Abstract
Automatic segmentation of cell nuclei is critical in several high-throughput cytometry applications whereas manual segmentation is laborious and irreproducible. One such emerging application is measuring the spatial organization (radial and relative distances) of fluorescence in situ hybridization (FISH) DNA sequences, where recent investigations strongly suggest a correlation between nonrandom arrangement of genes to carcinogenesis. Current automatic segmentation methods have varying performance in the presence of nonuniform illumination and clustering, and boundary accuracy is seldom assessed, which makes them suboptimal for this application. The authors propose a modular and model-based algorithm for extracting individual nuclei. It uses multiscale edge reconstruction for contrast stretching and edge enhancement as well as a multiscale entropy-based thresholding for handling nonuniform intensity variations. Nuclei are initially oversegmented and then merged based on area followed by automatic multistage classification into single nuclei and clustered nuclei. Estimation of input parameters and training of the classifiers is automatic. The algorithm was tested on 4,181 lymphoblast nuclei with varying degree of background nonuniformity and clustering. It extracted 3,515 individual nuclei and identified single nuclei and individual nuclei in clusters with 99.8 ± 0.3% and 95.5 ± 5.1% accuracy, respectively. Segmented boundaries of the individual nuclei were accurate when compared with manual segmentation with an average RMS deviation of 0.26 μm (~2 pixels). The proposed segmentation method is efficient, robust, and accurate for segmenting individual nuclei from fluorescence images containing clustered and isolated nuclei. The algorithm allows complete automation and facilitates reproducible and unbiased spatial analysis of DNA sequences.
Keywords: high-throughput segmentation, multiscale edge representation, nonuniform illumination, multiscale thresholding, watershed, region merging, cluster analysis
In recent years, investigations on nuclear architecture (spatial organization of nuclear organelles into well-defined compartments) and nonrandom gene positioning show significant impact on protein expression and cell functions (1–4). In these studies, the investigators used 2-D spatial distributions of relative and radial distances of fluorescence in situ hybridization (FISH)-labeled DNA sequences in interphase nuclei. They established the correlation between the spatial proximity of translocation prone genes and carcinogenesis (5–7). The motivation for this article stems from this interesting application in genomic organization. Our ultimate goal is to develop an exploratory data analysis system for studying the correlation between genomic organization and carcinogenesis (8). Naturally, such a system requires a segmentation method that can efficiently extract nuclei from fluorescence images and also possesses a high degree of segmentation accuracy (inaccurate segmentation can bias the spatial analysis of genes). The central theme of this article is to address these requirements for 2-D images.
Segmentation of cells or cell nuclei is a key component in most quantitative microscopic image analysis. Post segmentation, one can measure features relating to cell morphology, spatial organization of cells, and the distribution of specific molecules inside and on the surface of individual cells. On one hand, these quantitative measures can be used to do simple tasks, e.g., counting the number of cells (nuclei) and identify different cell types and cell-cycle phases. On the other hand, these measures can also be utilized in answering complex questions, such as the underlying mechanism of cell-cell communication processes (9) and spatial organization of subcellular structures (7,10). Additionally, the information from quantitative analysis can also serve as input(s) for testing data-driven mathematical models of the underlying physical processes (11). Such quantification can potentially shed insight into various cellular and molecular mechanisms and improve diagnosis and treatment of major human diseases, such as cancer.
In the past few years, numerous cell and cell nuclei segmentation algorithms have been proposed for a wide range of cytometry applications. For example, detection and enumeration of tumor cells (12–14), cell tracking and tracking individual fluorescent particles inside single cells (15–18), classification and identification of different mitotic phenotypes (19,20), and spatial statistical analysis of DNA sequences and nuclear structures (8,21). The most prominent segmentation techniques used for cell and nuclei delineation include gradient-curvature-driven methods, viz., active snakes, active contours, deformable models (19,22–24); levelsets (25–28); dynamic programming-based methods (29–33); graph-cut methods (34); and watershed and region-growing methods (35–37) combined with region-merging methods (38–41).
The energy minimization-based formulations, e.g., gradient-curvature-driven methods and levelsets-based methods, begin with a user-initiated boundary (contour or control points) and then calculate force fields, both stretching (repulsive force) and bending (attractive force), over the image domain using the generalized diffusion equation. The final object boundary is identified by “balancing” the stretching and bending forces extracted from the image data. Such approaches, however, are based on local optimization, either in a discrete setting or using partial differential equations and curve evolution. The main advantages of these methods are that they detect sharp changes in the object’s topology (concavities) and are easily extendable (at least levelsets-based methods) to handle higher-dimension images. However, these methods are sensitive to initialization since the energy minimization is subject to local minima and also to the force constants and the regularization terms.
Dynamic-programming and graph-cut methods also start with user-initiated control points, but they find a globally optimal path between two points in 2-D, based on an a priori figure of merit, typically the summation of weights of the pixels (edges) that are cut. Dynamic-programming-based methods have been successfully applied for 2-D segmentation whereas graph search techniques have been extended for 2-D and higher dimensions. Unlike gradient-curvature-based methods, conventional dynamic-programming and graph-cut methods are not iterative. These methods also have the particular advantage of being extremely robust, virtually insensitive to initialization and noise levels, and highly accurate (the user can interactively correct). But they come at the price of automation, and they also have “leaking” problem when an object has a weak boundary condition or is grouped together with another object having a similar intensity and also have a tendency to “bend” across sharp corners because of curvature limitations.
The most commonly used cell nuclei segmentation algorithms use the combination of region-growing and region-merging approach. The main advantage of these methods is the automation of the entire segmentation process, and this has been exploited to great success in several high-throughput applications for segmenting cells and cell nuclei from fluorescence images (15,20,42–48). However, region-growing (e.g., watershed algorithm) methods suffer from oversegmentation problem, especially, when the grey-level intensities of the objects in the fluorescence vary spatially. This can be circumvented, at least to a certain degree, by using region-merging methods based on adjacency-, size-, shape-, and intensity-based descriptors. But that makes the process ineffective when applied to new datasets because the user needs to carefully adjust (or guess) these parameters for merging. Additionally, these methods also have a tendency to inaccurately (over/under) segment (overlapping) nuclei with weak edge information. In most cases, the weak edges may be due to the intrinsic optical resolution limits of the acquisition system. Deconvolution methods (49) or contrast stretching methods (50,51) (e.g., windowing, histogram equalization) can be used to enhance the signal strength along the edges, but both these approaches also have their own demerits. For instance, deconvolution requires estimation of experimental point spread function, which can be extremely laborious and subjective, whereas (linear) contrast stretching can saturate pixels which fall outside the window and since the intensity values often drift across the image either prior knowledge or intensive human interaction is needed to choose the right window at each part of the image. In this aspect, multiscale techniques have shown promising results because of their ability to analyze the edges at different spatial scales (52–55) and will be explored in this article. Furthermore, most segmentation algorithms typically aim for segmenting all nuclei (high sensitivity) and at the cost of segmentation accuracy, which might not be optimal for some applications.
In this article, we address the aforementioned short comings commonly associated with 2-D high-throughput cell nuclei segmentation applications, namely, weak edge information, varying background intensities, and the requirement of input parameters. Consequently, we choose a model-based, region-growing and region-merging approach along with three novel concepts for our high-throughput cell nuclei segmentation. First, we use a modified multiscale (wavelet based) technique for enhancing the signal strength (intensity) at the boundaries of clustered nuclei (CN). Second, we propose and develop a multiscale thresholding technique based on entropy information from multiple scales for handling nonuniform background intensity variations. Last, we use statistical pattern recognition tools to automatically derive or estimate the input parameters required for segmenting CN. Furthermore, we also perform rigorous quantitative assessment using various measures (e.g., classification accuracy, segmentation accuracy) on approximately 4,200 cell nuclei from seven datasets of 2-D fluorescence images with varying degrees of clustering and background noise.
Methods and Materials
Overview of Segmentation
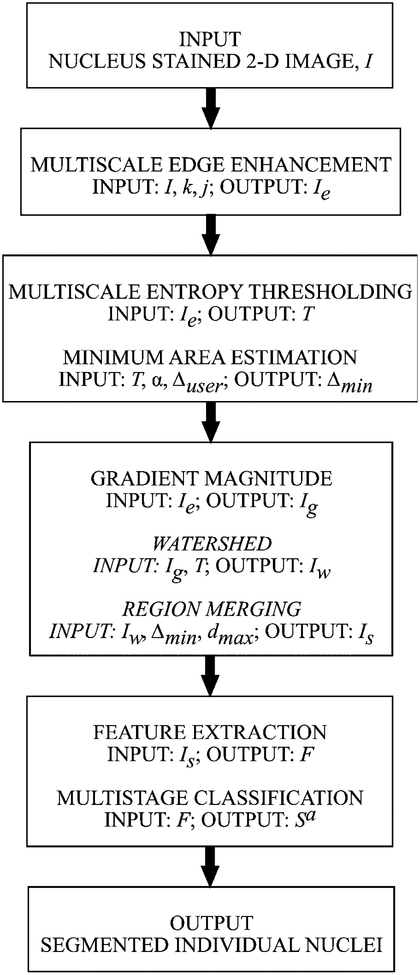
The algorithm starts with a 2-D fluorescence image with nuclear staining, for example, the DNA dye 4′, 6-diamidino-2-phenylindole (DAPI). In the first module, multiscale edge enhancement (MEE) technique is used for enhancing the boundaries of the nuclei in the nuclear staining channel. A particular advantage of this method is that it allows selective enhancement the edges of different sized objects and it also controls and suppresses the magnification of background noise. Next, the reconstructed image from MEE, Ie, is thresh-olded to extract nuclei (foreground objects) from the background using a multiscale entropy-based approach (MET). The entropy information measure, calculated over an adaptive window size, allows the proposed thresholding technique to extract foreground objects from background for nonuniformly illuminated samples. This thresholded image is used as a binary mask in all downstream operations. The third module of the algorithm estimates the minimum nuclear area (Δmin) from the thresholded objects using K-means clustering. Then, the nuclei are (over-) segmented using watershed, a morphological region-growing technique, from the gradient magnitude of the reconstructed image. This is followed by region merging, based on minimum area (Δmin) and maximum depth (dmax) constraints, to resolve the problem of oversegmentation. Finally, the algorithm calculates a 6-D feature vector for each segmented object and uses a multistage classifier to automatically classify the segmented objects (Sa) into clustered nuclei (Sa,CN) and single nuclei (Sa,SN). The flow diagram for the proposed modular high-throughput segmentation of clustered cell nuclei from 2-D fluorescence images is shown in Figure 1.
Figure 1.
Flow diagram illustrating the modular framework of the proposed high-throughput segmentation system.
Edge Enhancement Using Multiscale Edge Representation
The multiscale edge representation of signals was first described by Mallat (52), and since then it has evolved into two main formulations, namely, multiscale zero crossings and multiscale gradient maxima. The latter approach, which is used in this article, was originally developed by Mallat and Zhong (54) and expanded to a tree structured representation by Lu (56). The MEE technique comprises to two stages. In the first stage, the input signal is decomposed into several scales using wavelet transform (WT). The edges in each scale, identified using the gradient information (modulus maxima along the gradient direction), are enhanced using a user-defined stretching factor. In the second phase, the enhanced multiscale edges are used to reconstruct the image using an iterative alternating projection algorithm.
Decomposition.
The first step of multiscale edge representation involves applying a WT with a particular class of (non-orthogonal) separable spline wavelets (i.e., scaling and oriented functions) on the input image. The separable spline scaling function ϕ(x, y) plays the role of a smoothing filter, and the corresponding oriented wavelets are given by its partial derivatives:
| (1) |
The associated 2-D dyadic WT of intensity image I(x, y), size of I(x, y) is N × N and N = 2J, at scale 2j for position (x, y) and in orientation k is defined in the usual way:
| (2) |
The result is a representation of I(x, y) as a sequence of vector fields indexed by scale j, , where
| (3) |
An important feature of this representation is the information it provides about the gradient (edges) in I(x, y). From Eqs. (1)–(3), it can be easily shown that the 2-D WT gives the gradient of I(x, y) smoothed by ϕ(x, y) at dyadic scales. We call this representation multiscale gradient, and it is formally defined as
| (4) |
This multiscale gradient representation of I(x, y) is complete and redundant (the total number of coefficients in ∇2jI(x, y) is more than the original signal). These wavelets are not orthogonal; however, they do form a frame (53), and I(x, y) can be recovered (to be explained shortly), with minimal error, from Eq. (3) through the use of an associated family of synthesis wavelets. Furthermore, for discrete data (e.g., digital images), these calculations can be done in an efficient and stable manner (52,54).
For the intensity image, I(x, y), the positions of rapid variations are of interest, i.e., edge points; therefore we consider the local maxima of the gradient magnitude at various scales:
| (5) |
More precisely, a point (x, y) is considered a multiscale edge point at scale 2j if the magnitude of the gradient ρ2jI attains local maxima along the gradient direction θ2jI, which is given by:
| (6) |
For each scale, 2j, we collect the edge points along with the corresponding values of the gradient (i.e., the WT values) at that scale. The resulting local gradient maxima set at scale 2j are then given by
| (7) |
such that, ρ2jI(xi, yi) has local maxima at (xi, yi) along the direction θ2jI(xi, yi). For a J-level 2-D WT, the following set
| (8) |
is called the multiscale edge representation of image I(x, y). Here, S2JI(x, y) is the low-pass approximation of I(x, y) at the coarsest scale, 2J, and is equal to the global mean of I(x, y).
Since the magnitude of edge gradients characterizes the intensity difference between different objects and background regions in an image, we enhance the contrast between them by applying appropriate transformations on edge gradients. With the multiscale edge representation, different transformations of the edge gradients can be designed and used at different scales, thus enabling us to selectively enhance the edges of certain size objects. This is a simple consequence of the location information embedded in the multiscale edge representation.
Although several types of transformations can be applied to edge gradients in each scale, we choose to apply the simplest transformation on edge gradients by linearly stretching the gradient magnitudes at each scale by a user-defined stretching factor. Indeed, for the edge sets A2j(I) given by Eq. (7), we define the corresponding stretched edge set with a stretching factor, k ≥ 1, simply by multiplying each gradient maximum value by the scalar factor k and independent of scale index j, such that
| (9) |
Note that the stretching factor has the effect of scaling the length (magnitude) of the gradient vector at each edge point without affecting its direction. Logically, if we use a scale-dependant stretching factor, then we can selectively enhance the boundaries of different sized objects. (For instance, at higher scales, the multiscale edge representation, Eq. (9), will have fewer edge gradient locations corresponding to smaller objects than larger objects because the smaller objects will be blurred because of the smoothing function of wavelet decomposition.) However, since we intend to enhance the edges of (approximately) similar-sized nuclei in our images, that is, edge enhancement is limited to only one type of objects in the images; hence, we use a constant stretching factor, k, across all scales.
Reconstruction.
Once the multiscale edge representation (location of the gradient maxima, direction or gradient maxima) has been constructed from the input image, we can reconstruct a close approximation of the input image from this sparse multiscale edge representation [Eq. (8)] using an alternating projection algorithm developed by Mallat and Zhong (54). Briefly, Mallat-Zhong’s reconstruction algorithm attempts to find a inverse WT consistent with the multiscale scale edge representation while enforcing a constraint imposed by the reproducing property of WT. To enforce such constraint on a set of functions in the reconstruction process—denoted as h(x) in (54)—they project these functions onto the space characterized by the reproducing kernels. (We refer readers to Mallat-Zhong’s paper (54) for a detailed mathematical proof on the convergence of the iterative algorithm and for the derivation of the lower bound for convergence.)
Note that Mallat-Zhong’s original algorithm was designed for sparse representation (i.e., compact coding) of 2-D signals in terms of multiscale edges and not for edge enhancement. Hence, we made two modifications to their algorithm: (i) we start with a modified multiscale edge representation of Eq. (8), in which [A2j(I)] is replaced by Eq. (9); and (2) we allow for the S2JI(x, y) term to update during each cycle of the iteration in the alternating projection algorithm. These two modifications ensure the reconstruction process to not only enhance the intensity along edges but also improve the contrast within and around the regions enclosed by edges.
Multiscale Entropy Thresholding
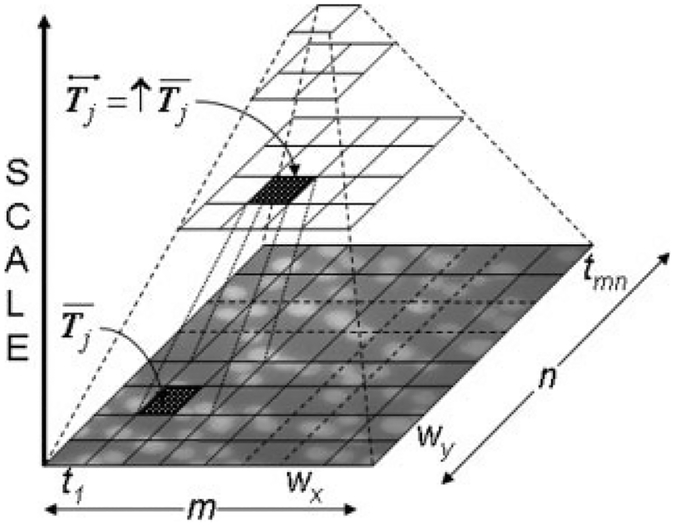
We apply a modified multiscale thresholding technique for extracting the nuclei from the background because of significant nonuniformity of nuclear brightness and background intensity. The technique uses a Laplacian pyramid data structure (Fig. 2) for selecting a window size based on the Shannon entropy (51). The proposed multiscale entropy-based thresholding method (MET) can be described as follows:
Figure 2.
Laplacian tree structure used in the multiscale entropybased thresholding technique.
Suppose the size of a discrete image I(x, y) is M × N, with l grey-levels. Let, pi denote the probability of the ith grey-level computed from the histogram of I(x, y). Then, the Shannon entropy (θ), is given by the following equation:
| (10) |
When θ attains a large value, it indicates that the image has more information (entropy) and it attains a maximal value when the pixel intensities in the image are uniformly distributed.
We start the multiscale thresholding process by dividing the image I(x, y) into small tiles, T = {t1, t2,…, tmn}, each of size wx × wy pixels, so that M = mwx and N = nwy. Then, we create a feature vector, F = {θi}, where θi is the Shannon entropy for tile ti ∈ T. Next, we calculate a threshold value θ′ for F using Otsu’s method (57) and identify the corresponding tiles, T′ ⊂ T with θi > θ′. After that, we threshold the pixels within each of the tiles in T′ by using Otsu’s method. Unlike the previous step, where the entropy value of each tile was used, in this step, the thresholding is done using the grey-value intensities of the pixels within each tile.
For the remaining tiles that were not thresholded, , we enlarge their window size (wx = 2wx and wy = 2wy) according to the Laplacian tree structure (Fig. 2). Let denote the set of all enlarged tiles, then for each tile, , we calculate its entropy value, append it to the feature vector, such that , and then calculate a new threshold valueθ′ for the updated feature vector using Otsu’s method. If , then we threshold only those pixels within , which have not been thresholded earlier using Otsu’s method, otherwise, will be enlarged until it is thresholded. Naturally, in some cases, this recursive enlargement will make the tile size equal to size of the input image, in which case the proposed multiscale thresholding method will be identical to global Otsu thresholding method.
Estimating Nuclear Area
Once the nuclei are extracted, we use that information to estimate the minimum nucleus area (Δmin) for the region-merging operation—a postprocessing step to merge oversegmented nuclei from watershed delineation. Since Δmin is a critical parameter that can affect segmentation, we have made it automatically adaptable to the size of the nuclei in the input image using statistical pattern recognition tools. This is instead of prompting the user to supply this parameter for each dataset. Moreover, such information might be unknown a priori unless manually estimated, which can be laborious when analyzing large volumes of data. The basis of this automated strategy is that, in most images, a fraction of nuclei will be isolated and the multiscale entropy-based thresholding will successfully extract them as individual objects from which nuclear size can be estimated. We realize that this assumption is not valid for all images and that this strategy will fail, particularly, when all the nuclei in the image are clustered. Under those circumstances, the algorithm defaults to the user-defined minimum area value, Δuser.
The minimum nuclear size is estimated as follows: First, we label the thresholded foreground objects and calculate the area of labeled objects, thus creating a 1-d feature space. The feature space is segregated into four classes to capture different sized foreground objects of approximately: (1) single nucleus; (2) small clusters up to 2 overlapping nuclei; (3) medium clusters with three to four nuclei; and (4) large range clusters with more than six nuclei. The 1-D feature space is classified using K-means clustering (51) and the minimum nuclear size Δmin is estimated as
| (11) |
where αuser is a user-defined parameter and μ(·) and σ(·) denote the mean and standard-deviation of the area for class 1, respectively. Note that it is not necessary to classify the feature space into exactly four classes; instead we can do the estimation process even when objects are categorized into two classes.
Watershed Segmentation and Region Merging
Although the multiscale thresholding method can successfully extract isolated nuclei, it will fail to delineate individual nuclei from clusters. For this purpose, we apply watershed algorithm (35,37) on the gradient magnitude (Ig) of edge enhanced image Ie and use the thresholded binary image from the previous step as a mask. In spite of the edge enhancement image, watershed algorithm will lead to severe oversegmentation of all the foreground objects into fragments, both isolated and CN alike, because of the intensity fluctuations within the objects and, thus, requiring region merging (15,47).
We apply two constraints: user-defined maximum depth (dmax) and minimum nuclear size (Δmin) calculated using Eq. (11), for merging oversegmented fragments from watershed delineation. In our implementation, we sort all the catchment basins (fragments) based on their depth and then on size. Next, a fragment is merged with one of its eight-connected neighbors either if the current fragment is at a depth d < dmax or has an area smaller than Δmin. The region-merging process is iterative and allows for only one mergence per oversegmented fragment in each iteration and does not allow mergence when (1) one of the two segments to be merged has a neighbor that has been merged in current iteration and (2) both segments exceed the minimum size. When all the basins have been processed, the final step of the region-merging process removes objects of size less than Δmin. This strategy ensures morphologically accurate merging of the oversegmented fragments of isolated nuclei to form individual nuclei. Similarly, the oversegmented fragments of nuclei in clusters will also merge to form bigger objects and in majority of the cases these merged objects will correspond to the individual nuclei within the clusters. Thus, allowing us to extract individual nuclei from clusters in addition to those that are already fairly well separated.
Classification of Segmented Nuclei
The segmentation process, described earlier, is not a perfect system and is bound to have some failures. For instance, closely packed or overlapping nuclei devoid of any perceptible boundary information will remain as clusters, and the region-merging process is also bound to generate morphologically inaccurate segmentation. The final step of our algorithm addresses these quality control issues by analyzing the feature measurements of the segmented nuclei and automatically segregating them into SN and CN. (Note that we use the term “clustered nuclei,” in a loose sense, to describe all those objects that do not belong to the SN class.)
The automatic classification is achieved using a multistage classifier system, where the first-stage classifier functions as a coarse-grain classifier and resulting classification is used as an input for the second-stage classifier for further refinement. The entire process involves three steps: features measurement, coarse-grain classifier, and fine-grain classifier. In the features measurement step, we label each of the segmented objects (both single and CN) and then compute a 6-D feature vector,
| (12) |
where imin = minor axis of inertia, imax = major axis of inertia, fmin = maximum Feret diameter, fmin = minimum Feret diameter, f⊥ = maximum perpendicular Feret diameter, and hc = Horton’s compactness factor or ratio of square of perimeter to four times the area. We selected these features based on our preliminary assessment of the nuclei’s morphology in our datasets. Next, we apply K-means algorithm to classify the feature space into two classes (SN and CN) by initiating the algorithm with a 50% prior-probability for each class. The output from the K-means algorithm will improve/adjust the prior probability estimate for each class-this depends entirely on the quality of the feature descriptors. Subsequently, we use a quadratic normal Bayes classifier, QDC (17), with the improved prior-probability estimates to refine the classification of the 6-D feature space into SN and CN classes.
Performance Assessment
To demonstrate the effectiveness of the MEE for CN we use the following quantitative measure of contrast between CN, EI:
| (13) |
where, i is the number of neighbors, μf and σf denote the mean and the standard deviation of the intensity values of the nucleus (f), respectively, and μe,i and σe,i denote the mean and standard deviation of the intensity values of the common edge (e) between two closely packed nuclei, respectively. To calculate the common edge between a nucleus and its neighbors within a cluster, first, we dilate one-time the binary region corresponding to the nucleus and calculate the “exclusive-OR” area of the nucleus and its dilated version, and then we find the intersection between the “exclusive-OR” and the binary region of its ith neighbor.
The effectiveness of multiscale entropy-based thresholding and watershed with region-merging steps was assessed using the following metrics:
percent nuclei missed by the multiscale entropy-based thresholding, MMET;
percent nuclei watershed and region-merging algorithm failed to delineate, MWRM;
percent CN that were not split and are still clustered after applying the proposed algorithm, PNCafter;
percent declustering, defined as
| (14) |
where PNCbefore denotes the percent of CN in the input image determined visually.
Additionally, we also do the accuracy assessment of multistage classifier for SN class and CN class using the following parameters: (1) TP, true positives, the total number of objects that were correctly identified; (2) FP, false positives, total number of incorrectly identified objects; and (3) PPV, positive predictive value,
| (15) |
as a measure of specificity. We also use the yield ratio, defined as
| (16) |
as a measure for assessing the algorithm’s sensitivity.
Segmentation accuracy of SN objects.
We use root-mean-square deviation, RMSD, to evaluate the segmentation accuracy of the SN objects only (32,58) because of their importance in further quantitative analysis. Briefly, our evaluation method computes RMSD by comparing pairs of segmented regions within an image, where one measurement is taken from the manual ground-truth segmentation (Sm,SN) and the other from the proposed segmentation (Sa,SN), such that and denote the boundaries of n segmented nuclei belonging to class SN, respectively. Let denote the distance transform, DT (51) calculated from the “binary-NOT” of the ground-truth segmentation boundary. (Hereafter, we will drop the SN superscript for notational convenience.) Then, we define the RMSD measure for all the SN within an image as
| (17) |
By this definition, each element of the RMSD vector reflects the amount of agreement between manual and automatic segmentation of a region, such that 0 ≤ RMSDi ≤ ∞. When RMSDi approaches 0, it indicates exact match between automatic and manual segmentation, and higher values of RMSD indicate (significant) difference between automatic and manual segmentation.
Samples and Image Acquisition
We used seven different datasets, D1 to D7, derived from four different lymphoblast cell lines, C1 to C4. For all datasets, we acquired 3-D stacks of 4% paraformaldehyde/0.3 phosphate buffered saline fixed nuclei, which had been stained with the DNA dye DAPI. (All cell lines were also stained for two genes using FISH but these channels were not used in this study.) To account for variations in nuclei density (total number of nuclei with in the field of view) and nuclei aggregation, we captured images of the same sample from different locations on the glass slide and under different confluence conditions, respectively. The images were acquired using a 60×, 1.4 NA, oil objective lens, on a Nikon fluorescent microscope. All images were 1,392 × 1,040 pixels with horizontal and axial resolution of 0.132 μm and 0.5 μm, respectively, and the 3-D stacks of the DAPI channel were collapsed to form a 2-D image using maximum intensity projection to satisfy the input requirements of our 2-D algorithm.
Table 1 summarizes the test datasets and lists the manual count of the total number of isolated nuclei, the total number of nuclei in clusters, and the percent (foreground) area occupied by nuclei. In general, nuclei from all cell lines were regular and flat, and in most datasets nuclei were mostly uniform in size. In addition, nuclei from all datasets also exhibited significant variation (based on visual inspection) in their textural property (DNA staining) across the field of view along with nonuniform variation in the background. The datasets D1 and D5, derived from cell line (C1), had similar nuclei aggregation (PNCbefore values 47.0 and 54.9%, respectively) but varied in terms of area occupied by nuclei (24.1 and 7.4% respectively). On the contrary, datasets D3 and D7, derived from cell line C3, differed mostly in terms of the degree of aggregation (PNCbefore values of 24.2% vs. 66.1%, respectively) but had similar nuclei density (~10%). In datasets derived from cell line C2, i.e., D2 and D6, both nuclei density and nuclei aggregation were lower in D2. The dataset D7 derived from cell line C4 had moderate amount of nuclei aggregation (PNCbefore value of 31.9%) and lower nuclei density (6.2%).
Table 1.
Summary of test datasets used for evaluating the proposed high-throughput algorithm
| DATASET | CELL LINE | IMAGES (N) | ISOLATED NUCLEI (N) | CLUSTERED NUCLEI (N) | PNCBEFORE (%) | FOREGROUND (%) |
|---|---|---|---|---|---|---|
| D1 | C1 | 16 | 865 | 767 | 47.0 | 24.1 |
| D2 | C2 | 18 | 217 | 78 | 26.4 | 5.2 |
| D3 | C3 | 12 | 380 | 121 | 24.2 | 12.7 |
| D4 | C4 | 16 | 261 | 122 | 31.9 | 6.2 |
| D5 | C1 | 17 | 120 | 146 | 54.9 | 7.4 |
| D6 | C2 | 10 | 175 | 368 | 67.8 | 20.4 |
| D7 | C3 | 15 | 190 | 371 | 66.1 | 9.8 |
| Sum | 104 | 2,208 | 1,973 |
PNCbefore = percentage of nuclei in clusters in the input image(s) and foreground = percentage area occupied by nuclei in the inputimage (calculated from the ground truth data).
Implementation
We developed a MATLAB (Version 7.1, Mathworks, Natick, MA)-based application along with DIP image (image processing toolbox, academic version, Delft University, Netherlands) and PRTOOLs (pattern recognition toolbox, academic/research version, Delft University) for implementing the multiscale thresholding, nuclear area estimation, segmentation, and classification steps of the algorithm. The MEE step was performed using an in-house script developed in Last-Wave (59).
For the MEE step, we used cubic spline wavelets as edge detectors (54,55), wavelet decomposition up to six scales, a stretching factor (k) of 4 for enhancing the multiscale edges, and 15 iterations for reconstruction (alternating projection algorithm). Since our input images were not dyadic, we created a dyadic version (2,048 × 2,048 pixels) of the input image by reflecting the input image along the right and bottom boundaries. For the multiscale thresholding, we used Δuser = 550 pixels, and the starting tile size (wx and wy) was automatically determined by calculating the best block size with dimensions at least . This resulted in wx = 16 pixels and wy = 16 pixels (calculated using “bestblk” function in MATLAB). Following the thresholding process, we retained only that portion corresponding to the original input image size (i.e., 1,392 × 1,040 pixels) and used that in all succeeding steps. We used a value of αuser = −1.5 for calculating the minimum nuclear size [Eq. (9)] and for the region-merging process we used dmax = 4. The ground truth data used for the assessment of segmentation accuracy was generated using an in-house, semiautomatic MATLAB application based on the 2-D dynamic programming (31).
Results
We batch-processed all input images with the aforementioned set of input parameters using the in-house LastWave script and the developed MATLAB application. The entire process took on average 65 seconds for a typical image with approximately 80 nuclei. At least two third of this processing time was spent on input and output operations (e.g., saving the segmented nuclei into individual files). All datasets were analyzed on a 32-bit workstation with two dual-core AMD Opteron (2.0-GHz processor) and 4 GBs of RAM. (Note: The LastWave and MATLAB applications utilized only one processor for computations.)
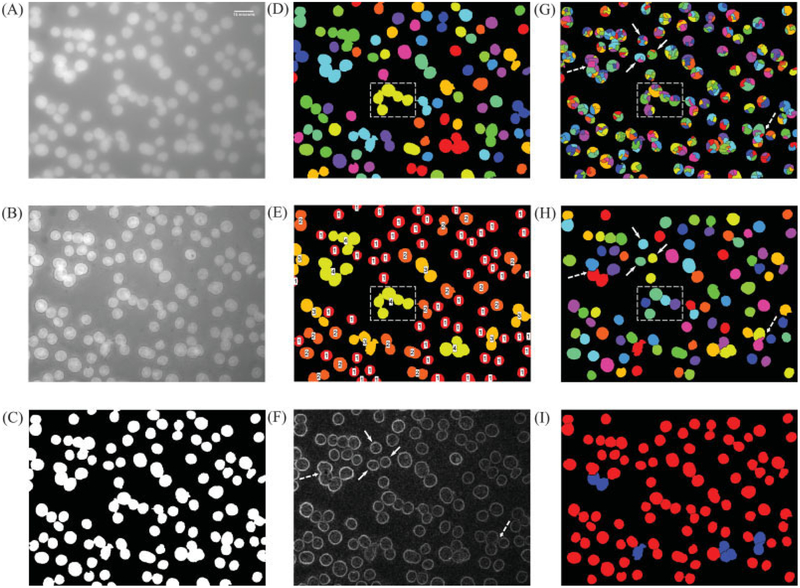
Figure 3 shows the step-by-step results from the proposed algorithm. Figure 3A shows the representative input image and was obtained by taking a maximum intensity projection of 3-D stack. Figure 3B shows the multiscale edge enhanced image (Ie) and Figure 3C shows the result of applying the MET on Ie. Clearly, the thresholding process does an excellent job at extracting all the nuclei present in the image. Figures 3D and 3E show the input (labeled thresholded objects) and the output (classification results from the K-means classifier) for the minimum nuclear size estimation step, respectively. Figure 3E also shows the class labels (1–4) overlaid on the thresholded objects and the minimum nuclear area was estimated from “class 1” objects (red colored objects). Figure 3G shows the results after applying watershed on the gradient magnitude of multiscale edge enhanced image (Fig. 3F). (Note: The thresholded binary image, Fig. 3B, was used a mask when applying watershed on the gradient magnitude of the MEE image.) In spite of the edge enhancement image, we can clearly notice the severe oversegmentation of all the foreground objects, both isolated (solid arrows in Figs. 3F-3G) and CN (dashed-rectangular box in Figs. 3D, 3E, and 3G). Figure 3H shows the results after applying the region-merging operation with size and depth constraints, and we can clearly notice that the oversegmented objects, both individual and CN, from the watershed operation are successfully merged into individual, well-separated objects (solid arrows and dashed-rectangular box in Fig. 3H, respectively). Figure 3I shows the final classification results from the multistage classifier, where the red- and blue-colored objects represent single/individual nuclei and CN, respectively.
Figure 3.
Results from various steps of the proposed algorithm. (A) 2-D image with DAPI stained nuclei, scale bar at top right corner = 15 μm; (B) reconstructed image after applying MEE; (C) output from the multiscale entropy-based thresholding of reconstructed image; (D) labeled objects from Figure 3B used for calculating the minimum nuclear area (objects along the image edges are ignored during the estimation process); (E) result of clustering the size of the labeled objects in Figure 3D into four classes using K-means; (F) gradient magnitude of the reconstructed image in Figure 2B; (G) result of watershed segmentation on the gradient magnitude image; (H) result of region merging based on maximum depth and minimum nuclear area constraints and after removing the edge nuclei; (I) result from multistage classifier (edge nuclei have been discarded), red color: single nuclei (Sa,SN); and blue color: clustered nuclei (Sa,CN). In Figures 3F–3H, solid-line arrow and dashed-line arrow point to an isolated nucleus and overlapping nuclei without any edge information between them and the effect of watershed and region merging, respectively. In Figures 3G–3H, dashed rectangle is used for illustrating the declustering of closely packed nuclei and corresponds to a clustered region in MET thresholded image (shown in Figs. 3D–3E).
Performance Assessment
Multiscale edge enhancement.
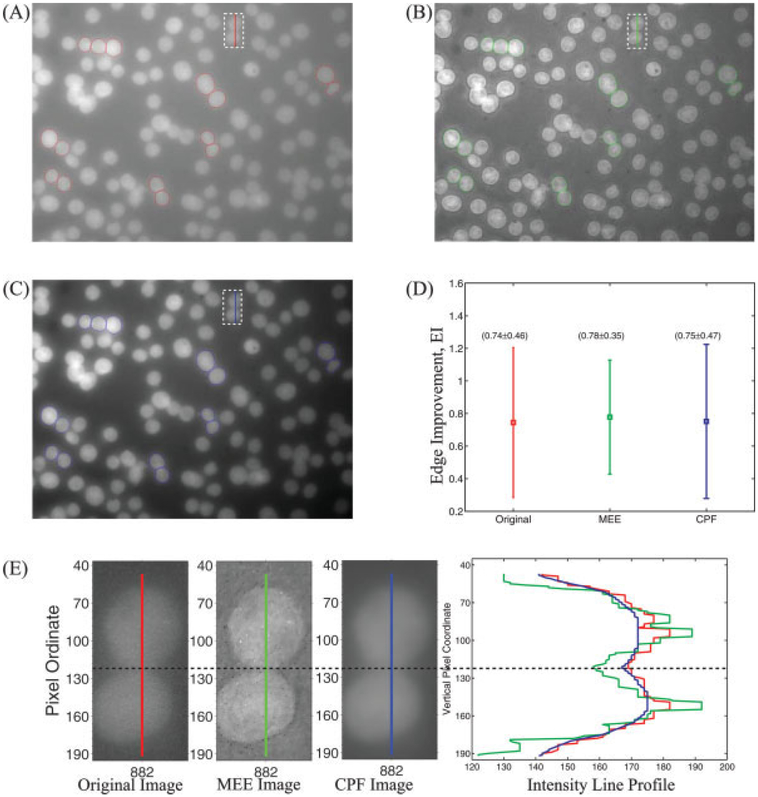
Figure 4 demonstrates the edge enhancement along the interface of clustered (closely packed) nuclei by comparing the edge improvement (EI) from Eq. (13) after MEE and corner-preserving filtering (CPF) (60). Figures 4A–4C show the selected nuclei (total of 15) overlaid on the original image (red color polygons), MEE image (green color polygons), and corner-preserving-filtered image (blue color polygons), respectively. Figure 4D shows the error bar (average ± standard deviation) for the EI calculated from original image, MEE image, and CPF image. We can clearly see that the EI values (average and standard deviation) for the original image and CPF image are similar (0.74 vs. 0.75), suggesting that CPF does not enhance the edges. On the contrary, for the MEE image the average value of EI increased 18% (from 0.74 for the original image to 0.78) along with a 24% decrease in the standard deviation (from 0.46 for the original image to 0.35), suggesting that that the MEE was able to enhance the contrast (intensity difference) between the foreground and the background. This can also be verified visually from Figure 4E, which compares the line profile across two (closely packed) nuclei from the original input image (vertical red line), the MEE image (vertical green line), and the CPF image (vertical blue line). The horizontal dashed line corresponds to region of separation between the upper and the lower nucleus. We can notice that the contrast (intensity difference) between background and foreground is best (at least visually) for the MEE image (green color profile in the right most graph in Fig. 4E).
Figure 4.
Comparison of edge enhancement from MEE and corner-preserving filtering (CPF). (A) Original image along with nuclei selected for analysis (red color polygons); (B) MEE image along with nuclei selected for analysis (green color polygons); (C) edge enhanced image after CPF along with nuclei selected for analysis (blue color polygons); (D) error-bar plot showing EI [Eq. (13)] for the nuclei selected from the original, MEE, and CPF images. (E) intensity line profiles from original, MEE, and CPF images across two closely packed nuclei selected from top-middle region (shown as a dashed rectangle with a vertical line in Figs. 4A–4C). The dashed horizontal line represents the approximate location where the two nuclei are separated.
Multiscale entropy thresholding and minimum nuclear area estimation.
Table 2 summarizes the performance of minimum nuclear size estimation, multiscale thresholding, and segmentation (watershed with region merging) steps. The proposed minimum nuclear size estimation methodology identified a Δmin value of 1,170 pixels (averaged over all seven datasets), which is twice the Δuser value of 550 pixels, suggesting that it was able to adaptively adjust the Δmin value, irrespective of the amount of clustering. For instance, datasets D3 and D6, which were derived from same cell line C3, have different extents of nuclei aggregation, but the average Δmin value for these datasets is almost identical (1,014 pixels and 1,023 pixels, respectively) because in Eq. (11) we use the size statistics of only “class 1” type objects, hence the extent of aggregation should have minimal effect on the estimation process. The MET on edge enhanced images appeared to perform consistently even in the presence of nonuniform illumination. The proposed multiscale entropy-based thresholding failed to extract 41 nuclei from the total available 4,181 nuclei with an unweighted average missing rate of 1.4 ± 1.01% per dataset (column MMET in Table 2).
Table 2.
Summary of results from estimation of minimum nuclear area and from watershed and region-merging algorithm
| DATASET | ΔMIN (PIXELS) | MMET (%) | MWRM (%) | MTOTAL (%) |
|---|---|---|---|---|
| D1 | 1,463 | 0.1 | 1.8 | 1.9 |
| D2 | 1,072 | 1.4 | 0.7 | 2.1 |
| D3 | 1,014 | 0.4 | 1.8 | 2.2 |
| D4 | 1,184 | 1.8 | 2.1 | 3.9 |
| D5 | 1,116 | 3.0 | 1.9 | 4.9 |
| D6 | 1,314 | 2.2 | 1.7 | 3.9 |
| D7 | 1,023 | 1.1 | 2.0 | 3.1 |
| Average | 1,169 | 1.4 | 1.7 | 3.1 |
Δmin = estimated minimum area for region merging; MMET = percent nuclei missed by multiscale entropy-based thresholding; MWRM = percent nuclei missed by watershed and region merging; and MTotal = percent nuclei missed by the proposed algorithm
Watershed and region merging.
The watershed with region-merging algorithm showed a total failure rate of 1.7% and performed consistently across all datasets with an average MWRM of 1.7 ± 0.47%. The segmentation step missed the least number of nuclei for dataset D2 with an MWRM of only 0.7% because majority of the input nuclei in this dataset were fairly well isolated (217 out of 295 nuclei). (Note that the MWRM failure rates indicates the nuclei missed by WRM only, thus the total failure rate is sum of MMET and MWRM.) When both the failure rates were combined, the proposed system failed to delineate 2.7% nuclei and had an average unweighted combined failure rate of 3.1 ± 1.1% per dataset.
In Table 3, we summarize the performance of declustering. The proposed algorithm successfully isolated 1,438 nuclei, i.e., declustering 73% of 1,973 CN. The algorithm’s best and worst performance, in terms of percent declustering, comes from datasets D3 and D5, respectively, in which it isolated 96 and 62% of CN. The algorithm isolated an unweighted average of 74.1 ± 10.8% CN per dataset and extracted 3,532 nuclei from a total of 4,181 available nuclei, resulting in a 0.84 yield ratio. The MEE certainly improved segmentation of CN when compared with our earlier work (8), in which, the algorithm declustered (unweighted average) 52.4 ± 36.6%. This indicates that the MEE renders the current approach insensitive to the degree of clustering and background intensity variations in the input image.
Table 3.
Algorithm’s declustering performance
| DATASET | PNCAFTER (%) | DECLUSTERING (%) |
|---|---|---|
| D1 | 12.3 | 73.8 |
| D2 | 6.8 | 74.4 |
| D3 | 1.0 | 95.9 |
| D4 | 8.9 | 72.1 |
| D5 | 21.1 | 61.6 |
| D6 | 16.9 | 75.0 |
| D7 | 22.6 | 65.8 |
| Average | 74.1 |
PNCafter = percent of nuclei that still are clustered after the segmentation [Eq. (14)].
Classification accuracy.
Table 4 summarizes the classification accuracy results for both classes, Sa,SN and Sa,CN, using three parameters: (a) the total number of objects that were correctly identified, TP; (b) total number of incorrectly identified objects, FP; and (c) positive predictive value, PPV. The multistage classifier identified SN and CN objects with an overall accuracy (PPV) of 99.8 and 93.6%, respectively, and with an average unweighted PPV of 99.8 ± 0.3% and 95.5 ± 5.1%, respectively.
Table 4.
Comparison of the output from automatic classification and manual assessment
| SN | CN | |||||
|---|---|---|---|---|---|---|
| DATASET | TP | FP | PPV | TP | FP | PPV |
| D1 | 1,390 | 4 | 99.7 | 96 | 10 | 90.6 |
| D2 | 268 | 0 | 100.0 | 9 | 1 | 90.0 |
| D3 | 485 | 0 | 100.0 | 2 | 0 | 100.0 |
| D4 | 334 | 3 | 99.1 | 14 | 0 | 100.0 |
| D5 | 197 | 0 | 100.0 | 24 | 0 | 100.0 |
| D6 | 425 | 0 | 100.0 | 44 | 5 | 89.8 |
| D7 | 416 | 1 | 99.8 | 59 | 1 | 98.3 |
| Sum = 3,515 | Sum = 8 | Average = 99.8 | Sum = 248 | Sum = 17 | Average = 95.5 | |
SN = class of single nucleus objects; CN = class of clustered nuclei objects; TP = true positives; FP = false positives; and PPV = positive predictive value [Eq. (15)].
Segmentation accuracy.
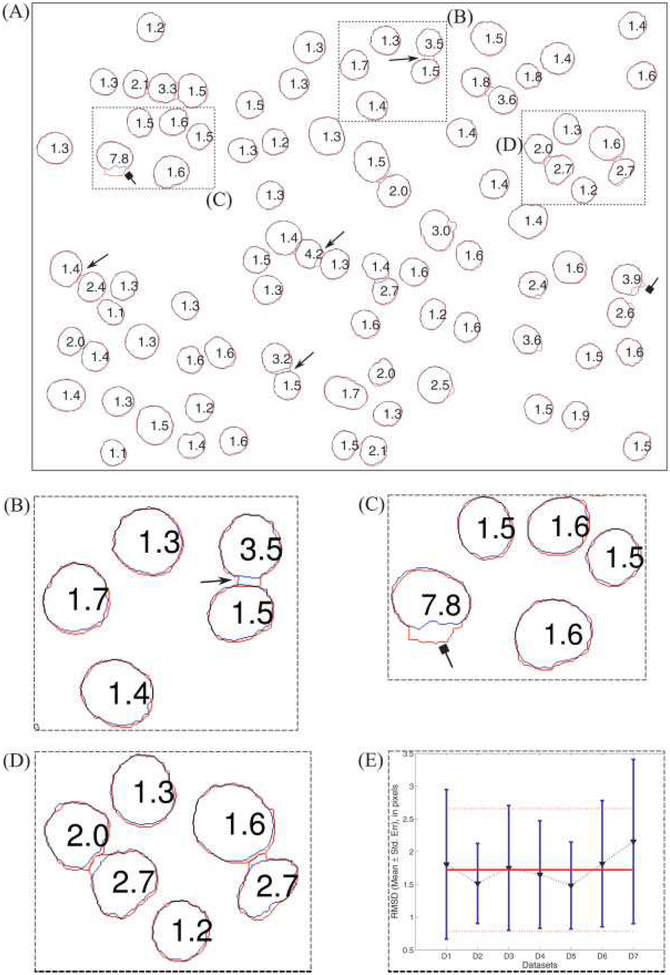
We analyzed the deviation between automatic and manual nuclei boundaries using the RMSD measure [Eq. (17)]. Figure 5A shows the overlay of the nuclei boundaries obtained from the proposed algorithm (Sa,SN, red color) and from the manual segmentation (Sm’SN, blue color) along with the RMSD value, in pixels, for each nucleus in the representative sample image (Fig. 3A). Figures 5B–5D shows the enlargement of regions B–D in Figure 5A. In general, segmentation of nuclei from the proposed method appears to be in good agreement with manual ground truth with approximately 2 pixels RMSD values. However, on close inspection, we found two cases resulting in larger RMSD values than the average RMSD value, viz. (1) when two nuclei are close to one another, then the boundary of one nucleus tends to “jump across” the interstitial space between the two (shown as lines with arrow heads in the Figs. 5A–5B); and (2) when more than two nuclei are closely packed and part of this cluster gets segmented into objects of both Sa,SN and Sa,CN (shown as lines with square heads in Figs. 5A and 5C). Figure 5E shows the average value of RMSD (mean ± standard deviation) for all Sa,SN nuclei in each dataset (blue lines) and the average value of RMSD (mean ± standard deviation) for all Sa,SN nuclei from all seven datasets (horizontal red lines). We can clearly see that algorithm works consistently across all datasets, irrespective of background noise and aggregation, and the average RMSD for all seven test datasets is 1.75 ± 0.94 pixels (i.e., 0.23 ± 0.13 μm). This suggests that the segmentation boundary from our method is acceptable and close to the limits of blurring that could occur because of the point spread function of the optical system.
Figure 5.
Quantitative comparison of the boundaries from automatic segmentation and manual segmentation. (A) The red and green color borders correspond to automatic and manual boundaries, respectively. The numbers inside each object is the RMS deviation (in pixels). (B)–(D) zoomed-in version of regions (B), (C), and (D) in Figure 5A. The arrows with triangular heads point to the “jump across” effect and the arrows with square heads point to nuclei with large RMSD because of improper breakage of a cluster. (E) RMSD (in pixels) for each dataset, triangular marker points are the mean values the blue bars denote ± 1 standard deviation. The solid red line is the average RMSD (1.75 pixels) for nuclei from datasets and the dashed red lines (above and below) the solid line denote the ± 1 standard deviation (0.94 pixels).
Sensitivity analysis of input parameters.
One of our design objectives was to automatically extract the input parameters. The proposed algorithm requires four main parameters, viz. maximum depth (dmax), minimum area (Δuser), number of DWT levels (j), and stretching factor (k). (The number of iterations used by the MEE algorithm is not an input parameter. In the Mallat-Zhong alternating projection algorithm (54), the number of iterations is used as a stopping criterion in the conjugate gradient method. Hence, it is only logical to use a reasonable number of iterations and results do not improve if more iterations are used.)
On the basis of our preliminary investigations, we found that after a certain depth, dmax, the final outcome from region-merging algorithm did not change when the area constraint was fixed (data not shown). Next, although the algorithm uses Δuser in certain operations (like, starting tile size for MET), it is mainly used as “fail safe” parameter for region-merging operation to handle those cases when the assumptions underlying our data-driven Δmin estimation approach are not met. Therefore, any reasonable values of Δuser and dmax should still generate reproducible results.
We analyzed the effect of number of DWT levels (j) and stretching factor (k) on downstream steps by keeping the remaining input parameters identical to those used for our test datasets. The results for a representative fluorescence image (Fig. 2A) are summarized in Table 5. This input image had a total of 96 nuclei (ignoring the nuclei along the edges), of which 41 nuclei were clustered. These nuclei occupied 20.9% of the total cross-sectional area. The starting tile size was 16 × 16 pixels and the MET successfully extracted almost all nuclei. Also, the Δmin value remained constant (approximately 1,725 pixels) across all six combinations of j and k. This is expected because MEE enhances the contrast between edges of closely packed objects (nuclei), and therefore should have minimal impact on Δmin estimation process. Thus, the number of nuclei that remain as clusters is the only basis for comparing the performance. The results indicate that the performance is only affected when either fewer dyadic scales (row 1, Table 5) or a smaller stretching factor (row 3, Table 5) is used for reconstruction. In the former case, using only the first few dyadic scales, the reconstruction process amplifies false edges along with the true boundaries. This, in turn, leads to oversegmentation of both isolated and CN. The region-merging step should be able to handle the oversegmentation of isolated nuclei; however, this will also adversely merge the oversegmented clustered objects. In the latter case, when a smaller stretching factor was used, the edges of objects in higher scale (external boundaries of CN) were enhanced similar to the edges in lower scale (boundaries of individual nuclei within the cluster) because MEE uniformly stretches the gradient magnitude of multiscale edges. Naturally, this will cause the watershed lines (catchment basins) to enclose the external boundary of the cluster. Thus, the algorithm will fail to effectively decluster the overlapping/CN.
Table 5.
Effect of the number of DWT levels (j) and stretching factor (k) used in multiscale edge enhancement on intermediate results for image shown in Figure 2A
| SN | CN | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| (j, k) | ΔMIN (PIXELS) | MMET (%) | MWRM (%) | TP | FP | NNC | TP | FP | NNC |
| (2,4) | 1,675 | 1 | 0 | 86 | 0 | 0 | 4 | 0 | 8 |
| (4,4) | 1,682 | 1 | 1 | 90 | 0 | 0 | 2 | 0 | 4 |
| (6,2) | 1,701 | 1 | 0 | 80 | 0 | 0 | 7 | 0 | 15 |
| (6,4) | 1,721 | 0 | 0 | 90 | 0 | 0 | 2 | 2 | 4 |
| (6,8) | 1,728 | 0 | 0 | 94 | 0 | 0 | 1 | 0 | 2 |
| (8,4) | 1,723 | 0 | 1 | 93 | 0 | 0 | 1 | 0 | 2 |
Row 4 (6 DWT levels and k = 4) represents the values used for analyzing all datasets: NNC = number of nuclei in clusters.
Discussion
Recent investigations of genomic organization show overwhelming evidence of nonrandom arrangement of subnuclear entities in human genome and its correlation to carcinogenesis (7,10). Current studies on genomic organization manually segment interphase cell nuclei with FISH-labeled DNA sequences and analyze the relative and radial distance distributions of the FISH signal. It is evident that this application requires development of automatic and highly accurate cell nuclei segmentation for a meaningful, unbiased, and reproducible analysis. Such automated techniques coupled with advanced statistical models can provide vital understanding of genomic organization and assist development of better diagnostic tools for detecting cancer (61).
In recent years, the availability of high-content and high-throughput image acquisition systems has prompted the development of several modular, model-based image analysis platforms for cytometry applications (17,39,46,62,63). Model-based segmentation methods, particularly region-growing and region-methods, which use a priori knowledge of the input images, have shown considerable promise for automatic nuclei segmentation. However, we observed that most region-growing and region-merging methods were ill suited for studying genomic organization because their performance suffered when the nuclei were severely clustered and had weak edge information. The presence of nonuniform background, like in our datasets, also affected their performance. Preprocessing the input images to restore/enhance the objects of interest using techniques like deconvolution and background correction may partially alleviate these problems. But such techniques are often time-consuming and complicated, and they do not always give satisfactory results.
Our main objective was to develop a high-throughput application for segmenting clustered and isolated nuclei from fluorescence images with extremely high accuracy and high specificity, if necessary at the cost of lower yield ratio (sensitivity). Given the advantages of the model-based approaches, we developed an automatic segmentation algorithm combining well-known, high-level image processing operations in a modular framework to make the segmentation process extremely flexible and suitable for batch-processing large volumes of datasets. In this work, we used wavelet-based edge enhancement for increasing the contrast along the edges of CN and multiscale entropy-based technique for thresholding the foreground from nonuniformly illuminated fluorescence images. Furthermore, we also used data-driven approaches from statistical pattern recognition to estimate certain input parameters, thus eliminating, at least partially, the need for user intervention.
The proposed algorithm’s modular framework is similar to the “pipeline” framework used in the CellProfiler by Carpenter et al. (62). CellProfiler’s generic image analysis platform allows the user to carry out quantitative image analysis using the “pipeline” strategy (a combination of various well-established image-processing steps) and test them on a representative image prior to batch-processing dataset(s). CellProfiler bundles existing (like, MATLAB Image Processing Toolbox, Mathworks, Natick, MA) and research-based image processing algorithms, thus permitting rapid prototyping. But our target application required statistical pattern recognition tools in addition to the modifications of several low-level image processing algorithms (e.g., MEE and multiscale thresholding), hence CellProfiler could not accomplish our task and thus we designed and built our application.
Our approach also shares some similarity to the multimodel approach described by Lin et al. (63). They also use similar steps (region-based segmentation, model-based object merging, model-selection, and classification) in their work to simultaneously segment and classify heterogeneous populations of cell nuclei. However, unlike our approach, Lin et al. incorporate user interaction and the tree-based region-merging approach to permit visual control of the quality of segmentation. Our method achieves better classification accuracy (PPV of 99.8% compared with 93.5%), and the segmentation accuracy (~1.75 pixels RMS deviation) is also within acceptable limits. Our approach can also segment and classify heterogeneous populations by changing the number of classes in our final multistage classification step.
Despite the problems of clustering and nonuniform illumination in our datasets, the proposed algorithm performed extremely well on all datasets. The combined failure rate from these steps was only 2.7% of total nuclei (114 out of 4,181), with an unweighted average failure rate of 3.1 ± 1.1% per dataset. Although the percentage of nuclei missed for datasets D4, D5, and D6 is slightly higher than the average failure rate, it should not be a major concern for our target application because it is relatively easy to acquire more images.
The proposed algorithm extracted 3,532 nuclei from a total of 4,181 available nuclei, resulting in a 0.84 yield ratio. Our yield ratio is relatively lower than the 0.95 yield ratio reported, independently, by Harder et al. (20) and Beaver et. al (64) for their adaptive thresholding methods. However, the former report showed no evidence of nuclear clustering and uneven background in their illustrated test image, and the latter assumed that the nuclei in their images are reasonably well separated. Our method makes no such assumptions and yet achieves a reasonable yield percentage even under nonuniform background and clustering conditions. Moreover, in our target application, accuracy of the segmented boundary and positive predictive value are more important than higher yield because, as mentioned earlier, it is relatively easy to acquire more images. The approach was also robust enough to extract large number of individual nuclei from clusters, mostly because of MEE. On an average, the algorithm was able to reduce the clustering by 74%, which is better than our previous effort of 52% (8) based on a similar framework except for the MEE.
The multistage classifier detected SN and CN with an overall accuracy (positive predictive value) of 99.8 and 93.8%, respectively. The PPV for SN class nuclei is comparable with the best results reported by Chen et al. (17). They obtained a similar overall PPV (~99%) for SN using a 28 feature measurements and by training their classifier with 300 sample nuclei. On the contrary, our algorithm achieves this high accuracy using only six features and requires no training datasets because we use a multistage classifier for refining the classification results. We get a (slightly) lower PPV for CN objects, but it is still reasonably high (>93%). Moreover, a slightly lower PPV for CN objects should not be a concern for two reasons: (1) these objects can be omitted from further analysis and (2) overall we are losing only 17 nuclei (sum of FP for CN) because of the misclassification.
Our algorithm, just as any other approaches, is not a perfect system and has some shortcomings. First, the MEE algorithm, albeit very effective at enhancing the edges, will fail to enhance the edges of overlapping nuclei with minimal or no edge information (dashed arrows in Fig. 3F). This can adversely affect the performance and effectiveness of downstream operations. For instance, in scenarios when the overlapping nuclei has no boundary information in the image (dashed arrows in Fig. 3F), the watershed operation can incorrectly label catchment basin(s) or fragment(s) by spanning them across more than one nucleus (dashed arrows in Fig. 3G) and the merging process will fail to yield visually and morphologically accurate results because of the increased likelihood of merging fragment(s) from one object to the fragments from its neighbor(s) (dashed arrows in Fig. 3H). In this context, it would be interesting to investigate the performance of rule-based and morphological constraint based region-merging approaches (15,42,43). Second, in the MEE, we use a constant stretching factor (scalar) across all the scales, and this was not a major problem for our datasets because most of our nuclei had similar morphology. However, if we were to segment objects of different sizes then using a scalar valued stretching factor will selectively enhance only one type of object. A better alternative would be to use different stretching factors for each scale. Third, our region-merging approach can lead to inaccurate segmentation of closely packed nuclei where the boundary can “jump across” the interstitial space between the nuclei (line with arrow heads in Figs. 5A and 5B). Conceivably, these discrepancies could be automatically identified and handled by using better rule-based merging constraints and dynamic-programming-based methods (31). Last, our proposed algorithm is not entirely parameter free, but the effect of each input parameter on the segmentation process is easy to understand and not particularly sensitive. We use simple pattern recognition tools for parameter estimation and relieve the user from extensive interaction. In future, we envision the new generation algorithms to incorporate similar or more advanced statistical pattern recognition techniques for prediction and estimation of input parameters.
In conclusion, the proposed high-throughput algorithm shows great potential for being an important image processing tool for analyzing the complex spatial organization of genes and its role in carcinogenesis. We expect to obtain useful biological results about the spatial organization of specific FISH-labeled DNA sequences in interphase nuclei using the methods described herein (results will be reported separately). Because of its modularity, parts of the algorithm can be either combined or replaced with other image processing techniques in a seamless fashion, thus, enabling the approach suitable for other dataset(s). Hence, in spite of designing the algorithm based on the requirements of a specific application, we anticipate other cytometry applications requiring accurate cell nuclei segmentation to benefit from this work. In future studies, we will analyze the effect of segmentation accuracy on the outcome of spatial analysis for DNA sequences. Furthermore, we plan to analyze the effect of scale-dependent stretching factor (k will be a vector of length equal to number scales) and to explore the potential of second generation wavelets, like edgelets, beamlets, and contourlets (65), for analyzing 3-D stacks of fluorescence images. We also plan to use this application for studying heterogeneous populations of cell nuclei and use deformable boundary-fitting algorithms (22) and dynamic-programming-based methods (31) for handling irregular shaped nuclei.
Offering the broadest portfolio of peripheral blood controls for immunophenotyping by flow cytometry.
Offering a wide portfolio of intracellular and surface marker controls for flow cytometry
Unique controls for normal and abnormal leukocyte populations
Ready to use controls with no dilution required
Reference values similar to those found in relevant blood types
Covering CD markers for TBNK, Stem Cells, and Leukemia and Lymphoma
Acknowledgments
We thank Ty Voss and Tatiana Karpova from LRBGE, NCI, for their help with image acquisition and the Delft University, Netherlands, for providing DIPImage and PRTools toolboxes.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was also supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Parts of the method described in this article were presented at the Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues V, Photonics West (SPIE) in San Jose, CA, USA, in January 2007.
This article is a US government work and, as such, is in the public domain in the United States of America.
Literature Cited
- 1.Gasser SM. Positions of potential: Nuclear organization and gene expression. Cell 2001;104:639–642. [DOI] [PubMed] [Google Scholar]
- 2.Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. Chromosome dynamics in the yeast interphase nucleus. Science 2001;294:2181–2186. [DOI] [PubMed] [Google Scholar]
- 3.Oliver B, Misteli T. A non-random walk through the genome. Genome Biol 2005;6: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pederson T Gene territories and cancer. Nat Genet 2003;34:242–243. [DOI] [PubMed] [Google Scholar]
- 5.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuc-lear compartmentalization of immunoglobulin loci during lymphocyte development. Science 2002;296:158–162. [DOI] [PubMed] [Google Scholar]
- 6.Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet 2003;34:287–291. [DOI] [PubMed] [Google Scholar]
- 7.Meaburn KJ, Misteli T, Soutoglou E. Spatial genome organization in the formation of chromosomal translocations. Semin Cancer Biol 2007;17:80–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudla PR, Collins J, Meaburn KJ, Misteli T, Lockett SJ. HiFLO: A high-throughput system for spatial analysis of FISH loci in interphase nuclei In: Farkas DL, Leif RC, Nicolau DV, editors. Proceedings of SPIE. Vol. 6441, Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues V. San Jose, CA: SPIE; 2007. p 644119 (1–9). [Google Scholar]
- 9.Knowles D, Ortiz de Solorzano C, Jones A, Lockett SJ.Analysis of the 3D spatial organization of cells and subcellular structures in tissue In: Farkas DL, Leif RC, editors. Optical Diagnostics of Living Cells III. Volume 3921, Proceedings of SPIE. San Jose, CA, USA: SPIE; 2000. p 66–73. [Google Scholar]
- 10.Meaburn KJ, Misteli T. Cell biology: Chromosome territories. Nature 2007;445:379–781. [DOI] [PubMed] [Google Scholar]
- 11.Gudla PR.Texture-based segmentation and finite element mesh generation for heterogeneous biological image data, Ph.D. dissertation. College Park: University of Maryland; 2005. [Google Scholar]
- 12.Lockett SJ, Herman B. Automatic detection of clustered, fluorescent-stained nuclei by digital image-based cytometry. Cytometry 1994;17:1–12. [DOI] [PubMed] [Google Scholar]
- 13.Fang B, Hsu W, Lee ML. On the accurate counting of tumor cells. IEEE Trans Nano-biosci 2003;2:94–103. [DOI] [PubMed] [Google Scholar]
- 14.Al-Awadhi F, Jennison C, Hurn M. Statistical image analysis for a confocal microscopy two-dimensional section of cartilage growth. J Roy Stat Soc Appl Stat 2004; 53:31–49. [Google Scholar]
- 15.Pinidiyaarachchi A, Wahlby C. Seeded watersheds for combined segmentation and tracking of cells ICIAP 2005: International Conference on Image Analysis and Processing. Volume 3617, Lecture notes in computer science. Cagliari, Italy: Springer Berlin/Heidelberg, 2005. p 336–343. [Google Scholar]
- 16.Sage D, Neumann FR, Hediger F, Gasser SM, Unser M. Automatic tracking of individual fluorescence particles: Application to the study of chromosome dynamics. IEEE Trans Image Process 2005;14:1372–1383. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Zhou X, Wong ST. Automated segmentation, classification, and tracking of cancer cell nuclei in time-lapse microscopy. IEEE Trans Biomed Eng 2006;53:762–766. [DOI] [PubMed] [Google Scholar]
- 18.Kellogg RA, Chebira A, Goyal A, Cuadra PA, Zappe SF, Minden JS, Kovacevic J.Towards an Image Analysis Toolbox for High-Throughput Drosophila Embryo RNAi Screens 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI 2007). Arlington, VA: IEEE; 2007. p 288–291. [Google Scholar]
- 19.Dow AI, Shafer SA, Kirkwood JM, Mascari RA, Waggoner AS. Automatic multiparameter fluorescence imaging for determining lymphocyte phenotype and activation status in melanoma tissue sections. Cytometry 1996;25:71–81. [DOI] [PubMed] [Google Scholar]
- 20.Harder N, Neumann B, Held M, Liebel U, Erfle H, Ellenberg J, Eils R, Rohr K. Auto-mated Recognition of Mitotic Patterns in Fluorescence Microscopy Images of Human Cells 3rd IEEE International Symposium on Biomedical Imaging: Nano to Macro. Arlington, VA: IEEE; 2006. p 1016–1019. [Google Scholar]
- 21.Gue M, Messaoudi C, Sun JS, Boudier T. Smart 3D-FISH: Automation of distance analysis in nuclei of interphase cells by image processing. Cytometry Part A 2005; 67A:18–26. [DOI] [PubMed] [Google Scholar]
- 22.Xu C, Prince JL. Snakes, shapes, and gradient vector flow. IEEE Trans Image Process 1998;7:359–369. [DOI] [PubMed] [Google Scholar]
- 23.Belien JA, van Ginkel HA, Tekola P, Ploeger LS, Poulin NM, Baak JP, van Diest PJ. Confocal DNA cytometry: A contour-based segmentation algorithm for automated three-dimensional image segmentation. Cytometry 2002;49:12–21. [DOI] [PubMed] [Google Scholar]
- 24.Adiga PS. Segmentation of volumetric tissue images using constrained active contour models. Comput Methods Programs Biomed 2003;71:91–104. [DOI] [PubMed] [Google Scholar]
- 25.Osher S, Sethian JA. Fronts propagation with curvature-dependent speed: Algorithms based on Hamilton-Jacobi formulations. J Comput Phys 1988;79:12–49. [Google Scholar]
- 26.Sethian JA. Level Set Methods: Evolving Interfaces in Geometry, Fluid Mechanics, Computer Vision, and Material Sciences. Cambridge, UK: Cambridge University Press; 1999. [Google Scholar]
- 27.Ortiz de Solorzano C, Malladi R, Lelieve SA, Lockett SJ. Segmentation of nuclei and cells using membrane related protein markers. J Microsc 2001;201:404–415. [DOI] [PubMed] [Google Scholar]
- 28.Zimmer C, Olivo-Marin JC. Coupled parametric active contours. IEEE Trans Pattern Anal Machine Intell 2005;27:1838–1842. [DOI] [PubMed] [Google Scholar]
- 29.Geusebroek J, Smeulders A, Geerts H. A minimum cost approach for segmenting networks oflines. Int J ComputVis 2001;43:99–111. [Google Scholar]
- 30.Appleton B, Talbot H. Globally minimal surfaces bycontinuous maximal flows. IEEE Trans Pattern Anal Machine Intell 2006;28:106–118. [DOI] [PubMed] [Google Scholar]
- 31.Baggett D, Nakaya MA, McAuliffe M, Yamaguchi TP, Lockett S. Whole cell segmentation in solid tissue sections. Cytometry Part A 2005;67A:137–143. [DOI] [PubMed] [Google Scholar]
- 32.McCullough DP, Gudla PR, Harris BS, Collins JA, Meaburn KJ, Nakaya M-A, Yama-guchi TP, Misteli T, Lockett SJ. Segmentation of whole cells and cell nuclei from 3D optical microscope images using dynamic programming. IEEE Trans Med Imaging 2007. doi: 10.1109/TMI.2007.913135 (published online). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartesaghi A, Sapiro G, Subramaniam S. An energy-based three-dimensional segmentation approach for the quantitative interpretation of electron tomograms. IEEE Trans Image Process 2005;14:1314–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boykov Y, Funka-Lea G. Graph cuts and efficient N-D image segmentation. Int J Comput Vis 2006;70:109–131. [Google Scholar]
- 35.Beucher S, Lantuejoul C.Use of watershed in contour detection International Workshop on Image Processing, Real-Time Edge and Motion Detection/Estimation. Rennes, France: 1979. [Google Scholar]
- 36.Meyer F, Beucher S. Morphological segmentation. J Vis Commun Image Represent 1990;1:21–46. [Google Scholar]
- 37.Vincent L, Soille P. Watersheds in digital spaces: An efficient algorithm based on immersion simulations. IEEE Trans Pattern Anal Machine Intell 1991;13:583–598. [Google Scholar]
- 38.Bleau Andre, Leon LJ. Watershed-based segmentation and region merging. Comput Vis Image Understanding 2000;77:317–370. [Google Scholar]
- 39.Raman S, Maxwell CA, Barcellos-Hoff MH, Parvin B. Geometric approach to segmentation and protein localization in cell culture assays. J Microsc 2007;225:22–30. [DOI] [PubMed] [Google Scholar]
- 40.Beare R A locally constrained watershed transform. IEEE Trans Pattern Anal Machine Intell 2006;28:1063–1074. [DOI] [PubMed] [Google Scholar]
- 41.Strasters KC, Jan JG. Three-dimensional image segmentation using a split, merge and group approach. Pattern Recognit Lett 1991;12:307–325. [Google Scholar]
- 42.Adiga U, Malladi R, Fernandez-Gonzalez R, Ortiz de Solorzano C. High-throughput analysis of multispectral images of breast cancer tissue. IEEE Trans Image Process 2006;15:2259–2268. [DOI] [PubMed] [Google Scholar]
- 43.Adiga PSU, Chaudhuri BB. An efficient method based on watershed and rule-based merging for segmentation of 3-D histopathological images. Pattern Recognit 2001;34:1449–1458. [Google Scholar]
- 44.Lin G, Adiga U, Olson K, Guzowski JF, Barnes CA, Roysam B. A hybrid 3D watershed algorithm incorporating gradient cues and object models for automatic segmentation of nuclei in confocal image stacks. Cytometry Part A 2003;56A:23–36. [DOI] [PubMed] [Google Scholar]
- 45.Tscherepanow M, Zollner F, Kummert F. Automatic segmentation of unstained living cells in bright-field microscope images In: Perner P, editor. Workshop proceedings: workshop on mass-data analysis of images and signals (MDA 2006). Leipzig, Germany: IBaI CD-Report (ISSN 1671–2671); 2006. p 86–95. [Google Scholar]
- 46.Lin G, Chawla MK, Olson K, Guzowski JF, Barnes CA, Roysam B. Hierarchical, model-based merging of multiple fragments for improved three-dimensional segmentation of nuclei. Cytometry Part A 2005;63A:20–33. [DOI] [PubMed] [Google Scholar]
- 47.Wahlby C, Sintorn IM, Erlandsson F, Borgefors G, Bengtsson E. Combining intensity, edge and shape information for 2D and 3D segmentation of cell nuclei in tissue sections. J Microsc 2004;215(Part 1):67–76. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen HT, Marcel W, Rein van den B. Watersnakes: Energy-driven watershed segmentation. IEEE Trans Pattern Anal Machine Intell 2003;25:330–342. [Google Scholar]
- 49.Cannell MB, McMorland A, Soeller C. Image enhancement by deconvolution In: Pawley JB, editor. Handbook of Biological Confocal Microscopy, 3rd ed. New York: Springer Science; 2006. p 488–500. [Google Scholar]
- 50.Chang SK. Principles of Pictorial Information Systems Designs. Upper Saddle River, NJ: Prentice-Hall; 1989. [Google Scholar]
- 51.Pitas I Digital Image Processing Algorithms and Applications. New York: Wiley; 2000. [Google Scholar]
- 52.Mallat S Zero-crossing of a wavelet transform. IEEE Trans Inf Theory 1991;37:1019–1033. [Google Scholar]
- 53.Mallat S A Wavelet Tour of Signal Processing. San Diego, CA: Academic Press; 1999. [Google Scholar]
- 54.Mallat SG, Zhong S. Characterization of signals from multiscale edges. IEEE Trans Pattern Anal Machine Intell 1992;14:710–732. [Google Scholar]
- 55.Zhong S Signal representation from wavelet transform maxima, Ph.D. dissertation. New York: New York University; 1990. [Google Scholar]
- 56.Lu J Signal recovery and noise reduction with wavelets, Ph.D. dissertation. Hanover, NH: Dartmouth College; 1993. [Google Scholar]
- 57.Otsu N A threshold selection method from gray-level histograms. IEEE Trans Syst ManCybern 1979;9:62–66. [Google Scholar]
- 58.Price JH, Hunter EA, Gough DA. Accuracy of least squares designed spatial FIR filters for segmentation of images of fluorescence stained cell nuclei. Cytometry 1996; 25:303–316. [DOI] [PubMed] [Google Scholar]
- 59.Bacry E LastWave. Available http://www.cmap.polytechnique.fr/bacry/LastWave/index.html. Accessed 2004.
- 60.El-Fallah AI, Ford GE. Mean curvature evolution and surface area scaling in image filtering. IEEE Trans Image Process 1997;6:750–753. [DOI] [PubMed] [Google Scholar]
- 61.Meaburn KJ, Misteli T. Locus-specific and activity-independent gene repositioning during early tumorigenesis. J Cell Biol 2008;180:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol 2006; 7:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin G, Chawla MK, Olson K, Barnes CA, Guzowski JF, Bjornsson C, Shain W, Roy-sam B. A multi-model approach to simultaneous segmentation and classification of heterogeneous populations of cell nuclei in 3D confocal microscope images. Cytometry Part A 2007;71A:724–736. [DOI] [PubMed] [Google Scholar]
- 64.Beaver W, Kosman D, Tedeschi G, Bier E, McGinnis W, Freund Y. Segmentation of Nuclei in Confocal Image Stacks Using Performance Based Thresholding 4th IEEE International Symposium on Biomedical Imaging: Nano to Macro. Arlington, VA: IEEE; 2007. pp 1016–1019. [Google Scholar]
- 65.Donoho DL, Huo X. Combined image representation using edgelets and wavelets In: Unser MA, Aldroubi A, Laine AF, editors. Proceedings of SPIE. Volume 3813, Wavelet Applications in Signal and Image Processing VII. Denver, CO: SPIE; 1999. p 468–476. [Google Scholar]