BSA.
Block Fc receptors on cells with Fc block (anti-mouse CD16/32; 0.5 μ g/1 × 106 cells) (see Note 10).
Incubate on ice for 20 min.
Wash cells once with ice-cold DPBS/0.02% BSA and resuspend again in DPBS/0.02% BSA and stain cells with: (1) IgG2a-FITC and IgG2a-PE isotype controls (0.5 μ g each/1 × 106 cells) and (2) CD45.2 FITC and CD45.1 PE (0.5 μ g each/1 × 106 cells) (see Note 11). In addition, prepare one tube with unstained cells for setting up the flow cytometer.
Incubate on ice for 20 min.
Wash cells once with ice-cold DPBS/0.02% BSA and resus-pend in 0.5 mL ice-cold DPBS/0.02% BSA for immediate analysis or ice-cold DPBS/0.02% BSA/1% paraformaldehyDefixing buffer if the samples will be analyzed later (do not store samples longer than 2–3 days).
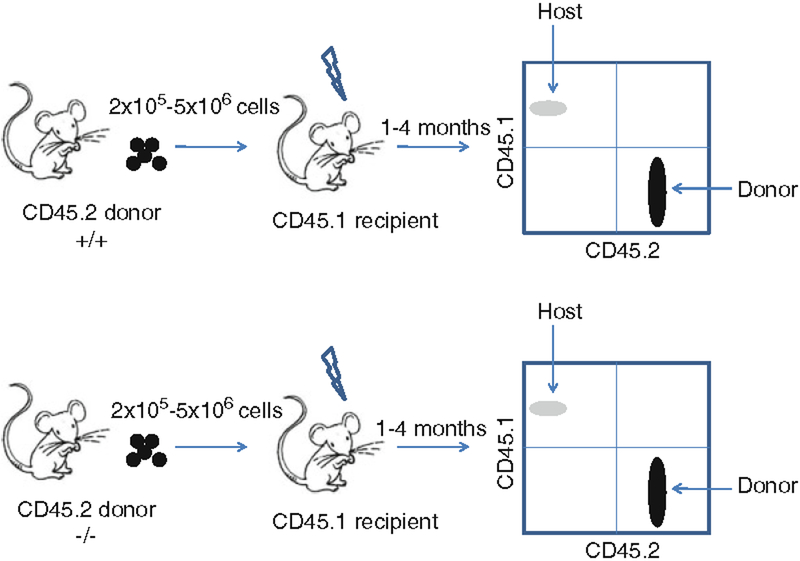
Run samples on a flow cytometer and analyze data using the appropriate software (e.g., FlowJo). Donor cells stain positive for CD45.2, whereas host cells are positive for CD45.1 (Fig. 2).
Fig. 2.
An example of a strategy for transplanting fetal liver cells from a wild-type (+/+) and a knockout (−/−) CD45.2 donor. Different numbers offetal liver cells (e.g., 2 × 105 −5 × 106) can be transplanted into a lethally irradiated CD45.1 recipient and analyzed for donor reconstitution 1–4 months posttransplantation. A CD45.1 × CD45.2 staining usually shows some CD45.1-positive cells (5–10%) in the wild type since residual hematopoietic stem and progenitor cells will start to contribute to hematopoiesis. The CD45.2 staining can be combined with markers for T cells, B cells, or myeloid cells to obtain information on the development of the different hematopoietic lineages.
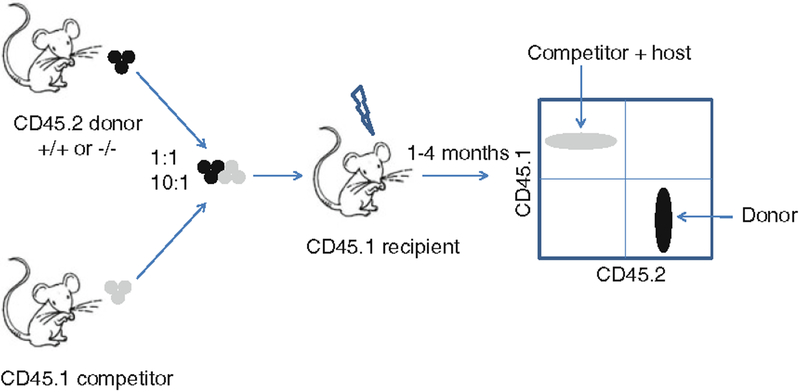
Fig. 3.
An example of a strategy for competitive transplantation between a CD45.2 donor and a CD45.1 competitor. Different combinations of donor and competitor cells can be mixed (e.g., 1:1 and 10:1) to test the function of CD45.2 hematopoietic stem cells and progenitors.
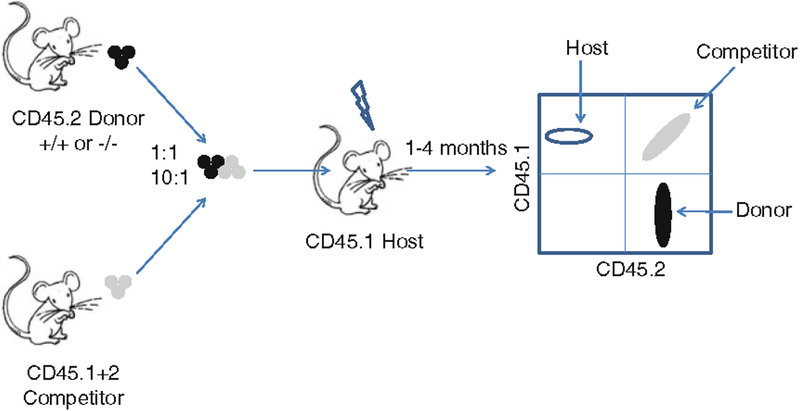
Fig. 4.
An example of a strategy for competitive transplantation between a CD45.2 donor and a CD45.1/CD45.2 competitor in a CD45.1 recipient. This strategy allows clear distinction of the transplanted CD45.2 or CD45.1/CD45.2 cells from the CD45.1 host.
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. This research was supported, in part, by the Intramural Research Program of National Institutes of Health, National Cancer Institute, and Center for Cancer Research. The content of this publication does not necessarily re flect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
4. Notes
Traditionally, as well as to save on cage numbers, one to three females are set up with each male.
For more information on the practical aspects of setting up timed breeding, see http://jaxmice.jax.org/jaxnotes/archive/501d.html.
Follow institutional guidelines in euthanizing embryos and adult mice. It is important to keep in mind that at E15, the embryo has developed perception of pain. Therefore, the embryos must be chilled on wet ice followed by decapitation. For more information, refer to http://web.ncifcrf.gov/rtp/lasp/intra/acuc/fred/guidelines/ACUC18.pdf.
In some instances, fetal liver cell numbers will be limited, and transplanting less than 0.5 × 106 cells/recipient will be necessary. This could lead to problems with short-term engraftment, but co-transplanting normal adult CD45.1 bone marrow cells as a support will prevent that and still allow tracking of CD45.2 donor cells. We have successfully transplanted 0.2 × 106 cells/recipient from an E14.5 donor without any support cells.
For further analysis of the function of speci fic HSC and/or progenitor cell populations, cell sorting can be performed, e.g., sorting and transplanting of Lin−, Mac1+, c-Kit+, and Sca1+ (LSK) fetal liver cells or highly puri fied Lin−, Mac1+, c-Kit+, Sca1+, CD34−, Flk2−, CD48−, and CD150+ HSCs. In those cases, few hundred or few thousand cells are obtained per fetal liver and support marrow needs to be co-transplanted to ensure short-term reconstitution. In addition, HSC function can be analyzed in a competitive repopulation assay, e.g., by transplanting knockout and wild-type fetal liver cells in different ratios, e.g., 1:1 and 10:1, and tracking by CD45.1 and CD45.2 staining (Fig. 3). Finally, intercrossing CD45.1 and CD45.2 mice to produce recipient mice expressing both CD45 allotypes can more accurately distinguish between donor and host cells in a competitive transplantation assay (Fig. 4).
We irradiate with 950 rads in one dose. Different protocols are used for irradiation, e.g., some laboratories irradiate with a total dose of 1,150 rads split into two doses 1–3 h apart. Make sure to include mice that are only injected with saline to control for the efficiency of the irradiation.
Get proper training for performing lateral tail vein injection from your institution veterinary services before setting up experiments.
Perform the procedure after proper training and with support from your institution veterinary services. For further guidelines on rodent bleeding, see http://oacu.od.nih.gov/ARAC/documents/Rodent_Bleeding.pdf.
Both injection and inhalation anesthesia can be used. However, inhalation anesthesia is preferred since it is easier to control and the animals recover faster compared to injection inhalation. Again, perform the procedure after proper training and with support from your institution veterinary services.
It is very important to block Fc receptors since they can bind the Fc part of the test antibodies and produce false positive signals in the flow analysis.
Other antibodies can be added to the CD45.1 and CD45.2 staining, e.g., antibodies against myeloid, T- and B-cell markers for analyzing the potential of the fetal liver cells to differentiate into these lineages.
References
- 1.Mikkola HK, Orkin SH (2006) The journey of developing hematopoietic stem cells. Development 133:3733–3744 [DOI] [PubMed] [Google Scholar]
- 2.Orkin SH, Zon LI (2008) Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132:631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ema H, Nakauchi H (2000) Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood 95:2284–2288 [PubMed] [Google Scholar]
- 4.Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ (2005) SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121:1109–1121 [DOI] [PubMed] [Google Scholar]
- 5.Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, Lio P, Macdonald HR, Trumpp A (2008) Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135:1118–1129 [DOI] [PubMed] [Google Scholar]
- 6.Kühn R, Schwenk F, Aguet M, Rajewsky K (1995) Inducible gene targeting in mice. Science 269:1427–1429 [DOI] [PubMed] [Google Scholar]
- 7.de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, Kioussis D (2003) Transgenic mice with hematopoietic and lymphoid speci fic expression of Cre. Eur J Immunol 33:314–325 [DOI] [PubMed] [Google Scholar]
- 8.Kim I, Saunders TL, Morrison SJ (2007) Sox17 dependence distinguishes the transcriptional regulation offetal from adult hematopoietic stem cells. Cell 130:470–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMahon KA, Hiew SY, Hadjur S, Veiga-Fernandes H, Menzel U, Price AJ, Kioussis D, Williams O, Brady HJ (2007) Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell 1:338–345 [DOI] [PubMed] [Google Scholar]
- 10.Kaufman M (1992) The atlas of mouse development. Academic, London [Google Scholar]
- 11.Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML (2000) Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29:52–54 [DOI] [PubMed] [Google Scholar]