Abstract
HD-Zip proteins are unique to plants, and contain a homeodomain closely linked to a leucine zipper motif, which are involved in dimerization and DNA binding. Based on homology in the HD-Zip domain, gene structure and the presence of additional motifs, HD-Zips are divided into four families, HD-Zip I–IV. Phylogenetic analysis of HD-Zip genes using transcriptomic and genomic datasets from a wide range of plant species indicate that the HD-Zip protein class was already present in green algae. Later, HD-Zips experienced multiple duplication events that promoted neo- and sub-functionalizations. HD-Zip proteins are known to control key developmental and environmental responses, and a growing body of evidence indicates a strict link between members of the HD-Zip II and III families and the auxin machineries. Interactions of HD-Zip proteins with other hormones such as brassinolide and cytokinin have also been described. More recent data indicate that members of different HD-Zip families are directly involved in the regulation of abscisic acid (ABA) homeostasis and signaling. Considering the fundamental role of specific HD-Zip proteins in the control of key developmental pathways and in the cross-talk between auxin and cytokinin, a relevant role of these factors in adjusting plant growth and development to changing environment is emerging.
Keywords: Arabidopsis, developmental pathways, environmental responses, HD-Zip transcription factors, hormones
1. The HD-Zip Class of Proteins
The homeodomain-leucine zipper (HD-Zip) class of proteins appears to be present exclusively in the plant kingdom and is characterized by the presence of a homeodomain closely linked to a leucine zipper motif [1]. The Arabidopsis genome codes for 48 HD-Zip proteins that, on the basis of sequence homology in the HD-Zip domain, the presence of additional conserved motifs, and specific intron and exon positions, have been grouped into four families: HD-Zip I (17 members), HD-Zip II (10 members), HD-Zip III (5 members) and HD-Zip IV (16 members) [2,3,4,5,6,7].
HD-Zip genes are evolutionary highly conserved and there is evidence that they were already present in green algae [8,9,10,11]. Later in evolution, the HD-Zip class experienced multiple duplication events that promoted neo- and sub-functionalizations for terrestrial life [11].
Experimental work has demonstrated that the HD-Zip domain, but not the HD by itself, interacts with DNA [12], and it has been shown that a correct spatial relationship between the HD and the leucine zipper motif is crucial for DNA binding [12]. Binding-site selection analysis and subsequent chromatin immunoprecipitation sequencing (ChIP-seq) experiments have determined that the HD-Zip proteins recognize pseudo-palindromic DNA elements [3,12,13,14]. HD-Zip I proteins interact with the CAAT(A/T)ATTG motif [12,15,16] whereas HD-Zip II proteins preferentially bind the CAAT(C/G)ATTG motif [12,13]. Binding-site selection analysis identified GTAAT(G/C)ATTAC as the sequence preferentially recognized by HD-Zip III proteins [3]; however, more recent genome-wide binding-site experiments suggest that the AT(G/C)AT central core is sufficient for DNA binding [14]. For HD-Zip IV proteins, the CATT(A/T)AATG motif was shown to be required for DNA binding [13] and found in the promoters of true target genes [17,18,19]. Interestingly, the identified cis-elements are very similar, particularly the HD-Zip II and III binding sites which share the same core sequence [AAT(G/C)ATT] [3,12], thus suggesting that members of the different families of HD-Zip proteins may regulate common target genes [20,21].
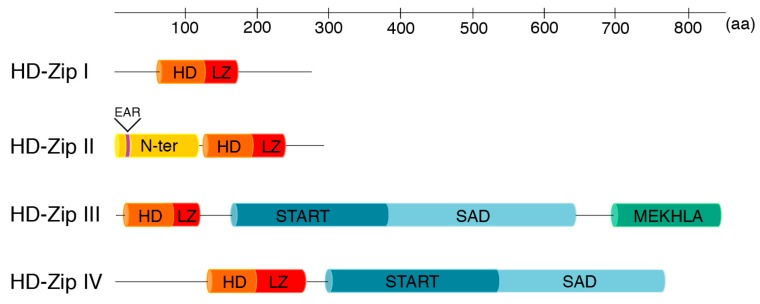
Beside the homeodomain-leucine zipper motif, HD-Zip I proteins have no other established functional domain; conversely, most of the HD-Zip II transcription factors contain an LxLxL type of ERF-associated amphiphilic repression (EAR) motif [7,22] (Figure 1), and there is evidence that they function as negative regulators of gene expression [7,20,23,24,25]. Furthermore, it was recently shown that HOMEOBOX ARABIDOPSIS THALIANA (HAT) 1 and HAT22, two members of the HD-Zip II protein family, interact with the TOPLESS (TPL) co-repressor protein via the EAR motif [26]. In addition to the HD-Zip domain, HD-Zip III and HD-Zip IV proteins contain a steroidogenic acute regulatory protein-related lipid-transfer (START) domain motif with putative lipid-binding capability and a Small body size–mothers against decapentaplegic homolog 4 (Smad4) activation domain (SAD) [27,28] (Figure 1). Finally, HD-Zips III, and not HD-Zips IV, share a MEKHLA domain (Figure 1). A region within this domain contains a region homologous to the PAS (Per-Arnt-Sim)-domain known to act as intracellular sensor of light, oxygen, or redox-potentials [28]. Consistently, it has been reported that REVOLUTA (REV), a member of the HD-Zip III family, acts as a redox-sensitive transcription factor [29]. Furthermore, it has also been shown that the MEKHLA domain is involved in the dimerization of HD-Zip III proteins with DORNROSCHEN (DRN) and DORNROSCHEN-like (DNRL), two APETALA2 (AP2) transcription factors involved in embryo patterning [30].
Figure 1.
Schematic representation of the protein domains possessed by each HD-Zip family. ATHB1, ATHB2, ATHB8, and GLABRA2 were chosen as representative members of the HD-Zip I, II, III and IV families, respectively. N-term, N-terminus consensus; EAR, LxLxL type of ERF-associated amphiphilic repression; HD, Homeodomain; Zip, Leucine zipper; START, steroidogenic acute regulatory protein-related lipid-transfer domain; SAD, Small body size–mothers against decapentaplegic homolog 4 (Smad4) activation domain; MEKHLA, named after the identification of the highly conserved amino acids Met, Glu, Lys, His, Leu, Ala. Adapted from Ariel et al (2007) [6].
HD-Zip proteins are known to control key developmental and environmental responses [21,31,32,33]. In particular, a large body of evidence indicates that HD-Zip I and HD-Zip II proteins are involved in environmental responses whereas HD-Zip III and HD-Zip IV proteins act as core developmental factors. However, several recent studies have led to review this conclusion. Indeed, evidence is accumulating that, on one hand, key developmental regulators (members of HD-Zip III and IV families) play an important role in abiotic stress responses and, on the other hand, environmental factors (members of the HD-Zip I and II families) have relevant functions in developmental pathways, thus suggesting that HD-Zip transcription factors may be crucial in adjusting development to changing environment. Here we report recent advances on the understanding of the complex interactions of HD-Zip transcription factors between themselves and with hormone signaling networks, including those involved in abiotic and biotic stress responses.
2. HD-Zips I
Sequence comparison and phylogenetic analysis indicated that members of the HD-Zip I protein family can be classified into six different clades, I to VI, and that the presence and the position of conserved sequences may be related to specific function(s) [4,34]. Genome-wide expression studies revealed that several Arabidopsis HD-Zip I genes show transcriptional changes in response to treatments with abscisic acid (ABA) [4], and there is evidence that members of clades I and II of the HD-Zip I family have roles related to drought stress and ABA signaling in different plant species [35,36,37,38,39,40,41]. For example, ARABIDOPSIS THALIANA HOMEOBOX (ATHB) 7 and ATHB12 (belonging to the clade I) [34] are both strongly induced by water deficit and ABA. Chromatin immunoprecipitation (ChIP) and gene expression analyses have demonstrated that ATHB7 and ATHB12 positively regulate the expression of five genes encoding clade A protein phosphatases type 2C (PP2C), acting as central negative regulators of ABA signaling [42,43,44]. Furthermore, it has also been shown that ATHB7 and ATHB12 act to repress the transcription of two members of the PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL) gene family, encoding the ABA receptors [45,46,47]. Together the data indicate that ATHB7 and ATHB12 function as negative regulators of the ABA response in Arabidopsis [48]. Evidence exists that HOMEOBOX (HB) 6 (also known as ATHB6, clade II) may also act as a negative regulator of ABA signaling [49].
The HELIANTHUS ANNUUS HOMEOBOX4 (HAHB4) gene, encoding a protein homologous to ATHB7 and ATHB12, is also regulated by ABA and drought, as well as by methyl-jasmonic acid (MeJa) or ethylene (ET) or biotic stresses [50,51]. The ectopic expression of HAHB4 in Arabidopsis negatively affects the synthesis of ET and resulted in plants more resistant to drought [50]. In addition, functional analysis of transgenic Arabidopsis and maize plants constitutively expressing HAHB4 suggested that this HD-Zip I protein acts as an integrator of MeJa and ET pathways [51].
It is worth mentioning that in some plants there is evidence that ABA synthesis and signaling is relevant to fully activate defense responses against insect herbivores and, in general, hormonal interactions are important for regulating plant responses to abiotic stresses and growth-defense tradeoffs [52]. Of interest is the finding that the ATHB13 (clade V) gene, positively regulated by low temperature, drought, and salinity, can confer cold, drought and broad-spectrum disease resistance when overexpressed [39,53,54]. The results point to a role of some HD-Zip I proteins as integrators of internal and external signals in the regulation of abiotic and biotic stresses.
Together with the effects on stresses described above, the overexpression of HD-Zip I proteins very often resulted also in alterations of the shape and growth of the plant, including cotyledon, leaf and supporting organs [55,56], suggesting a role of some HD-Zip I proteins in specific growth and/or developmental pathways. It is worth mentioning that at least ATHB12 and some ABA signaling components are regulated by KANADI1 (KAN1) [57], a factor controlling organ polarity, including the patterning of leaf primordia in Arabidopsis [58]. Furthermore, evidence strongly suggests that ATHB12 acts as a positive regulator of endoreduplication and cell growth during leaf development [59]. In addition, it has been reported that the Medicago truncatula HB1, highly related to ATHB7 and ATHB12, interacts with both ABA and auxin signaling in the regulation of organ development. The MtHB1 gene is strongly regulated by salt, osmotic and ABA stresses. There is evidence that MtHB1 controls the emergence of lateral roots likely by repressing the auxin-regulated LOB-Binding Domain 1 (LBD1) gene [60], a member of a family of plant-specific transcription factors involved in lateral organ development [61].
In the post-embryonic development, ATHB5 (a.k.a. HB5, clade II) behaves as a growth-promoting transcription factor of the hypocotyl. In particular, it promotes the gibberellin acid (GA)-mediated expansion of the epidermal and cortex cells by a positive direct modulation of the expression of EXPANSIN3 (EXP3), a gene involved in cell wall extension [62]. ATHB1 (Clade III) is a direct target of PHYTOCHROME INTERACTING FACTOR 1 (PIF1), a basic helix-loop-helix (bHLH) transcription factor involved in the regulation of light responses downstream of phytochromes [63] and plays a role in hypocotyl growth under short-day regime likely through a positive regulation of genes involved in cell elongation [64]. Interestingly, ATHB1 expression is positively regulated by ethylene in Arabidopsis [65], known to regulate the elongation of the hypocotyl in low light and shade [66], whereas the tomato ortholog HB1 directly regulates the expression of 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE (ACO) gene encoding a key enzyme in ethylene biosynthesis [67].
Very recent work implicated three related HD-Zip I proteins belonging to clade VI [34] in the control of shoot branching [68]. Indeed, it was found that TEOSINTE BRANCHED1, CYCLOIDEA, PCF (TCP) transcription factor BRANCHED1 (BRC1), that functions inside axillary buds to prevent constitutive branch outgrowth [69], binds to and positively regulates the transcription of the ATHB21, ATHB40 and ATHB53 (a.k.a. HB21, HB40 and HB53) genes, all belonging to clade VI. These HD-Zip I proteins are necessary and sufficient to enhance the expression of 9-CIS-EPOXICAROTENOID DIOXIGENASE 3 (NCED3), a key ABA biosynthesis gene, and for ABA accumulation inside axillary buds in conditions of low Red/Far-Red (R/FR) ratio light or short photoperiod. This, in turn, causes suppression of bud development. Relevantly, the BRC1/ATHB21/40/53 regulatory module appears to be conserved in monocot and dicot species [69].
Besides the multiple links found between HD-Zip I proteins and ABA, evidence of a direct interaction between HD-Zips I and auxin also exists [70]. Auxin has a central role during embryogenesis and post-embryonic development. The transcriptional auxin response is regulated by AUXIN RESPONSE FACTOR (ARF) transcription factors and AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) proteins. In the absence of auxin, AUX/IAAs act as repressors by forming heterodimers with ARFs; the auxin-mediated degradation of AUX/IAAs releases the inhibition on ARF transcription factors [71,72]. MONOPTEROS (MP)/ARF5 and its interacting AUX/IAA partner BODENLOS (BDL)/IAA12 play an important role during the embryonic and post-embryonic development. Both a dominant mutant of (BDL)/IAA12 and a loss-of-function mutant of MP/ARF5 lack a seedling root and display cotyledon defects [73,74,75,76]. Interestingly, it was found that the ATHB5 protein (clade II) directly negatively regulates BDL/IAA12 expression. Overexpression of ATHB5 during embryogenesis transcriptionally suppresses the expression of BDL/IAA12 and rescues the rootless phenotype of the bdl/iaa12 dominant mutant. Together the data lead to the hypothesis that ATHB5 may contribute to spatially restrict BDL/IAA12 expression during embryogenesis [70]. Evidence that ATHB6, a close homolog of ATHB5, may act redundantly with ATHB5 in the negative regulation of BDL/IAA12 have also been provided [70].
Finally, the cross-talk between ethylene and auxin in the control of root elongation mediated by ATHB52 has been recently uncovered. It is very well established that root elongation is inhibited by ethylene in Arabidopsis and other species through the action of auxin [77,78,79,80]. The ATHB52 gene is positively regulated by ETHYLENE-INSENSITIVE3 (EIN3), a key transcription factor of the ethylene signal transduction pathway. A molecular and genetic analysis has shown that ATHB52 binds the promoters of PIN FORMED2 (PIN2), coding for a polar auxin carrier, and of WAVY ROOT GROWTH1 (WAG1) and WAG2, encoding PIN polarity regulators. The positive modulation by ethylene of the PIN2/WAG1/WAG2 module exerted through ATHB52 could affect the local polar auxin transport in the root tip resulting in the inhibition of primary root elongation [81].
3. HD-Zips II
The HD-Zip II protein family contains ten members which can be divided into four clades (α-δ) [7]. Remarkably, all the HD-Zip II γ (ATHB2, HAT1, HAT2) and δ (HAT3, ATHB4) genes are rapidly induced by changes in the R/FR ratio light that promote shade avoidance in the Angiosperms [7,82] and several evidence exist that HD-Zip II γ and δ proteins act as positive regulators of this response [7,23,25,83,84,85,86,87].
Interestingly, evidence demonstrates that, besides their function in shade avoidance, HD-Zip II γ and δ transcription factors play a crucial role in embryo apical development and essential developmental processes in sunlight, including shoot apical meristem (SAM) activity, organ polarity and gynoecium development [20,31,88,89,90,91].
Several links have been established between HD-Zip II γ and δ proteins and auxin. Plants with elevated levels of ATHB2 display a constitutive shade avoidance response, and it was shown that ATHB2-induced elongation of the hypocotyl depends on the auxin transport system, as it is abolished by auxin transport inhibitors [23]. Furthermore, the lateral root phenotype observed in ATHB2 overexpressing seedlings is rescued by IAA [23]. Finally, it was found that both auxin synthesis and transport are affected in hat3 athb4 and hat3 athb4 athb2 mutant embryos [20,31].
Recently, HAT1 has been linked to brassinosteroid (BR) signaling pathway. BRs signal through a plasma membrane-localized receptor kinase to modulate the BES1/BZR1 (BRI1-EMS SUPPRESSOR 1/BRASSINAZOLE RESISTANT1) family of transcription factors that positively and negatively regulate a large number of genes [92,93,94,95,96,97]. Relevantly, it was recently found through ChIP experiments that HAT1 is a direct target of BES1 [98]. HAT1 functions redundantly with its close homolog HAT3, as the double loss-of-function mutant hat1 hat3 displayed a reduced BR response stronger than that of the hat1 and hat3 single mutants. Expression levels of several BR-repressed genes are increased in hat1 hat3 double mutant and reduced in HAT1 overexpressing lines, thus strongly suggesting that HAT1 functions to repress the expression of a subset of BR target genes. Consistently, it was found that HAT1 binds to DNA elements in BR-repressed gene promoters and functions as a BES1 corepressor [98]. Furthermore, it was shown that GSK3 (GLYCOGEN SYNTHASE KINASE 3)-like kinase BIN2 (BRASSINOSTEROID-INSENSITIVE 2), a negative regulator of the BR pathway, increases the stability of HAT1 [98,99,100,101] (Figure 2).
Figure 2.
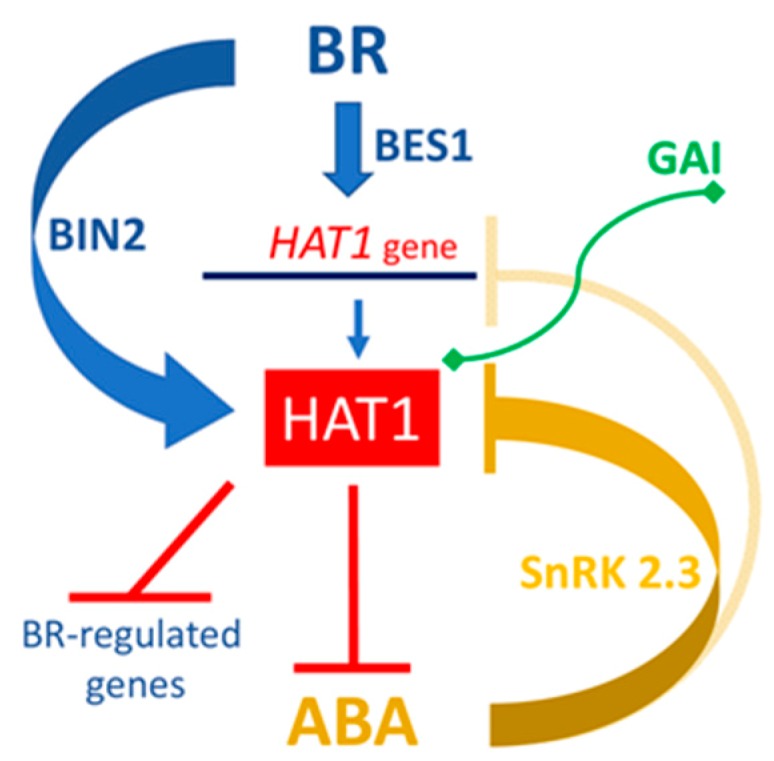
Positive and negative hormonal pathways regulated by HAT1. BR positively regulates the expression of HAT1 through the BES1 transcription factor and stabilizes HAT1 protein through the BIN2 kinase. HAT1 acts together with BES1 as a transcriptional repressor of BR-regulated genes. HAT1, whose expression is negatively regulated by ABA at the transcriptional and post-transcriptional levels, represses the expression of NCED3 and ABA3 resulting in a reduction of ABA synthesis. The SnRK 2.3 kinase, positively acting in the ABA signaling, affects both HAT1 protein stability and DNA binding activity. The GAI interaction with HAT1 is also indicated.
Furthermore, HAT1 was identified among the transcription factors interacting with the GIBBERELLIN INSENSITIVE (GAI) DELLA protein, a master negative regulator in gibberellin (GA) signaling [102,103] (Figure 2). Further work is needed to establish the specific GA-response(s) in which HAT1 is involved.
Recent work has demonstrated that HAT1, apart from its role in BR-mediated growth responses [98], in GA signaling, and in viral defense response in a manner dependent on salicylic acid (SA) [104], it also negatively regulates, redundantly with HAT3, ABA-mediated drought responses through suppression of ABA biosynthesis and signaling [105]. The expression of both HAT1 and HAT3 is indeed repressed by ABA. Evidence have been provided that HAT1 can bind to specific DNA sequences on the promoters of NCED3 and ABA DEFICIENT (ABA) 3, two key ABA biosynthesis genes, and negatively regulate their expression, thus resulting in a reduction of ABA synthesis. In addition, it was observed that HAT1 overexpressing plants display reduced sensitivity to ABA and less tolerance to drought stress, whereas the double loss-of-function hat1 hat3 mutant show opposite phenotypes. Finally, it was found that Sucrose non-fermenting 1-related protein kinase (SnRK) 2.3, a positive component of ABA signaling, physically interacts with and phosphorylates HAT1, decreasing its protein stability and binding activity [105] (Figure 2).
Relevantly, at least other two HD-Zip II proteins, ATHB17 and HAT22/ABA-INSENSITIVE GROWTH 1 (ABIG1), are linked to ABA [106,107]. ATHB17 expression is induced by ABA, and evidence have been provided that athb17 loss-of-function mutants are ABA-insensitive and drought-sensitive whereas lines overexpressing ATHB17 display opposite phenotypes. Interestingly, the effect of ATHB17 on seedling growth in the presence of ABA is stage-specific. Indeed, it is observed exclusively during the post-germination seedling establishment stage [106]. Recent work identified HAT22/ABIG1 as a transcription factor required for ABA-mediated growth inhibition, but not for seed dormancy and stomatal closure. It has been proposed that drought acts through ABA to increase HAT22/ABIG1 transcription which, in turn, inhibits new shoot growth and promotes leaf senescence [107].
4. HD-Zips III
The HD-Zip III family contains five members: ATHB8, CORONA (CNA), PHABULOSA (PHB), PHAVOLUTA (PHV), and REV. It is well established that HD-Zip III proteins act as master regulators of embryonic apical fate [108], are required to maintain an active SAM and to establish lateral organ polarity [109,110,111] and are necessary for xylem formation and specification [112,113,114,115,116,117]. Recent work has also implicated HD-Zip III proteins in the regulation of the shade avoidance response [14,21,32].
The pattern of HD-Zip III expression largely coincides with that of auxin distribution [8,9,115,118,119,120,121,122]. Furthermore, HD-Zip III genes are regulated at the post-transcriptional level by the microRNAs miR165/166, which negatively affect their expression through mRNA cleavage [110,123]. Relevantly, REV directly positively regulates the HD-Zip II genes HAT3, ATHB4, ATHB2, and HAT2, and evidence exists that PHB and PHV are involved in the control of HAT3 expression [14,20]. It has been recently shown that REV physically interacts with HAT3 and ATHB4 to directly repress MIR165/166 expression in the adaxial side of the leaf [91].
The interconnection between HD-Zip III transcription factors and auxin began to be clarified especially through the molecular-genetic analysis of the vascular system [124]. REV directly positively regulates the auxin biosynthetic genes TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1) and YUCCA5 (YUC5) [14,125]. Furthermore, it has been demonstrated that genes implicated in auxin transport, including the influx carriers LIKE AUXIN RESISTANT 2 (LAX2) and LAX3, and response are also direct targets of REV [122,125,126]. Interestingly, REV also upregulates the expression of NAKED PINS IN YUC MUTANTS 1 (NPY1) and WAG1, encoding an AGC protein kinase highly similar to PINOID (PID) [127]. NPY genes encode proteins with a Broad-Complex, Tramtrack, and Bric-a-brac (BTB)-Poxvirus and Zinc Finger (POZ) domain that together with AGC kinases determine the subcellular polar targeting of the PIN efflux carriers, thus establishing the direction of auxin transport [128,129,130,131].
A recent study reinforces the interconnection between HD-Zip III transcription factors and auxin signaling [132]. HD-Zip III proteins are known to determine xylem patterning in the Arabidopsis root [116], and it has been shown that PHB directly interacts with the promoter of both MP/ARF5, a transcription factor gene playing a major role in vascular development, and IAA20, encoding an IAA protein that is stable in the presence of auxin and able to interact with MP. The double mutant of IAA20 and its closest homolog IAA30 forms ectopic protoxylem, whereas elevated levels of IAA30 result in discontinuous protoxylem, analogous to a weak mp mutant. It has therefore proposed a mechanism in which PHB stabilizes the auxin response within the xylem axis by activating both MP and its repressors IAA20 and IAA30 to ensure correct vascular patterning and differentiation of xylem cells [132].
Cross-talk between auxin and cytokinin (CK) is crucial during several developmental processes, including vascular development. Several studies have indicated that ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6 (AHP6), an inhibitory pseudophosphotransfer protein, is positively regulated by auxin and counteracts CK signaling, allowing protoxylem formation in the root. Conversely, CK signaling negatively regulates the spatial domain of AHP6 expression [133,134]. Interestingly, it has been shown that PHB acts redundantly with other HD-Zip III transcription factors to downregulate AHP6 expression, either directly or by alteration of auxin signaling [116,133].
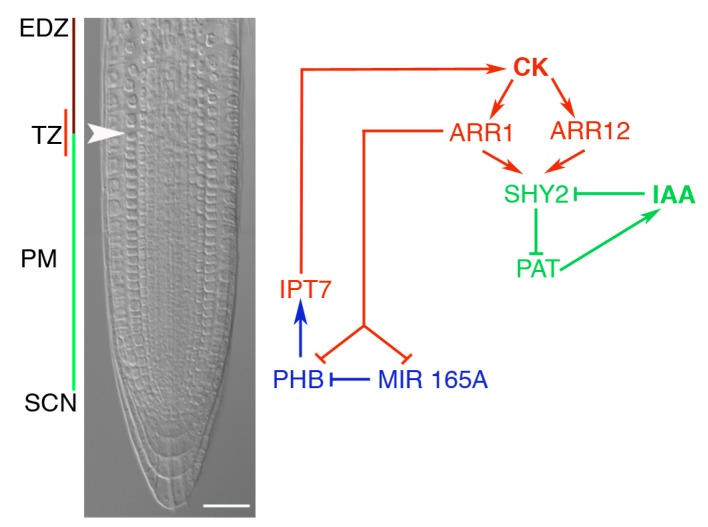
The interaction between HD-Zip III proteins and cytokinin network is further strengthened by the finding that PHB directly activates ISOPENTENYLTRANSFERASE 7 (IPT7), a gene coding for a rate-limiting component of the cytokinin biosynthesis pathway. This in turn promotes cell differentiation and regulates root length [135]. These results, together with the finding that CK is transported in the phloem [136], suggests that CK is synthesized in the meristem vasculature through the activity of PHB and then delivered to the transition zone (TZ) to promote differentiation. Consistently, the expression of SHORT HYPOCOTYL 2 (SHY2), a CK primary target necessary and sufficient to promote cell differentiation at the TZ [137], is weaker in phb phv double loss-of-function mutants with respect to wild type but is reestablished after CK treatment. In addition, the root meristem phenotype of the dominant shy2-2 mutant is suppressed in phb phv, further confirming the hypothesis that PHB-dependent CK biosynthesis in the distal part of the root influences cell differentiation at the proximal TZ [135]. Relevantly, it was also shown that CK represses both PHB and miR165 [135]. The authors referred to these interactions as an incoherent regulatory loop in which CK represses both its activator and a repressor of its activator and proposed that this circuit might provide robustness against CK fluctuations [135] (Figure 3).
Figure 3.
Interplay between the HD-Zip III protein PHB and CKs in the regulation of root meristem size. DIC image of the Arabidopsis root tip with different developmental zones indicated, stem cell niche (SCN), proximal meristem (PM), transition zone (TZ), and elongation and differentiation zone (EDZ), together with a schematic representation of the molecular interactions involved in the determination of RAM size. PHB induces CK biosynthesis in the PM of the root; cytokinin is delivered to the transition zone where activates ARABIDOPSIS RESPONSE REGULATOR (ARR) 1. ARR1 represses the expression of PHB at the TZ, thus restricting PHB expression to the distal part of the PM. Remarkably, ARR1 also represses the transcription of MIR165A [135]. In addition, ARR1 induces the expression of SHY2, a CK primary target necessary and sufficient to promote cell differentiation at the TZ. White arrowhead indicates the TZ of the cortex tissue, placed at the boundary between the last meristematic cell and the first differentiating cell [137]. A color code has been used to indicate different pathways: CK pathway, red; IAA pathway, green; PHB/miR165, blue. Scale bar, 20 μm.
Furthermore, recent work has shown that PHB also regulates cell activities at the TZ by repressing B-ARRs, positive regulators of CK signaling [138,139]. Such a repressive effect of PHB on B-ARR activities is enhanced by high cytokinin [139].
A recent study on the mechanism underlying shoot regeneration in Arabidopsis provided an additional link between HD-Zip III transcription factors and CK signaling [140], somehow predicted also on the basis of protein-protein interaction studies between HD-Zip III and DNR/DNRL transcription factors [30,141,142] and the analysis of the regeneration capacity of a cna mutant encoded by the hoc locus [143]. Four B-ARR transcription factors, ARR1, ARR2, ARR10, and ARR12, have essential roles in shoot regeneration. Indeed, the shoot regenerative capacity is impaired in the arr1 arr10 arr12 mutant with respect to wild type [144,145,146]. The A-type ARRs play opposite roles in shoot regeneration, and it has been shown that overexpression of ARR7 or ARR15 results in a marked reduction of the regeneration capacity [147]. Remarkably, it was found that ARR1, ARR2, ARR110 and ARR12 interact with PHB, PHV and REV HD-Zip III transcription factors, and that these complexes in turn activates the expression of WUSCHEL (WUS), a gene essential in maintaining SAM activity [140,148].
Beside the evidence of molecular interactions between HD-Zip III proteins and key components of the auxin and cytokinin networks, it has been recently reported that REV positively directly regulates the expression of the gene encoding the ABA receptor protein PYL6 [125,126]. Remarkably, PYL6 is oppositely directly regulated by KAN1, a key determinant of abaxial cell fate in the leaf [59,127,149]. Furthermore, microarray data revealed that the expression of REV, PHB and PHV significantly decreases upon ABA application, as a consequence of ectopic induction of miR165 expression [21,126]. It has been therefore proposed that the connection between ABA perception and signaling and HD-Zip III transcription factors may be required to adapt leaf development to alterations in water availability [21,126].
Finally, it is worth mentioning that recent work has revealed that the expression of miR165/166 is regulated by a complex hormonal cross-talk during root development in Arabidopsis [150]. Evidence have been provided that miR 165/166 has important functions in ABA and abiotic stress responses. It was indeed found that reduction in the expression of miR165/166 results in drought and cold resistance phenotypes and hypersensitivity to ABA during seed germination and seedling development. Furthermore, it was shown that miR165/166-mediated regulatory module is linked with ABA responses likely through a direct regulation by HD-Zip III proteins of ABA INSENSITIVE4 (ABI4), a regulator of ABA signaling, and β-glucosidase 1 (BG1), known to hydrolyze glucose-conjugated, biologically inactive ABA to produce active ABA [151]. In addition, there is also evidence that the cross-talk between miR165 and ABA is involved in root xylem formation and vascular acclimation to water deficit. It was indeed shown that under limited water availability endodermal ABA signaling activates MIR165A, thus leading to increased miR165 levels that repress HD-Zip III transcription factors in the stele. Together the data nicely show how a pathway known to control core developmental processes is used as a mean to adjust xylem formation under conditions of abiotic stress [152].
5. HD-Zips IV
Many of the HD-Zip IV genes were shown to be specifically or preferentially expressed in the epidermis of developing embryos and/or other plant organs [153]. It is worth mentioning that the epidermis plays a critical role also in plant defense against pathogens and in protection from environmental stresses [154,155].
GLABRA2 (GL2), the first identified member of the HD-Zip IV protein family, promotes trichome differentiation [156], and suppresses root hair formation in the root epidermis [19,156]. In particular, GL2 controls cell fate determination of N-cells (non-hair cells; atrichoblast) [157], through a negative regulation of the phospholipase D zeta 1 (PLDz1) [18], and of several bHLH transcription factors including ROOT HAIR DEFECTIVE 6 (RDH6) involved in root hair initiation [156]. There is strong evidence that BRs are required to maintain position-dependent cell fate specification in root epidermis, as loss of BR signaling results in loss of H-cells (hair cells; trichoblast). Indeed, BRs are required for a correct expression of WEREWOLF (WER) and GL2, master regulators of epidermal patterning, and of CAPRICE (CPC), a direct downstream target of WER [158].
Recent work has also established a link between HD-Zip IV proteins and GA signaling [159]. It was indeed shown that DELLA proteins interact directly with MERISTEM LAYER 1 (ATML1) and its paralogue PLANT DEFENSIN 2 (PDF2), two HD-Zip IV proteins required for epidermis specification and binding to the L1 box present in the promoters of epidermis-specific genes [17,160]. Silencing of both ATML1 and PDF2 inhibits epidermis-specific gene expression and delays germination [157]. Evidence were provided that, upon seed imbibition, increased GA levels reduce DELLA protein levels, thus releasing ATML1/PDF2 to activate epidermis-specific expression and promote seed germination [159].
Apart from their role in development, interactions between HD-Zip IV transcription factors and environmental responses have also been reported [161]. For example, HOMEODOMAIN GLABRA 11 (HDG11) was identified via activation tagging as a gene involved in drought tolerance. The mutant has higher levels of ABA than the wild type and displays enhanced root growth with more lateral roots and reduced stomatal density. Overexpression of HDG11 also conferred drought tolerance associated with augmented lateral roots and reduced leaf stomatal density in both Arabidopsis and tobacco [162]. Higher level of ABA and improved drought tolerance have been observed also in cotton, poplar and rice transgenic plants overexpressing HDG11 [161,163]. It has been suggested that HDG11 positively regulates the expression of cell-wall-loosening protein genes, including EXP5, resulting in a well-developed root system [164].
Intriguingly, HDG11 acts as a maternal regulator of zygote asymmetry through a direct activation of the WUSCHEL RELATED HOMEOBOX 8 (WOX8) gene whose product leads to asymmetric division of the zygote [165].
6. Conclusions
A fundamental question in plant biology is how plants integrate environmental signals with intrinsic developmental programs and how coordinate the growth of different organs depending on resource availability. Over recent years remarkable progress has been made, and the molecular mechanisms controlling these processes are being elucidated. The functional analysis of the HD-Zip proteins revealed that they are part of complex networks involved in the integration of external signals through the regulation of hormonal pathways involved in the control of fundamental developmental processes. Although many factors belonging to each HD-Zip family are implicated in specific processes, it is interesting to note that the simple overexpression of ATHB2 (and other members of the HD-Zip II family) and HDG11 is sufficient to generate plants looking for optimal light and water for growth, respectively. This could be explained by the existence of organ- and/or tissue-specific hubs that stimulated by external and/or internal (hormonal) signals converge to coherently adjust the development and growth to a specific environmental signal. These hubs may have been generated during evolution through multiple duplication events that have promoted neo and sub-functionalization of factors operating on specific pathways. The identification of these hubs it has the potential to lead to a more unified vision of the development and growth of the plant according to environmental stresses that could be applied for the improvement of the cultivated plants.
Acknowledgments
We thank all our collaborators who made the work on the HD-Zips an exciting and gratifying experience. Our apologies to the many researchers whose work or original publications has not been cited here because of space limitations.
Abbreviations
| ABA | Abscisic acid |
| ABA | ABA deficient |
| ABI4 | ABA INSENSITIVE4 |
| ABIG1 | ABA-INSENSITIVE GROWTH 1 |
| ACCO | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE |
| AHP6 | HISTIDINE PHOSPHOTRANSFER PROTEIN 6 |
| AP2 | APETALA2 |
| AUX/IAA | AUXIN/INDOLE-3-ACETIC ACID |
| ARF | AUXIN RESPONSE FACTOR |
| ARR | ARABIDOPSIS RESPONSE REGULATOR |
| ATHB | ARABIDOPSIS THALIANA HOMEOBOX |
| ATML1 | MERISTEM LAYER 1 |
| BDL | BODENLOS |
| BES1/BZR1 | BRI1-EMS SUPPRESSOR 1/BRASSINAZOLE RESISTANT1 |
| BG1 | β-glucosidase 1 |
| b-HLH | basic Helix-Loop-Helix |
| BIN2 | BRASSINOSTEROID-INSENSITIVE 2 |
| BR | Brassinosteroid |
| BRC1 | BRANCHED1 |
| ChIP | Chromatin Immunoprecipitation |
| ChIP-seq | Chromatin Immunoprecipitation sequencing |
| CK | Cytokinin |
| CNA | CORONA |
| CPC | CAPRICE |
| DRN | DORNROSCHEN |
| DNRL | DORNROSCHEN-like |
| EAR | ERF-Associated Amphiphilic Repression |
| EDZ | Elongation and Differentiation Zone |
| EIN3 | ETHYLENE-INSENSITIVE 3 |
| EXP3 | EXPANSIN 3 |
| ET | Ethylene |
| H cell | Hair cell; trichoblast |
| HAHB4 | HELIANTHUS ANNUUS HOMEOBOX 4 |
| HB | HOMEOBOX |
| HAT1 | HOMEOBOX ARABIDOPSIS THALIANA 1 |
| HDG11 | HOMEODOMAIN GLABRA 11 |
| HD-Zip | Homeodomain-leucine zipper |
| IPT7 | ISOPENTENYLTRANSFERASE 7 |
| GA | Gibberellin Acid |
| GAI | GIBBERELLIN INSENSITIVE |
| GL2 | GLABRA2 |
| GSK3 | GLYCOGEN SYNTHASE KINASE 3 |
| KAN1 | KANADI 1 |
| LAX2 | LIKE AUXIN RESISTANT 2 |
| LBD1 | LOB-Binding Domain 1 |
| MeJa | Methyl-Jasmonic acid |
| MP | MONOPTEROS |
| N cell | Non-hair cell; atrichoblast |
| NCED3 | 9-CIS-EPOXICAROTENOID DIOXIGENASE 3 |
| NPY1 | NAKED PINS IN YUC MUTANTS 1 |
| PDF2 | PLANT DEFENSIN 2 |
| PHB | PHABULOSA |
| PHV | PHAVOLUTA |
| PID | PINOID |
| PIF1 | PHYTOCHROME INTERACTING FACTOR 1 |
| PIN2 | PIN FORMED2 |
| PLDz1 | Phospholipase D zeta 1 |
| PM | Proximal Meristem |
| POZ | Broad-Complex, Tramtrack, and Bric-a-brac (BTB)-Poxvirus and Zinc Finger |
| PP2C | Protein Phosphatases type 2C |
| PYL | PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE |
| RDH6 | ROOT HAIR DEFECTIVE 6 |
| R/FR | Red/Far-Red |
| REV | REVOLUTA |
| SAD | Small body size–mothers against decapentaplegic homolog 4 (Smad4) activation domain |
| SAM | Shoot Apical Meristem |
| SCN | Stem Cell Niche |
| SHY2 | SHORT HYPOCOTYL 2 |
| SnRK | Sucrose non-fermenting 1-related protein kinase |
| START | Steroidogenic acute regulatory protein-related lipid-transfer |
| TAA1 | TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 |
| TCP | TEOSINTE BRANCHED1, CYCLOIDEA, PCF |
| TPL | TOPLESS |
| TZ | Transition Zone |
| WAG1 | WAVY ROOT GROWTH 1 |
| WER | WEREWOLF |
| WOX8 | WUSCHEL RELATED HOMEOBOX 8 |
| WUS | WUSCHEL |
| YUC5 | YUCCA5 |
Author Contributions
G.S., M.C. and M.P. performed the review of the literature; G.M. and I.R. wrote the manuscript. All authors approved the final draft.
Funding
This research was funded by the Italian Ministry of Education, University and Research, PRIN Program (https://www.researchitaly.it/), grant number 2010HEBBB8_004 and Italian Ministry of Agricultural, Food, Forestry and Tourism Policies, CISGET-BIOTECH Project n. 15924 18/05/2018.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ruberti I., Sessa G., Lucchetti S., Morelli G. A novel class of plant proteins containing a homeodomain with a closely linked leucine zipper motif. EMBO J. 1991;10:1787–1791. doi: 10.1002/j.1460-2075.1991.tb07703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sessa G., Carabelli M., Ruberti I., Baima S., Lucchetti S., Morelli G. Identification of distinct families of HD-Zip proteins in Arabidopsis thaliana. In: Puigdomenech P., Coruzzi G., editors. Plant Molecular Biology: Molecular-Genetic Analysis of Plant Development and Metabolism. Volume 81. Springer; Berlin/Heidelberg, Germany: 1994. pp. 411–426. (NATO-ASI Series). [Google Scholar]
- 3.Sessa G., Steindler C., Morelli G., Ruberti I. The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-Zip proteins. Plant Mol. Biol. 1998;38:609–622. doi: 10.1023/A:1006016319613. [DOI] [PubMed] [Google Scholar]
- 4.Henriksson E., Olsson A.S., Johannesson H., Johansson H., Hanson J., Engstrom P., Soderman E. Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 2005;139:509–518. doi: 10.1104/pp.105.063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura M., Katsumata H., Abe M., Yabe N., Komeda Y., Yamamoto K.T., Takahashi T. Characterization of the class IV homeodomain leucine zipper gene family in Arabidopsis. Plant Physiol. 2006;141:1363–1375. doi: 10.1104/pp.106.077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ariel F.D., Manavella P.A., Dezar C.A., Chan R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007;12:419–426. doi: 10.1016/j.tplants.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Ciarbelli A.R., Ciolfi A., Salvucci S., Ruzza V., Possenti M., Carabelli M., Fruscalzo A., Sessa G., Morelli G., Ruberti I. The Arabidopsis homeodomain-leucine zipper II gene family: Diversity and redundancy. Plant Mol. Biol. 2008;68:465–478. doi: 10.1007/s11103-008-9383-8. [DOI] [PubMed] [Google Scholar]
- 8.Floyd S.K., Bowman J.L. Distinct developmental mechanisms reflect the independent origins of leaves in vascular plants. Curr. Biol. 2006;16:1911–1917. doi: 10.1016/j.cub.2006.07.067. [DOI] [PubMed] [Google Scholar]
- 9.Floyd S.K., Zalewski C.S., Bowman J.L. Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics. 2006;173:373–388. doi: 10.1534/genetics.105.054239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu R., Chi X., Chai G., Kong Y., He G., Wang X., Shi D., Zhang D., Zhou G. Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa) PLoS ONE. 2012;7:e31149. doi: 10.1371/journal.pone.0031149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romani F., Reinheimer R., Florent S.N., Bowman J.L., Moreno J.E. Evolutionary history of HOMEODOMAIN LEUCINE ZIPPER transcription factors during plant transition to land. New Phytol. 2018;219:408–421. doi: 10.1111/nph.15133. [DOI] [PubMed] [Google Scholar]
- 12.Sessa G., Morelli G., Ruberti I. The Athb-1 and -2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J. 1993;12:3507–3517. doi: 10.1002/j.1460-2075.1993.tb06025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tron A.E., Bertoncini C.W., Palena C.M., Chan R.L., Gonzalez D.H. Combinatorial interactions of two amino acids with a single base pair define target site specificity in plant dimeric homeodomain proteins. Nucl. Acids Res. 2001;29:4866–4872. doi: 10.1093/nar/29.23.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brandt R., Salla-Martret M., Bou-Torrent J., Musielak T., Stahl M., Lanz C., Ott F., Schmid M., Greb T., Schwarz M., et al. Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 2012;72:31–42. doi: 10.1111/j.1365-313X.2012.05049.x. [DOI] [PubMed] [Google Scholar]
- 15.Sessa G., Morelli G., Ruberti I. DNA-binding specificity of the homeodomain-leucine zipper domain. J. Mol. Biol. 1997;274:303–309. doi: 10.1006/jmbi.1997.1408. [DOI] [PubMed] [Google Scholar]
- 16.Palena C.M., Gonzalez D.H., Chan R.L. A monomer-dimer equilibrium modulates the interaction of the sunflower homeodomain leucine-zipper protein Hahb-4 with DNA. Biochem. J. 1999;341:81–87. [PMC free article] [PubMed] [Google Scholar]
- 17.Abe M., Takahashi T., Komeda Y. Identification of a cis regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 2001;26:487–494. doi: 10.1046/j.1365-313x.2001.01047.x. [DOI] [PubMed] [Google Scholar]
- 18.Ohashi Y., Oka A., Rodrigues-Pousada R., Possenti M., Ruberti I., Morelli G., Aoyama T. Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science. 2003;300:427–430. doi: 10.1126/science.1083695. [DOI] [PubMed] [Google Scholar]
- 19.Lin Q., Ohashi Y., Kato M., Tsuge T., Gu H., Qu L.J., Aoyama T. GLABRA2 directly suppresses Basic Helix-Loop-Helix transcription factor genes with diverse functions in root hair development. Plant Cell. 2015;27:2894–2906. doi: 10.1105/tpc.15.00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turchi L., Carabelli M., Ruzza V., Possenti M., Sassi M., Peñalosa A., Sessa G., Salvi S., Forte V., Morelli G., et al. Arabidopsis HD-Zip II transcription factors control embryo development and meristem function. Development. 2013;140:2118–2129. doi: 10.1242/dev.092833. [DOI] [PubMed] [Google Scholar]
- 21.Brandt R., Cabedo M., Xie Y., Wenkel S. Homeodomain leucine zipper proteins and their role in synchronizing growth and development with the environment. J. Int. Plant Biol. 2014;56:518–526. doi: 10.1111/jipb.12185. [DOI] [PubMed] [Google Scholar]
- 22.Kagale S., Links M.G., Rozwadowski K. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 2010;152:1109–1134. doi: 10.1104/pp.109.151704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steindler C., Matteucci A., Sessa G., Weimar T., Ohgishi M., Aoyama T., Morelli G., Ruberti I. Shade avoidance responses are mediated by the ATHB-2 HD-Zip protein, a negative regulator of gene expression. Development. 1999;125:4235–4245. doi: 10.1242/dev.126.19.4235. [DOI] [PubMed] [Google Scholar]
- 24.Ohgishi M., Oka A., Morelli G., Ruberti I., Aoyama T. Negative autoregulation of the Arabidopsis homeobox gene ATHB-2. Plant J. 2001;25:389–398. doi: 10.1046/j.1365-313x.2001.00966.x. [DOI] [PubMed] [Google Scholar]
- 25.Sawa S., Ohgishi M., Goda H., Higuchi K., Shimada Y., Yoshida S., Koshiba T. The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J. 2002;32:1011–1022. doi: 10.1046/j.1365-313X.2002.01488.x. [DOI] [PubMed] [Google Scholar]
- 26.Causier B., Ashworth M., Guo W., Davies B. The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 2012;158:423–438. doi: 10.1104/pp.111.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schrick K., Nguyen D., Karlowski W.M., Mayer K.F. START lipid/sterolbinding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 2004;5:R41. doi: 10.1186/gb-2004-5-6-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukherjee K., Burglin T.R. MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain-leucine zipper III proteins. Plant Physiol. 2006;140:1142–1150. doi: 10.1104/pp.105.073833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie Y., Huhn K., Brandt R., Potschin M., Bieker S., Straub D., Doll J., Drechsler T., Zentgraf U., Wenkel S. REVOLUTA and WRKY53 connect early and late leaf development in Arabidopsis. Development. 2014;141:4772–4783. doi: 10.1242/dev.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandler J.W., Cole M., Flier A., Grewe B., Werr W. The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development. 2007;134:1653–1662. doi: 10.1242/dev.001016. [DOI] [PubMed] [Google Scholar]
- 31.Turchi L., Baima S., Morelli G., Ruberti I. Interplay of HD-Zip II and III transcription factors in auxin-regulated plant development. J. Exp. Bot. 2015;66:5043–5053. doi: 10.1093/jxb/erv174. [DOI] [PubMed] [Google Scholar]
- 32.Merelo P., Paredes E.B., Heisler M.G., Wenkel S. The shady side of leaf development: The role of the REVOLUTA/KANADI1 module in leaf patterning and auxin-mediated growth promotion. Curr. Opin. Plant Biol. 2017;35:111–116. doi: 10.1016/j.pbi.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 33.Roodbarkelari F., Groot E.P. Regulatory function of homeodomain-leucine zipper (HD-ZIP) family proteins during embryogenesis. New Phytol. 2017;213:95–104. doi: 10.1111/nph.14132. [DOI] [PubMed] [Google Scholar]
- 34.Arce A.L., Raineri J., Capella M., Cabello J.V., Chan R.L. Uncharacterized conserved motifs outside the HD-Zip domain in HD-Zip subfamily I transcription factors; a potential source of functional diversity. BMC Plant Biol. 2011;11:42. doi: 10.1186/1471-2229-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Söderman E., Mattsson J., Engström P. The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J. 1996;10:375–381. doi: 10.1046/j.1365-313X.1996.10020375.x. [DOI] [PubMed] [Google Scholar]
- 36.Dezar C.A., Gago G.M., González D.H., Chan R.L. Hahb-4, a sunflower homeobox-leucine zipper gene, is a developmental regulator and confers drought tolerance to Arabidopsis thaliana plants. Transgenic Res. 2005;14:429–440. doi: 10.1007/s11248-005-5076-0. [DOI] [PubMed] [Google Scholar]
- 37.Deng X., Phillips J., Bräutigam A., Engström P., Johannesson H., Ouwerkerk P.B.F., Ruberti I., Salinas J., Vera P., Iannacone R., et al. A homeodomain leucine zipper gene from Craterostigma plantagineum regulates abscisic acid responsive gene expression and physiological responses. Plant Mol. Biol. 2006;61:469–489. doi: 10.1007/s11103-006-0023-x. [DOI] [PubMed] [Google Scholar]
- 38.Agalou A., Purwantomo S., Övernäs E., Johannesson H., Zhu X., Estiati A., de Kam R.J., Engström P., Slamet-Loedin I.H., Zhu Z., et al. A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 2008;66:87–103. doi: 10.1007/s11103-007-9255-7. [DOI] [PubMed] [Google Scholar]
- 39.Ebrahimian-Motlagh S., Ribone P.A., Thirumalaikumar V.P., Allu A.D., Chan R.L., Mueller-Roeber B., Balazadeh S. JUNGBRUNNEN1 Confers Drought Tolerance Downstream of the HD-Zip I Transcription Factor AtHB13. Front. Plant Sci. 2017;8:2118. doi: 10.3389/fpls.2017.02118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen W., Li H., Teng R., Wang Y., Wang W., Zhuang J. Genomic and transcriptomic analyses of HD-Zip family transcription factors and their responses to abiotic stress in tea plant (Camellia sinensis) Genomics. 2018 doi: 10.1016/j.ygeno.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 41.Yang Q., Niu Q., Li J., Zheng X., Ma Y., Bai S., Teng Y. PpHB22, a member of HD-Zip proteins, activates PpDAM1 to regulate bud dormancy transition in “Suli” pear (Pyrus pyrifolia White Pear Group) Plant Physiol. Biochem. 2018;127:355–365. doi: 10.1016/j.plaphy.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 42.Merlot S., Gosti F., Guerrier D., Vavasseur A., Giraudat J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001;25:295–303. doi: 10.1046/j.1365-313x.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- 43.Saez A., Apostolova N., González-Guzmán M., González-García M.P., Nicolás C., Lorenzo O., Rodríguez P.L. Gain-of-function and loss-of-function phenotypes of the protein phosphatases 2C HAB1 reveal its role as a negative regulator of abscisic acid signaling. Plant J. 2004;37:354–369. doi: 10.1046/j.1365-313X.2003.01966.x. [DOI] [PubMed] [Google Scholar]
- 44.Kuhn J.M., Boisson-Dernier A., Dizon M.B., Maktabi M.H., Schroeder J.I. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 2006;140:127–139. doi: 10.1104/pp.105.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyazono K., Mikayawa T., Sawano Y., Kubota K., Kang H.-J., Asano A., Miyauchi Y., Takahashi M., Zhi Y., Fujita K., et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 46.Park S.Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T.F., et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santiago J., Rodrigues A., Saez A., Rubio S., Antoni R., Dupeux F., Park S.-Y., Márquez J.A., Cutler S.R., Rodríguez P.L. Modulation of drought resistance by the abscisic acid-receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009;60:575–588. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- 48.Valdes A.E., Overnas E., Johansson H., Rada-Iglesias A., Engstrom P. The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol. Biol. 2012;80:405–418. doi: 10.1007/s11103-012-9956-4. [DOI] [PubMed] [Google Scholar]
- 49.Himmelbach A., Hoffmann T., Leube M., Höhener B., Grill E. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 2002;21:3029–3038. doi: 10.1093/emboj/cdf316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manavella P.A., Arce A.L., Dezar C.A., Bitton F., Renou J.P., Crespi M., Chan R.L. Cross-talk between ethylene and drought signalling pathways is mediated by the sunflower Hahb-4 transcription factor. Plant J. 2006;48:125–137. doi: 10.1111/j.1365-313X.2006.02865.x. [DOI] [PubMed] [Google Scholar]
- 51.Manavella P.A., Dezar C.A., Bonaventure G., Baldwin I.T., Chan R.L. HAHB4, a sunflower HD-Zip protein, integrates signals from the jasmonic acid and ethylene pathways during wounding and biotic stress responses. Plant J. 2008;56:376–388. doi: 10.1111/j.1365-313X.2008.03604.x. [DOI] [PubMed] [Google Scholar]
- 52.Nguyen D., Rieu I., Mariani C., van Dam N.M. How plants handle multiple stresses: Hormonal interactions underlying responses to abiotic stress and insect herbivory. Plant Mol. Biol. 2016;91:727–740. doi: 10.1007/s11103-016-0481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cabello J.V., Arce A.L., Chan R.L. The homologous HD-Zip I transcription factors HaHB1 and AtHB13 confer cold tolerance via the induction of pathogenesis-related and glucanase proteins. Plant J. 2012;69:141–153. doi: 10.1111/j.1365-313X.2011.04778.x. [DOI] [PubMed] [Google Scholar]
- 54.Gao D., Appiano M., Huibers R.P., Chen X., Loonen A.E.H.M., Visser R.G.F., Wolters A.-M.A., Bai Y. Activation tagging of ATHB13 in Arabidopsis thaliana confers broad-spectrum disease resistance. Plant Mol. Biol. 2014;86:641–653. doi: 10.1007/s11103-014-0253-2. [DOI] [PubMed] [Google Scholar]
- 55.Aoyama T., Dong C.H., Wu Y., Carabelli M., Sessa G., Ruberti I., Morelli G., Chua N.H. Ectopic expression of the Arabidopsis transcriptional activator Athb-1 alters leaf cell fate in tobacco. Plant Cell. 1995;7:1773–1785. doi: 10.1105/tpc.7.11.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hanson J., Johannesson H., Engström P. Sugar-dependent alterations in cotyledon and leaf development in transgenic plants expressing the HDZhdip gene ATHB13. Plant Mol. Biol. 2001;45:247–262. doi: 10.1023/A:1006464907710. [DOI] [PubMed] [Google Scholar]
- 57.Merelo P., Xie Y., Brand L., Ott F., Weigel D., Bowman J.L., Heisler M.G., Wenkel S. Genome-Wide Identification of KANADI1 Target Genes. PLoS ONE. 2013;8:e77341. doi: 10.1371/journal.pone.0077341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kerstetter R.A., Bollman K., Taylor R.A., Bomblies K., Poethig R.S. KANADI regulates organ polarity in Arabidopsis. Nature. 2001;411:706–709. doi: 10.1038/35079629. [DOI] [PubMed] [Google Scholar]
- 59.Hur Y.S., Um J.H., Kim S., Kim K., Park H.J., Lim J.S., Kim W.Y., Jun S.E., Yoon E.K., Lim J., et al. Arabidopsis thaliana homeobox 12 (ATHB12), a homeodomain-leucine zipper protein, regulates leaf growth by promoting cell expansion and endoreduplication. New Phytol. 2015;205:316–328. doi: 10.1111/nph.12998. [DOI] [PubMed] [Google Scholar]
- 60.Ariel F., Diet A., Verdenaud M., Gruber V., Frugier F., Chan R., Crespi M. Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell. 2010;22:2171–2183. doi: 10.1105/tpc.110.074823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu C., Luo F., Hochholdinger F. LOB Domain Proteins: Beyond Lateral Organ Boundaries. Trends Plant Sci. 2016;21:159–167. doi: 10.1016/j.tplants.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 62.Stamm P., Topham A.T., Mukhtar N.K., Jackson M.D.B., Tomé D.F.A., Beynon J.L., Bassel G.W. The Transcription Factor ATHB5 Affects GA-Mediated Plasticity in Hypocotyl Cell Growth during Seed Germination. Plant Physiol. 2017;173:907–917. doi: 10.1104/pp.16.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leivar P., Quail P.H. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Capella M., Ribone P.A., Arce A.L., Chan R.L. Arabidopsis thaliana HomeoBox 1 (AtHB1), a Homedomain-Leucine Zipper I (HD-Zip I) transcription factor, is regulated by PHYTOCHROME-INTERACTING FACTOR 1 to promote hypocotyl elongation. New Phytol. 2015;207:669–682. doi: 10.1111/nph.13401. [DOI] [PubMed] [Google Scholar]
- 65.Zhong G.Y., Zhong G.V., Burns J.K. Profiling ethylene-regulated gene expression in Arabidopsis thaliana by microarray analysis. Plant Mol. Biol. 2003;53:117–131. doi: 10.1023/B:PLAN.0000009270.81977.ef. [DOI] [PubMed] [Google Scholar]
- 66.Pierik R., Djakovic-Petrovic T., Keuskamp D.H., de Wit M., Voesenek L.A. Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and DELLA proteins in Arabidopsis. Plant Physiol. 2009;149:1701–1712. doi: 10.1104/pp.108.133496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin Z., Hong Y., Yin M., Li C., Zhang K., Grierson D. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 2008;55:301–310. doi: 10.1111/j.1365-313X.2008.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.González-Grandío E., Pajoro A., Franco-Zorrilla J.M., Tarancón C., Immink R.G., Cubas P. Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. USA. 2017;114:E245–E254. doi: 10.1073/pnas.1613199114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aguilar-Martínez J.A., Poza-Carrión C., Cubas P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell. 2007;19:458–472. doi: 10.1105/tpc.106.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Smet I., Lau S., Ehrismann J.S., Axiotis I., Kolb M., Kientz M., Weijers D., Jürgens G. Transcriptional repression of BODENLOS by HD-ZIP transcription factor HB5 in Arabidopsis thaliana. J. Exp. Bot. 2013;64:3009–3019. doi: 10.1093/jxb/ert137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lavy M., Estelle M. Mechanisms of auxin signaling. Development. 2016;143:3226–3229. doi: 10.1242/dev.131870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leyser O. Auxin signaling. Plant Physiol. 2018;176:465–479. doi: 10.1104/pp.17.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berleth T., Jurgens G. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development. 1993;118:575–587. [Google Scholar]
- 74.Hardtke C.S., Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hamann T., Mayer U., Jurgens G. The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development. 1999;126:1387–1395. doi: 10.1242/dev.126.7.1387. [DOI] [PubMed] [Google Scholar]
- 76.Hamann T., Benkova E., Bäurle I., Kientz M., Jurgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Růzicka K., Ljung K., Vanneste S., Podhorská R., Beeckman T., Friml J., Benková E. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell. 2007;19:2197–2212. doi: 10.1105/tpc.107.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stepanova A.N., Yun J., Likhacheva A.V., Alonso J.M. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell. 2007;19:2169–2185. doi: 10.1105/tpc.107.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Swarup R., Perry P., Hagenbeek D., Van Der Straeten D., Beemster G.T., Sandberg G., Bhalerao R., Ljung K., Bennett M.J. Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell. 2007;19:2186–2196. doi: 10.1105/tpc.107.052100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Markakis M.N., De Cnodder T., Lewandowski M., Simon D., Boron A., Balcerowicz D., Doubbo T., Taconnat L., Renou J.P., Höfte H., et al. Identification of genes involved in the ACC-mediated control of root cell elongation in Arabidopsis thaliana. BMC Plant Biol. 2012;12:208. doi: 10.1186/1471-2229-12-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miao Z.-Q., Zhao P.-X., Mao J., Yu L., Yuan Y., Tang H., Liu Z.-B., Xiang C. Arabidopsis HB52 mediates the crosstalk between ethylene and auxin by transcriptionally modulating PIN2, WAG1, and WAG2 during primary root elongation. Plant Cell. 2018 doi: 10.1105/tpc.18.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carabelli M., Morelli G., Whitelam G., Ruberti I. Twilight-zone and canopy shade induction of the ATHB-2 homeobox gene in green plants. Proc. Natl. Acad. Sci. USA. 1996;93:3530–3535. doi: 10.1073/pnas.93.8.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sorin C., Salla-Martret M., Bou-Torrent J., Roig-Villanova I., Martínez-García J.F. ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J. 2009;59:266–277. doi: 10.1111/j.1365-313X.2009.03866.x. [DOI] [PubMed] [Google Scholar]
- 84.Carabelli M., Turchi L., Ruzza V., Morelli G., Ruberti I. Homeodomain-Leucine Zipper II family of transcription factors to the limelight: Central regulators of plant development. Plant Signal. Behav. 2013;8:e25447. doi: 10.4161/psb.25447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ruberti I., Sessa G., Ciolfi A., Possenti M., Carabelli M., Morelli G. Plant adaptation to dynamically changing environment: The shade avoidance response. Biotechnol. Adv. 2012;30:1047–1058. doi: 10.1016/j.biotechadv.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 86.Ruzza V., Sessa G., Sassi M., Morelli G., Ruberti I. Auxin coordinates shoot and root development during shade avoidance response. In: Zažímalová E., Petrasek J., Benková E., editors. Auxin and Its Role in Plant Development. Springer; Berlin/Heidelberg, Germany: 2014. [Google Scholar]
- 87.Carabelli M., Possenti M., Sessa G., Ruzza V., Morelli G., Ruberti I. Arabidopsis HD-Zip II proteins regulate the exit from proliferation during leaf development in canopy shade. J. Exp. Bot. 2018;69:5419–5431. doi: 10.1093/jxb/ery331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bou-Torrent J., Salla-Martret M., Brandt R., Musielak T., Palauquim J.C., Martinez-Garcia J.F., Wenkel S. ATHB4 and HAT3, two class II HD-ZIP transcription factors, control leaf development in Arabidopsis. Plant Signal. Behav. 2012;7:1382–1387. doi: 10.4161/psb.21824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reymond M.C., Brunoud G., Chauvet A., Martínez-Garcia J.F., Martin-Magniette M.L., Monéger F., Scutt C.P. A light-regulated genetic module was recruited to carpel development in Arabidopsis following a structural change to SPATULA. Plant Cell. 2012;24:2812–2825. doi: 10.1105/tpc.112.097915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zúñiga-Mayo V.M., Marsch-Martínez N., de Folter S. JAIBA, a class-II HD-ZIP transcription factor involved in the regulation of meristematic activity, and important for correct gynoecium and fruit development in Arabidopsis. Plant J. 2012;71:314–326. doi: 10.1111/j.1365-313X.2012.04990.x. [DOI] [PubMed] [Google Scholar]
- 91.Merelo P., Ram H., Pia Caggiano M., Ohno C., Ott F., Straub D., Graeff M., Cho S.K., Yang S.W., Wenkel S., et al. Regulation of MIR165/166 by class II and class III homeodomain leucine zipper proteins establishes leaf polarity. Proc. Natl. Acad. Sci. USA. 2016;113:11973–11978. doi: 10.1073/pnas.1516110113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goda H., Sawa S., Asami T., Fujioka S., Shimada Y., Yoshida S. Comprehensive comparison of auxin-regulated and brassinosteroid regulated genes in Arabidopsis. Plant Physiol. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nemhauser J.L., Mockler T.C., Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guo H., Li L., Ye H., Yu X., Algreen A., Yin Y. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2009;106:7648–7653. doi: 10.1073/pnas.0812346106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li L., Ye H., Guo H., Yin Y. Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc. Natl. Acad. Sci. USA. 2010;107:3918–3923. doi: 10.1073/pnas.0909198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sun Y., Fan X.Y., Cao D.M., Tang W., He K., Zhu J.Y., He J.X., Bai M.Y., Zhu S., Oh E., et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P., et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65:634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- 98.Zhang D., Ye H., Guo H., Johnson A., Zhang M., Lin H., Yin Y. Transcription factor HAT1 is phosphorylated by BIN2 kinase and mediates brassinosteroid repressed gene expression in Arabidopsis. Plant J. 2014;77:59–70. doi: 10.1111/tpj.12368. [DOI] [PubMed] [Google Scholar]
- 99.Li J., Nam K.H., Vafeados D., Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li J., Nam K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 101.Yan Z., Zhao J., Peng P., Chihara R.K., Li J. BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 2009;150:710–721. doi: 10.1104/pp.109.138099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Daviere J.M., Achard P. Gibberellin signaling in plants. Development. 2013;140:1147–1151. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- 103.Marín-de la Rosa N., Sotillo B., Miskolczi P., Gibbs D.J., Vicente J., Carbonero P., Oñate-Sánchez L., Holdsworth M.J., Bhalerao R., Alabadí D., et al. Large-scale identification of gibberellin-related transcription factors defines group VII ETHYLENE RESPONSE FACTORS as functional DELLA partners. Plant Physiol. 2014;166:1022–1032. doi: 10.1104/pp.114.244723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zou L.J., Deng X.G., Han X.Y., Tan W.R., Zhu L.J., Xi D.-J., Zhang D.-W., Lin H.-H. Role of Transcription Factor HAT1 in Modulating Arabidopsis thaliana Response to Cucumber Mosaic Virus. Plant Cell Physiol. 2016;57:1879–1889. doi: 10.1093/pcp/pcw109. [DOI] [PubMed] [Google Scholar]
- 105.Tan W., Zhang D., Zhou H., Zheng T., Yin Y., Lin H. Transcription factor HAT1 is a substrate of SnRK2.3 kinase and negatively regulates ABA synthesis and signaling in Arabidopsis responding to drought. PLoS Genet. 2018;14:e1007336. doi: 10.1371/journal.pgen.1007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Park M.Y., Kim S.A., Lee S.J., Kim S.Y. ATHB17 is a positive regulator of abscisic acid response during early seedling growth. Mol. Cells. 2013;35:125–133. doi: 10.1007/s10059-013-2245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu T., Longhurst A.D., Talavera-Rauh F., Hokin S.A., Barton M.K. The Arabidopsis transcription factor ABIG1 relays ABA signaled growth inhibition and drought induced senescence. Elife. 2016;5:e13768. doi: 10.7554/eLife.13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smith Z.R., Long J.A. Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature. 2010;464:423–426. doi: 10.1038/nature08843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McConnell J.R., Emery J., Eshed Y., Bao N., Bowman J., Barton M.K. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 110.Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 111.Prigge M.J., Otsuga D., Alonso J.M., Ecker J.R., Drews G.N., Clark S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhong R., Ye Z.H. IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell. 1999;11:2139–2152. doi: 10.1105/tpc.11.11.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Baima S., Possenti M., Matteucci A., Wisman E., Altamura M., Ruberti I., Morelli G. The Arabidopsis ATHB8 HD-Zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 2001;126:643–655. doi: 10.1104/pp.126.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ohashi-Ito K., Fukuda H. HD-Zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol. 2003;44:1350–1358. doi: 10.1093/pcp/pcg164. [DOI] [PubMed] [Google Scholar]
- 115.Ohashi-Ito K., Kubo M., Demura T., Fukuda H. Class III homeodomain leucine-zipper proteins regulate xylem cell differentiation. Plant Cell Physiol. 2005;46:1646–1656. doi: 10.1093/pcp/pci180. [DOI] [PubMed] [Google Scholar]
- 116.Carlsbecker A., Lee J.-Y., Roberts C.J., Dettmer J., Lehesranta S., Zhou J., Lindgren O., Moreno-Risueno M.A., Vatén A., Thitamadee S., et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ilegems M., Douet V., Meylan-Bettex M., Uyttewaal M., Brand L., Bowman J.L., Stieger P.A. Interplay of auxin, KANADI and Class III HD-ZIP transcription factors in vascular tissue formation. Development. 2010;137:975–984. doi: 10.1242/dev.047662. [DOI] [PubMed] [Google Scholar]
- 118.Baima S., Nobili F., Sessa G., Lucchetti S., Ruberti I., Morelli G. The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development. 1995;121:4171–4182. doi: 10.1242/dev.121.12.4171. [DOI] [PubMed] [Google Scholar]
- 119.Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 120.Ursache R., Miyashima S., Chen Q., Vatén A., Nakajima K., Carlsbecker A., Zhao Y., Helariutta Y., Dettmer J. Tryptophan-dependent auxin biosynthesis is required for HD-ZIP III-mediated xylem patterning. Development. 2014;141:1250–1259. doi: 10.1242/dev.103473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Donner T.J., Sherr I., Scarpella E. Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development. 2009;136:3235–3246. doi: 10.1242/dev.037028. [DOI] [PubMed] [Google Scholar]
- 122.Baima S., Forte V., Possenti M., Peñalosa A., Leoni G., Salvi S., Felici B., Ruberti I., Morelli G. Negative feedback regulation of auxin signaling by ATHB8/ACL5-BUD2 transcription module. Mol. Plant. 2014;7:1006–1025. doi: 10.1093/mp/ssu051. [DOI] [PubMed] [Google Scholar]
- 123.Tang G., Reinhart B.J., Bartel D.P., Zamore P.D. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ruonala R., Ko D., Helariutta Y. Genetic Networks in Plant Vascular Development. Annu. Rev. Genet. 2017;51:335–359. doi: 10.1146/annurev-genet-120116-024525. [DOI] [PubMed] [Google Scholar]
- 125.Huang T., Harrar Y., Lin C., Reinhart B., Newell N.R., Talavera-Rauh F., Hokin S.A., Barton M.K., Kerstetter R.A. Arabidopsis KANADI1 acts as a transcriptional repressor by interacting with a specific cis-element and regulates auxin biosynthesis, transport, and signaling in opposition to HD-ZIPIII factors. Plant Cell. 2014;26:246–262. doi: 10.1105/tpc.113.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Reinhart B.J., Liu T., Newell N.R., Magnani E., Huang T., Kerstetter R., Michaels S., Barton M.K. Establishing a framework for the Ad/abaxial regulatory network of Arabidopsis: Ascertaining targets of class III homeodomain leucine zipper and KANADI regulation. Plant Cell. 2013;25:3228–3249. doi: 10.1105/tpc.113.111518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu T., Reinhart B.J., Magnani E., Huang T., Kerstetter R., Barton M.K. Of blades and branches: Understanding and expanding the Arabidopsis Ad/Abaxial regulatory network through target gene identification. Cold Spring Harb. Symp. Quant. Biol. 2013;77:31–35. doi: 10.1101/sqb.2013.77.014480. [DOI] [PubMed] [Google Scholar]
- 128.Cheng Y., Qin G., Dai X., Zhao Y. NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2007;104:18825–18829. doi: 10.1073/pnas.0708506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cheng Y., Qin G., Dai X., Zhao Y. NPY genes and AGC kinase genes define two key steps in auxin-mediated cotyledon organogenesis. Proc. Natl. Acad. Sci. USA. 2008;105:21017–21022. doi: 10.1073/pnas.0809761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dhonukshe P., Huang F., Galvan-Ampudia C.S., Mähönen A.P., Kleine-Vehn J., Xu J., Quint A., Prasad K., Friml J., Scheres B., et al. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development. 2010;137:3245–3255. doi: 10.1242/dev.052456. [DOI] [PubMed] [Google Scholar]
- 131.Furutani M., Sakamoto N., Yoshida S., Kajiwara T., Robert H.S., Friml J., Tasaka M. Polar-localized NPH3-like proteins regulate polarity and endocytosis of PIN-FORMED auxin efflux carriers. Development. 2011;138:2069–2078. doi: 10.1242/dev.057745. [DOI] [PubMed] [Google Scholar]
- 132.Müller C.J., Valdés A.E., Wang G., Ramachandran P., Beste L., Uddenberg D., Carlsbecker A. PHABULOSA Mediates an Auxin Signaling Loop to Regulate Vascular Patterning in Arabidopsis. Plant Physiol. 2016;170:956–970. doi: 10.1104/pp.15.01204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mähönen A.P., Bishopp A., Higuchi M., Nieminen K.M., Kinoshita K., Törmäkangas K., Ikeda Y., Oka A., Kakimoto T., Helariutta Y. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311:94–98. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 134.Bishopp A., Help H., El-Showk S., Weijers D., Scheres B., Friml J., Benková E., Mähönen A.P., Helariutta Y. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr. Biol. 2011;21:917–926. doi: 10.1016/j.cub.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 135.Dello Ioio R., Galinha C., Fletcher A.G., Grigg S.P., Molnar A., Willemsen V., Scheres B., Sabatini S., Baulcombe D., Maini P.K., et al. A PHABULOSA/cytokinin feedback loop controls root growth in Arabidopsis. Curr. Biol. 2012;22:1699–1704. doi: 10.1016/j.cub.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 136.Bishopp A., Lehesranta S., Vatén A., Help H., El-Showk S., Scheres B., Helariutta K., Mähönen A.P., Sakakibara H., Helariutta Y. Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr. Biol. 2011;21:927–932. doi: 10.1016/j.cub.2011.04.049. [DOI] [PubMed] [Google Scholar]
- 137.Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M.T., Aoyama T., Costantino P., Sabatini S. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322:1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- 138.Argyros R.D., Mathews D.E., Chiang Y.H., Palmer C.M., Thibault D.M., Etheridge N., Argyros D.A., Mason M.G., Kieber J.J., Schaller G.E. Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell. 2008;20:2102–2116. doi: 10.1105/tpc.108.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sebastian J., Ryu K.H., Zhou J., Tarkowská D., Tarkowski P., Cho Y.H., Yoo S.D., Kim E.S., Lee J.Y. PHABULOSA Controls the Quiescent Center-Independent Root Meristem Activities in Arabidopsis thaliana. PLoS Genet. 2015;11:e1004973. doi: 10.1371/journal.pgen.1004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang T.Q., Lian H., Zhou C.M., Xu L., Jiao Y., Wang J.W. A Two-Step Model for de Novo Activation of WUSCHEL during Plant Shoot Regeneration. Plant Cell. 2017;29:1073–1087. doi: 10.1105/tpc.16.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Banno H., Ikeda Y., Niu Q.W., Chua N.H. Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell. 2001;13:2609–2618. doi: 10.1105/tpc.13.12.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ikeda Y., Banno H., Niu Q.-W., Howell S.H., Chua N.-H. The ENHANCER OF SHOOT REGENERATION 2 gene in Arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol. 2006;47:1443–1456. doi: 10.1093/pcp/pcl023. [DOI] [PubMed] [Google Scholar]
- 143.Duclercq J., Assoumou Ndong Y.P., Guerineau F., Sangwan R.S., Catterou M. Arabidopsis shoot organogenesis is enhanced by an amino acid change in the ATHB15 transcription factor. Plant Biol. 2011;13:317–324. doi: 10.1111/j.1438-8677.2010.00363.x. [DOI] [PubMed] [Google Scholar]
- 144.Mason M.G., Mathews D.E., Argyros D.A., Maxwell B.B., Kieber J.J., Alonso J.M., Ecker J.R., Schaller G.E. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell. 2005;17:3007–3018. doi: 10.1105/tpc.105.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ishida K., Yamashino T., Yokoyama A., Mizuno T. Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 2008;49:47–57. doi: 10.1093/pcp/pcm165. [DOI] [PubMed] [Google Scholar]
- 146.Zhang T.Q., Lian H., Tang H., Dolezal K., Zhou C.M., Yu S., Chen J.H., Chen Q., Liu H., Ljung K., et al. An intrinsic microRNA timer regulates progressive decline in shoot regenerative capacity in plants. Plant Cell. 2015;27:349–360. doi: 10.1105/tpc.114.135186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Buechel S., Leibfried A., To J.P., Zhao Z., Andersen S.U., Kieber J.J., Lohmann J.U. Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur. J. Cell Biol. 2010;89:279–284. doi: 10.1016/j.ejcb.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 148.Mayer K.F.X., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/S0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 149.Eshed Y., Baum S.F., Perea J.V., Bowman J.L. Establishment of polarity in lateral organs of plants. Curr. Biol. 2001;11:1251–1260. doi: 10.1016/S0960-9822(01)00392-X. [DOI] [PubMed] [Google Scholar]
- 150.Singh A., Roy S., Singh S., Das S.S., Gautam V., Yadav S., Kumar A., Singh A., Samantha S., Sarkar A.K. Phytohormonal crosstalk modulates the expression of miR166/165s, target Class III HD-ZIPs, and KANADI genes during root growth in Arabidopsis thaliana. Sci. Rep. 2017;7:3408. doi: 10.1038/s41598-017-03632-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yan J., Zhao C., Zhou J., Yang Y., Wang P., Zhu X., Tang G., Bressan R.A., Zhu J.K. The miR165/166 Mediated Regulatory Module Plays Critical Roles in ABA Homeostasis and Response in Arabidopsis thaliana. PLoS Genet. 2016;12:e1006416. doi: 10.1371/journal.pgen.1006416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ramachandran P., Wang G., Augstein F., de Vries J., Carlsbecker A. Continuous root xylem formation and vascular acclimation to water deficit involves endodermal ABA signalling via miR165. Development. 2018;145:dev159202. doi: 10.1242/dev.159202. [DOI] [PubMed] [Google Scholar]
- 153.Chew W., Hrmova M., Lopato S. Role of Homeodomain leucine zipper (HD-Zip) IV transcription factors in plant development and plant protection from deleterious environmental factors. Int. J. Mol. Sci. 2013;14:8122–8147. doi: 10.3390/ijms14048122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Javelle M., Vernoud V., Rogowsky P.M., Ingram G.C. Epidermis: The formation and functions of a fundamental plant tissue. New Phytol. 2011;189:17–39. doi: 10.1111/j.1469-8137.2010.03514.x. [DOI] [PubMed] [Google Scholar]
- 155.Rerie W.G., Feldmann K.A., Marks M.D. The glabra 2 gene encodes a homeodomain protein required for normal trichome development in Arabidopsis. Genes Dev. 1994;8:1388–1399. doi: 10.1101/gad.8.12.1388. [DOI] [PubMed] [Google Scholar]
- 156.Di Cristina M., Sessa G., Dolan L., Linstead P., Baima S., Ruberti I., Morelli G. The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 1996;10:393–402. doi: 10.1046/j.1365-313X.1996.10030393.x. [DOI] [PubMed] [Google Scholar]
- 157.Masucci J.D., Rerie W.G., Foreman D.R., Zhang M., Galway M.E., Marks M.D., Schiefelbein J.W. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development. 1996;122:1253–1260. doi: 10.1242/dev.122.4.1253. [DOI] [PubMed] [Google Scholar]
- 158.Kuppusamy K.T., Chen A.Y., Nemhauser J.L. Steroids are required for epidermal cell fate establishment in Arabidopsis roots. Proc. Natl. Acad. Sci. USA. 2009;106:8073–8076. doi: 10.1073/pnas.0811633106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rombolá-Caldentey B., Rueda-Romero P., Iglesias-Fernández R., Carbonero P., Oñate-Sánchez L. Arabidopsis DELLA and two HD-ZIP transcription factors regulate GA signaling in the epidermis through the L1 box cis-element. Plant Cell. 2014;26:2905–2919. doi: 10.1105/tpc.114.127647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Abe M., Takahashi T., Komeda Y. Cloning and characterization of an L1 layer-specific gene in Arabidopsis thaliana. Plant Cell Physiol. 1999;40:571–580. doi: 10.1093/oxfordjournals.pcp.a029579. [DOI] [PubMed] [Google Scholar]
- 161.Yu L., Chen X., Wang Z., Wang S., Wang Y., Zhu Q., Li S., Xiang C. Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 2013;162:1378–1391. doi: 10.1104/pp.113.217596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yu H., Chen X., Hong Y.Y., Wang Y., Xu P., Ke S.D., Liu H.Y., Zhu J.K., Oliver D.J., Xiang C.B. Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell. 2008;20:1134–1151. doi: 10.1105/tpc.108.058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yu L.-H., Wu S.-J., Peng Y.-S., Liu R.-N., Chen X., Zhao P., Xu P., Zhu J.-B., Jiao G.-L., Pei Y., et al. Arabidopsis EDT1/HDG11 improves drought and salt tolerance in cotton and poplar and increases cotton yield in the field. Plant Biotechnol. J. 2016;14:72–84. doi: 10.1111/pbi.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Xu P., Cai X.-T., Wang Y., Xing L., Chen Q., Xiang C.-B. HDG11 upregulates cell-wall-loosening protein genes to promote root elongation in Arabidopsis. J. Exp. Bot. 2014;65:4285–4295. doi: 10.1093/jxb/eru202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ueda M., Aichinger E., Gong W., Groot E., Verstraeten I., Vu L.D., De Smet I., Higashiyama T., Umeda M., Laux T. Transcriptional integration of paternal and maternal factors in the Arabidopsis zygote. Genes Dev. 2017;31:617–627. doi: 10.1101/gad.292409.116. [DOI] [PMC free article] [PubMed] [Google Scholar]