Abstract
In this study, 15 methanol-inducible and 9 constitutive promoters were used to drive the expression of Thermomyces dupontii lipase (TDL) in Pichia pastoris. Of the 15 methanol-inducible promoters, formaldehyde dehydrogenase promoter (PFLD1) showed the highest efficiency in driving lipase production, followed by alcohol oxidase 1 (PAOX1) and dihydroxyacetone synthase (PDAS1) promoters. The maximum lipase activity of transformants with PFLD1, PAOX1 and PDAS1 promoters in 5-l bioreactor was 27,076, 24,159 and 22,342 U/ml, respectively. For the nine constitutive promoters, glycosyl phosphatidyl inositol-anchored protein promoter (PGCW14) produced the highest amount of lipases in a medium containing glucose or glycerol as the only carbon source, followed by mitochondrial alcohol dehydrogenase isozyme (P0472) and glyceraldehyde-3-phosphate dehydrogenase (PGAP) promoters. The maximum lipase yields in 5-l bioreactors under the control of PGCW14, P0472 and PGAP promoters were 17,353, 15,046 and 14,276 U/ml, respectively. The result of this study not only identifies a few highly efficient promoters for the heterologous expression of TDL in P. pastoris, but also casts some insight into the optimization of protein production in heterologous systems.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1531-5) contains supplementary material, which is available to authorized users.
Keywords: Lipase, Pichia pastoris, Promoter, Thermomyces dupontii
Introduction
Lipases (triacylglycerol acylhydrolases EC 3.1.1.3) can be defined as a class of hydrolase. They can hydrolyze long-chain acylglycrols in aqueous medium. Meanwhile, they catalyze synthesis of esters and fine chemicals through direct esterification and transesterification in no-aqueous medium. Nowadays, lipases are widely used in industries of feed additives, detergent, food processing, fine chemicals, and biodiesel production (Jaeger and Eggert 2002; Singh et al. 2016).
The lipase from Thermomyces dupontii (formerly classified as Talaromyces thermophilus) is a thermo-active and alkaline enzyme (named TDL). Previous studies revealed TDL with great potential values in many industrial applications. TDL presented good stability in alkaline pH and different surfactants, which is helpful for its application as additive in detergents (Romdhane et al. 2010). Meanwhile, the high transesterification activity of TDL is very useful for its application in biodiesel production (Romdhane et al. 2013). Furthermore, TDL can be used as a biocatalyst for the synthesis of chiral intermediate of Pregablin through kinetic resolution of 2-carboxyethyl-3-cyano-5-methylhexanoic acid ethyl ester (CNDE) (Ding et al. 2018). Although TDL shows a potential value for commercial application, the low production of TDL and secretion of TDL with other hydrolytic enzymes by T. dupontii are the main hurdles limiting its further application (Romdhane et al. 2012). Heterologous expression of TDL in P. pastoris is an effective method to overcome these hurdles. Previous study demonstrated that the production of TDL in P. pastoris was 260% higher than that of T. dupontii at flask shake culture. Furthermore, SDS–PAGE showed that the recombinant TDL was almost free from contaminating proteins, since the recombinant strain secretes few native proteins, which facilitates downstream processing (Zhang et al. 2015).
Promoter plays an important role in the expression of heterologous lipases in P. pastoris. Generally, the promoters for the expression of lipases in P. pastoris include methanol-inducible and constitutive promoters. Until now, the most commonly used methanol-inducible and constitutive promoters are alcohol oxidase 1 promoter (PAOX1) and glyceraldehyde-3-phosphate dehydrogenase promoter (PGAP), respectively. PAOX1 is extensively employed for the expression of lipases in P. pastoris (Borrelli and Trono 2015). Compared to PAOX1, PGAP is used to drive some lipase expression (Wang et al. 2012a, b). Besides PAOX1 and PGAP, few promoters are employed for the expression of lipase in P. pastoris. In this study, TDL is expressed in P. pastoris under the control of 15 methanol-inducible and 9 constitutive promoters. The results of this study will provide some suitable promoters for heterologous expression of TDL and lay a foundation for further improvement of this lipase in P. pastoris.
Materials and methods
Strains, plasmids and reagents
The Escherichia coli strain Top 10 was conserved in our laboratory and used to propagate plasmids. P. pastoris X33 was purchased from Invitrogen (Carlsbad, CA, USA) and used as host for the expression of TDL. The vector pPICZαA was also purchased from Invitrogen. DNA polymerase (PrimeSTARTMHS), restriction enzymes (EcoRI, NotI and SacI), in-fusion cloning kit and T4-DNA ligase were purchased from Takara Biotechnology (Dalian, China). The gene of TDL (GenBank: JF414585.1) without signal sequence was optimized according to the preference of P. pastoris and synthesized by the Genewiz (Suzhou, China). Sodium hydroxide was purchased from Merck (Darmstadt, Germany).
Medium
Media for E. coli Top 10 and P. pastoris X33 included LBZ (LB with 25 µg/ml zeocin), YPD (yeast extract peptone dextrose medium), YPDZ (yeast extract peptone dextrose medium with 100 µg/ml zeocin), BMGY (buffered glycerol complex medium), BMMY (buffered methanol complex medium) and BMDY (buffered dextrose complex medium). LBZ, YPDZ, BMGY, BMDY and BMMY were prepared according to the protocol provided by Invitrogen (https://www.thermofisher.com).
Promoter cloning and construction expression vectors
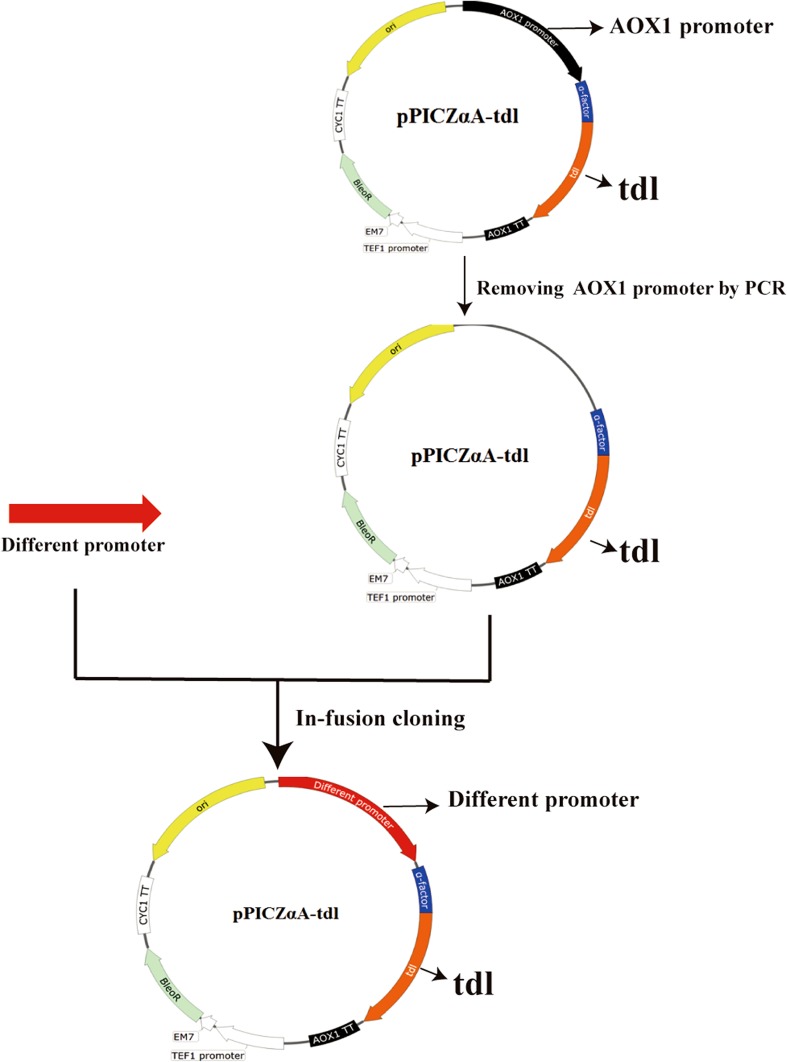
Based on the results of previous studies (De Schutter et al. 2009; Xu et al. 2018), 15 methanol-inducible and 9 constitutive promoters were selected for heterologous expression of TDL. The optimized gene (tdl-opt) without the predicted signal sequence was digested by EcoRI and NotI, and ligated into pPICZαA to form pPICZαA–tdl-opt, for the expression of TDL under promoter PAOX1. The following 23 expression vectors with different promoters were constructed based on the pPICZαA–tdl-opt. The 3.6-kb fragment of pPICZαA–tdl-opt without promoter PAOX1 was amplified from pPICZαA–tdl-opt using the specific primers. And then, the 23 different promoters were ligated to 3.5-kb fragment by in-fusion cloning to form 23 expression vectors with different promoters. All the expression vectors contained the saccharomyces cerevisiae α-factor secretion signal downstream of different promoters. The details of these 24 promoters are listed in Table 1. The primers used in this study are listed in supplementary Table 1.
Table 1.
List of selected promoters for the heterologous expression of TDL
| Promoters | Corresponding genes | Locus ID |
|---|---|---|
| PAOX1 | Alcohol oxidase 1 | PAS_chr4_0821 |
| PAOX2 | Alcohol oxidase 2 | PAS_chr4_0152 |
| PDAS1 | Dihydroxyacetone synthase 1 | FJ752551.1 |
| PDAS2 | Dihydroxyacetone synthase 2 | FJ752552.1 |
| PFLD1 | Formaldehyde dehydrogenase | AF066054.1 |
| PTPI | Triose phosphate isomerase | PAS_chr3_0951 |
| PCAT | Catalase A | PAS_chr2-2_0131 |
| PFDH | NAD(+)-dependent formate dehydrogenase | PAS_chr3_0932 |
| PFBP | Fructose 1,6-bisphosphatase | PAS_chr3_0868 |
| PFBA2 | Fructose 1,6-bisphosphate aldolase | PAS_chr1-1_0319 |
| P0547 | Peroxiredoxin | PAS_chr1-4_0547 |
| P0043 | Mitochondrial aldehyde dehydrogenase | PAS_chr4_0043 |
| PFGH | Non-essential intracellular esterase | PAS_chr3_0867 |
| P0374 | Hypothetical protein | PAS_chr3_0374 |
| P0099 | Mitochondrial NAD+ transporter | PAS_chr3_0099 |
| PGAP | Glyceraldehyde 3-phosphate dehydrogenase | PAS_chr2-1_0437 |
| PGCW14 | Glycosyl phosphatidyl inositol-anchored protein | PAS_chr1-4_0586 |
| P0472 | Mitochondrial alcohol dehydrogenase isozyme | PAS_chr2-1_0472 |
| P0769 | Pyruvate kinase | PAS_chr2-1_0769 |
| P0188 | Pyruvate decarboxylase isozymes | PAS_chr3_0188 |
| P0090 | Polyamine transport protein specific for spermine | PAS_chr1-4_0090 |
| P0065 | Plasma membrane multidrug transporter of the major facilitator superfamily | PAS_chr2-2_0065 |
| P0016 | 2-Deoxyglucose-6-phosphate phosphatase | PAS_chr2-1_0016 |
| PTEF | Translation elongation factor 1 | M10992.1 |
Transformation of P. pastoris and isolation of recombinant clones
The recombinant E. coli Top 10 containing different expression plasmids were grown in LBZ at 37 °C and 220 rpm for 18 h. The expression plasmids were isolated and purified using TIANprep Midi Plasmid Kit (TIANGEN, Beijing, China). The expression vectors with different promoters were linearized with SacI and the concentration of SacI-linearized plasmids were analyzed by NanoDrop 2000 (Thermo Scientific). 80 ng SacI-linearized plasmids were transformed into P. pastoris X33 competent cell. The transformants were plated and screened on YPDZ plates. The detailed protocol for the selection of recombinant strains is provided in supplementary material. The gene copy numbers of recombinant strain with different promoters were detected by Quantitative real-time PCR (qPCR). The single-copied recombinant strains were isolated for shake flask fermentation.
Shake flask cultures
For the expression of TDL under methanol-inducible promoters, the single-copied recombinant strains were inoculated into 10 ml BMGY in a 150-ml flask and incubated at 30 °C and 200 rpm for 24 h. Then the recombinant cells were harvested by centrifugation, re-suspended in BMMY, and transferred to 50 ml BMMY (OD600 is 1.0) in a 250-ml flask, incubated at 30 °C and 200 rpm. 0.75% (v/v) methanol was added to the culture at every 24 h, and 0.5 ml culture was harvested every 24 h for lipase and biomass concentration assay. The recombinant strains showing higher lipase activity were isolated for high cell density fermentation.
For the expression of TDL under constitutive promoters, the single-copied recombinant strains were also inoculated into 10 ml YPD in a 150-ml flask and cultured overnight at 30 °C and 200 rpm. 1 ml of overnight culture was transferred to 250-ml flask containing 50 ml BMGY or BMDY, and incubated at 30 °C and 200 rpm for 72 h. 1 ml culture was harvested every 24 h for lipase and biomass concentration assay. The recombinant strains with higher lipase activity were isolated for high cell density fermentation.
High cell density fermentation
High cell density fermentation was carried out in 5-l bioreactor. The detailed protocol of high cell density fermentation is provided in supplementary material. For the expression of TDL under methanol-inducible promoters, the recombinant strains were cultivated for about 23 h in batch on glucose medium. When the glucose was exhausted, it was then changed to methanol induction phase. For the expression of TDL under constitutive promoters, the recombinant strains were cultivated on basal salt medium and using glucose or glycerol as sole carbon source. The enzyme activity, total protein concentration and dry cell weight (DCW) were monitored throughout the fermentation.
Detection methods
The lipase activity was detected by the pH-stat (Metrohm, Herisau, Switzerland) method using olive oil as substrate according to the previous method (Wang et al. 2013). The pH and temperature for lipase activity determination were set at 9.5 and 60 °C, respectively. The amount of enzyme that liberates 1 µmol fatty acid per minute was defined as one unit (U) of the activity. The concentration of total protein was detected by Bradford method using BSA as standard. DCW was determined according to the previous method (Wang et al. 2013). The qPCR experiments were performed according to the previous method (Sha et al. 2013). The glyceraldehyde 3-phosphate dehydrogenase gene (GAP) was selected as the reference gene.
Results
Construction of expression vectors with different promoters
15 methanol-inducible and 9 constitutive promoters were selected for the heterologous expression of TDL in P. pastoris (Table 1). These 15 methanol-inducible promoters included PAOX1, PFLD1 and PDAS1, etc. The nine constitutive promoters included PGAP, PGCW14, mitochondrial alcohol dehydrogenase isozyme promoter (P0472), etc. Those methanol-inducible and constitutive promoters were cloned from P. pastoris X33, which were about 800 bp upstream of the start codon ATG. The process for construction of different expression vectors is depicted in Fig. 1.
Fig. 1.
Construction of the recombinant vectors containing different promoters
Heterologous expression of TDL under methanol-inducible promoters
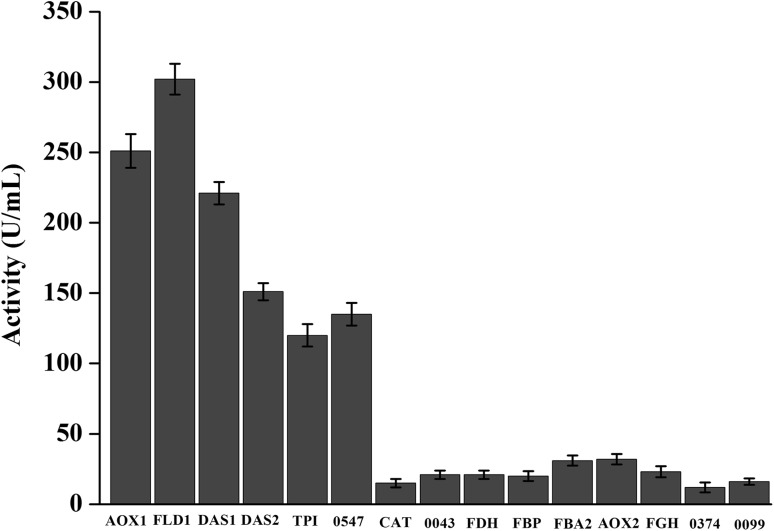
Those 15 expression vectors with different methanol-inducible promoters were linearized and transformed to P. pastoris X33. The recombinant strain with classic promoter PAOX1 was used as control. Due to a small amount (80 ng) of linearized plasmids were transformed into P. pastoris X33, only two to four clones were isolated for each promoter. The mean relative lipase activity of recombinant strains with different methanol-inducible promoters are mostly distributed from 4.8 to 120.3% compared to the recombinant strain with promoter PAOX1 (Fig. 2). Meanwhile, the gene copy numbers of recombinant strains with different methanol-inducible promoters were determined by qPCR. The results of qPCR showed that these recombinant strains were single-copied clone. The lipase activities of recombinant strain using PAOX1 (named X33–PAOX1–tdl), PDAS1 (named X33–PDAS1–tdl) and PFLD1 (named X33–PFLD1–tdl) were much higher than other promoters (Fig. 2). So X33–PAOX1–tdl, X33–PDAS1–tdl and X33–PFLD1–tdl were isolated for shake flask fermentation. After 120 h methanol induction, the maximum lipase activity of X33–PAOX1–tdl, X33–PDAS1–tdl and X33–PFLD1–tdl were 542, 602 and 585 U/ml, respectively (supplementary Fig. 1). The OD600 values of these three recombinant strains were similar during the process of cultivation.
Fig. 2.
Lipase activities of the transformant representatives controlled by different promoters
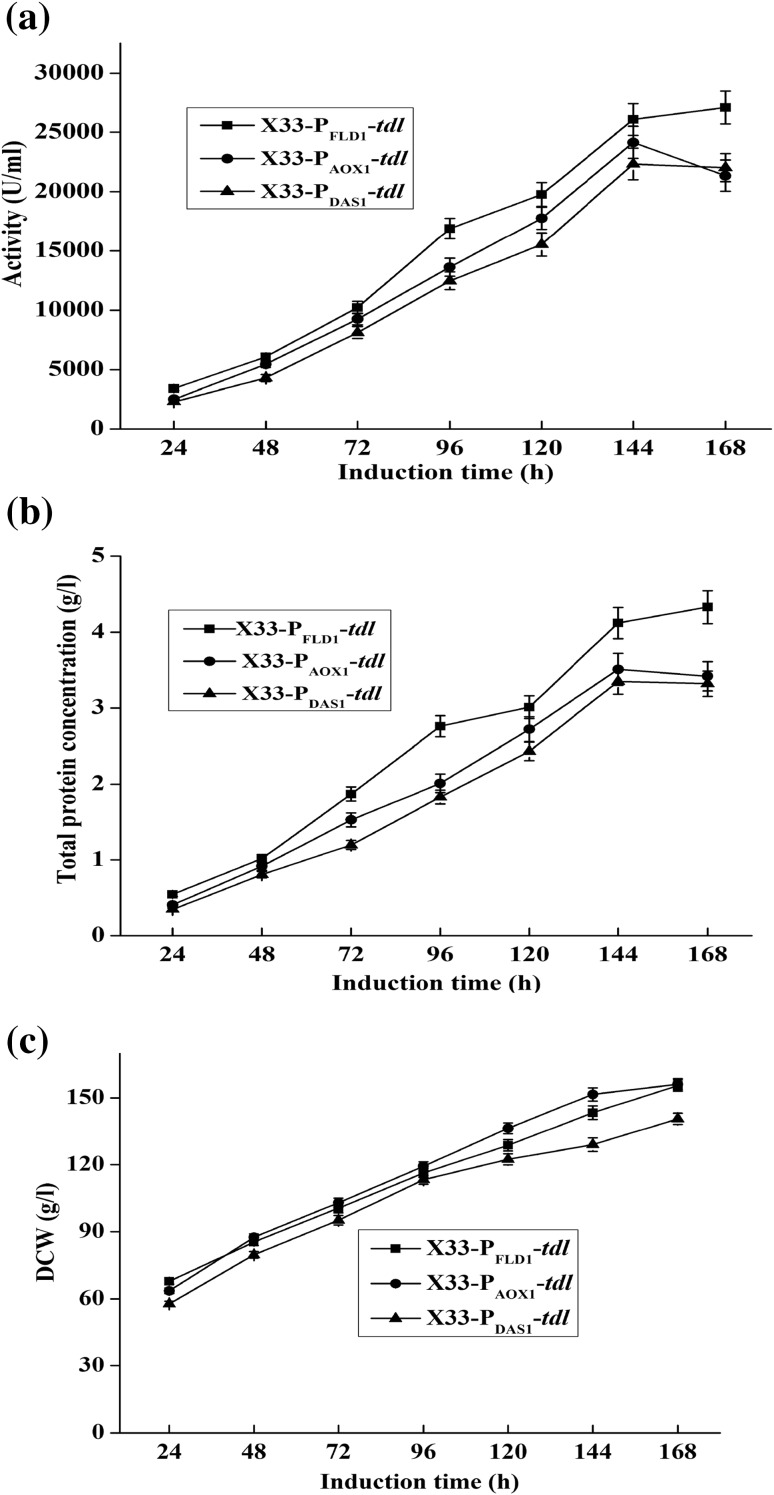
Furthermore, X33–PAOX1–tdl, X33–PDAS1–tdl and X33–PFLD1–tdl were cultivated in 5-l bioreactor. The lipase activity, total protein concentration and DCW of these three recombinant strains are shown in Fig. 3. X33–PFLD1–tdl also exhibited the highest activity among these three recombinants in 5-l bioreactor. The maximum lipase activity of X33–PFLD1–tdl reached 27,076 U/ml after 168 h induction, which was 1.11- and 1.21-fold higher than those of the recombinant strains X33–PAOX1–tdl and X33–PDAS1–tdl, respectively (Fig. 3a). The maximum total protein concentration of X33–PAOX1–tdl, X33–PDAS1–tdl and X33–PFLD1–tdl were 3.65, 3.35, and 4.63 g/l, respectively (Fig. 3b). The maximum DCW of X33–PFLD1–tdl, X33–PAOX1–tdl and X33–PDAS1–tdl were 147, 146 and 141 g/l, respectively (Fig. 3c).
Fig. 3.
Lipase activity (a), total protein concentration (b) and DCW (c) of X33–PFLD1–tdl, X33–PAOX1–tdl and X33–PDAS1–tdl in 5-l bioreactor. The lipase activity was detected by pH-stat method (pH 9.5 and 60 °C) using olive oil as substrate. Total protein concentration was detected by Bradford method. DCW was obtained by centrifuging 10 ml samples in a pre-weighted 20 ml centrifuge tube and discarding supernatant, and then allowing the pellet to constant weight at 100 °C. Measurements are the average value ± standard error from independently duplicate fermentations
The heterologous expression of TDL under constitutive promoters
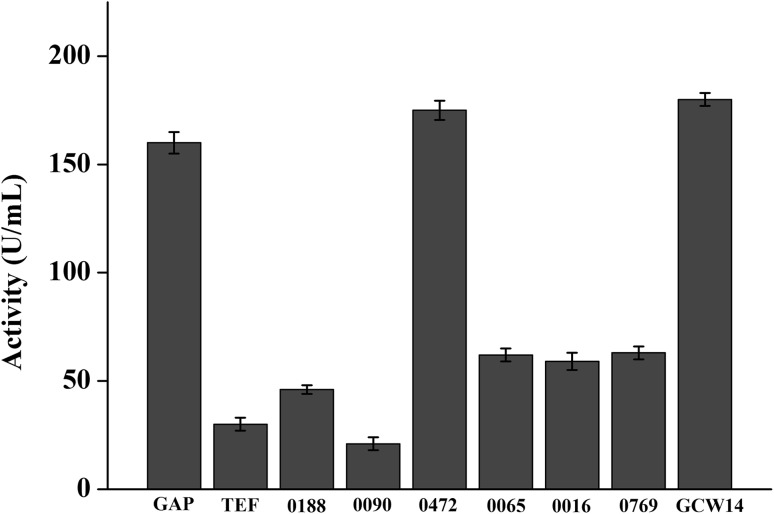
Those nine constitutive expression vectors were transformed to P. pastoris X33 by electro-transformation. The recombinant strains were cultured in YPD medium and lipase activity was determined after 36 h cultivation at 30 °C and 200 rpm. The recombinant strain with PGCW14 showed the highest lipase activity, followed by P0472 and PGAP transformant (Fig. 4). Meanwhile, the results of qPCR revealed that these recombinant strains were single-copied clone. The PGCW14, P0472 and PGAP transformant (named X33–PGAP–tdl, X33–PGCW14–tdl and X33–P0472–tdl) were selected for shake flask fermentation. These three recombinant strains were cultured in BMGY or BMDY medium for 72 h at 30 °C and 200 rpm. The maximum lipase activity of X33–PGAP–tdl, X33–PGCW14–tdl and X33–P0472–tdl in BMGY medium were 241, 251 and 265 U/ml, respectively. The maximum lipase activities of these three recombinant strains in BMDY medium were 261, 273 and 286 U/ml, respectively (supplementary Fig. 2).
Fig. 4.
Lipase activities of different constitutive promoter transformants during initial isolation
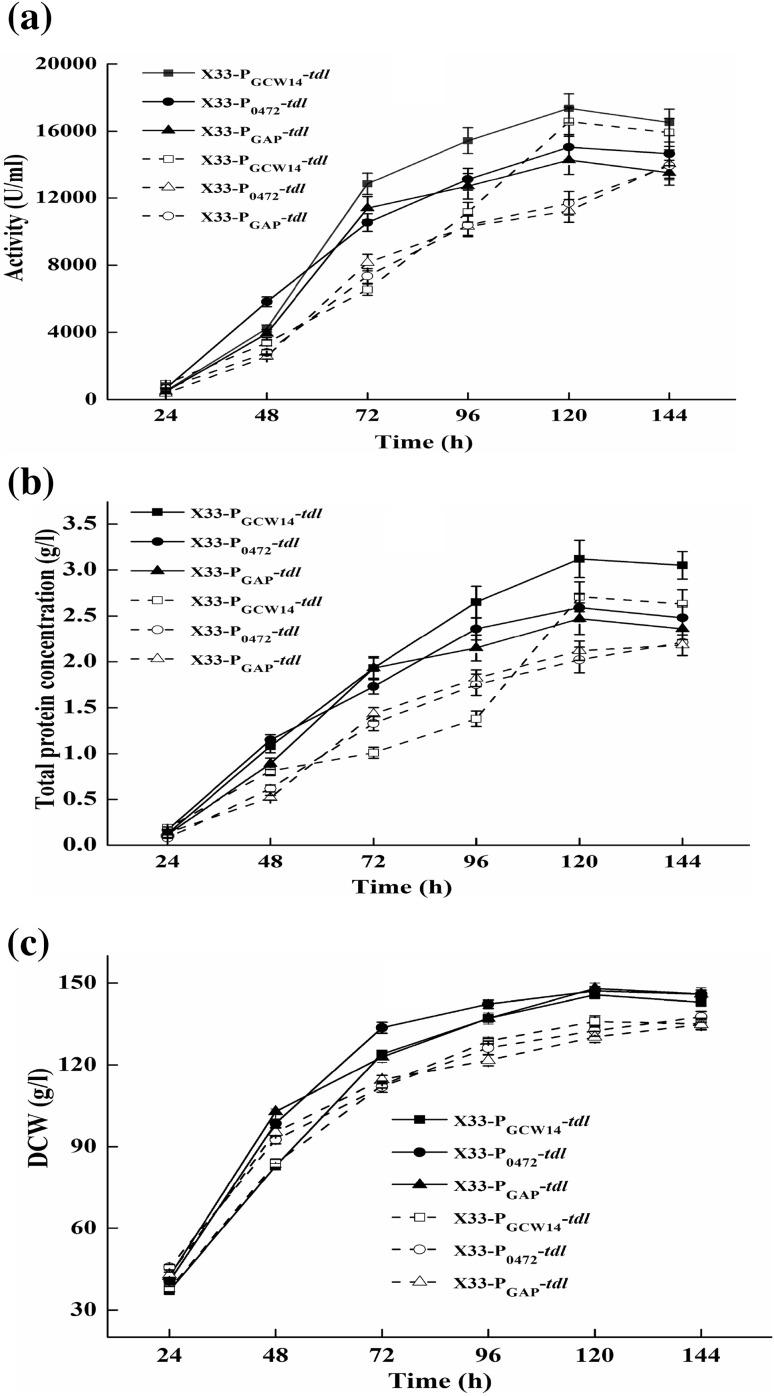
The expression level of TDL under PGAP, PGCW14 and P0472 was further evaluated in 5-l bioreactor. As seen in Fig. 5, the maximum lipase activity and total protein concentration produced by X33–PGCW14–tdl in medium using glucose as sole carbon source reached 16,569 U/ml and 2.71 g/l, respectively. Compared with X33–PGAP–tdl, the maximum lipase activity and total protein concentration were increased by 18.8 and 24.3%, respectively. Meanwhile, the maximum lipase activity and total protein concentration of TDL driven by PGCW14 were 1.18- and 1.23-folds higher than that by P0472. The DCW of X33–PGAP–tdl, X33–PGCW14–tdl and X33–P0472–tdl were almost same during the high cell density fermentation (Fig. 5c). The lipase activity, total protein concentration and DCW of these three recombinant strains in medium using glycerol as sole carbon source are depicted in Fig. 5. The maximum lipase activity of X33–PGCW14–tdl was 17,353 U/ml, which was 115.3 and 121.5% to that of X33–P0472–tdl and X33–PGAP–tdl, respectively (Fig. 5a). The maximum total protein concentration of X33–PGCW14–tdl reached 3.12 g/l at 120 h, which was 120.4 and 126.3% to that of X33–P0472–tdl and X33–PGAP–tdl, respectively (Fig. 5b). The maximum DCW of X33–PGAP–tdl, X33–PGCW14–tdl and X33–P0472–tdl was 148, 145 and 147 g/l, respectively (Fig. 5c).
Fig. 5.
Lipase activity (a), total protein concentration (b) and DCW (c) of X33–PGCW14–tdl, X33–P0472–tdl and X33–PGAP–tdl in 5-l bioreactor. Solid line represents glycerol as sole carbon source and dashed line represents the glucose as sole carbon source. Measurements are the average value ± standard error from independently duplicate fermentations
Discussion
As a thermo-active and alkaline lipase, TDL shows a great potential value in many industrial applications. The production of TDL from the wild-type strain T. dupontii is less than satisfactory, which limits its further applications. For commercial exploitation of TDL, it is crucial to achieve high yield of this protein. Previous study demonstrated that heterologous expression of TDL in P. pastoris was an effective method to improve the production of TDL (Zhang et al. 2015). As one of the most commonly used expression systems, P. pastoris has been used extensively for heterologous protein production. P. pastoris has many advantages such as powerful secretion ability, ease of genetic manipulation, and mature fermentation process (Ahmad et al. 2014; Li et al. 2017).
Promoter is a key factor for heterologous lipase production in P. pastoris (Vogl and Glieder 2013; Arruda et al. 2016). Until now, PAOX1 is the most commonly used promoter for the efficient expression of heterologous lipase. Besides PAOX1, there are few methanol promoters which have been used to drive the expression of lipase. In this study, TDL was expressed under the control of 15 methanol promoters. To circumvent gene copy number on the production of TDL, only single-copied clone was selected for study. X33–PFLD1–tdl showed the highest lipase activity at flask shake (602 U/ml) and 5-l fermentor (27,076 U/ml) among all the 15 promoter transformants, followed by X33–PAOX1–tdl and X33–PDAS1–tdl. Alcohol oxidase 1 (AOX1), formaldehyde dehydrogenase (FLD1) and dihydroxyacetone synthase (DAS) are key enzymes of the methanol utilization pathway. These genes are strongly induced when methanol is only carbon source (Yurimoto et al. 2011). The lipase activity from PFLD1 was 1.06-fold higher than PAOX1 which agreed with previous research that the expression level of Yarrowia lipolytica lipase 2 under control of PFLD1 was comparable with PAOX1 (Wang et al. 2012a, b). Meanwhile, the production of TDL under PDAS1 was 22,342 U/ml, which was 90.9% of PAOX1. These results indicate that PFLD1 and PDAS1 are alternative methanol-inducible promoters to PAOX1 for the TDL production in P. pastoris.
Expression under the control of constitutive promoters is also an effective method to improve the production of TDL. Compared to methanol-inducible promoters, constitutive promoters have several advantages such as omits the use of methanol as inducer, simplifies cultivation efforts, and shortens fermentation time (Vogl and Glieder 2013). In the present study, nine constitutive promoters were chosen for the expression of TDL based on literature data. These nine promoters were tested on glycerol and glucose as carbon sources. PGCW14 transformant showed the highest lipase activity among all test promoters, followed by P0472 and PGAP transformant, when glucose is only carbon source. The change to glycerol as only carbon source, PGCW14 transformant also exhibited the highest lipase activity. These results are similar to previous studies, the expression level of EGFP under control of PGCW14 was ten and five times higher than PGAP when carbon sources were glucose and glycerol, respectively (Liang et al. 2013). Previous study showed that the maximum amylase activity from P0472 recombinant strain was 1.25-fold higher than PGAP strain during fed batch cultivation at 5-l bioreactor (Xu et al. 2018). These results suggest that PGCW14 and P0472 are an attractive alternative to PGAP for the expression of TDL in P. pastoris.
The production of TDL under the control of other promoters was lower than above methanol-inducible (PFLD1, PDAS1 and PAOX1) and constitutive promoters (PGAP, PGCW14 and P0472), which means they are not the ideal promoters for the heterologous expression of TDL. However, these different promoters will become a toolbox in our further research. The effects of co-expression of chaperones on the production of TDL in P. pastoris will be investigated in our further study. Different chaperones will be expressed under the control of these promoters. The results of this study demonstrated that PFLD1, PDAS1, PGCW14 and P0472 were an attractive alternative to classic promoter PAOX1 and PGAP for the expression of TDL in P. pastoris. These 24 promoters will lay a foundation for our further study and also give some clues for the heterologous expression of recombinant protein in P. pastoris.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (DOCX 11069 KB)
Acknowledgements
This work was supported by the National High Technology Project of People’s Republic of China (No. 2014AA093514).
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
References
- Ahmad M, Hirz M, Pichler H, Schwab H. Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol. 2014;98:5301–5317. doi: 10.1007/s00253-014-5732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda A, Reis VC, Batista VD, Daher BS, Piva LC, De Marco JL, de Moraes LM, Torres FA. A constitutive expression system for Pichia pastoris based on the PGK1 promoter. Biotechnol Lett. 2016;38:509–517. doi: 10.1007/s10529-015-2002-2. [DOI] [PubMed] [Google Scholar]
- Borrelli GM, Trono D. Recombinant lipases and phospholipases and their use as biocatalysts for industrial applications. Int J Mol Sci. 2015;16:20774–20840. doi: 10.3390/ijms160920774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter K, Lin YC, Tiels P, Van Hecke A, Glinka S, Weber-Lehmann J, Rouzé P, Van de Peer Y, Callewaert N. Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol. 2009;27:561–566. doi: 10.1038/nbt.1544. [DOI] [PubMed] [Google Scholar]
- Ding X, Zheng RC, Tang XL, Zheng YG. Engineering of Talaromyces thermophilus lipase by altering its crevice-like binding site for highly efficient biocatalytic synthesis of chiral intermediate of Pregablin. Bioorg Chem. 2018;77:330–338. doi: 10.1016/j.bioorg.2018.01.018. [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Eggert T. Lipases for biotechnology. Curr Opin Biotechnol. 2002;13:390–397. doi: 10.1016/S0958-1669(02)00341-5. [DOI] [PubMed] [Google Scholar]
- Li J, Sun C, Chen L, Sun L, Duan L, Zheng Q, Hu X. Optimization of the secretory expression of recombinant human C-reactive protein in Pichia pastoris. 3 Biotech. 2017;7:291. doi: 10.1007/s13205-017-0917-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Zou C, Lin Y, Zhang X, Ye Y. Identification and characterization of PGCW14: a novel, strong constitutive promoter of Pichia pastoris. Biotechnol Lett. 2013;35:1865–1871. doi: 10.1007/s10529-013-1265-8. [DOI] [PubMed] [Google Scholar]
- Romdhane IB, Fendri A, Gargouri Y, Gargouri A, Belghith H. A novel thermoactive and alkaline lipase from Talaromyces thermophilus fungus for use in laundry detergents. Biochem Eng J. 2010;53:112–120. doi: 10.1016/j.bej.2010.10.002. [DOI] [Google Scholar]
- Romdhane IB, Frikha F, Maalej-Achouri I, Gargouri A, Belghith H. Gene cloning and molecular characterization of the Talaromyces thermophilus lipase catalyzed efficient hydrolysis and synthesis of esters. Gene. 2012;494:112–118. doi: 10.1016/j.gene.2011.11.059. [DOI] [PubMed] [Google Scholar]
- Romdhane IB, Romdhane ZB, Bouzid M, Gargouri A, Belghith H. Application of a chitosan-immobilized Talaromyces thermophilus lipase to a batch biodiesel production from waste frying oils. Appl Biochem Biotechnol. 2013;171:1986–2002. doi: 10.1007/s12010-013-0449-y. [DOI] [PubMed] [Google Scholar]
- Sha C, Yu XW, Li F, Xu Y. Impact of gene dosage on the production of lipase from Rhizopus chinensis CCTCC M201021 in Pichia pastoris. Appl Biochem Biotechnol. 2013;169:1160–1172. doi: 10.1007/s12010-012-0050-9. [DOI] [PubMed] [Google Scholar]
- Singh R, Kumar M, Mittal A, Mehta PK. Microbial enzymes: industrial progress in 21st century. 3 Biotech. 2016;6:174. doi: 10.1007/s13205-016-0485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl T, Glieder A. Regulation of Pichia pastoris promoters and its consequences for protein production. N Biotechnol. 2013;30:385–404. doi: 10.1016/j.nbt.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Wang X, Sun Y, Ke F, Zhao H, Liu T, Xu L, Liu Y, Yan Y. Constitutive expression of Yarrowia lipolytica lipase LIP2 in Pichia pastoris using GAP as promoter. Appl Biochem Biotechnol. 2012;166:1355–1367. doi: 10.1007/s12010-011-9524-4. [DOI] [PubMed] [Google Scholar]
- Wang XF, Shen XG, Sun YC, Zhao HY, Xu L, Liu Y, Yan YJ. Production of Yarrowia lipolytica lipase LIP2 in Pichia pastoris using the nitrogen source-regulated FLD1 promoter. J Chem Technol Biot. 2012;87:553–558. doi: 10.1002/jctb.2749. [DOI] [Google Scholar]
- Wang JR, Li YY, Xu SD, Li P, Liu JS, Liu DN. High-level expression of pro-form lipase from Rhizopus oryzae in Pichia pastoris and its purification and characterization. Int J Mol Sci. 2013;15:203–217. doi: 10.3390/ijms15010203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Zhu J, Zhu Q, Xing Y, Cai M, Jiang T, Zhou M, Zhang Y. Identification and characterization of novel promoters for recombinant protein production in yeast Pichia pastoris. Yeast. 2018;35:379–385. doi: 10.1002/yea.3301. [DOI] [PubMed] [Google Scholar]
- Yurimoto H, Oku M, Sakai Y (2011) Yeast methylotrophy: metabolism, gene regulation and peroxisome homeostasis. Int J Microbiol. 2011:101298 [DOI] [PMC free article] [PubMed]
- Zhang X, Li X, Xia L. Expression of a thermo-alkaline lipase gene from Talaromyces thermophilus in recombinant Pichia pastoris. Biochem Eng J. 2015;103:263–269. doi: 10.1016/j.bej.2015.08.011. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (DOCX 11069 KB)