Abstract
Advances in amyloid research rely on improved access to the β-amyloid peptide, Aβ. N-terminal methionine extended Aβ, Aβ(M1–42), is a readily expressed and widely used form of Aβ with comparable properties to the natural Aβ(1–42) peptide. Expression of Aβ(M1–42) is simple to execute and avoids an expensive and often difficult enzymatic cleavage step associated with expression and isolation of Aβ(1–42). This paper reports an efficient method for expression and purification of Aβ(M1–42) and 15N-labeled Aβ(M1–42). This method affords the pure peptide at about 19 mg per liter of bacterial culture through simple and inexpensive steps in three days. This paper also reports a simple method for construction of recombinant plasmids, and the expression and purification of Aβ(M1–42) peptides containing familial mutations. We anticipate that these methods will enable experiments that would otherwise be hindered by insufficient access to Aβ.
Graphical Abstact
Introduction
The β-amyloid peptide, Aβ, is central to the pathology of Alzheimer’s disease.1-3 The 40- and 42-amino acid alloforms of Aβ aggregate to form fibrils and oligomers in Alzheimer’s disease. The 42-amino acid alloform, Aβ(1–42) aggregates more rapidly and is more toxic than Aβ(1–40).4
A plentiful source of pure Aβ peptides, including isotopically labeled Aβ and various mutants associated with familial Alzheimer’s disease, is essential to progress in research in Aβ aggregation and Alzheimer’ s disease. Early efforts to generate Aβ focused on chemical synthesis of the peptide. Chemical synthesis of Aβ can lead to impurities, such as amino acid deletion products, that are difficult to eliminate during purification.5 Within the past fifteen years, expression has emerged as a useful alternative for preparing Aβ of superior purity. Expressed Aβ has been reported to aggregate three times faster and be significantly more toxic toward neuronal cells than synthetic Aβ.5 Expressed Aβ is typically generated as a fusion protein that is cleaved after expression using a protease.5-7 This approach requires expression and purification of the protease and an affinity purification step, which can make the preparation of Aβ costly and time-intensive.
Walsh and co-workers have introduced an Aβ expression system that circumvents the need for protease cleavage and affinity chromatography. In this expression system, Aβ(1–40) and Aβ(1–42) are expressed as variants Aβ(M1–40) and Aβ(M1–42) that contain an N-terminal methionine residue that originates from the translational start codon (Figure 1).8 Aβ(M1–40) and Aβ(M1–42) behave almost identically to the native peptides in aggregation and toxicity assays, and the additional N-terminal methionine has little impact on the fibril structure.9, 10 Because of these characteristics, Aβ(M1–40) and Aβ(M1–42) have emerged as widely used alternatives to the native Aβ peptides.11-16 Although this expression system provides ready access to Aβ(M1–40) with yields of 5-20 mg per liter of bacterial culture, the preparation of Aβ(M1–42) gives substantially lower yields.8
Figure 1.
Sequences of Aβ(1–40), Aβ(1–42), and Aβ(M1–42).
Here we report an efficient method for expression and purification of Aβ(M1–42) and associated homologues, including the uniformly 15N-labeled peptide and familial mutants. Our method simplifies the construction of plasmids containing mutant Aβ sequences and bypasses cumbersome steps in previously reported purification procedures. Our approach offers several major advantages over previous procedures: (1) a short preparation time of only three days, (2) minimal expense, (3) easier laboratory techniques, and (4) production of substantial amounts of highly pure Aβ peptides at about 19 mg per liter of bacterial culture.
Material and Methods
All chemicals were used as received unless otherwise noted. Deionized water (18 MΩ) was obtained from a Thermo Scientific Barnstead Genpure Pro water purification system. The pET-Sac-Aβ(M1–42) was a gift from Dominic Walsh (Addgene plasmid # 71875).8 DNA sequences that encode Aβ(M1–42) familial mutants were purchased in 500 ng quantities from Genewiz. NdeI and SacI restriction enzymes, CutSmart buffer, and shrimp alkaline phosphatase (rSAP) were purchased from New England Biolabs (NEB). TOP10 Ca2+-competent E. coli and BL21 DE3 PLysS Star Ca2+-competent E. coli, T4 ligase, and ethidium bromide were purchased from Thermo Fisher Scientific. Zymo ZR plasmid miniprep kit was purchased from Zymo Research. Zymoclean Gel DNA Recovery Kit was purchased from Zymo Research. Carbenicillin and chloramphenicol were purchased from RPI Research Products. The carbenicillin was added to culture media as a 1000X stock solution (50 mg/mL) in water. The chloramphenicol was added to culture media as a 1000X stock solution (34 mg/mL) in EtOH. 15NH4Cl was purchased from Cambridge Isotope Laboratories.
The concentration of DNA was measured using a Thermo Scientific NanoDrop spectrophotometer. E. coli were incubated in a Thermo Scientific MaxQ Shaker 6000. E. coli were lysed using a QSonica Q500 ultrasonic homogenizer. Analytical reverse-phase HPLC was performed on an Agilent 1200 instrument equipped with a Phenomonex Aeris PEPTIDE 2.6u XBC18 column with a Phenomonex SecurityGuard ULTRA cartridges guard column for C18 column. Preparative reverse-phase HPLC was performed on a Rainin Dynamax instrument SD-200 equipped with an Agilent ZORBAX 300SB-C8 semi-preparative column (9.4 × 250 mm) with a ZORBAX 300SB-C3 preparative guard column (9.4 × 15 mm). During purifications, the C8 column and the guard column were heated to 80 °C in a Sterlite plastic bin equipped with a Kitchen Gizmo Sous Vide immersion circulator. (Any water heater large enough to submerge a HPLC column should be sufficient.) HPLC grade acetonitrile and deionized water (18 MΩ), each containing 0.1% trifluoroacetic acid (TFA), were used for analytical and preparative reverse-phase HPLC. MALDI-TOF mass spectrometry was performed using an AB SCIEX TOF/TOF 5800 System. 1H-15N HSQC NMR was performed using a Bruker DRX500 500 MHz spectrometer equipped with a cryogenic probe.
For details of the expression and purification of Aβ(M1–42) and 15N-labeled Aβ(M1–42), the construction of recombinant plasmids, and the expression and purification of mutant Aβ(M1–42) peptides, see the Supporting Information.
Results and Discussion
The following describes the procedures that we have developed for the preparation of the Aβ(M1–42) wild-type and mutant peptides: For expression of the wild-type Aβ(M1–42) peptide, we use the commercially available plasmid, pET-Sac-Aβ(M1–42).8 For expression of mutant Aβ(M1–42) peptides, we construct recombinant plasmids containing mutant Aβ(M1–42) gene sequences using standard cloning techniques. Upon expression in E. coli, the peptides form inclusion bodies. The inclusion bodies are subjected to multiple rounds of washing, followed by solubilization in urea buffer. The resulting solution is filtered using a hydrophilic syringe filter and then immediately applied to a reverse-phase HPLC column. Pure HPLC fractions are then combined and lyophilized to give the peptide as a white powder. For biophysical and biological studies, the purified peptide is further treated with NaOH and then re-lyophilized. Yields are assessed both gravimetrically and by UV absorption. The composition and purity of the peptides are assessed by analytical HPLC, MALDI-MS, and SDS-PAGE with silver staining.
Expression of Aβ(M1–42)
To express Aβ(M1–42), pET-Sac-Aβ(M1–42) plasmid is transformed into BL21(DE3)pLysS competent E. coli. Expression is induced by isopropyl β-D-1-thiogalactopyranoside (IPTG). The expressed peptide is pelleted with the inclusion bodies, which are washed several times and then solubilized with 8 M urea. The yield of Aβ(M1–42) depends on the extent of cell growth prior to IPTG induction, with an OD600 of ca. 0.45 proving optimal for wild-type Aβ(M1–42) production. Growth to substantially higher or lower OD600 values gives lower yields of peptide.
Purification of Aβ(M1–42) by preparative HPLC
At this point in the procedure, the expressed peptide is handled like a synthetic peptide, and HPLC is used to purify it. The solution of the inclusion bodies in 8 M urea is filtered to prevent damaging the HPLC column. Initially, a 0.22 μm nylon syringe filter was used, but doing so resulted in substantial loss of peptide. The hydrophobicity and propensity of Aβ to aggregate appear to make Aβ particularly prone to loss in filters. We screened several types of syringe filters to optimize peptide recovery, monitoring the relative concentrations of peptide by UV absorbance at 280 nm (Table 1).17 We found that a 0.22 μm hydrophilic filter, such as hydrophilic polyvinylidene fluoride (PVDF) or polyethersulfone (PES), provided satisfactory peptide recovery.
Table 1.
The effect of different syringe filters on Aβ(M1–42) recovery.
| Filter type | UV absorbance at 280 nm |
Peptide recovery |
|---|---|---|
| non-filtered Aβ sample | 0.7279 ± 0.0052 | N/A |
| Millex-HV PES (0.22 μm) | 0.6294 ± 0.0001 | 86.5% |
| Fisher hydrophilic PVDF (0.22 μm) | 0.6279 ± 0.0009 | 86.3% |
| Millex-GV hydrophilic PVDF (0.22 μm) | 0.5703 ± 0.0003 | 78.3% |
| Fisher nylon (0.22 μm) | 0.2907 ± 0.0001 | 39.9% |
| Millex-GV MCE (0.22 μm) | 0.2273 ± 0.0001 | 31.2% |
A typical HPLC trace of unpurified Aβ(M1–42) shows three major peaks (Figure 2A). The first peak is the largest and contains mostly monomer, and the second and the third peaks appear to be oligomers (Figure S1). For preparative HPLC, a reverse-phase silica-based C8 column is used as the stationary phase, and a gradient of water and acetonitrile containing 0.1% trifluoroacetic acid is used as the mobile phase. To enhance resolution and reduce peak tailing, it was necessary to heat the column. Without heating, the resolution and yield of peptide are substantially lower. Aβ(M1–42) peptide monomer generally elutes at around 34% acetonitrile when the C8 column is heated to 80 °C in a water bath. HPLC fractions containing pure peptide were combined, and the purity was confirmed by analytical HPLC (Figure 2B). Acetonitrile was removed by rotary evaporation, and the aqueous solution of pure peptide was then frozen and lyophilized. These procedures typically yield about 19 mg of Aβ(M1–42) as the trifluoroacetate salt from one liter of bacterial culture.
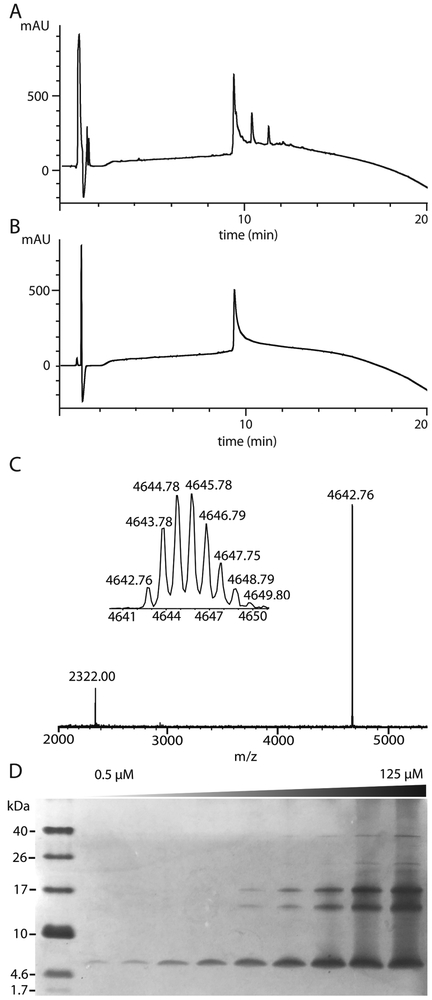
Figure 2.
Purification and characterization of Aβ(M1–42). (A) Typical analytical HPLC trace of filtered crude Aβ(M1–42) sample. (B) Typical analytical HPLC trace of purified Aβ(M1–42). (C) MALDI mass spectrum of purified Aβ(M1–42). (D) Silver-stained SDS-PAGE gel (16% polyacrylamide) of increasing concentrations of Aβ(M1–42) from 0.5 to 125 μM. A 12-μL aliquot was loaded in each lane of the gel.
This purification procedure does not require specialized equipment or costly reagents and is not time-consuming. It avoids the use of specialized and costly columns, such as cation-exchange chromatography columns and size-exclusion chromatography columns. Another advantage of this procedure is that it yields lyophilized powder as the final peptide product. Working with lyophilized peptide is convenient for subsequent studies as it can be dissolved in any appropriate buffer at a desired concentration.
The purity and composition of the Aβ(M1–42) peptide were further assessed through MALDI-MS, and SDS-PAGE with silver staining. MALDI-MS confirms that the observed mass of Aβ(M1–42) matches the expected mass (Figure 2C). The silver-stained SDS-PAGE gel shows that at low concentrations, the Aβ(M1–42) peptide exists as a monomer. At higher concentrations, Aβ(M1–42) begins to form oligomers with molecular weights consistent with trimers and tetramers (Figure 2D).
Sample preparation for biophysical and biological studies
The propensity of Aβ to aggregate necessitates the preparation of monomeric Aβ for subsequent studies.18 Without any sample preparation, studies are reported to be irreproducible.19 Fezoui and co-workers reported that treatment of Aβ with NaOH disrupts aggregates and generates Aβ that is monomeric or nearly monomeric.20 This NaOH-treated Aβ is widely used in subsequent aggregation studies.18
We applied this procedure to each batch of expressed Aβ to generate aliquots for further studies. Thus, the lyophilized powder was dissolved in 2 mM NaOH, and the pH was adjusted, if necessary, by addition of 0.1 M NaOH, to give a pH 10.5 solution. The solution was sonicated for one minute, the concentration was determined by UV absorbance at 280 nm, and the yield of Aβ(M1–42) was calculated. The solution was then aliquoted in 0.0055 or 0.020 micromole portions into small tubes, and these samples were frozen and lyophilized. The lyophilized aliquots are stored in a desiccator at −20 °C.
Expression of 15N-labeled Aβ(M1–42)
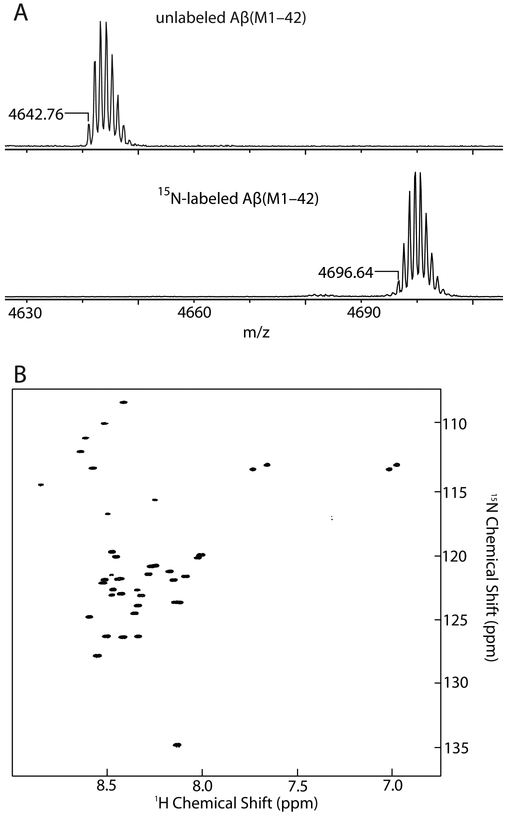
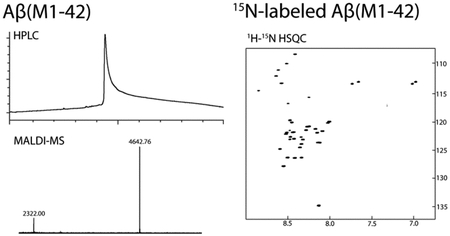
15N-labeled Aβ peptides are useful tools for structural studies by NMR and for studying binding profiles of Aβ. For expression of 15N-labeled Aβ(M1–42), E. coli are grown to an OD600 of ca. 0.45 in LB media, then the LB media is exchanged to M9 minimal media containing 15NH4Cl. Expression is induced in the 15N-enriched M9 media for 16 hours with IPTG. Purification and sample preparation of 15N-labeled Aβ(M1–42) is performed identically to unlabeled Aβ(M1–42). The composition of the 15N-labeled Aβ(M1–42) was assessed by MALDI-MS (Figure 3A). A 1H-15N HSQC NMR spectrum of 160 μM 15N-labeled Aβ(M1–42) in 50 mM potassium phosphate buffer in 10% D2O was recorded at 5 °C with a 500 MHz NMR spectrometer equipped with a cryogenic probe (Figure 3B). This spectrum matches the NMR spectrum reported by Macao and co-workers.9
Figure 3.
(A) MALDI spectra of unlabeled Aβ(M1–42) and 15N-labeled Aβ(M1–42) peptides. (B) 1H-15N HSQC NMR spectrum of 160 μM 15N-labeled Aβ(M1–42) peptide at 5 °C at 500 MHz equipped with a cryogenic probe.
The yield of the 15N-labeled Aβ(M1–42) peptide is comparable to that of the unlabeled Aβ(M1–42) peptide, at around 19 mg per liter of bacterial culture. Access to such amounts of the 15N-labeled peptide at low cost is enabling for performing experiments such as SAR by NMR.21
Construction of recombinant plasmids for expression of mutant Aβ(M1–42) peptides
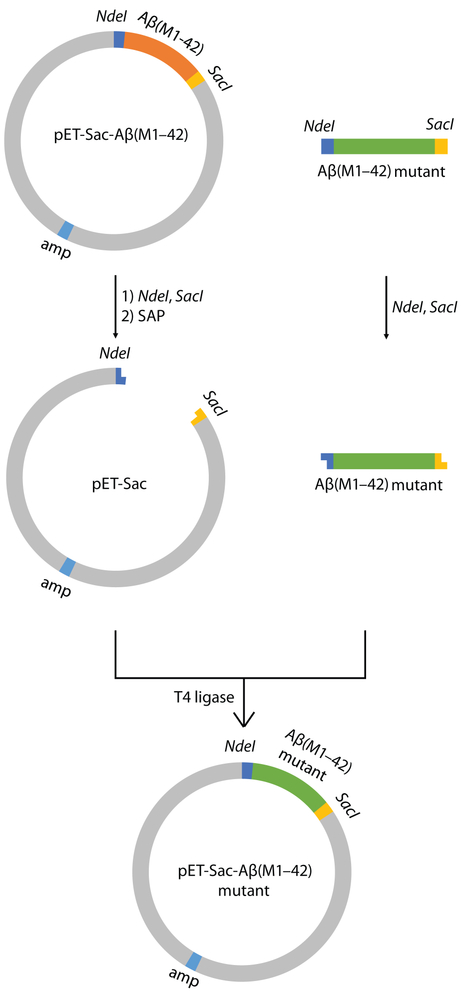
To express Aβ(M1–42) peptides containing familial mutations, we construct recombinant plasmids by ligating enzymatically digested pET-Sac-Aβ(M1–42) and DNA sequences that encode Aβ(M1–42) mutants (Figure 4). In this procedure, pET-Sac-Aβ(M1–42) is first digested with NdeI and SacI restriction enzymes to remove the wild-type Aβ(M1–42) sequence. Next, the digested pET-Sac vector is treated with shrimp alkaline phosphatase (rSAP) to remove the terminal phosphate groups. The digested vector is isolated by agarose gel electrophoresis purification using a commercially available kit. Synthetic DNA encoding each mutant Aβ(M1–42) is purchased and then digested with NdeI and SacI to generate the insert. The vector and insert are ligated using T4 ligase and then transformed into TOP 10 competent E. coli. E. coli transformed with ligated plasmid form colonies on agar containing carbenicillin. Plasmids are isolated from colonies, and the sequences are verified by DNA sequencing. For this paper, we constructed five plasmids with familial mutations: A21G, E22G, E22K, E22Q, and D23N.
Figure 4.
Molecular cloning strategy to construct recombinant plasmids of Aβ(M1–42) containing familial mutatations.
This cloning strategy is inexpensive and is simpler to execute than site-directed mutagenesis. The entire cloning procedure takes two days, and many mutants can be generated concurrently. Another advantage of this strategy is that Aβ(M1–42) plasmids containing multiple point mutations can be prepared as easily as plasmids containing single point mutations.
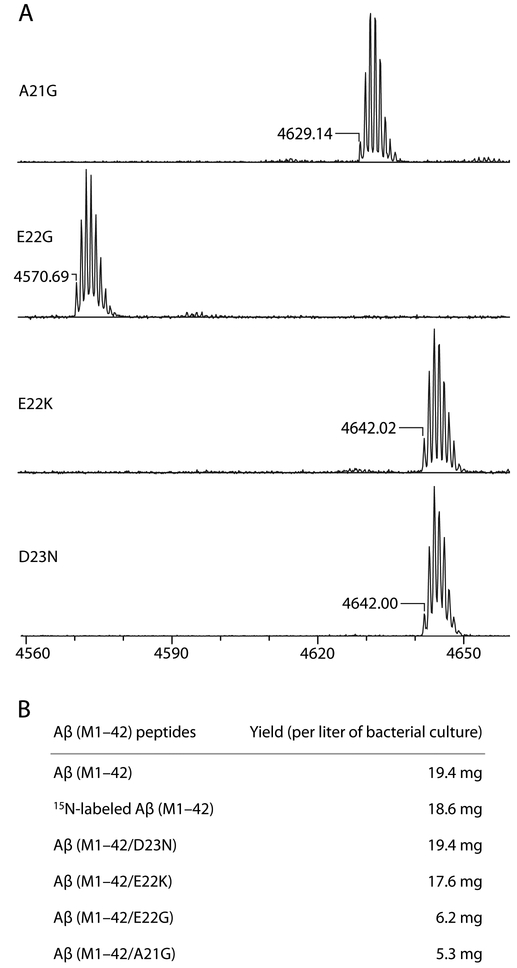
The purification and preparation of Aβ(M1–42) containing familial mutations is performed identically to that of Aβ(M1–42). The composition of familial mutant Aβ(M1–42) peptides was assessed using MALDI-MS (Figure 5A). The expression levels and yields of the Aβ(M1–42) familial mutants varied due to different aggregation propensities of the peptides. Analytical HPLC traces of crude samples of the A21G and E22Q mutants showed smaller first peaks and larger second and third peaks, suggesting that more oligomers are formed after dissolving the inclusion bodies. Figure 5B shows typical yields of the peptides. Our expression and purification procedures proved unsuitable for the E22Q mutant, which showed very little monomer in the HPLC trace.
Figure 5.
(A) MALDI mass spectra of Aβ(M1–42) peptides with A21G, E22G, E22K, and D23N mutations. (B) Typical yields of Aβ(M1–42), 15N-labeled Aβ(M` 1–42), and mutant Aβ(M1–42) peptides.
Conclusion
The procedures described herein provide an efficient method for expression and purification of Aβ(M1–42), 15N-labeled Aβ(M1–42), and Aβ(M1–42) containing several familial mutations. Our method employs the most convenient features of protein expression and peptide purification to provide ready access to good quantities of the pure peptides. We anticipate that our method will provide new opportunities to pilot experiments that require large amounts of Aβ. We also anticipate that this method can be adjusted for the expression and purification of other amyloidogenic proteins.
Supplementary Material
Acknowledgments
We thank Phil Dennison at the UCI NMR facility and Felix Grun and Ben Katz at the UCI Mass Spectrometry facility for their assistance and discussions. We also thank members of Martin, Spitale, Weiss, and Prescher laboratories for providing helpful advice and equipment.
Funding
This work was supported by the National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS), Grant GM097562.
Footnotes
Supporting Information
The Supporting Information contains procedures for the expression, purification, sample preparation, and characterization of Aβ(M1–42) and 15N-labeled Aβ(M1–42), and Aβ(M1–42) peptides with familial mutations.
The Supporting Information is available free of charge on the ACS Publications website at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
References
- 1.Benilova I, Karran E, and De Strooper B (2012) The toxic Abeta oligomer and Alzheimer's disease: an emperor in need of clothes, Nature neuroscience 15, 349–357. [DOI] [PubMed] [Google Scholar]
- 2.Haass C, and Selkoe DJ (2007) Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide, Nature reviews. Molecular cell biology 8, 101–112. [DOI] [PubMed] [Google Scholar]
- 3.Chiti F, and Dobson CM (2017) Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade, Annual review of biochemistry 86, 27–68. [DOI] [PubMed] [Google Scholar]
- 4.Jarrett JT, Berger EP, and Lansbury PT (2002) The carboxy terminus of the .beta. amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer's disease, Biochemistry 32, 4693–4697. [DOI] [PubMed] [Google Scholar]
- 5.Finder VH, Vodopivec I, Nitsch RM, and Glockshuber R (2010) The recombinant amyloid-beta peptide Abeta1-42 aggregates faster and is more neurotoxic than synthetic Abeta1-42, Journal of molecular biology 396, 9–18. [DOI] [PubMed] [Google Scholar]
- 6.Hortschansky P, Schroeckh V, Christopeit T, Zandomeneghi G, and Fandrich M (2005) The aggregation kinetics of Alzheimer's beta-amyloid peptide is controlled by stochastic nucleation, Protein science : a publication of the Protein Society 14, 1753–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao YH, and Chen YR (2015) A novel method for expression and purification of authentic amyloid-beta with and without (15)N labels, Protein expression and purification 113, 63–71. [DOI] [PubMed] [Google Scholar]
- 8.Walsh DM, Thulin E, Minogue AM, Gustavsson N, Pang E, Teplow DB, and Linse S (2009) A facile method for expression and purification of the Alzheimer's diseaseassociated amyloid beta-peptide, The FEBS journal 276, 1266–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macao B, Hoyer W, Sandberg A, Brorsson AC, Dobson CM, and Hard T (2008) Recombinant amyloid beta-peptide production by coexpression with an affibody ligand, BMC biotechnology 8, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silvers R, Colvin MT, Frederick KK, Jacavone AC, Lindquist S, Linse S, and Griffin RG (2017) Aggregation and Fibril Structure of AbetaM01-42 and Abeta1-42, Biochemistry 56, 4850–4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandberg A, Luheshi LM, Sollvander S, Pereira de Barros T, Macao B, Knowles TP, Biverstal H, Lendel C, Ekholm-Petterson F, Dubnovitsky A, Lannfelt L, Dobson CM, and Hard T (2010) Stabilization of neurotoxic Alzheimer amyloid-beta oligomers by protein engineering, Proc Natl Acad Sci U S A 107, 15595–15600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen SI, Linse S, Luheshi LM, Hellstrand E, White DA, Rajah L, Otzen DE, Vendruscolo M, Dobson CM, and Knowles TP (2013) Proliferation of amyloidbeta42 aggregates occurs through a secondary nucleation mechanism, Proc Natl Acad Sci U S A 110, 9758–9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertini I, Gonnelli L, Luchinat C, Mao J, and Nesi A (2011) A new structural model of Abeta40 fibrils, Journal of the American Chemical Society 133, 16013–16022. [DOI] [PubMed] [Google Scholar]
- 14.Colvin MT, Silvers R, Ni QZ, Can TV, Sergeyev I, Rosay M, Donovan KJ, Michael B, Wall J, Linse S, and Griffin RG (2016) Atomic Resolution Structure of Monomorphic Abeta42 Amyloid Fibrils, Journal of the American Chemical Society 138, 9663–9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meisl G, Yang X, Hellstrand E, Frohm B, Kirkegaard JB, Cohen SI, Dobson CM, Linse S, and Knowles TP (2014) Differences in nucleation behavior underlie the contrasting aggregation kinetics of the Abeta40 and Abeta42 peptides, Proc Natl Acad Sci U S A 111, 9384–9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munke A, Persson J, Weiffert T, De Genst E, Meisl G, Arosio P, Carnerup A, Dobson CM, Vendruscolo M, Knowles TPJ, and Linse S (2017) Phage display and kinetic selection of antibodies that specifically inhibit amyloid self-replication, Proc Natl Acad Sci U S A 114, 6444–6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Syringe filters with large pore sizes (0.45 μm) result in incomplete filtration of the peptide and risk damaging HPLC columns.
- 18.Teplow DB (2006) Preparation of Amyloid β-Protein for Structural and Functional Studies, 413, 20–33. [DOI] [PubMed] [Google Scholar]
- 19.Wood SJ, Maleeff B, Hart T, and Wetzel R (1996) Physical, morphological and functional differences between ph 5.8 and 7.4 aggregates of the Alzheimer's amyloid peptide Abeta, Journal of molecular biology 256, 870–877. [DOI] [PubMed] [Google Scholar]
- 20.Fezoui Y, Hartley DM, Harper JD, Khurana R, Walsh DM, Condron MM, Selkoe DJ, Lansbury PT, Fink AL, and Teplow DB (2009) An improved method of preparing the amyloid β-protein for fibrillogenesis and neurotoxicity experiments, Amyloid 7, 166–178. [DOI] [PubMed] [Google Scholar]
- 21.Shuker SB, Hajduk PJ, Meadows RP, and Fesik SW (1996) Discovering High-Affinity Ligands for Proteins: SAR by NMR, Science 274, 1531–1534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.