Abstract
Yak is one of the largest native mammalian species at the Himalayas, the highest plateau area in the world with an average elevation of >4,000 m above the sea level. Yak is well adapted to high altitude environment with a set of physiological features for a more efficient blood flow for oxygen delivery under hypobaric hypoxia. Yet, the genetic mechanism underlying its adaptation remains elusive. We conducted a cross-tissue, cross-altitude, and cross-species study to characterize the transcriptomic landscape of domestic yaks. The generated multi-tissue transcriptomic data greatly improved the current yak genome annotation by identifying tens of thousands novel transcripts. We found that among the eight tested tissues (lung, heart, kidney, liver, spleen, muscle, testis, and brain), lung and heart are two key organs showing adaptive transcriptional changes and >90% of the cross-altitude differentially expressed genes in lung display a nonlinear regulation. Pathways related to cell survival and proliferation are enriched, including PI3K-Akt, HIF-1, focal adhesion, and ECM–receptor interaction. These findings, in combination with the comprehensive transcriptome data set, are valuable to understanding the genetic mechanism of hypoxic adaptation in yak.
Keywords: yak, high altitude adaptation, hypoxia, transcriptome
Introduction
Tibetan plateau is the highest plateau in the world with an average elevation of 4,000 m above the sea level, and is thus an ideal natural laboratory for studying the biological mechanism of physiological adaptions to high altitude hypoxia in human and animals. Currently there are ∼5 million native Tibetans permanently reside on the Tibetan Plateau as agro-pastoralists managing yak pastoralism and barley cultivation. Therefore, Tibetans and domestic yaks are two ideal models native to the highland for high altitude adaptation studies given their long-term colonization and widespread distribution across the plateau.
Yak (Bos grunniens) is one of the largest native highland mammals on the Tibetan Plateau with a population size of over 14 million, and distributes alongside the Tibetan pastoralists across the plateau with elevations ranging from 3,000 to 5,000 m (Zhang 1989; Wiener et al. 2003). The unearthed fossil records suggest a wide distribution of the progenitor of yak across north-eastern Eurasia ∼2 Ma (Zhang 1989; Wiener et al. 2003), while the domestication of yak is believed to be a relatively recent event that likely took place subsequent to the introduction of domestication technology into the plateau by the Neolithic agriculturalists ∼10,000–7,000 years (Qi et al. 2013), and since then the domestic yak became the most important livestock for Tibetan highlanders and dispersed across the plateau following the migration of native Tibetans (Qi 2004; Guo et al. 2006; Qi et al. 2008, 2013, 2014, 2015). Currently, the great majority of yaks raised on the plateau are domestic whereas the wild ones are confined to remote areas of the plateau with a rather small population size of <20,000 (Schaller and Liu 1996).
Compared with lowland cattle, domestic yaks have developed physiological features adapted to high altitude hypoxia such as larger pulmonary alveolar area per unit area, thinner alveolar septum, thinner blood–air barrier (Wei and Yu 2008), and smooth muscles within the arteriole wall of the micro-artery with diameter of <50 µm whereas lowland cattle do not have such a structure (Zhou et al. 2015). The native highlander Tibetans also acquired similar physiological features such as greater hypoxic and hypercapnic ventilatory responsiveness, larger lungs, better lung function, greater lung diffusion capacity, minimal hypoxic pulmonary hypertension, higher levels of exhaled nitric oxide, and lower levels of hemoglobin concentration (Wu and Kayser 2006). These physiological features may facilitate a more efficient blood flow for oxygen delivery under hypobaric hypoxia. In addition, both domestic yak and Tibetans maintain a normal blood hemoglobin level in comparison with lowland cattle (Zhang et al. 1994) and lowland Han population (Peng et al. 2017), respectively, suggesting that the highland Tibetans and domestic yak likely took similar strategies for protecting them from the high altitude polycythemia (red cell overproduction).
Despite the importance of domestic yak in studying high altitude adaptation, few comparative studies have been conducted in part due to the lack of lowland controls because domestic yaks have been confined to a relatively small zones with altitude ranging from 3,000 to 5,000 m above the sea level. For this reason, published studies have been mainly focusing on the functional analyses of genome-wide adaptive changes identified by genomic sequence comparison between highland Tibetans and lowland Han populations (Beall et al. 2010; Bigham et al. 2010; Simonson et al. 2010; Yi et al. 2010; Peng et al. 2011; Xu et al. 2011), and successfully identified EPAS1 and EGLN1 as two key genes involved in the maintenance of normal hemoglobin concentration of Tibetans under a high altitude hypoxic environment (Xiang et al. 2013; Peng et al. 2017), while genes underlying the adaptation to hypoxia in yaks seem different from humans (Qiu et al. 2015). Compared with yaks, humans are much recent inhabitants on the Tibetan Plateau based on genetic and archaeological evidence (13,000–30,000 years ago) (Zhao et al. 2009; Peng et al. 2011; Qi et al. 2013; Meyer et al. 2017). It is thus likely that natural selection had acted on a different set of genes/pathways in yaks, which is yet to be uncovered.
In addition to genome-wide adaptive changes at the DNA sequence level, gene expression as an intermediate phenotype connecting DNA sequences and physiological traits is highly informative in revealing molecular pathways/networks involved in genetic adaptation. However, the contribution of the genome-wide transcriptomic changes to high altitude adaptation remain undetermined due to the lack of highlander native human tissue samples. In the case of unavailability of highland Tibetan tissue samples, domestic yak may serve as an ideal natural animal model and the resulted transcriptomic profiles could provide insights to understanding the adaptive evolution in other highland species including humans (e.g., Tibetans).
In this study, we collected multiple tissue samples from domestic yaks permanently living at 3,400 m, 4,200 m, and 5,000 m. As yaks are native highland species and only found at highland area with elevation of over 3,000 m, there is no lowland controls available. Phylogenetically the taurine cattle (Bos taurus) are the most closely related lowland Bovidae species to yaks and can serve as lowland controls although they diverged at least 1 Ma (Chen et al. 2018; Wu et al. 2018). Here, we choose Zaosheng cattle, a Chinese native species living at 1,500 m and geographically adjacent to the Tibetan Plateau as the lowland control. We performed RNA sequencing (RNA-Seq) and depicted a detailed transcriptomic landscape across three different altitudes and eight major organs. The comparison between highland yaks and lowland cattle leads to the identification of a set of genes and molecular pathways that may explain genetic adaptation of domestic yak to high altitude hypoxia. By leveraging information from multiple altitudes, we also identified genes and pathways with possible adaptive changes which may explain the genetic mechanism of hypoxic adaptation in yaks.
Materials and Methods
Sample Collection and Transcriptome Sequencing
We collected tissue samples from 18 indigenous adult male yaks with a gross body weight of 200 kg and an age of 4–6 years old, from five pastoral areas representing three major altitudes (3,400 m, 4,200 m, 5,000m) for yak pastoralism on the Tibetan Plateau, and four indigenous adult male cattle with a gross body weight of 400 kg and an age of 4–6 years old from 1,500 m as low altitude control (fig. 1A). These samples include six yaks from Maqu county in Gannan Tibetan Autonomous Prefecture of Gansu Province representing altitude of 3,400 m, six from Dangxiong County and Linzhou County for altitude of 4,200 m, six from Anduo County and Bange County for altitude of 5,000 m in Tibetan Autonomous Region, and four indigenous Zaosheng cattle from Ningxian county in Gansu Province. Tissue samples from eight major organs including heart, liver, spleen, lung, kidney, testis, muscle, and brain were taken within 2 h after animals were sacrificed on site where animals were located, and tissue samples were stored immediately in the liquid nitrogen during field and transportation. After transporting to the laboratory, all tissue samples were transferred to the –80 °C freezer for long-term storage. Total RNA was isolated using TRIzol reagent (Biotopped). Libraries from the resulting total RNA were prepared using the TruSeq paired-end mRNA-Seq kit and sequenced on the Illumina HiSeq 2500 platform. Because lung and heart are the major organs for oxygen exchange and blood transportation and likely play important roles in hypoxic response, we therefore generated RNA-seq data of lung and heart for all 18 animals, while for other tissues, we generated RNA-seq data for at least three individuals from each altitude. The protocol of this study was reviewed and approved by the Internal Review Board of Kunming Institute of Zoology, Chinese Academy of Sciences.
Fig. 1.
—Refinement of yak genome annotation and general expression pattern. (A) Experimental design and sampling strategy. (B) Refined annotation discovers more isoforms for each gene in comparison with the reference set. (C) Hierarchical clustering of pairwise Spearman correlations for the 120,409 coding exons highlights a tissue-specific transcriptional pattern. Samples are color-coded according to tissue type, sampling altitude, and sequencing batch. (D) A small number of genes dominate yak transcriptome in all tested tissues.
For each sample, we generated 100-bp paired-end reads and the raw sequencing data were deposited to the Genome Sequence Achieve (http://gsa.big.ac.cn), under accession number PRJCA000530.
Annotation Refinement by Reference-Assisted Assembly
The yak genome sequence (GCF_000298355.1_BosGru_v2.0) and annotation information were downloaded from NCBI database (ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq/vertebrate_mammalian/Bos_mutus/all_assembly_versions/GCF_000298355.1_BosGru_v2.0; last accessed December 25, 2018). RNA-Seq reads were preprocessed following our previous protocol (Peng et al. 2017), which removes reads with adapters or with >10% ambiguous bases or with >50% low quality bases (Q-score ≤ 5). The retained reads were then mapped to the reference genome using STAR release 2.4.0h (Dobin et al. 2013) with following parameters “–outFilterMultimapNmax 20 –outFilterTypeBySJout –outSAMunmapped Within –alignSJoverhangMin 8 –alignSJDBoverhangMin 1 –outFilterMismatchNmax 999 –outSAMattributes NH HI AS NM MD –alignIntronMin 20 –alignIntronMax 1000000 –outSAMtype BAM SortedByCoordinate –chimSegmentMin 20 –outSAMstrandFieldintronMotif –outFilterIntronMotifsRemoveNoncanonical.” Resulted bam files were feed to cufflinks v.2.2.1 (Trapnell et al. 2010) to improve the current annotation. For each sample, a transcript annotation in gtf format was generated, and cuffmergewas used to merge all files to obtain the final annotation for yak transcriptome.
We further applied a series of filters to verify newly assembled transcripts. First, the new yak annotation file was compared with the reference annotation by cuffcompare, resulting in three categories of transcripts: 1) matched transcripts (transfrag class code “=”), where exon/intron coordinates completely match that of reference; 2) novel isoforms (transfrag class code “j”), where at least one splice junction is identical to a reference junction; and 3) novel transcripts, where none of the exons matches any reference exon nor within any annotated genic regions. All other transfrag classes represent more complex scenarios and were ignored.
To verify novel isoforms of annotated genes or novel transcripts, we calculated the number of supporting reads for novel exon–exon junctions. A supporting read is defined as spanning the junction with a 20-bp region centered on the junction point, and no mismatch could be found in this region. To consider a junction as validated, we required at least five supporting reads from at least two yak samples. Isoforms or transcripts with ≥30% of their novel junctions validated will be defined as verified for downstream analysis.
Coding Potential Prediction for Novel Transcripts
For novel transcripts absent in the reference set, we applied Coding Potential Assessment Tools or CPAT (Wang et al. 2013) to calculate their coding probability. To choose a reasonable threshold for coding transcripts, we curated 25,380 coding and 4,127 noncoding transcripts from yak reference annotation and computed respective CPAT scores. The highest prediction accuracy was achieved at a threshold of 0.52, which was adopted to determine potentially coding transcripts.
Expression Estimate of Constitutive Exons
To identify constitutive exons in our data set, we filtered overlapping exons from two different transcripts using our refined gene annotation. Then, HTSeq tool (Anders et al. 2015) was applied to count the number of RNA-Seq reads that were mapped to constitutive exons. Read pairs were removed if they span a region oversized the range of target locus, or were mapped to multiple loci, or showed improper mapping, or were mapped to multiple locations.
Identification of Orthologous Exons in Cattle
Whole genome sequence and annotation for cattle (GCF_000003205.5_Btau_4.6.1) were downloaded from NCBI database (ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq/vertebrate_mammalian/Bos_taurus/all_assembly_versions/suppressed/GCF_000003205.5_Btau_4.6.1; last accessed December 25, 2018). Constitutive exons identified above were mapped to cattle genomes using BLAT (Kent 2002). An exon that mapped to the genome with a sequence similarity of 0.95 and an aligned fraction of 0.99 is retained. If there were more than one hit for an exon sequence (paralogues) or hits from two distinct exons overlap, those exons were excluded from further analysis.
DEG Analysis
To identify differential expression, we applied voom (Law et al. 2014) embedded in the R package limma, which is a Bayesian method and accounts for mean-variance relationship. We performed analyses on individual exons in different tissues, only exons with one transcript per million reads (TPM) > 1 (Li and Dewey 2011) in at least 1/3 of total samples were included. The linear model we used is Exp ∼ Altitude + Batch. Multiple tests correction were performed using Benjamini–Hochberg approach (Benjamini and Hochberg 1995). The widely used P-value threshold of 0.05 is applied to detect differential expression. Furthermore, we classified differentially expressed exons (DEEs) into four categories based the expression pattern: 1) high group, where the expression level is higher in animals from 5,000 m compared with those from 3,400 m and 4,200 m; 2) up group, where the expression level increases as altitude elevates; 3) low group, where the expression level is lower at 5,000 m in comparison to 3,400 m and 4,200 m; 4) down group, where the expression level negatively correlates with altitudes.
Functional Enrichment Analysis
We applied TopGO package (Alexa et al. 2006) in R to identify enriched gene ontology (Ashburner et al. 2000) categories for various gene sets. To determine pathway enrichment, we downloaded KEGG pathway annotation (Kanehisa and Goto 2000) and performed hypergeometric test. All tests were adjusted by Benjamini–Hochberg approach.
Construction of Gene Co-Expression Network
WGCNA (Langfelder and Horvath 2008) was used to perform weighted co-editing network analysis. Briefly, pair-wise correlations were first calculated between all possible pairs of expressed genes across all samples, and then were used to construct the adjacency matrix. Next topological overlap matrix (TOM) was constructed using the above adjacency matrix, and co-expressed genes were clustered together by average linkage hierarchical clustering. The resulting clusters were cut by the dynamic hybrid tree cut algorithm and branches under the cut were defined as different modules (Langfelder et al. 2008). For each module, a module eigengene (ME) was derived as the first principle component. Highly correlated modules (in terms of ME) were further merged using mergeCutHeight of 0.25.
We next calculated ME-based connectivity kME or module membership (MM). For a given gene i, MMq(i) = cor[x(i), Eq], where q is the module, x(i) denotes the expression level, and Eq is the ME for module q (Horvath and Dong 2008; Xue et al. 2013). Hub genes were defined as those with highest MM values (MM > 0.9 for a module).
Identification of Tissue Dominant Genes
For each gene, we first calculated the first quartile, the third quartile, and the interquartile range of its expression in each tissue, and then define it as tissue dominant if its first quartile expression in one tissue is higher than the sum of the third quartile expression and two times of the inter quartile range in any other tissues.
Results
We generated RNA-seq data from eight main organs including heart, liver, spleen, lung, kidney, testis, muscle, and brain in domestic yaks permanently living at three representative altitude areas on the Tibetan Plateau (fig. 1A), including six animals at 3,400 m, six at 4,200 m, and six at 5,000 m (see details in Materials and Methods). In total, we generated RNA-seq data for 18 lungs (lg), 17 hearts (ht), 9 kidneys (kd), 9 livers (lv), 9 muscles (ms), 9 spleens (sp), 8 testes (ts), 6 parietal lobes (pl), and 3 prefrontal cortices (pfc). We also generated RNA-seq data for matched tissues from four Chinese native male cattle (B.taurus) permanently living at an elevation of 1,500 m close to the Tibetan Plateau (fig. 1A). In total, ∼5.4 billion short reads have been generated after removing adapter-embedded or low quality reads. On average, each sample contains ∼24 million reads, and 91% of them can be unambiguously mapped to the yak reference genome.
Refinement of Yak Annotation
Despite the intriguing role of yak in understanding genetic adaptation, genomic studies on yak are still sparse (Qiu et al. 2012), mainly due to difficulties in sample acquisition for transcriptome analysis. As a consequence, the current annotation of the yak genome is far from complete compared with other well studied species. We thus seek to refine the yak annotation using newly generated RNA-seq data, which to the best of our knowledge, is the largest transcriptome profiles for yak to date. By applying cufflinks and cuffmerge (Trapnell et al. 2010), we conducted reference-assisted assembling, resulting in a comprehensive set of 241,829 transcripts from 57,370 loci, which covered all of 31,084 previously annotated transcripts including 25,393 protein-coding, 3,819 pseudogenes, 1,564 tRNAs, and 308 misc_RNAs. This result suggests the current reference may only represent a minimal set of all yak transcripts. Thus, a refined annotation will give us a broader picture regarding transcriptional regulation in yak.
In the reference set, there are 18,447 protein coding genes with 25,393 coding transcripts in total. Through our refined efforts, we identified 18,742 novel isoforms of those coding genes with high confidence (see Materials and Methods, supplementary table S1, Supplementary Material online), where 5,034 genes have at least one new isoform, 3,066 genes have at least two new isoforms and 2,073 have at least three new isoforms (fig. 1B). This finding largely expands the current catalog of splicing forms of protein coding genes in yak, and it should be noted that these transcripts may represent a subset of all possible isoforms due to the stringent criteria we applied. Similarly, we also verified 19,128 novel transcripts with at least two exons (supplementary table S2, Supplementary Material online), and predicted their coding probability using CPAT (Wang et al. 2013). The distribution shows the majority of the novel transcripts has low coding probability and are likely noncoding (data not shown), but 3,874 (20.3%) could be potentially coding using a threshold learned from known coding and noncoding transcripts in yak (Materials and Methods).
Tissue Expression Pattern in Yak
To develop an extensive understanding of yak transcriptome, we focused on constitutive exons from all known and new coding transcripts, exons shorter than 50 base pairs or partially overlapped exons from two transcripts were excluded to avoid uncertainty. The number of reads uniquely mapped to these exons were counted by using HTSeq tools (Anders et al. 2015). In total, 120,409 exons from 17,571 protein coding genes were retained for downstream analysis.
Consistent with findings from GTEx project (GTEx Consortium 2015), we found each tissue presented a distinct transcriptional signature and all samples are unambiguously grouped by tissue origin (fig. 1C). The primary separation, as observed in previous studies (Soumillon et al. 2013; Yu et al. 2015), is between testis and other tissues, which may reflect the hyperactive transcriptional machinery in testis. Prefrontal cortex and parietal lobeare are indistinguishable as expected and were thus categorized as a single tissue (brain) in downstream analyses. Similar to humans (GTEx Consortium 2015), heart and muscle samples were clustered together, which is consistent with the common understanding that a large proportion of heart is composed of cardiac muscle tissue.
As observed in our study and others (GTEx Consortium 2015), a small number of genes dominated tissue transcription, and on average, at least 50% of reads aligned to coding genes were from 311 genes (fig. 1D). The level of dominance varies among tissues, as <150 genes in heart, muscle and liver contribute to >50% of aligned reads, and the number of genes in kidney, lung and spleen was around 400, whereas testis showed the lowest level of dominance with 50% of reads coming from >600 genes. When focusing on the top 100 genes with the most abundant reads, we found different tissues shared a large proportion of common genes, but they also represent tissue specific transcriptions relevant to their distinct functions (supplementary table S3, Supplementary Material online). For example, several actin and myosin genes including ACTN2 and MYH7 were among the top genes in muscular tissues such as heart and muscle (supplementary fig. S1, Supplementary Material online). With high data quality, the yak transcriptome data provide an important resource for future genomic and functional studies.
Transcriptional Changes in Response to Hypoxia
Yak is a native mammalian species on the Tibetan Plateau and its progenitor colonized the plateau at least 1 Ma (Zhang 1989), though domestication of yak is believed to be a rather recent event no earlier than 10,000 years ago (Qi et al. 2013). Yak has developed a number of physiological traits to cope with hypobaric hypoxia (Zhang et al. 1994; Wei and Yu 2008; Zhou et al. 2015). We therefore asked how altitude change could shape transcriptional regulation in yak. To concurrently capture the potential influence of alternative splicing, we limited our analysis to nonoverlapping exons which are expressed over one TPM in at least two thirds of samples. Tissues were analyzed separately to retain information from tissue-specific or -dominant genes. Because brain materials were only available from two elevations, we excluded brain samples from this analysis.
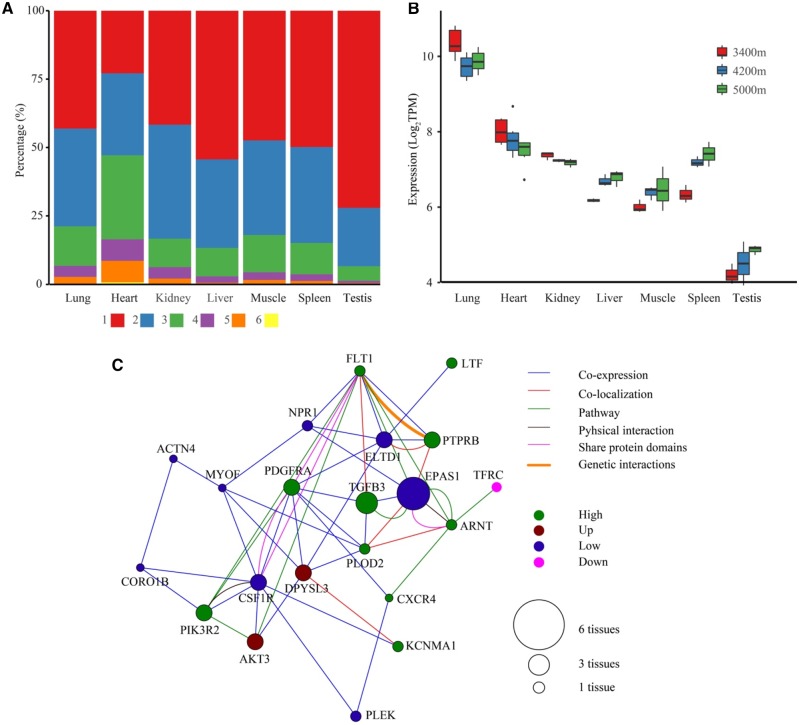
We detected various DEEs among altitudes within 48–2,724 (median 821) coding genes in those tissues (fig. 2A), where kidney has the fewest DEEs (118 in 48 coding genes) and testis has the most (8,907 DEEs in 2,724 genes). A majority of these DEEs (16,318 of 18,984 or 85.96%) was found in only one tissue, suggesting that the transcriptional rewiring is distinct among tissues. If we exclude testis which has at least 4,000 more DEEs than any other tissues, the pattern still holds as 9,982 of 11,523 (86.63%) DEEs are tissue-specific. At gene level, tissue-dominated differential expression is prominent as well, as 3,692 of 4,845 (76.20%) differentially expressed genes (DEGs) are from one single tissue. In particular, we found EPAS1 (encodes hypoxia-induced-factor 2α, HIF-2α) is differentially expressed in all tissues except for kidney (fig. 2B), but the expression level and pattern vary across tissues. In particular, lung has the highest expression of EPAS1 in general, and lung samples from 4,200 m show lower expression compared with those from 3,400 m or 5,000 m. In contrast, heart presents the second highest expression of EPAS1, but the expression level decreases along with altitude. While in other tissues such as liver, muscle, spleen or testis, EPAS1 expression increases with altitude. These observations indicate that the response to hypoxia in yak is highly complicated and involves fine tuning of transcription in multiple genes and tissues. There are 13 additional genes differentially expressed in at least five tissues (supplementary table S4, Supplementary Material online), and five of them are collagen genes (COL1A2, COL3A1, COL5A2, COL14A1, and COL15A1), highlighting the crucial role of collagen involved pathways in high altitude adaptation.
Fig. 2.
—Transcriptional changes in response to high altitude. (A) Distribution of DEEs across tissues. A majority of DEEs is restricted to one tissue type (the red portion). (B) Expression level of EPAS1 at different altitudes among different tissues. Only lung show a nonlinear change with the lowest expression at 4,200 m, whereas all other tissues show a linear change (increase or decrease linearly with altitude). (C) Network of differentially expressed hypoxic genes in lung. The colored solid circles indicate different expression patterns with altitude.
To further understand the molecular basis of adaptation to hypoxia, we collected a set of 473 hypoxia-related genes (HRGs) from hypoxia-related pathways identified in previous literature (Peng et al. 2017). Of them, nearly one third (144/473) are differentially expressed in at least one tissue (supplementary table S5, Supplementary Material online). At the tissue level, heart and kidney have the highest proportion of HRGs in their DEG sets (7.86%, 11/140 and 6.25%, 3/48, respectively), then are muscle (5.24%, 43/821), lung (4.04%, 21/520), spleen (3.91%, 45/1152), liver (2.77%, 25/902), and testis (2.46%, 67/2, 724).
Compared with lowland cattle, yak has enlarged lung and heart (Zhang 1989), enabling them to improve the efficacy of oxygen utilization in a hypoxic environment. Interestingly, we found lung harbors the highest proportion of differentially expressed HRGs (17/21, or 80.1%) with nonlinear (putatively adaptive) expression pattern among all tested tissues: (1/11, or 9.1% for heart, 1/3, or 33.3% for kidney, 22/43, or 51.2% for muscle, 11/45, or 24.4% for spleen, 16/25, or 64.0% for liver, and 19/67, or 28.4% for testis) (fig. 2C). We further performed in-depth analyses for these two organs to draw a clearer picture of the potential correlation between genetic factors and phenotypic changes, which were described below.
Transcriptomic Profiles of Lung
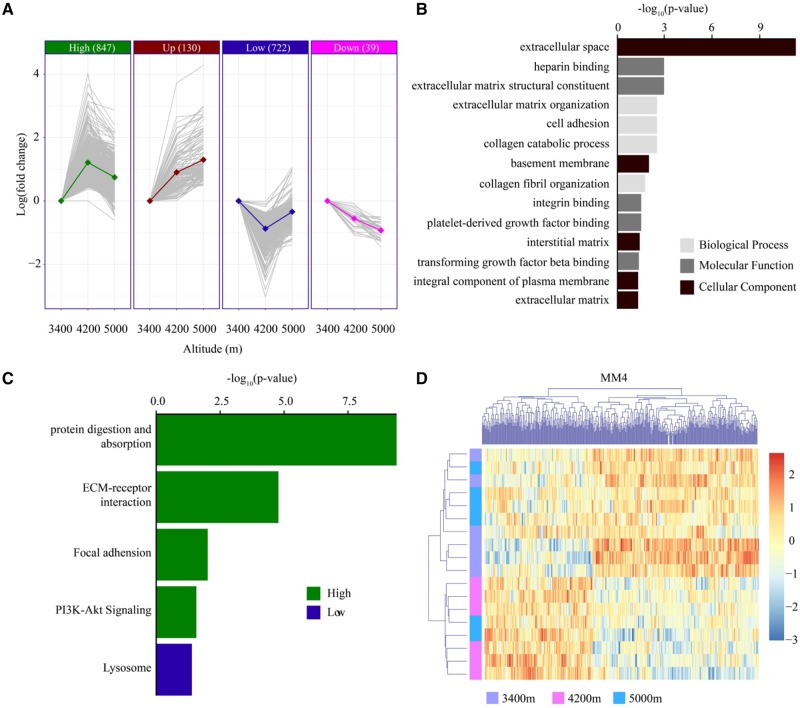
We collected 18 lung samples in total with six from each of the three elevations, and there are 70,516 qualified exons from 9,077 coding genes subject to analysis. After correcting for batch effect, we identified 1,738 (2.46%) DEEs representing 520 (5.73%) genes (DEGs). When breaking down by pairwise comparison, we found 1,626 DEEs from 470 genes are between 3,400 m and 4,200 m, 264 DEEs from 101 genes are between 3,400 m and 5,000 m and only 19 DEEs of 11 DEGs are between 4,200 m and 5,000 m. Importantly, we found overwhelmingly more DEEs (1,626) between 3,400 m and 4,200 m compared with 264 DEEs between 3,400 m and 5,000 m, implying a generally nonlinear correlation between expression level and elevation in lung as the example of EPAS1 we discussed above.
On the basis of expression pattern, we grouped DEEs into four classes, High, Low, Up, and Down (see Materials and Methods, supplementary table S6, Supplementary Material online). Notably, the dominance of High and Low groups confirms the nonlinear correlation observed above (fig. 3A), contributing to >90% of DEEs, highlighting the critical role of long-term adaptation in lung transcriptional regulation. In the High group, 14 GO categories are enriched and some of these categories have known roles in hypoxia. In particular, cell adhesion is reduced under an increasing intensity of hypoxia which affects biological processes such as vascular endothelium and extracellular matrix (Kaiser et al. 2012). Platelet-derived growth factor (PDGF) binding plays a pivotal role in the pathology of pulmonary hypertension, and hypoxia enhances PDGF signaling in the pulmonary vasculature (ten Freyhaus et al. 2011). Extracellular matrix functions and collagen can influence the behavior of vascular smooth muscle cells which are sensitive to hypoxia (Shami et al. 2014) (fig. 3B). In terms of pathway, protein digestion and absorption (bom04974), ECM–receptor interaction (bom04512), focal adhesion (bom04510), and PI3K-Akt signaling (bom04151) are enriched in the High group, whereas lysosome (bom04142) is enriched in the Low group (fig. 3C).
Fig. 3.
—Transcriptional patterns in lung. (A) The majority of differentially expressed exons in lung shows a nonlinear correlation (“High” or “Low”) with altitude. (B) Gene ontology enrichment for lung DEGs. (C) Result of pathway analysis for DEGs. (D) A co-expression module found in lung is enriched for nonlinear DEEs.
In addition to individual DEEs/DEGs, a co-expression network gives a systems-level view on how transcriptional changes respond to hypoxia. In general, 8,888 of 9,077 (97.92%) genes expressed in lung were assigned to 47 co-expression modules, and the number of genes range from 59 to 605 per module. The first principal component of each module known as modular eigengene (ME) was also calculated. By overlapping with DEGs, we found seven modules are enriched for DEGs. Of them, module 4 is the most significant one (P = 1.4 × 10−40 after BH correction), and 110 of 450 (24.44%) module genes are found to be DEGs (fig. 3D). Further analysis suggests lysosome pathway (bom04142) is overrepresented, though it is not statistically significant (adjusted P = 0.125). More intriguingly, DEGs have significantly higher intramodular connectivity than nonDEGs (median of 0.80 vs 0.57, P = 4.49 × 10−29, Wilcoxon's ranksum test), and all top 10 connected genes are DEGs (supplementary table S7, Supplementary Material online).
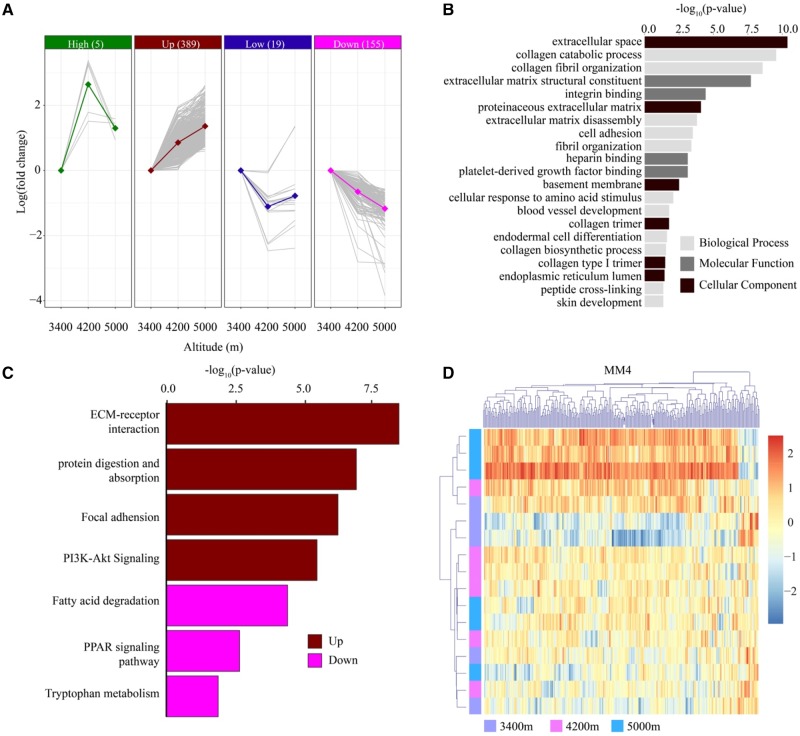
Transcriptomic Profiles of Heart
With the same experimental design, we tested 17 heart samples (one sample from the 3,400 m group was degraded), and identified 568 DEEs among three altitudes. In contrast to lung (>90%), only 4.2% (24 of 568) DEEs show nonlinear expression pattern consistent with the High or Low groups (fig. 4A and supplementary table S8, Supplementary Material online), suggesting a different pattern of gene expression regulation in heart. Gene ontology analysis discovered 21 functional categories enriched in the Up group, including blood vessel development (GO: 0001568), endodermal cell differentiation (GO: 0035987), as well as terms related to extracellular matrix and collagen functions (fig. 4B). The same four pathways enriched in lung were also found enriched in the Up group of heart (fig. 4C), implying they are important mechanisms in coping with hypoxia. Additionally, three other pathways (fatty acid degradation, PPAR signaling and tryptophan metabolism) are enriched in the Down group. As oxygen concentration decreases with elevation, down-regulation of these pathways will reduce hypoxia-induced metabolism shift from mitochondrial fatty acid β-oxidation to glycolysis (Galzio et al. 2012; Liu et al. 2014; Zhirov et al. 2014).
Fig. 4.
—Transcriptional patterns in heart. (A) The majority of differentially expressed exons in heart shows a linear (“Up” or “Down”) correlation with altitude. (B) Gene ontology enrichment for heart DEGs. (C) Result of pathway analysis for heart DEGs. (D) The co-expression module found in heart.
We further investigated 193 exons from 25 genes (supplementary table S9, Supplementary Material online) that are differentially expressed in both lung and heart. Of them, only 12 exons belong to the same group in both organs, and 177 (91.7%) switched from the High group (nonlinear) in lung to the Up group (linear) in heart, including 20 exons from FN1 and 42 exons from FBN1. It was reported that FN1 is hypoxia responsive in murine and human melanoma cells (Olbryt et al. 2011), and FBN1 is involved in heart disease, such as aortic aneurysm and aortic dissection (Lesauskaite et al. 2015).
In terms of co-expression, we grouped 7,490 of 7,743 (96.7%) heart genes into 27 modules, ranging from 62 to 2,965 genes per module. Among them, module 4 with 288 genes shows a significant enrichment (P = 1.09 × 10−21) for heart DEGs (fig. 4D). Similarly, DEGs in this module present higher intramodular connectivity than nonDEGs (0.87 vs 0.76, P = 2.45 × 10−8, Wilcoxon's rank sum test), and 6 of the 10 most connected genes are DEGs (supplementary table S10, Supplementary Material online), which is unlikely caused by chance considering 38 DEGs in this module (P = 5.12 × 10−4, Fisher's exact test).
Expression Pattern of Orthologous Exons
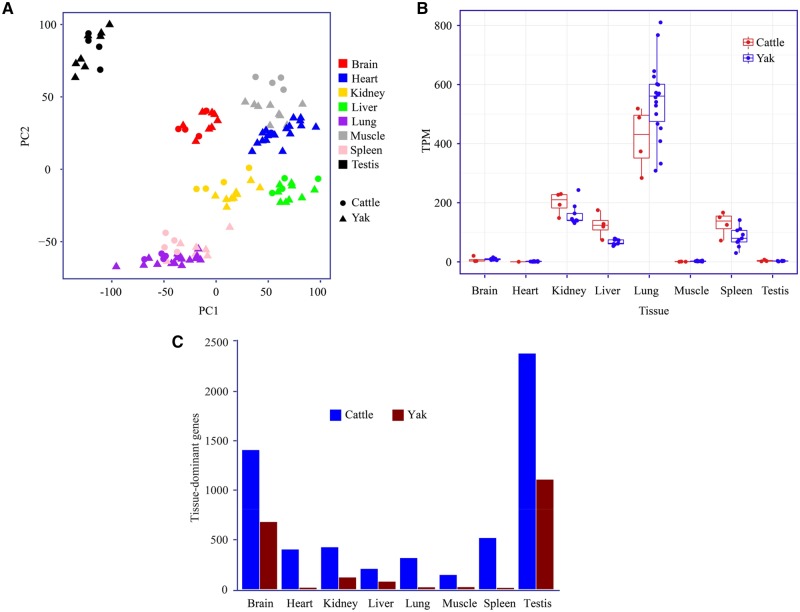
Another question of particular interest is whether there are expression changes specific to yak that are responsible for hypoxic adaptation. To address this, we chose Zaosheng cattle, a Chinese native cattle living at 1,500 m and geographically next to the Tibetan Plateau as lowland control. As described before, yak is a native highland species and only found at highland area with elevation of over 3,000 m, therefore there is no lowland controls available. Phylogenetically the taurine cattle (B.taurus) is the most closely related lowland Bovidae species to yak and can serve as lowland controls although they diverged at least 1 Ma (Chen et al. 2018; Wu et al. 2018). We sequenced transcriptome of the corresponding organs from four cattle samples (see Materials and Methods). We first investigated 100,841 yak-cattle orthologous exons with ≥ 95% sequence similarity and ≤ 1% size difference and found that intertissue variation explains most of the total variance, and then interspecies variations (fig. 5A). Considering the long-term phylogenetic divergence between yak and cattle, the majority of transcriptomic variances between yak and cattle probably resulted from between-species difference, therefore, the common assumption of transcriptome analysis might be violated that the expression of a majority of genes is similar across groups. To address this concern, we compared the normalization factors calculated using all exons and that from orthologs of 4,299 curated human housekeeping genes (Eisenberg and Levanon 2013). A high correlation between these two sets was observed (Pearson's r = 0.891), suggesting the common assumption still holds.
Fig. 5.
—Expression pattern of orthologous genes. (A) PCA analysis suggests tissue accounts for the majority of expression variance. (B) Expression pattern of a tissue-dominant gene (CDH1) for lung as an example. (C) Distribution of tissue-preferred genes across different tissues.
Given the rich tissue information embedded in our data set, we further investigated whether there is any interspecific difference in tissue expression pattern by studying genes highly or specifically expressed in one tissue (tissue-preferred genes, TPGs), which are determined using interquantile range (IQR) of expression in different tissues (fig. 5B). Among the 14,950 yak-cattle orthologous genes, 1,117 (7.5%) are TPGs in yak testis (fig. 5C and supplementary table S11, Supplementary Material online), which is the highest across tissues. Brain has the second abundant TPGs (804% or 5.4%), whereas the other tissues have small portion of TPGs (<1%). This observation is highly consistent with the common understanding that genes expressed in testis and brain execute distinct functions (Soumillon et al. 2013; GTEx Consortium 2015). A similar pattern is also found in cattle, with more TPGs in each tissue, which may reflect the small sample size in general. Furthermore, we sought to seek yak-specific TPGs which may harbor additional information for high altitude adaptation. Of 25 yak lung TPGs, three were not presented as cattle TPGs using a relaxed threshold, including CDH1, LOC102280947, and RASEF. Genes that are specifically repressed in individual tissues were also interrogated but their incidences are relatively rare and thus not discussed here.
Comparison with Results from the Yak Genome Study
Previously, a list of positively selected genes (PSGs) were identified by comparing yak genome with other species, among which, several genes are involved in the response to hypoxia including ADAM17 and ARG2 (Qiu et al. 2012). However, none of the 80 yak-specific PSGs showed differential expression in lung among three altitudes. Similarly, only two PSGs, ATP1A2 and MAPK12, were found differentially expressed in heart, suggesting changes of protein coding sequences and transcriptions may act independently in yak adaptation.
Pathway Regulation and High Altitude Adaptation
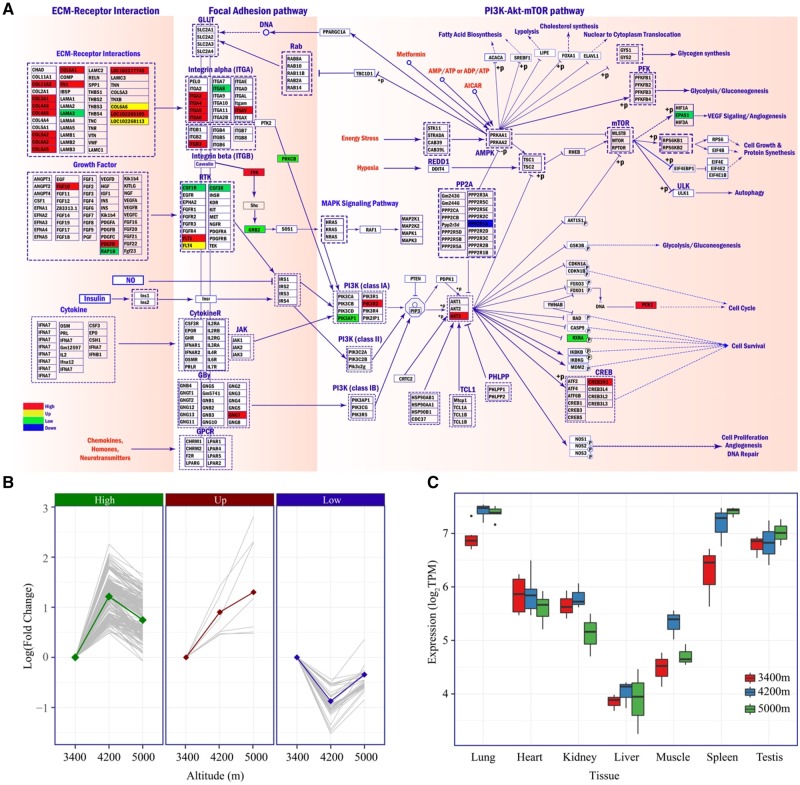
In our results, we found activation of three pathways (ECM–receptor interaction, focal adhesion and PI3K-AKT, fig. 6A) are explicitly associated with changed elevation in lung, heart and spleen. Collagens that encode cell-extracellular matrix (ECM) and interact with integrins are widely represented in ECM–receptor interaction and focal adhesion pathways, including six DEG (COL1A2, COL3A1, COL6A2, COL6A5, Loc102265105, and Loc102277745). We also identified four integrin genes (ITGA6, ITGA8, ITGAV, and ITGB3) in focal adhesion and PI3K-AKT pathways with significantly different expression among elevations. Those genes function as mechanoreceptors and provide a force-transmitting physical link between the EMC and cytoskeleton. These interactions between collagens and integrins lead to a direct or indirect control of multiple cellular activities such as adhesion, migration, differentiation, proliferation and apoptosis of cells.
Fig. 6.
—Expression pattern of key pathways. (A) Three functional pathways with enriched DEGs are tightly connected and activated at high altitudes. Genes filled with different colors represent differential expression pattern in at least one tissue. (B) A majority of differentially expressed genes of PI3K-AKT pathway in lung belongs to the “High” group. (C) Expression pattern of the AKT gene across different tissues.
The PI3K-AKT pathway is a well-studied intracellular signaling pathway with crucial roles in a number of biological processes, such as metabolism, cell growth and proliferation, and cancer development. In our analysis, we found this pathway is overrepresented in high- or up-regulated genes in multiple tissues (fig. 6B), including lung, heart, muscle, and spleen. Particularly among the adaptively regulated genes is AKT3 (fig. 6C), the central player of PI3K-AKT pathway and a proto-oncogene activated in multiple cancers to promote cell survival and proliferation. The stress of low oxygen level at high altitude mimics the tumor cell micro environment, which is alleviated by activating AKT3 and thus the PI3K-AKT pathway, preventing cells from apoptosis. Collectively, our observation of activated genes in ECM–receptor, focal adhesion and PI3K-AKT pathways highlights the potential molecular mechanism to overcome hypobaric hypoxia.
Discussion
Attribute to the decreasing cost of next generation sequencing, a large number of whole genome sequencing studies have been performed to explore the molecular mechanism of high-altitude adaptation, with a focus on Tibetan mammals (Qiu et al. 2012; Ai 2014). Despite the identification of candidate signals for adaptation, the connection between genotype and adaptive phenotype remains poorly understood. Transcriptome, on the other side, provides us a valuable resource to expand the genomic information by focusing on gene regulation, which may be more functionally relevant. Recently, a few transcriptomic studies have been conducted in Tibetan mammals (Qiu et al. 2012; Ai 2014; Lan et al. 2014; Shan et al. 2014; Wang et al. 2014; Jia et al. 2016; Li et al. 2017); however, the interpretation of their findings is constrained as only one or few tissues were sampled, and/or from a single altitude. Here, we generated comprehensive transcriptional profiles, including multiple tissues and multiple altitudes of domestic yaks. The comparisons among yaks from different altitudes revealed genes and pathways with possible adaptive changes in response to high altitude hypoxia. Notably, the yak lung displayed a characteristic gene expression regulation with a majority of the DEGs showing a nonlinear change. For example, as an important hypoxia-inducible factor, HIF-2 alpha (encoded by EPAS1) usually accumulate under chronic hypoxia (Bigham and Lee 2014). However, in yaks, the EPAS1 expression in lung at 4,200 m was down-regulated compared with the level at both 3,400 m and 5,000 m (fig. 2B), implying blunted hypoxic response.
Pathways are key components to carry out a variety of critical tasks in cells, such as gene regulation and signal transduction. In this study, we have identified several pathways enriched in DEGs among altitudes and/or between-species, including the PI3K-AKT signaling pathway and its downstream mTOR signaling pathway, both of which are connected to the HIF pathway in regulating expression of multiple genes and inducing downstream pathways under hypoxic conditions.
The PI3K-AKT pathway is an intracellular signaling pathway and plays an important role in regulating cell cycle, proliferation and survival. Under hypoxia, cells are exposed to environmental stresses and apoptosis is induced by hyper-permeability of the inner mitochondrial membrane, the generation of reactive oxygen species (ROS) or the activation of stress activated protein kinase (SAPK) (Greijer and van der Wall 2004). It has been widely studied that activating PI3K-AKT as well as downstream mTOR could protect cells from apoptosis in rats (Alvarez-Tejado et al. 2001) and multiple cancers (Kilic-Eren et al. 2013). Apparently, the same mechanism has been utilized in yaks to respond to hypoxic stress. Different from malignant tumors, however, the growth and division of yak cells is still under control, which may provide insights for biomedical studies to treat cancer.
In summary, we comprehensively profiled gene regulation in lung and heart, two key organs for adaptation to high altitude hypoxia, and we identified several molecular pathways showing adaptive changes in yak, which may explain the genetic mechanism of hypoxic adaptation. The reported transcriptomic profiles of yaks provide further insights to understanding adaptive evolution in other highland species including humans (e.g., Tibetans).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Mr. Xichao Yang (Gansu Agricultural University), Yan Guo, and Hui Zhang (Kunming Institute of Zoology, CAS) for field sampling and technical help. This study was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13010000 to B.S., XDA20040102 to X.Q.), and grants from the National 973 program (grant number 2013CB835203 to X.Q.), the National Natural Science Foundation of China (30870295, 31671329, and 31371269 to X.Q., 31621062 to B.S., 31460287 to Ouzhuluobu.), the West Light Doctoral Program (to X.Z.), and the Zhufeng Scholar Program of Tibetan University (to Ouzhuluobu).
Literature Cited
- Ai H. 2014. Population history and genomic signatures for high-altitude adaptation in Tibetan pigs. BMC Genomics 15(1):834.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A, Rahnenfuhrer J, Lengauer T.. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22(13):1600–1607. [DOI] [PubMed] [Google Scholar]
- Alvarez-Tejado M, et al. 2001. Hypoxia induces the activation of the phosphatidylinositol 3-kinase/Akt cell survival pathway in PC12 cells: protective role in apoptosis. J Biol Chem. 276(25):22368–22374. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W.. 2015. HTSeq – a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall CM, et al. 2010. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci U S A. 107(25):11459–11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 57:12. [Google Scholar]
- Bigham A, et al. 2010. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 6(9):e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham AW, Lee FS.. 2014. Human high-altitude adaptation: forward genetics meets the HIF pathway. Genes Dev. 28(20):2189–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, et al. 2018. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat Commun. 9(1):2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY.. 2013. Human housekeeping genes, revisited. Trends Genet. 29(10):569–574. [DOI] [PubMed] [Google Scholar]
- Galzio R, et al. 2012. Hypoxia modulation of peroxisome proliferator-activated receptors (PPARs) in human glioblastoma stem cells. Implications for therapy. J Cell Biochem. 113(11):3342–3352. [DOI] [PubMed] [Google Scholar]
- Greijer AE, van der Wall E.. 2004. The role of hypoxia inducible factor 1 (HIF-1) in hypoxia induced apoptosis. J Clin Pathol. 57(10):1009–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium 2015. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, et al. 2006. Origin of mitochondrial DNA diversity of domestic yaks. BMC Evol Biol. 6(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Dong J.. 2008. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 4(8):e1000117.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia C, et al. 2016. Gene co-expression network analysis unraveling transcriptional regulation of high-altitude adaptation of Tibetan pig. PLoS One 11(12):e0168161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser R, Friedrich D, Chavakis E, Böhm M, Friedrich EB.. 2012. Effect of hypoxia on integrin-mediated adhesion of endothelial progenitor cells. J Cell Mol Med. 16(10):2387–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S.. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1):27–30. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT – the BLAST-like alignment tool. Genome Res. 12(4):656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic-Eren M, Boylu T, Tabor V.. 2013. Targeting PI3K/Akt represses Hypoxia inducible factor-1alpha activation and sensitizes Rhabdomyosarcoma and Ewing's sarcoma cells for apoptosis. Cancer Cell Int. 13(1):36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan D, et al. 2014. RNA-Seq analysis of yak ovary: improving yak gene structure information and mining reproduction-related genes. Sci China Life Sci. 57(9):925–935. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S.. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9(1):559.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, Horvath S.. 2008. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24(5):719–720. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK.. 2014. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15(2):R29. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesauskaite V, et al. 2015. FBN1 polymorphisms in patients with the dilatative pathology of the ascending thoracic aorta. Eur J Cardiothorac Surg. 47(4):e124–e130. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12(1):323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, et al. 2017. Generation and characterization of the blood transcriptome of Macaca thibetana and comparative analysis with M. mulatta. Mol Biosyst. 13(6):1121–1130. [DOI] [PubMed] [Google Scholar]
- Liu Y, et al. 2014. HIF-1alpha and HIF-2alpha are critically involved in hypoxia-induced lipid accumulation in hepatocytes through reducing PGC-1alpha-mediated fatty acid beta-oxidation. Toxicol Lett. 226(2):117–123. [DOI] [PubMed] [Google Scholar]
- Meyer MC, et al. 2017. Permanent human occupation of the central Tibetan Plateau in the early Holocene. Science 355(6320):64–67. [DOI] [PubMed] [Google Scholar]
- Olbryt M, et al. 2011. Melanoma-associated genes, MXI1, FN1, and NME1, are hypoxia responsive in murine and human melanoma cells. Melanoma Res. 21(5):417–425. [DOI] [PubMed] [Google Scholar]
- Peng Y, et al. 2011. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol. 28:1075–1081. [DOI] [PubMed] [Google Scholar]
- Peng Y, et al. 2017. Down-regulation of EPAS1 transcription and genetic adaptation of tibetans to high-altitude hypoxia. Mol Biol Evol. 34:818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Cui C, Ouzhuluobu, Wu T, Su B.. 2014. Prehistoric colonization and demographic history of modern humans on the Tibetan plateau In: The encyclopedia of life sciences (eLS). John Wiley & Sons, Ltd, The Atrium, Southern Gate, Chichester, West Sussex, PO19 8SQ, the United Kingdom; p. 1–10. [Google Scholar]
- Qi X, et al. 2013. Genetic evidence of Paleolithic colonization and Neolithic expansion of modern humans on the Tibetan Plateau. Mol Biol Evol. 30:1761–1778. [DOI] [PubMed] [Google Scholar]
- Qi X, Han J, Blench R, Rege JEO, Hanotte O.. 2008. Understanding the yak pastoralism in Central Asian Highlands: genetic evidence for origin, domestication and dispersion of domestic yak In: Sanchez-Mazas A, Blench R, Ross M, Peiros I, Lin M, editors. Past human migrations in East Asia and Taiwan: matching archaeology, linguistics and genetics. London: RoutledgeCurzon; p. 428–443. [Google Scholar]
- Qi X. 2004. Genetic diversity, differentiation and relationship of domestic yak populations – a microsatellite and mitochondrial DNA study [PhD thesis]. [Lanzhou, P.R. China]: Lanzhou University.
- Qiu Q, et al. 2012. The yak genome and adaptation to life at high altitude. Nat Genet. 44(8):946–949. [DOI] [PubMed] [Google Scholar]
- Qiu Q, et al. 2015. Yak whole-genome resequencing reveals domestication signatures and prehistoric population expansions. Nat Commun. 6:10283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GB, Liu W.. 1996. Distribution, status, and conservation of wild yak Bos grunniens. Biol Conserv. 76(1):1–8. [Google Scholar]
- Shami A, Goncalves I, Hultgardh-Nilsson A.. 2014. Collagen and related extracellular matrix proteins in atherosclerotic plaque development. Curr Opin Lipidol. 25(5):394–399. [DOI] [PubMed] [Google Scholar]
- Shan L, et al. 2014. Transcriptome profiling identifies differentially expressed genes in postnatal developing pituitary gland of miniature pig. DNA Res. 21(2):207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson TS, et al. 2010. Genetic evidence for high-altitude adaptation in Tibet. Science 329(5987):72–75. [DOI] [PubMed] [Google Scholar]
- Soumillon M, et al. 2013. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 3(6):2179–2190. [DOI] [PubMed] [Google Scholar]
- ten Freyhaus H, et al. 2011. Hypoxia enhances platelet-derived growth factor signaling in the pulmonary vasculature by down-regulation of protein tyrosine phosphatases. Am J Respir Crit Care Med. 183(8):1092–1102. [DOI] [PubMed] [Google Scholar]
- Trapnell C, et al. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28(5):511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, et al. 2013. CPAT: coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 41(6):e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. 2014. Transcriptome profile analysis of adipose tissues from fat and short-tailed sheep. Gene 549(2):252–257. [DOI] [PubMed] [Google Scholar]
- Wei Q, Yu H.. 2008. Comparison of histological structure of pulmonary alveoli between 180 days old yak and plain cattle. J Qinghai Univ (Nat Sci). 26:36–39. (In Chinese with English Abstract). [Google Scholar]
- Wiener G, Jianlin H, Ruijun L.. 2003. The Yak. Bangkok, Thailand: The Regional Office for Asia and the Pacific, Food and Agriculture Organization of the United Nations. [Google Scholar]
- Wu D-D, et al. 2018. Pervasive introgression facilitated domestication and adaptation in the Bos species complex. Nat Ecol Evol. 2:1139–1145. [DOI] [PubMed] [Google Scholar]
- Wu T, Kayser B.. 2006. High altitude adaptation in Tibetans. High Alt Med Biol. 7(3):193–208. [DOI] [PubMed] [Google Scholar]
- Xiang K, et al. 2013. Identification of a Tibetan-specific mutation in the hypoxic gene EGLN1 and its contribution to high-altitude adaptation. Mol Biol Evol. 30:1889–1898. [DOI] [PubMed] [Google Scholar]
- Xu S, et al. 2011. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol Evol. 28(2):1003–1011. [DOI] [PubMed] [Google Scholar]
- Xue Z, et al. 2013. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500(7464):593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, et al. 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329:75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu NY, et al. 2015. Complementing tissue characterization by integrating transcriptome profiling from the Human Protein Atlas and from the FANTOM5 consortium. Nucleic Acids Res. 43(14):6787–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RC. 1989. China Yak. Lanzhou, P.R. China: Gansu Scientific & Technology Press. [Google Scholar]
- Zhang X, et al. 1994. Report on the physiological and biochemical index for Chinese yak. China Yak. 3:12–25. (In Chinese with English Abstract). [Google Scholar]
- Zhao M, et al. 2009. Mitochondrial genome evidence reveals successful Late Paleolithic settlement on the Tibetan Plateau. Proc Natl Acad Sci U S A. 106:21230–21235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhirov IV, Zaseeva AV, Masenko VP, Tereshchenko SN.. 2014. Peroxisome proliferator-activated receptors-alpha (PPAR-alpha) and chronic heart failure: is there a reason to discuss the metabolic strategy of treatment? Ter Arkh. 86:78–82. [PubMed] [Google Scholar]
- Zhou J, Yu S, He J, Cui Y.. 2015. Comparative structural organization and morphological analyses of intrapulmonary artery of adult yak and cattle. Chin J Vet Sci. 35:1840–1844. (In Chinese with English Abstract). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.