Abstract
Rice (Oryza sativa L.) is an important food crop providing energy and nutrients for more than half of the world population. It produces vast amounts of secondary metabolites. At least 276 secondary metabolites from rice have been identified in the past 50 years. They mainly include phenolic acids, flavonoids, terpenoids, steroids, alkaloids, and their derivatives. These metabolites exhibit many physiological functions, such as regulatory effects on rice growth and development, disease-resistance promotion, anti-insect activity, and allelopathic effects, as well as various kinds of biological activities such as antimicrobial, antioxidant, cytotoxic, and anti-inflammatory properties. This review focuses on our knowledge of the structures, biological functions and activities, biosynthesis, and metabolic regulation of rice secondary metabolites. Some considerations about cheminformatics, metabolomics, genetic transformation, production, and applications related to the secondary metabolites from rice are also discussed.
Keywords: rice, Oryza sativa, secondary metabolites, phytoalexins, biological functions, biosynthesis, elicitation, metabolic regulation
1. Introduction
Rice (Oryza sativa L.), which belongs to the Gramineae family, has been consumed by humans for almost 5000 years. Rice is a widely diffuse staple food, providing energy and nutrients for more than half of the world population, especially in Asia [1]. The most common rice consumed by humans is white rice (about 85%), and the rest is pigmented rice [2]. Furthermore, rice is a model plant for molecular studies of monocotyledonous species [3]. There are diverse secondary metabolites produced in rice. These metabolites are organ- and tissue-specific. For example, diterpenoid phytoalexins are mainly present in the leaves, whereas phenolic acids, flavonoids, sterols, and triterpenoids are mainly present in the bran [4]. Rice secondary metabolites play roles either as defense agents, by providing disease resistance and exerting anti-nematodal, anti-insect, and allelopathic ativities against biotic and abiotic stresses, or as plant growth regulators. They also show various kinds of biological activities, such as antimicrobial, antioxidant, cytotoxic, and anti-inflammatory properties, which are implicated in various health-promoting and disease-preventive effects. Rice metabolites mainly include phenolic acids, flavonoids, terpenoids, steroids, alkaloids. Some metabolites such as phenolic acids and flavonoids are also distributed in other plant species [5,6]. To our knowledge, many reviews have discussed a specific topic of rice secondary metabolites [7,8,9,10,11,12,13,14], but no review has focused on describing the whole variety of secondary metabolites of rice so far. Furthermore, significant advances on rice secondary metabolism have been made recently based on genomic, biosynthesis regulation, and metabolomic approaches [15,16,17,18]. In this review, we summarize and discuss the developments from studies on the structural diversity, biological functions, biosynthesis, and metabolic regulations of rice secondary metabolites.
2. Structural Diversity and Roles of Rice Secondary Metabolites
Rice can accumulate a large number of secondary metabolites, such as phenolic acids, flavonoids, terpenoids, steroids, and alkaloids. These molecules play various physiological and ecological roles (i.e., antimicrobial, insecticidal, growth regulatory, and allelopathic activities). They also exhibit features beneficial to humans, including cytotoxic, anti-tumor, anti-inflammatory, antioxidant, and neuroprotective properties. For example, many phenolic acids, flavonoids, tocopherols, tocotrienols, γ-oryzanol, and phytic acid from rice exhibit antioxidant activities [10].
2.1. Phenolic Acids and Their Biological Functions
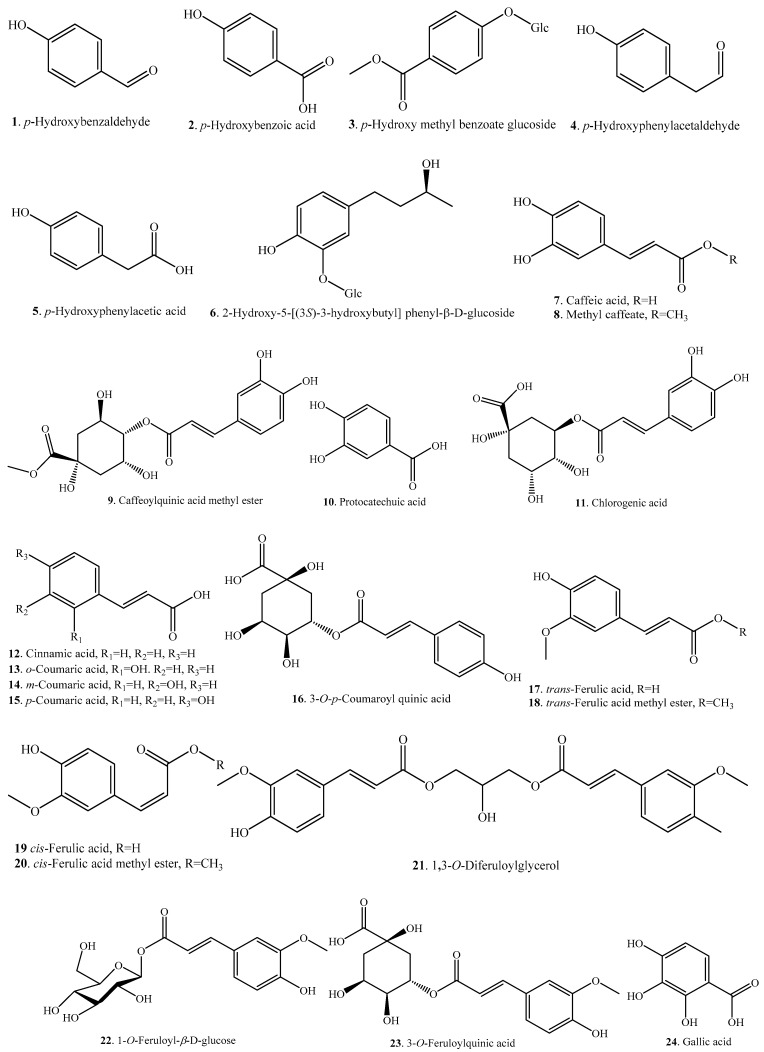
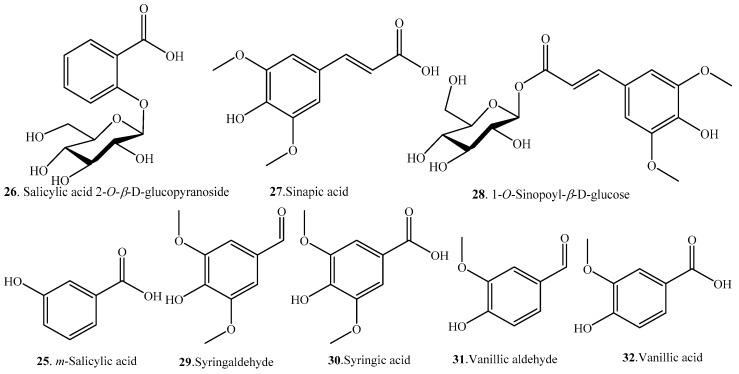
Rice phenolic acids can be classified as soluble-free, soluble-conjugated, and insoluble-bound forms. The insoluble-bound phenolic acids covalently boind to structural components of cells like cellulose, hemicellulose, lignin, pectin, rod-shaped structural proteins, etc. [19]. The distribution of rice phenolic acids exhibits varietal differences, and rice bran has the highest total phenolic acid content among four different fractions of whole rice grain [20,21]. Overall, p-hydroxybenzoic acid (2), caffeic acid (7), protocatechuic acid (10), ferulic acid (17/19), sinapic acid (27), syringic acid (30), and vanillic acid (32) are present in the whole rice grain, and ferulic acid (17/19) is the most abundant phenolic acid in the insoluble-bound fraction [22]. Normally, the pigmented rice contains phenolic acids with a larger structural diversity and in higher content than the non-pigmented rice [2,13]. About 32 phenolic acid analogues have be identified in rice. Rice phenolic acids and their biological activities are listed in Table 1. The structures of rice phenolic acids are shown in Figure 1. Most rice phenolic acids have antioxidant activities, though some of them have not been evaluated individually but only mixed with other rice phenolic acids [20].
Table 1.
Phenolic acids and their biological activities.
| Name | Rice Part Used for Isolation | Biological Activity and Function | Ref. |
|---|---|---|---|
| p-Hydroxybenzaldehyde (1) | Husk and bran | - | [24] |
| Bran | Antioxidant activity | [31] | |
| p-Hydroxybenzoic acid (2) | Root exudate | Allelopathic effect | [30] |
| Husk and bran | - | [24] | |
| p-Hydroxy methyl benzoate glucoside (3) | Bran | Antioxidant activity | [31] |
| p-Hydroxy phenyl acetaldehyde (4) | Husk and bran | - | [24] |
| p-Hydroxy phenyl acetic acid (5) | Husk and bran | - | [24] |
| 2-Hydroxy 5-[(3S)-3-hydroxybutyl] phenyl β-d-glucoside (HHPG) (6) | Brans of purple rice | Inhibitory activity on tunicamycin-induced retinal damage | [32] |
| Caffeic acid (7) | Endosperm and bran/embryo of indica variety | Antioxidant activity | [20] |
| Root exudate | Allelopathic effect | [30] | |
| Husk and bran | - | [24] | |
| Methyl caffeate (8) | Bran | Antioxidant activity | [31] |
| Caffeoyl quinic acid methyl ester (9) | Grains of brown rice | - | [33] |
| Protocatechuic acid (10) | Endosperm and bran/embryo of indica variety | Antioxidant activity | [20] |
| Chlorogenic acid (11) | Endosperm and bran/embryo of indica variety | Antioxidant activity | [20] |
| Cinnamic acid (12) | Husk and bran | - | [24] |
| o-Coumaric acid (13) | Endosperm and bran/embryo of indica variety | Antioxidant activity | [20] |
| Root exudate | Allelopathic effect | [30] | |
| m-Coumaric acid (14) | Grains | - | [23] |
| p-Coumaric acid (15) | Grains | - | [23] |
| Grains of brown rice | - | [33] | |
| 3-O-p-Coumaroyl quinic acid (16) | Grains of brown rice | - | [33] |
| Leaves | - | [34] | |
| trans-Ferulic acid (17) | Endosperm and bran/embryo of indica variety | Antioxidant activity | [20] |
| Grains | - | [23] | |
| Black rice bran | Antioxidant activity | [25] | |
| Husk and bran | - | [24] | |
| Bran | Antioxidant activity | [31] | |
| trans-Ferulic acid methyl ester (18) | Bran | Antioxidant activity | [31] |
| cis-Ferulic acid (19) | Bran | Antioxidant activity | [31] |
| cis-Ferulic acid methyl ester (20) | Bran | Antioxidant activity | [31] |
| 1,3-O-Diferuloylglycerol (21) | Leaves | - | [34] |
| 1-O-Feruloyl-β-d-glucose (22) | Leaves | - | [34] |
| 3-O-Feruloylquinic acid (23) | Leaves | - | [34] |
| Gallic acid (24) | Endosperm and bran/embryo of indica variety | Antioxidant activity | [20] |
| Husk and bran | - | [24] | |
| m-Salicylic acid (25) | Grains of brown rice | - | [33] |
| Salicylic acid 2-O-β-d-glucopyranoside (26) | Leaves | - | [34] |
| Sinapic acid (27) | Grains | - | [23] |
| 1-O-Sinapoyl-β-d-glucose (28) | Leaves | - | [34] |
| Syringaldehyde (29) | Grains of brown rice | - | [33] |
| Syringic acid (30) | Endosperm and bran/embryo of indica variety | Antioxidant activity | [20] |
| Root exudate | Allelopathic effect | [30] | |
| Husk and bran | - | [24] | |
| Grains of brown rice | - | [33] | |
| Vanillic aldehyde (31) | Bran | Antioxidant activity | [31] |
| Vanillic acid (32) | Root exudate | Allelopathic effect | [30] |
| Husk and bran | - | [24] | |
| Grains of brown rice | - | [33] |
Figure 1.
Structures of the phenolic acids isolated from rice.
Zaupa et al. revealed that the main rice phenolic acids are protecatechuic acid (10), p-coumaric acid (15), ferulic acid (17/19), sinapic acid (27), and vanillic acid (32) [23]. Ding et al. investigated eight rice varieties of O. sativa sp. japonica and O. sativa sp. indica planted in different areas of China for their phenolic acids distribution by using UPLC-MS method. A total of 12 phenolic compounds were identified in all rice varieties. Protocatechuic acid (10), ferulic acid (17/19), gallic acid (24), and syringic acid (30) were the dominant phenolic compounds in rice bran, while p-hydroxybenzaldehyde (1) was the main phenolic acid in rice husk. Bran and husk fractions provide more than 90% of phenolic acids and antioxidant activity of the whole rice plant. In addition, the rice subspecies japonica has significant higher phenolic acids content and antioxidant activity than the indica subspecies [24]. Ferulic acid (17/19) has also been found as the major phenolic compound in black rice bran, indicating the potential use of black rice bran as a natural source of antioxidants [25].
Phenolic acids are considered to be natural antioxidants, being able to scavenge free radicals that may increase oxidative stress and potentially damage large biological molecules such as lipids, proteins, and nucleic acids [26]. Therefore, the phenolic acid content was positively correlated with rice antioxidant capacity [27]. The development and utilization of phenolic acid analogues from rice husk and bran are important for improving the functionality of rice by-products.
Some phenolic acids are released from rice roots as allelochemicals. The main phenolic acids in the root exudates were identified as p-hydroxybenzoic (2), caffeic (7), p-coumaric (15), syringic (30), and vanillic (32) acids [28]. As these phenolic acids are released at relatively low concentrations in the soil and other plant species have a high level of tolerance against phenolic acids, they are considered the least important allelochemicals in rice [8,29,30].
2.2. Flavonoids and Their Biological Functions
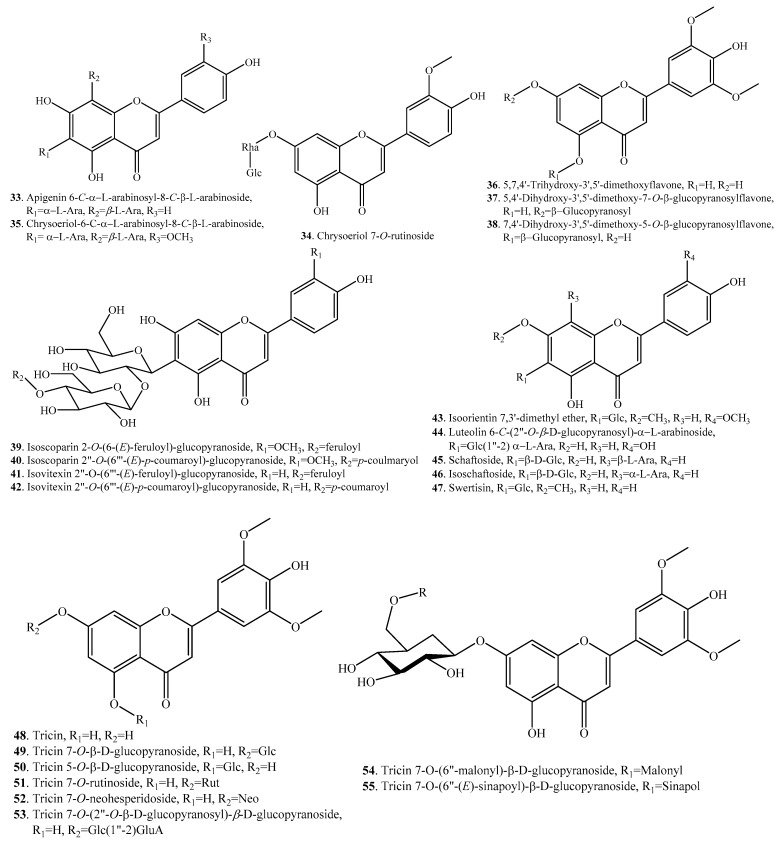
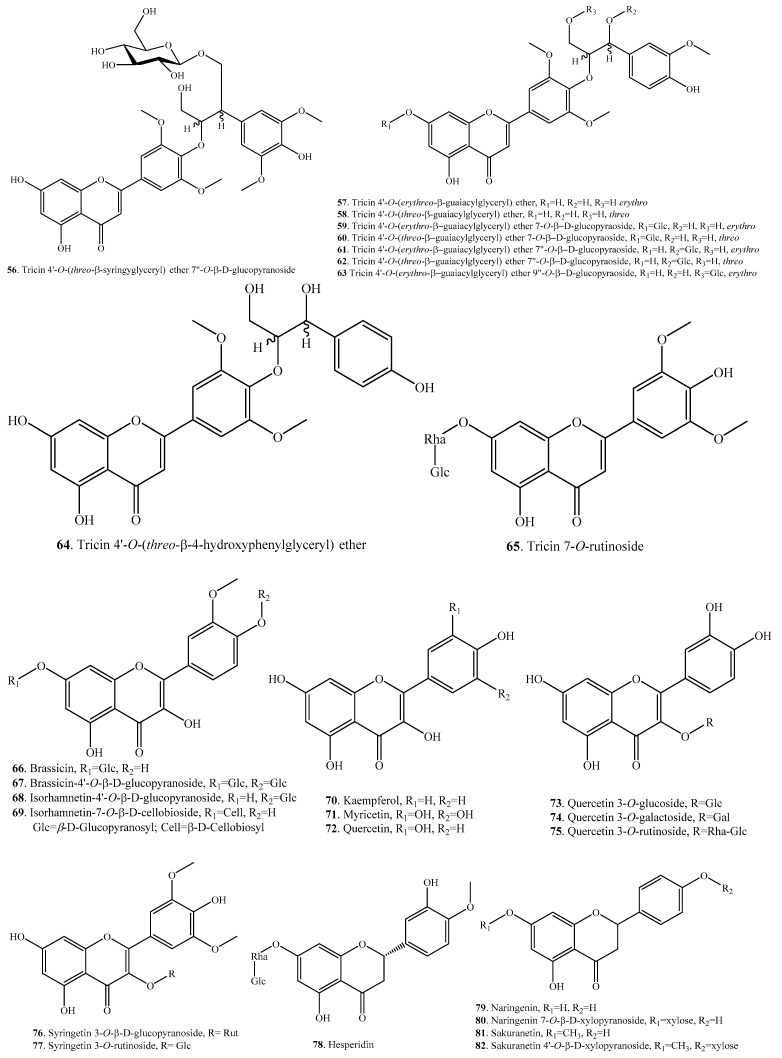
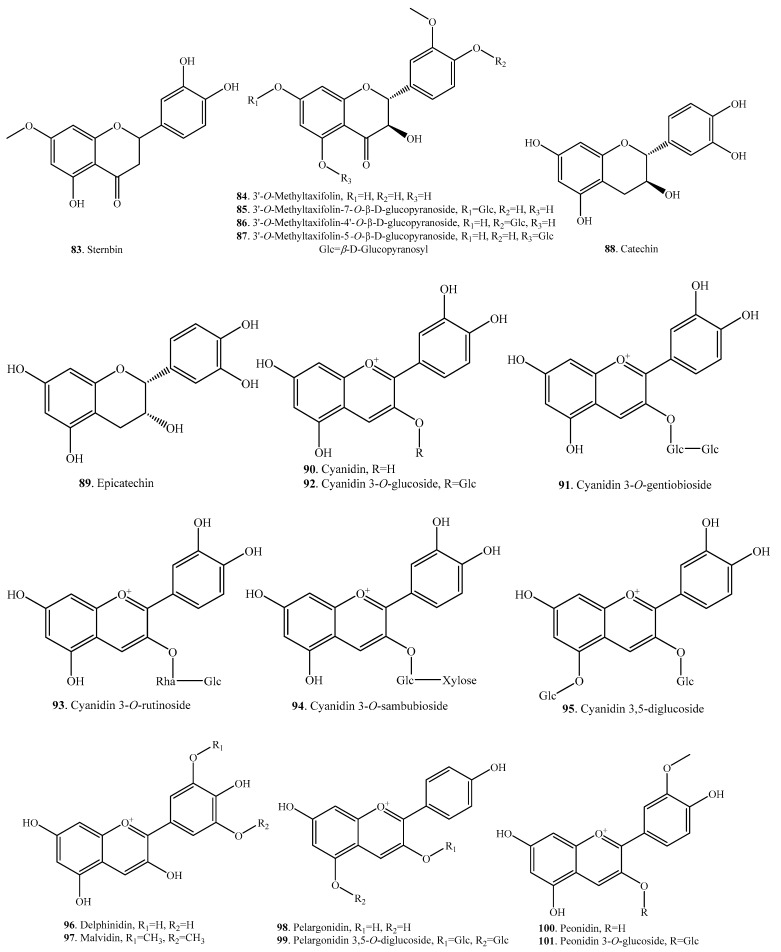
According to the structural features, rice flavonoids can be classified as flavones (33~65), flavonols (66~77), flavanones (or dihydroflavones, 78~83), flavanonols (84~87), flavanols (88, 89), and anthocyanins (90~101), along with their glycosides. Rice flavonoids mainly have antioxidant properties, though some of them have not been evaluated for their antioxidant activities [10]. Among them, anthocyanins are mainly distributed in pigmented rice plants [2]. Rice flavonoids and their biological activities are listed in Table 2. Their structures are shown in Figure 2.
Table 2.
Flavonoids and their biological activities.
| Name | Rice Part Used for Isolation | Biological Activity and Function | Ref. |
|---|---|---|---|
| Flavones | |||
| Apigenin 6-C-α-l-arabinosyl-8-C-β-l-arabinoside (33) | Leaves | - | [53] |
| Chrysoeriol 7-O-rutinoside (34) | Grains of brown rice | - | [33] |
| Chrysoeriol 6-C-α-l-arabinosyl-8-C-β-l-arabinoside (35) | Leaves | - | [53] |
| 5,7,4′-Trihydroxy-3′,5′-dimethoxyflavone (36) | Leaves | Allelopathic activity; antifungal activity | [54,55] |
| Seedlings | Allelopathic activity | [41] | |
| 5,4′-Dihydroxy-3′,5′-dimethoxy-7-O-β-glucopyranosylflavone (37) | Seedlings | - | [41] |
| 7,4′-Dihydroxy-3′,5′-dimethoxy-5-O-β-glucopyranosylflavone (38) | Seedlings | - | [41] |
| Isoscoparin 2-O-(6-(E)-feruloyl)-glucopyranoside (39) | Leaves | - | [53] |
| Isoscoparin 2″-O-(6‴-(E)-p-coumaroyl)-glucopyranoside (40) | Leaves | - | [53] |
| Isovitexin 2″-O-(6‴-(E)-feruloyl)-glucopyranoside (41) | Leaves | - | [53] |
| Isovitexin 2″-O-(6‴-(E)-p-coumaroyl)-glucopyranoside (42) | Leaves | - | [53] |
| Isoorientin 7,3′-dimethyl ether (43) | Leaves | - | [53] |
| luteolin 6-C-(2″-O-β-d-glucopyranosyl)-α-l-arabinoside (44) | Leaves | - | [53] |
| Schaftoside (45) | Leaves | Antifeedant activity | [35] |
| Isoschaftoside (46) | Leaves | Antifeedant activity | [35] |
| Swertisin (47) | Leaves | - | [53] |
| Tricin (48) | Bran | DPPH radical scavenging activity | [56] |
| Tricin 7-O-β-d-glucopyranoside (49) | Leaves | - | [53] |
| Tricin 5-O-β-d-glucopyranoside (50) | Leaves | - | [53] |
| Tricin 7-O-rutinoside (51) | Leaves | - | [53] |
| Tricin 7-O-neohesperidoside (52) | Leaves | - | [53] |
| Tricin 7-O-(2″-O-β-d-glucopyranosyl)-β-d-glucuronopyranoside (53) | Leaves | - | [53] |
| Tricin 7-O-(6″-O-malonyl)-β-d-glucopyranoside (54) | Leaves | - | [53] |
| Tricin 7-O-(6″-(E)-sinapoyl)-β-d-glucopyranoside (55) | Leaves | - | [53] |
| Tricin 4′-O-(threo-β-syringylglyceryl) ether 7″-O-β-d-glucopyranoside (56) | Leaves | - | [53] |
| Tricin 4′-O-(erythro-β-guaiacylglyceryl) ether (57) | Bran | DPPH radical scavenging activity | [56] |
| Bran | Cytotoxicity and apoptosis induction in multiple tumor cells | [38] | |
| Tricin 4′-O-(threo-β-guaiacylglyceryl) ether (58) | Bran | DPPH radical scavenging activity | [56] |
| Bran | Cytotoxicity and apoptosis induction in multiple tumor cells | [38] | |
| Tricin 4′-O-(erythro-β-guaiacylglyceryl) ether 7-O-β-d-glucopyranoside (59) | Leaves | - | [53] |
| Tricin 4′-O-(threo-β-guaiacylglyceryl) ether 7-O-β-d-glucopyranoside (60) | Leaves | - | [53] |
| Tricin 4′-O-(erythro-β-guaiacylglyceryl) ether 7″-O-β-d-glucopyranoside (61) | Leaves | - | [53] |
| Tricin 4′-O-(threo-β-guaiacylglyceryl) ether 7″-O-β-d-glucopyranoside (62) | Leaves | - | [53] |
| Tricin 4′-O-(erythro-β-guaiacylglyceryl) ether 9″-O-β-d-glucopyranoside (63) | Leaves | - | [53] |
| Tricin 4′-O-(threo-β-4-hydroxyphenylglyceryl) ether (64) | Leaves | - | [53] |
| Tricin 7-O-rutinoside (65) | Grains of brown rice | - | [33] |
| Flavonols | |||
| Brassicin (66) | Grains of transgenic japonica | Radical scavenging activity | [39] |
| Brassicin-4′-O-β-d-glucopyranoside (67) | Grains of transgenic japonica | Radical scavenging activity | [39] |
| Isorhamnetin-4′-O-β-d-glucopyranoside (68) | Grains of transgenic japonica | Radical scavenging activity | [39] |
| Isorhamnetin-7-O-β-d-cellobioside (69) | Grains of transgenic japonica | Radical scavenging activity | [39] |
| Kaempferol (70) | Husk and bran | - | [24] |
| Myricetin (71) | Rice flour | - | [57] |
| Quercetin (72) | Rice flour | - | [57] |
| Quercetin 3-O-glucoside (73) | Rice flour | - | [57] |
| Quercetin 3-O-galactoside = Hyperoside (74) | Rice flour | - | [57] |
| Qucertin 3-O-rutinoside = Rutin (75) | Rice flour | - | [57] |
| Syringetin 3-O-β-d-glucopyranoside (76) | Leaves | - | [53] |
| Syringetin 3-O-rutinoside (77) | Leaves | - | [53] |
| Flavanones | |||
| Hesperidin (78) | Rice flour | - | [57] |
| Naringenin (79) | Leaves | - | [42] |
| - | Antifungal activity | [43,44] | |
| Rice flour | - | [57] | |
| Rice fungal pathogen | - | [44,45] | |
| Naringenin 7-O-β-d-xylopyranoside (80) | Rice fungal pathogen | - | [45] |
| Sakuranetin (81) | Leaves | Antifungal activity | [43,44] |
| Leaves | Antibacterial and antifungal activities | [58] | |
| Leaves | Anti-Helicobacter pylori activity | [49] | |
| - | Antileishmanial and antitrypanosomal activities | [50] | |
| - | Antioxidant activity | [42] | |
| - | Anti-inflammatory activity | [47] | |
| - | Anti-mutagenic activity | [48] | |
| - | Induction of adipogenesis in 3T3-L1 cells | [46] | |
| - | Induction of melanogenesis in B16BL6 melanoma cells | [51] | |
| Sakuranetin 4′-O-β-d-xylopyranoside (82) | Rice fungal pathogen | - | [45] |
| Sternbin (83) | Rice fungal pathogen | - | [44] |
| Flavanonols | |||
| 3′-O-Methyltaxifolin (84) | Grains of transgenic japonica | Radical scavenging activity | [39] |
| 3′-O-Methyltaxifolin-7-O-β-d-glucopyranoside (85) | Grains of transgenic japonica | Radical scavenging activity | [39] |
| 3′-O-Methyltaxifolin-4′-O-β-d-glucopyranoside (86) | Grains of transgenic japonica | Radical scavenging activity | [39] |
| 3′-O-Methyltaxifolin-5-O-β-d-glucopyranoside (87) | Grains of transgenic japonica | Radical scavenging activity | [39] |
| Flavanols | |||
| Catechin (88) | Rice flour | - | [57] |
| Epicatechin (89) | Rice flour | - | [57] |
| Anthocyanins | |||
| Cyanidin (90) | Bran | - | [59] |
| Black rice kernels | Antioxidant activity | [52] | |
| Cyanidin 3-O-gentiobioside (91) | Bran | - | [60] |
| - | Inhibitory activity on tunicamycin-induced retinal damage | [32] | |
| Cyanidin 3-O-glucoside (92) | Bran | - | [60] |
| Inhibitory activity on tunicamycin-induced retinal damage | [32] | ||
| Cyanidin 3-O-rutinoside (93) | Kernels | - | [61] |
| Cyanidin 3-O-sambubioside (94) | Black rice kernels | Antioxidant activity | [52] |
| Cyanidin 3,5-O-diglucoside (95) | Kernels | - | [61] |
| Delphinidin (96) | Bran | - | [59] |
| Malvidin (97) | Bran | - | [59] |
| Pelargonidin (98) | Bran | - | [59] |
| Pelargonidin 3,5-O-diglucoside (99) | Pigmented rice | Antioxidant activity | [2] |
| Peonidin (100) | Black rice kernels | Antioxidant activity | [52] |
| Peonidin 3-O-glucoside (101) | Bran | - | [60] |
| - | Inhibitory activity on tunicamycin-induced retinal damage | [32] | |
| Black rice kernels | Antioxidant activity | [52] |
Figure 2.
Structures of the flavonoids isolated from rice.
Two apigenin C-glycosides schaftoside (45) and isoschaftoside (46) were identified in whole rice leaves [35] and phloem [36]. The contents of both flavones were higher in the phloem of an insect-resistant rice variety than in a susceptible variety, which suggested that schaftoside (45) and isoschaftoside (46) in rice act as an antifeedant against brown planthopper (Nilaparvata lugens) [37].
Flavones 56~64 belong to flavonolignans. Both tricin 4′-O-(erythro-β-guaiacylglyceryl) ether (57) and tricin 4′-O-(threo-β-guaiacylglyceryl) ether (58) from Njavara rice bran had cytotoxic activity and induced apoptosis in multiple tumor cells by themitochondrial pathway, which indicated their possible role as potential cytotoxic agents against cancer cells [38].
The flavonoids in rice include aglycones (i.e., quercetin, kaempferol and tricin) and their glycosides. Eight flavonoids, i.e., brassicin (66), isorhamnetin-4′-O-β-d-glucopyranoside (67), brassicin-4′-O-β-d-glucopyranoside (68), isorhamnetin-7-O-β-d-cellobioside (69), 3′-O-methyltaxifolin (84), 3′-O-methyltaxifolin-7-O-β-d-glucopyranoside (85), 3′-O-methyltaxifolin-4′-O-β-d-glucopyranoside (86), and 3′-O-methyltaxifolin-5-O-β-d-glucopyranoside (87), were isolated from Oryza sativa sp. japonica c.v. Hwa-Young. This cultivar has a high flavonoid content in the seeds and, particularly, in the endosperm tissue [39].
Two flavones, i.e., O-glycosides 5,4′-dihydroxy-3′,5′-dimethoxy-7-O-β-glucopyranosylflavone (37) and 7,4′-dihydroxy-3′,5′-dimethoxy-5-O-β-glucopyranosylflavone (38), were identified in allelopathic rice seedlings. Only their aglycone, 5,7,4′-trihydroxy-3′,5′-dimethoxyflavone (36), was found in the soil. These two flavone O-glycosides were exuded from the rice roots to the rhizosphere and were then transformed into their aglycone forms, which showed an allelopathic effect on associated weeds and microbes [40,41].
Sakuranetin (81) is a flavanone-type phytoalexin in rice active against plant pathogens. Naringenin (79) is considered the biosynthetic precursor of sakuranetin (81) in rice. The bioconversion of naringenin (79) into sakuranetin (81) is catalyzed by naringenin 7-O-methyltransferase (OsNOMT) in rice leaves [42]. The antifungal activity of sakuranetin (81) was found to be higher than that of naringenin (79) [43,44]. Very interestingly, sakuranetin (81) can be detoxificated into naringenin (79) and sternbin (83) by the rice blast pathogen Magnaporthe oryzae [44] and can also be detoxificated into naringenin (79), naringenin 7-O-β-d-xylopyranoside (80), and sakuranetin 4′-O-β-d-xylopyranoside (82) by the rice sheath blight pathogen Rhizoctonia solani [45].
Sakuranetin (81) is not only a plant antibiotic but also a potential pharmaceutical agent that induces adipogenesis in 3T3-L1 cells through enhanced expression of peroxisome proliferator-activated receptor γ2, contributing to the maintenance of glucose homeostasis in animals [46] and exhibits anti-inflammatory activity by inhibiting 5-lipoxygenase, which is involved in arachidonic acid metabolism in animal cells [47], anti-mutagenic activity [48], anti-Helicobacter pylori activity by inhibiting β-hydroxyacylacyl carrier protein dehydration [49], and antileishmanial and antitrypanosomal activities [50]. Sakuranetin (81) strongly stimulated melanogenesis in B16BL6 melanoma cells via the ERK1/2 and PI3K–AKT signaling pathways, which led to the upregulation of Tyr family genes, TRP1 and TRP2 [51].
Anthocyanins are widely distributed in black rice. Eight anthocyanins, i.e., cyanidin (90), cyanidin 3-O-gentiobioside (91), cyanidin 3-O-glucoside (92), cyanidin 3-O-rutinoside (93), cyanidin 3-O-sambubioside (94), cyanidin 3,5-O-diglucoside (95), peonidin (100), and peonidin 3-O-glucoside (101) were identified from the kernels of black rice by UPLC-Q-TOF-MS [52]. They showed obviously antioxidant activities. The protective effects were mainly due to their free radical scavenging capacity [52].
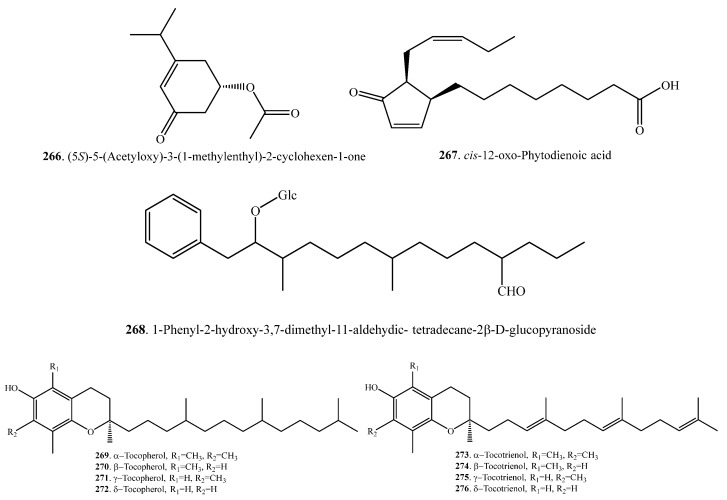
2.3. Terpenoids and Their Biological Functions
Rice terpenoids include monoterpenoids, sesquiterpenoids, diterpenoids, and triterpenoids. Some monoterpenoids and sesquiterpenoids are volatile components and are often distributed in rice leaves. Rice diterpenoids play roles as phytohormones and phytoalexins. The triterpenoids are usually distributed in rice bran. The monoterpenoids, sesquiterpenoids, and triterpenoids usually play functions as allelochemicals.
2.3.1. Monoterpenoids and Their Biological Functions
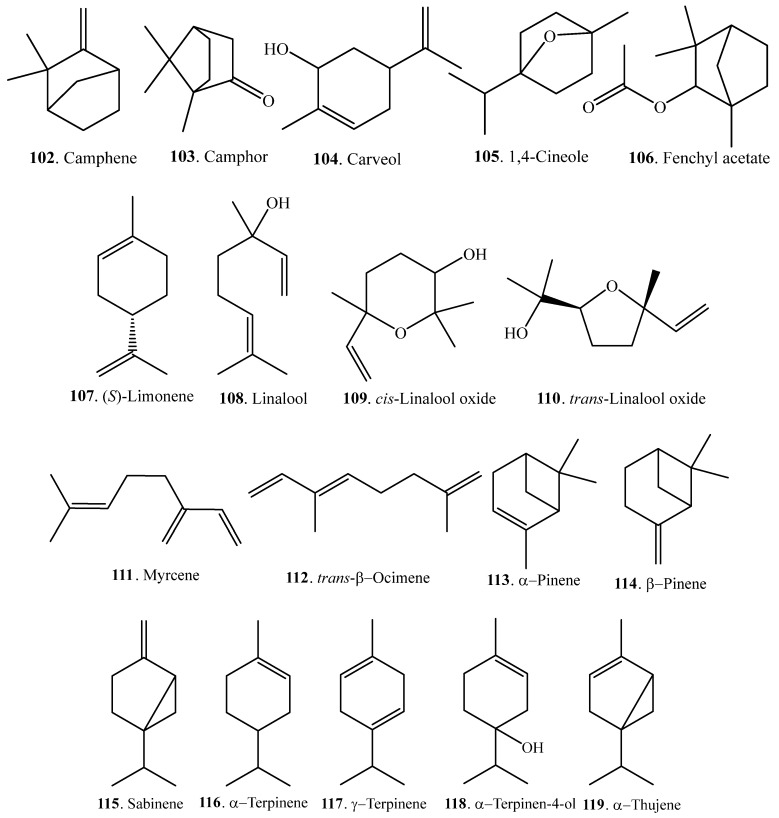
Monoterpenoids are mainly volatile compounds which confer rice its good aroma character. They can be extracted from the headspace of some rice bran samples by solid-phase microextraction (SPME). At least 18 monoterpenoids have been identified in rice. Their names and biological activities are listed in Table 3. Their structures are shown in Figure 3.
Table 3.
Monoterpenoids and their biological activities.
| Name | Rice Part Used for Isolation | Biological Activity and Function | Ref. |
|---|---|---|---|
| Camphene (102) | Bran | - | [65] |
| Camphor (103) | Bran | - | [65] |
| Carveol (104) | Bran | - | [65] |
| 1,4-Cineol (105) | Bran | - | [65] |
| Fenchyl acetate (106) | Bran | - | [65] |
| (S)-Limonene (107) | Leaves | - | [66] |
| Bran | - | [65] | |
| Seedlings | Antibacterial activity on Xoo | [63] | |
| Linalool (108) | Leaves | - | [66] |
| Leaves | Resistance induction to Xoo | [64] | |
| cis-Linalool oxide (109) | Bran | - | [65] |
| trans-Linalool oxide (110) | Bran | - | [65] |
| Myrcene (111) | Seedlings | - | [63] |
| Bran | - | [65] | |
| trans-β-Ocimene (112) | Bran | - | [65] |
| α-Pinene (113) | Seedlings | - | [63] |
| β-Pinene (114) | Bran | - | [65] |
| Sabinene (115) | Seedlings | - | [63] |
| Bran | - | [65] | |
| α-Terpinene (116) | Seedlings | - | [63] |
| γ-Terpinene (117) | Leaves | Antibacterial activity on Xoo | [62] |
| Terpinen-4-ol (118) | Bran | - | [65] |
| α-Thujene (119) | Seedlings | - | [63] |
Figure 3.
Structures of the monoterpenoids identified in rice.
Rice monoterpenoids are synthesized by various types of terpene synthases (TPSs), such as OsTPS20 and OsTPS24. These TPSs contain a transit peptide for localization in the chloroplasts where monoterpenes are biosynthesized from geranyl diphosphate (GPP) by TPSs via the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway. TPSs can be induced by jasmonic acid (JA). The amount of γ-terpinene (117) increased after JA treatment. γ-Terpinene (117) had significant antibacterial activity against Xoo. However, it did not show significant antifungal activity against the rice blast pathogen. The antibacterial mechanism of γ-terpinene (117) against Xoo involved damage to bacterial cell membranes [62].
Monoterpenes (S)-limonene (107), myrcene (111), α-pinene (113), sabinene (115), α-terpinene (116), and α-thujene (119) were detected from one-week-old Xoo-infected rice seedlings by the method of solid-phase microextraction-GC-MS. However, only (S)-limonene (107) severely inhibited Xoo growth, which suggests that (S)-limonene (107) plays a significant role in suppressing Xoo growth in rice seedlings [63].
Many volatile monoterpenoids including linalool (108) were accumulated in response to the exogenous application of JA. The xpression of linalool synthase gene was upregulated by JA. Vapour treatment with linalool (108) induced resistance to Xoo. The transgenic rice plants overexpressing linalool synthase gene were more resistant to Xoo, which suggests that linalool (108) plays an important role in JA-induced resistance to Xoo [64].
2.3.2. Sesquiterpenoids and Their Biological Functions
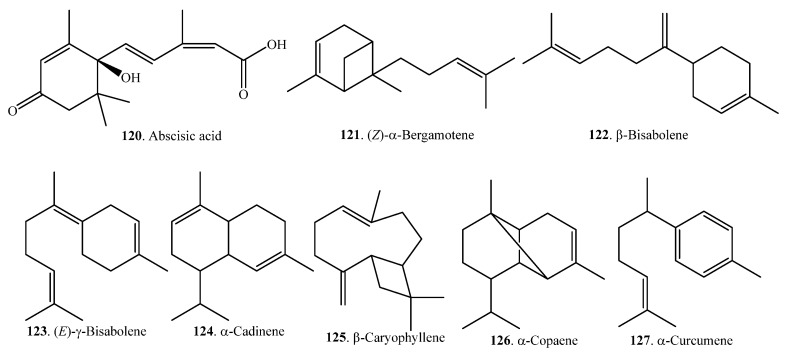
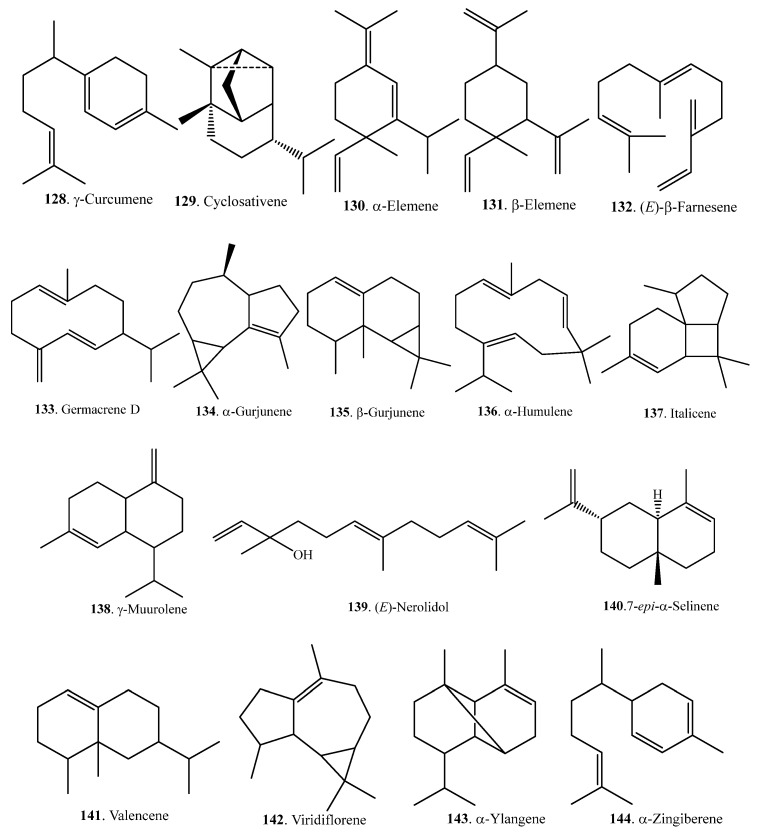
Sesquiterpenoids are also volatile components which contribute to the aroma quality of rice. They can be analyzed and identified by GC and GC-MS. The relative content of sesquiterpenoids was much lower, on average, than that of monoterpenoids in rice. Sesquiterpenoids are usually produced and released from wounds or microbe-infected sites. They act as signaling molecules that induce defense against tissue damage caused by herbivores or plant pathogens [67]. Rice sesquiterpenoids and their biological activities are listed in Table 4. Their structures are shown in Figure 4.
Table 4.
Sesquiterpenoids and their biological activities and functions.
| Name | Rice Part Used for Isolation | Biological Activity and Function | Ref. |
|---|---|---|---|
| Abscisic acid (120) | Whole rice plant | Regulation of growth and development | [69] |
| (Z)-α-Bergamotene (121) | Leaves | - | [66] |
| β-Bisabolene (122) | Bran | - | [65] |
| (E)-γ-Bisabolene (123) | Leaves | - | [66] |
| α-Cadinene (124) | Leaves | - | [66] |
| β-Caryophyllene (125) | Leaves | - | [66] |
| Bran | - | [65] | |
| α-Copaene (126) | Leaves | - | [66] |
| Bran | - | [65] | |
| Seedlings | - | [63] | |
| α-Curcumene (127) | Leaves | [66] | |
| γ-Curcumene (128) | Leaves | [66] | |
| Cyclosativene (129) | Seedlings | - | [63] |
| α-Elemene (130) | Bran | - | [65] |
| β-Elemene (131) | Seedlings | - | [63] |
| (E)-β-Farnesene (132) | Leaves | - | [68] |
| Germacrene D (133) | Leaves | [66] | |
| α-Gurjunene (134) | Bran | - | [65] |
| β-Gurjunene (135) | Leaves | [66] | |
| α-Humulene (136) | Leaves | [66] | |
| Italicene (137) | Leaves | [66] | |
| γ-Muurolene (138) | Leaves | [66] | |
| (E)-Nerolidol (139) | Leaves | Antibacterial activity against Xoo | [68] |
| 7-epi-α-Selinene (140) | Bran | - | [65] |
| Valencene (141) | Leaves | - | [66] |
| Viridiflorene (142) | Leaves | - | [66] |
| α-Ylangene (143) | Bran | - | [65] |
| α-Zingiberene (144) | Leaves | - | [66] |
Figure 4.
Structures of the sesquiterpenoids isolated from rice.
Sesquiterpenes are biosynthesized from farnesyl diphosphate (FPP) by TPSs via the mevalonate (MVA) pathway in the cytoplasm. Rice terpene synthase 18 was found to localize in the cytoplasm and synthesized the sesquiterpenes (E)-nerolidol (139) and (E)-β-farnesene (132), whose amounts increased after JA treatment. (E)-Nerolidol (139) had significant antibacterial activity against Xoo [68]. Rice sesquiterpenoids and their biological activities are listed in Table 4.
2.3.3. Diterpenoids and Their Biological Functions
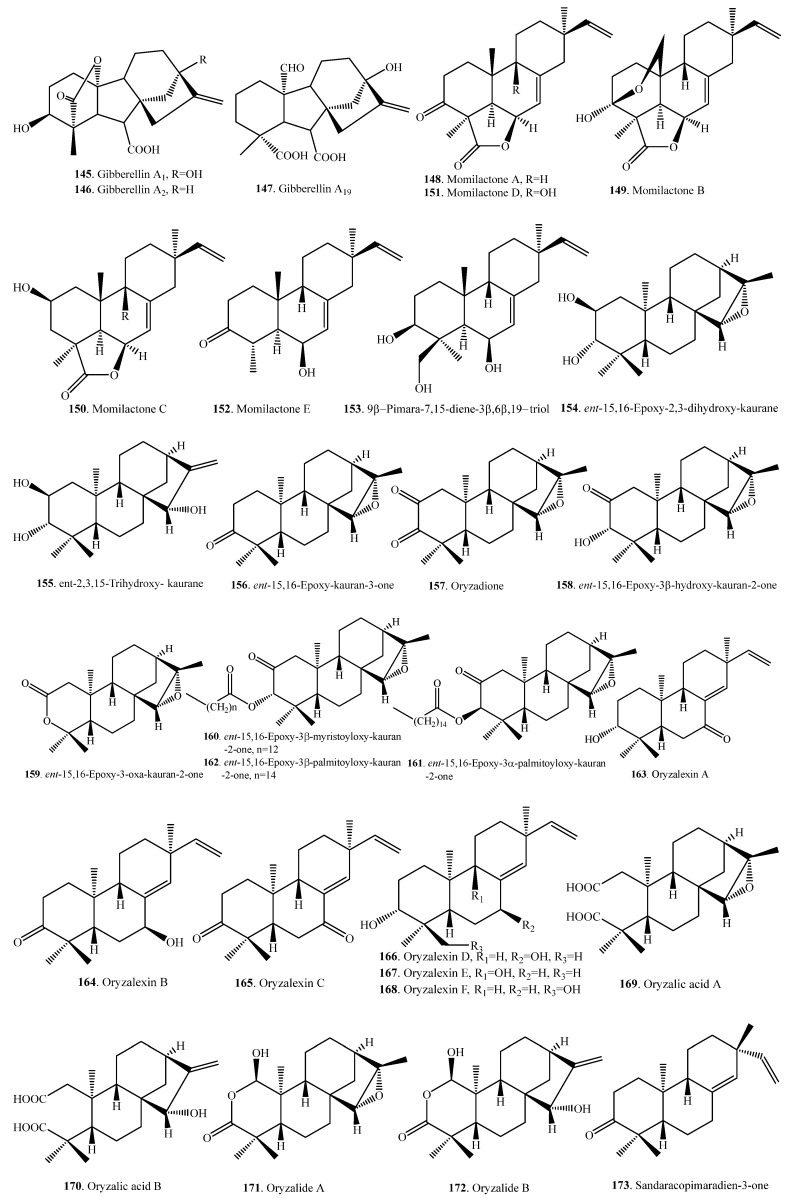
Almost all rice diterpenoids are members of the labdane-related superfamily, which includes not only phytohormone gibberellins (GAs) but also phytoalexins (i.e., phytocassanes, oryzalides, and oryzalexins), participate in the defense against pathogens, and are allelochemicals (i.e., momilactone B) inhibiting the growth of other plant species. Rice diterpenoids and their biological activities are listed in Table 5. Their structures are shown in Figure 5.
Table 5.
Diterpenoids and their biological activities.
| Name | Rice Part Used for Isolation | Biological Activity and Function | Ref. |
|---|---|---|---|
| Phytohormone gibberellins | |||
| Gibberellin A1 (145) | Whole plant | Growth-promoting activity | [70] |
| Gibberellin A4 (146) | Whole plant | Growth-promoting activity | [70] |
| Gibberellin A19 (147) | Whole plant | Growth-promoting activity | [70] |
| Pimaradiene-type diterpenoids | |||
| Momilactone A (148) | Coleoptiles | Antifungal activity | [71] |
| Bran | Growth inhibitory activity on rice roots | [90] | |
| Bran | Inhibitory activities on seed germination and growth of barnyard grass | [93] | |
| Root exudates | Allelopathy effect | [91] | |
| Momilactone B (149) | Coleoptiles | Antifungal activity | [71] |
| Seedlings | Growth inhibitory activity on rice roots | [90,92] | |
| Seedlings | Allelopathic effects | [40] | |
| Bran | Inhibitory activities on seed germination and growth of barnyard grass | [93] | |
| Root exudates | Allelopathy effect | [91] | |
| Momilactone C (150) | Bran | Weak growth inhibitory activity | [94] |
| Momilactone D (151) | Roots | - | [95] |
| Momilactone E (152) | Roots | - | [95] |
| 9β-Pimara-7,15-diene-3β,6β,19-triol (153) | Leaves | Weak antifungal activity | [72] |
| ent-Sandaracopimaradiene-type diterpenoids | |||
| ent-15,16-Epoxy-2,3-dihydroxy- kaurane (154) | Leaves of a bacterial leaf blight-resistant cultivar | Antibacterial activity | [85] |
| ent-2,3,15-Trihydroxy- kaurane (155) | Leaves of a bacterial leaf blight-resistant cultivar | Antibacterial activity | [85] |
| ent-15,16-Epoxy-kauran-3-one (156) | Leaves of a bacterial leaf blight-resistant cultivar | Antibacterial activity | [85] |
| ent-15,16-Epoxy-kauran-2,3-dione = Oryzadione (157) | Leaves of a bacterial leaf blight-resistant cultivar | Antibacterial activity | [86] |
| ent-15,16-Epoxy-3β-hydroxy-kauran-2-one (158) | Leaves of a bacterial leaf blight-resistant cultivar | Antibacterial activity | [86] |
| ent-15,16-Epoxy-3-oxa-kauran-2-one (159) | Leaves of a bacterial leaf blight-resistant cultivar | Antibacterial activity | [86] |
| ent-15,16-Epoxy-3β-myristoyloxy-kauran-2-one (160) | Leaves of a bacterial leaf blight-resistant cultivar | Antibacterial activity | [86] |
| ent-15,16-Epoxy-3α-palmitoyloxy-kauran-2-one (161) | Leaves of a bacterial leaf blight-resistant cultivar | Antibacterial activity | [86] |
| ent-15,16-Epoxy-3β-palmitoyloxy-kauran-2-one (162) | Leaves of a bacterial leaf blight-resistant cultivar | Antibacterial activity | [86] |
| Oryzalexin A (163) | Leaves | Inhibitory activity on spore germination and germ tube growth of Ochrobactrum oryzae | [73,76] |
| Roots | - | [95] | |
| Oryzalexin B (164) | Leaves | Inhibitory activity on spore germination and germ tube growth of O. oryzae | [75,76] |
| Oryzalexin C (165) | Leaves | Inhibitory activity on spore germination and germ tube growth of O. oryzae | [75,76] |
| Oryzalexin D(166) | Leaves | Inhibitory activity on spore germination of Magnaporthe Oryzae | [77] |
| Oryzalexin E (167) | Leaves | Inhibitory activity on spore germination of M. Oryzae | [78] |
| Oyzalexin F (168) | Leaves | Antimicrobial activity | [79] |
| Oryzalic acid A (169) | Leaves of a bacterial leaf blight-resistant cultivar | Antibacterial activity | [88] |
| Oryzalic acid B = ent-15-Hydroxy-2,3-secokauren- 2,3-dioic acid (170) | Leaves of a bacterial leaf blight-resistant cultivar | Antibacterial activity | [85] |
| Oryzalide A = ent-15,16-Epoxy-1α-hydroxy-2-oxa-kauran-3-one (171) | Leaves of a bacterial leaf blight-resistant cultivar | Antibacterial activity | [87,88] |
| Oryzalide B (172) | Leaves of a bacterial leaf blight-resistant cultivar | Antibacterial activity | [88] |
| Sandaracopimaradien-3-one (173) | Roots | - | [95] |
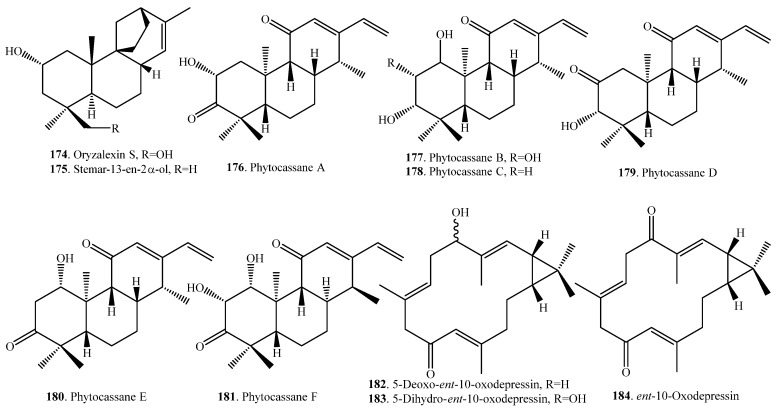
| Stemarene-type diterpenoids | |||
| Oryzalexin S (174) | Leaves | Antifungal activity | [96,97] |
| Stemar-13-en-2α-ol (175) | Leaves | Weak antifungal activity | [72] |
| ent-Cassadiene-type diterpenoids | |||
| Phytocassane A (176) | Leaves infected with M. oryzae; stems infected with Rhizoctonia Solani | Antifungal activity | [80] |
| Phytocassane B (177) | Leaves infected with M. oryzae; stems infected with R. Solani | Antifungal activity | [80] |
| Phytocassane C (178) | Leaves infected with M. oryzae; stems infected with R. Solani | Antifungal activity | [80] |
| Phytocassane D (179) | Leaves infected with M. oryzae; stems infected with R. Solani | Antifungal activity | [80] |
| Phytocassane E (180) | Cultured rice cells | Inhibition activity on spore germination and germ tube growth of M. oryzae | [81] |
| Phytocassane F (181) | Leaves | Antifungal activity | [72] |
| Casbene-type diterpenoids | |||
| 5-Deoxo-ent-10-oxodepressin (182) | Leaves | Antifungal activity | [83] |
| 5-Dihydro-ent-10-oxodepressin (183) | Leaves | Antifungal activity | [83] |
| ent-10-Oxodepressin (184) | Leaves | Antifungal activity | [84] |
Figure 5.
Structures of the diterpenoids isolated from rice.
The major endogenous GA in rice was identified as GA19 (147). Other GA analogs are GA1 (145) and GA4 (146). The level of active Gas, such as GA1 (145), may be regulated by the rate of biosynthesis of GA19 (147) or its metabolic conversion [70].
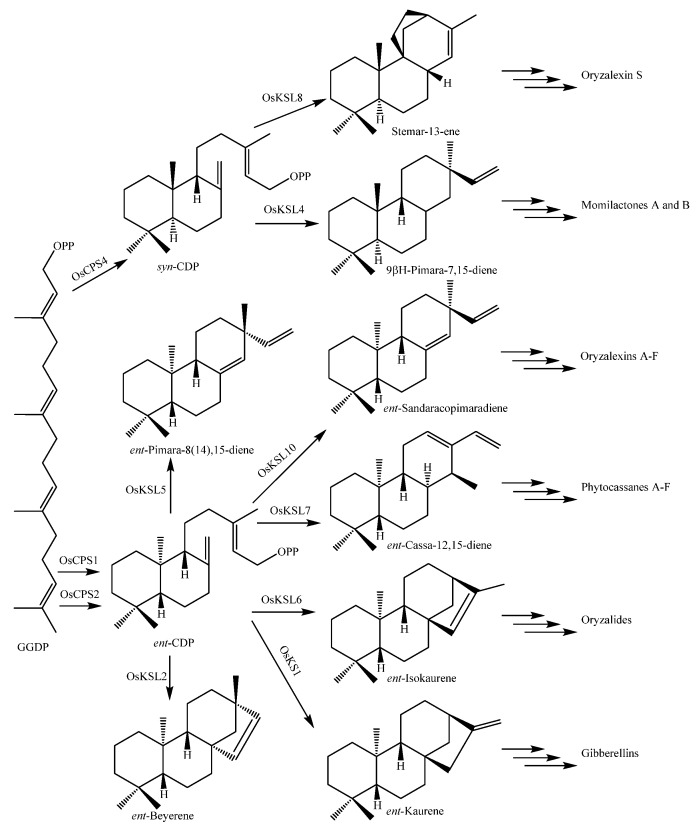
Up to now, 37 diterpenoid-type phytoalexin analogues have been identified from rice plants. They have been further classified into five subtypes according to their biosynthetic pathways and structural characters [17]. The first one (148~153) is the pimaradiene type which mainly includes momilactones A (148) and B (149) [71] and 9β-pimara-7,15-diene-3β,6β,19-triol (153) [72]. The second subtype (154~173) is the ent-sandaracopimaradiene type which mainly includes oryzalexins A~F (163~168) [73,74,75,76,77,78,79]. The third one is the stemarene type that contains oryzalexin S (174) [44] and stemar-13-en-2α-ol (175) [72]. The fourth one is the ent-cassadiene type, containing phytocassanes A~F (176~181) [72,80,81,82]. The fifth one is the casbene type, including 5-deoxo-ent-10-oxodeprssin (182) [83], 5-dihydro-ent-10-oxodepressin (183) [83], and ent-10-oxodepressin (184) [84].
A few oryzalide-related compounds were isolated from the leaves of a cultivar resistant to the Xoo. They were identified as ent-15,16-epoxy-2,3-dihydroxy-kaurane (154) [85], ent-2,3,15-trihydroxy-kaurane (155) [85], ent-15,16-epoxy-kauran-3-one (156) [85], oryzadione (157) [86], ent-15,16-epoxy-3β-hydroxy-kauran-2-one (158) [86], ent-15,16-epoxy-3-oxa-kauran-2-one (159) [86], ent-15,16-epoxy-3β-myristoyloxy-kauran-2-one (160) [86], ent-15,16-epoxy-3α-palmitoyloxy-kauran-2-one (161) [86], ent-15,16-epoxy-3β-palmitoyloxy-kauran-2-one (162) [86], oryzalide A (163) [87,88], oryzalide B (164) [88], oryzalic acid A (169) [88], and oryzalic acid B (170) [85]. In contrast to typical diterpene phytoalexins, the accumulation of oryzalide-related comounds is only moderately induced by Xoo infection [89].
Three compounds, i.e., 9β-pimara-7,15-diene-3β,6β,19-triol (153), stemar-13-en-2α-ol (175), and phytocassane F (181) were accumulated following an infection by the rice blast pathogen M. oryzae. 9β-pimara-7,15-diene-3β,6β,19-triol (153) and stemar-13-en-2α-ol (175) exhibited weak antifungal activity and may be the biosynthetic intermediates of rice phytoalexins momilactones and oryzalexin S (174), respectively. Phytocassane F (181) exhibited relatively high inhibitory activity against the mycelial growth of M. oryzae, to the same extent as the known phytoalexin phytocassane A (176) [72].
Some diterpenoids such as momilactones A (148) and B (149) have their obvious allelopathic effects. Momilactones A (148) and B (149) mainly distribute in rice husks, leaves, seedlings, and straw. They function as either rice defense systems against pathogens and insects or growth inhibitors in seed dormancy [90]. Both momilacontes A (148) and B (149) inhibited the growth of barnyard grass (Echinochloa crus-galli) and Echinochloa colonum, the most noxious weeds in rice field, at concentrations greater than 1 and 10 μM, respectively. Momilactone B (149) exhibited greater growth inhibitory activity than momilactone A (148) [91]. Momilactone B (149) was preferentially secreted from the rice roots into the neighboring environment over the entire life cycle at phytotoxic levels. Momilaconte B (149) seems to account for the majority of rice allelopathy, while momilactone A (148) accumulates to higher levels in the plant upon infection. Interestingly, both momilactones A (148) and B (149) inhibited root and shoot growth of rice seedlings only at concentrations greater than 100 μM and 300 μM, respectively. Therefore, the ability of momilactones A (148) and B (149) to suppress the growth of rice seedlings was much lower than their effect on E. crus-galli and E. colonum, with no visible damage to rice seedlings exerted by momilactones A (148) and B (149) at levels that were cytotoxic to other plant species [91]. Selective removal of the momilactones from the complex mixture of rice root exudates significantly reduced allelopathy, which demonstrated that momilactones served as allelochemicals [8,92].
2.3.4. Triterpenoids and Their Biological Functions
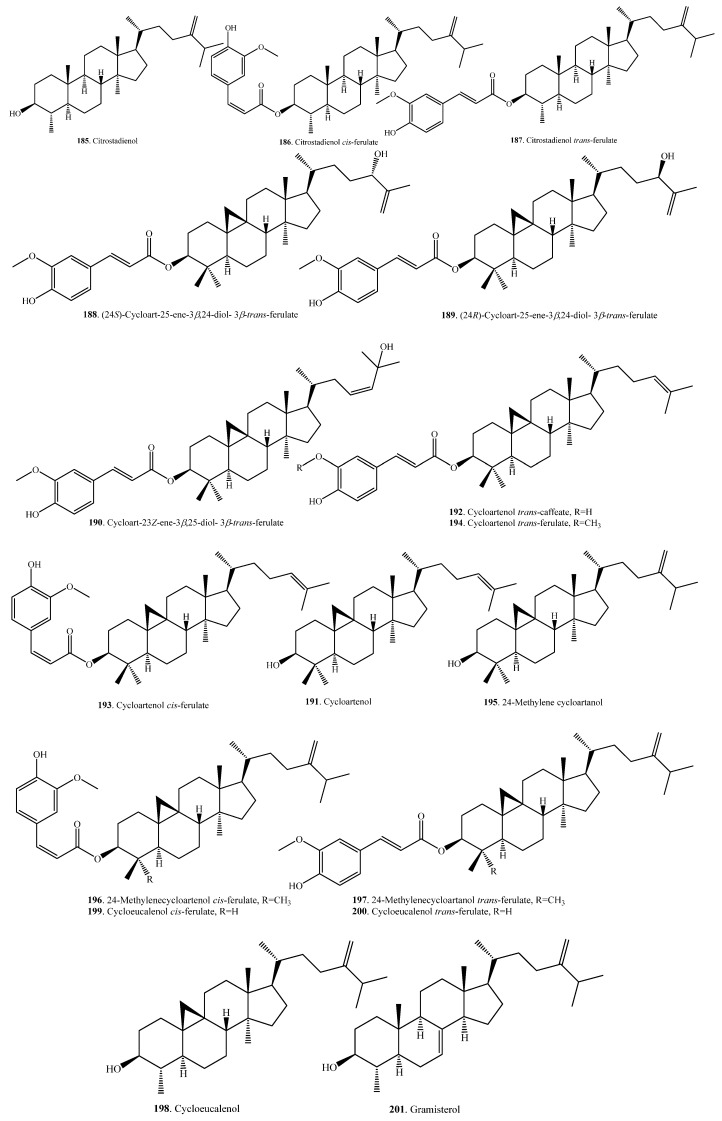
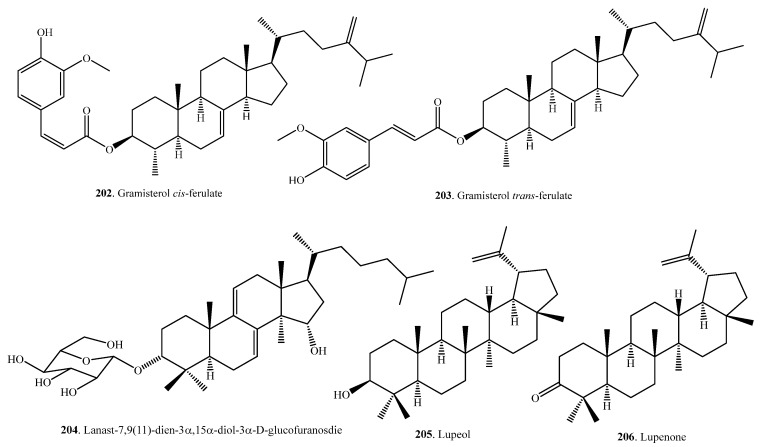
Triterpenoids are usually distributed in rice bran. Eight hydroxylated triterpene alcohol ferulates (188~190, 193, 194, 196, 197, 200) were isolated from rice bran. They showed moderate cytotoxic activity [98,99]. The seed coats (or bran) usually contain large amounts of bioactive metabolites. This was also observed for the seed coats of quinoa (Chenopodium quinoa), where there were various triterpenoids distributed. Quinoa triterpenoids showed antimicrobial and molluscicidal activities [100]. There are few reports about the physiological and ecological functions of rice triterpenoids. The aglycones of rice triterpenoids are citrostadienol (185), cycloartenol (191), cycloeucalenol (198), gramisterol (201), and lupeol (205). On the basis of the biosynthetic pathway, citrostadienol (185), cycloeucalenol (198), gramisterol (201), and their derivatives are considered nortriterpenoids. Rice triterpenoids and their biological activities are listed in Table 6. Their structures are shown in Figure 6.
Table 6.
Triterpenoids and their biological activities.
| Name | Rice Part Used for Isolation | Biological Activity and Function | Ref. |
|---|---|---|---|
| Citrostadienol (185) | Bran | - | [105] |
| Bran | Anti-inflammatory activity | [104] | |
| Citrostadienol cis-ferulate (186) | Bran | Anti-inflammatory activity | [104] |
| Citrostadienol trans-ferulate (187) | Bran | Anti-inflammatory activity | [104] |
| (24S)-Cycloart-25-ene-3β,24-diol-3β-trans-ferulate (188) | Bran | Moderate cytotoxic activity | [99] |
| (24R)-Cycloart-25-ene-3β,24-diol-3β-trans-ferulate (189) | Bran | Moderate cytotoxic activity | [99] |
| Cycloart-23Z-ene-3β,25-diol-3β-trans-ferulate (190) | Bran | Moderate cytotoxic activity | [99] |
| Cycloartenol (191) | Bran | - | [105] |
| Bran | Lowering postpradial hyperglyceimia | [103] | |
| Cycloartenol trans-caffeate (192) | Seeds | - | [106] |
| Cycloartenol cis-ferulate (193) | Bran | - | [98] |
| Cycloartanol trans-ferulate (194) | Bran | - | [98] |
| Bran | Moderate cytotoxic activity | [99] | |
| 24-Methylene cycloartanol (195) | Bran of black non-glutinous rice | Anti-cancer activity | [107] |
| Bran | Lowering postpradial hyperglyceimia | [103] | |
| 24-Methylene cycloartanol cis-ferulate (196) | Bran | - | [98] |
| Bran | Anti-inflammatory activity | [104] | |
| 24-Methylene cycloartanol trans-ferulate (197) | Bran | - | [98] |
| Bran | Moderate cytotoxic activity | [99] | |
| Cycloeucalenol (198) | Bran of black non-glutinous rice | Anti-cancer activity | [107] |
| Cycloeucalenol cis-ferulate (199) | Bran | Antioxidant activity | [31] |
| Cycloeucalenol trans-ferulate (200) | Bran | - | [98] |
| Bran | Anti-inflammatory activity | [104] | |
| Bran | Antioxidant activity | [31] | |
| Gramisterol (201) | Bran of black non-glutinous rice | Anti-cancer activity | [107] |
| Gramisterol cis-ferulate (202) | Bran | Anti-inflammatory activity | [104] |
| Gramisterol trans-ferulate (203) | Bran | Anti-inflammatory activity | [104] |
| Lanast-7,9(11)-dien-3α,15α-diol-3α-d-glucofuranoside (204) | Hulls | Herbicidal activity | [108] |
| Lupeol (205) | Bran of black non-glutinous rice | Anti-cancer activity | [107] |
| Lupenone (206) | Bran of black non-glutinous rice | Anti-cancer activity | [107] |
Figure 6.
Structures of the triterpenoids isolated from rice.
γ-Oryzanol is a mixture of triterpene and sterol ferulates extracted from rice bran [101]. In addition to its antioxidant activity, γ-oryzanol is often associated with cholesterol-lowering, anti-inflammatory, anti-cancer, and anti-diabetic properties [102]. The mixture of triterpene alcohols and sterols, with its components such as cycloartenol (191) and 24-methylene cycloartanol (195) from rice bran, can lower postpradial hyperglyceimia in mice and humans [103].
The main triterpene ferulates are cycloartenol ferulate (193/194) and 24-methylenecycloartanol ferulate (196/197). Both cycloeucalenol trans-ferulate (194) and 24-methylenecycloartanol cis-ferulate (196) showed anti-inflammatory activity in mice with inflammation induced by 12-O-tetradecanoylphorbol-13-acetate [104].
2.4. Steroids and Their Biological Functions
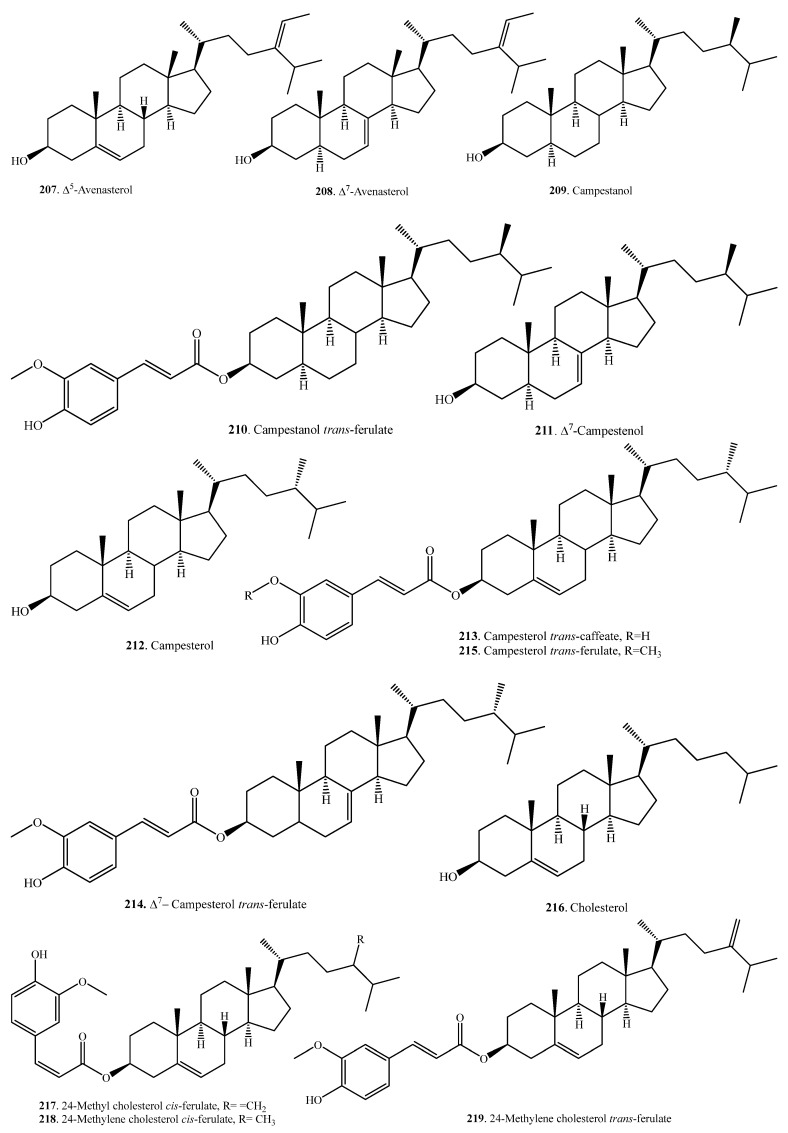
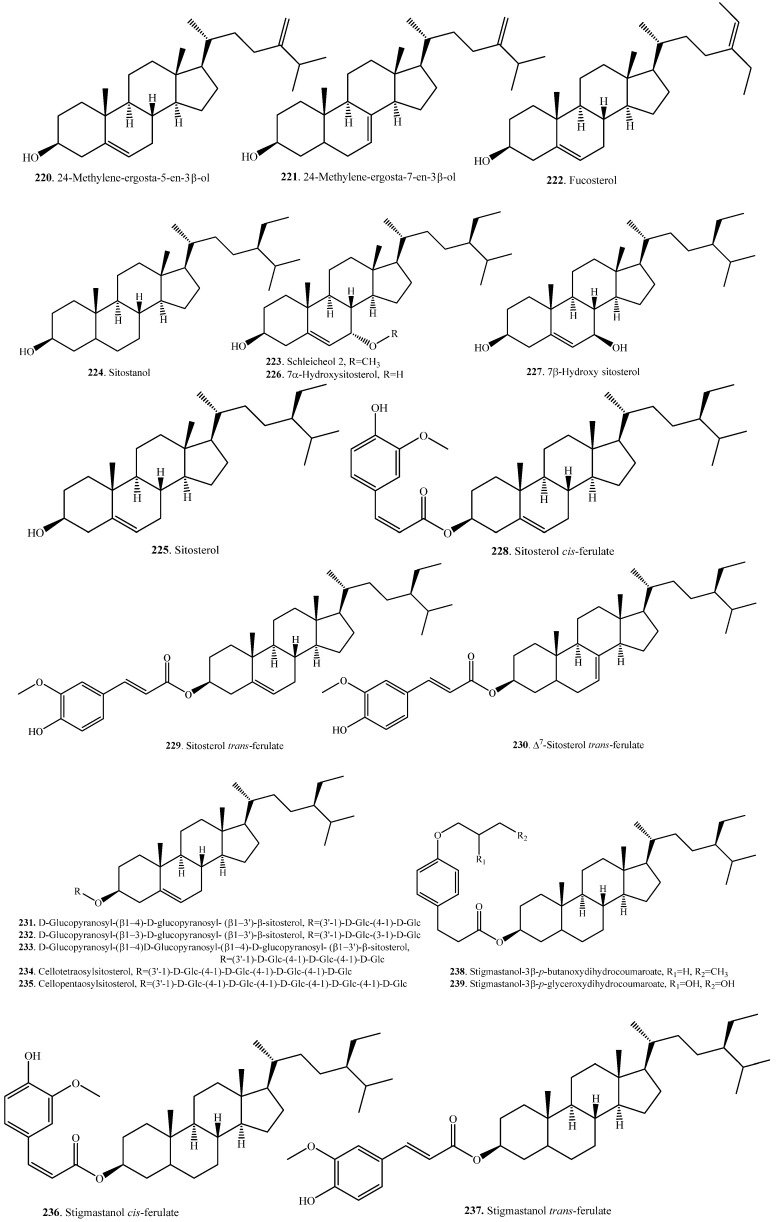
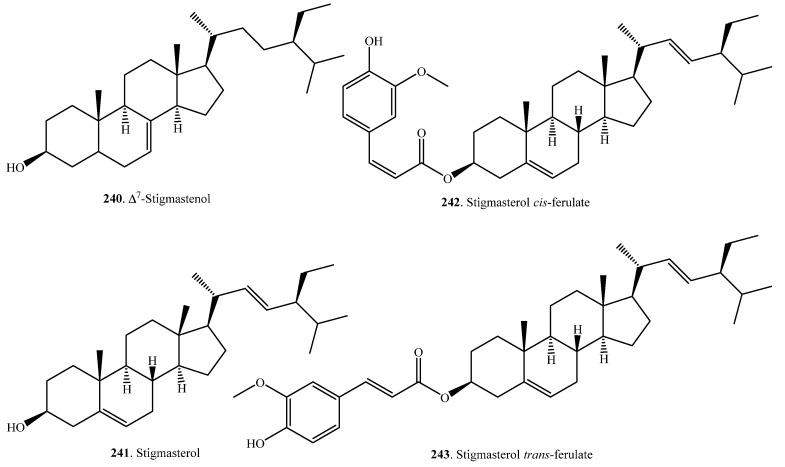
Plant steroids, generally termed phytosterols, are integral components of the membrane lipid bilayer in plants. They regulate membrane fluidity, influencing membrane’s properties, functions, and structure. An increase in the accumulation of sterols, namely, campesterol (209), β-sitosterol (225), and stigmasterol (241) was observed in rice as seedlings matured. These molecules are considered to have a role in drought stress tolerance in rice [109]. Steroids are usually distributed in the rice bran [107,110]. To date, 37 steroids have been identified from rice plants. Their names and biological activities are listed in Table 7. Their structures are shown in Figure 7.
Table 7.
Steroids and their biological activities.
| Name | Rice Part Used for Isolation | Biological Activity and Function | Ref. |
|---|---|---|---|
| ∆5-Avenasterol (207) | Germinating seeds | - | [106] |
| ∆7-Avenasterol (208) | Germinating seeds | - | [106] |
| Campestanol (209) | Germinating seeds | - | [106] |
| Campestanol trans-ferulate (210) | Bran | - | [98] |
| ∆7-Campestenol (211) | Germinating seeds | - | [106] |
| Campesterol (212) | Bran | - | [107] |
| Seedlings | Drought stress tolerance | [109] | |
| Campesterol trans-caffeate (213) | Bran | - | [98] |
| ∆7-Campesterol trans-ferulate (214) | Bran | - | [99] |
| Campesterol trans-ferulate (215) | Bran | - | [99] |
| Cholesterol (216) | Germinating seeds | - | [106] |
| 24-Methyl cholesterol cis-ferulate (217) | Bran | Anti-inflammatory activity | [105] |
| 24-Methylene cholesterol cis-ferulate (218) | Bran | Anti-inflammatory activity | [104] |
| 24-Methylene cholesterol trans-ferulate (219) | Bran | - | [98] |
| Bran | Anti-inflammatory activity | [104] | |
| 24-Methylene ergosta-5-en-3β-ol (220) | Bran | - | [107] |
| 24-Methylene ergosta-7-en-3β-ol (221) | Bran | - | [107] |
| Fucosterol (222) | Bran | - | [107] |
| Schleicheol 2 (223) | Bran | - | [110] |
| Sitostanol (224) | Germinating seeds | - | [106] |
| Sitosterol = β-Sitosterol (225) | Bran | - | [105,107] |
| Seedlings | Drought stress tolerance | [109] | |
| 7α-Hydroxy sitosterol (226) | Bran | - | [110] |
| 7β-Hydroxy sitosterol (227) | Bran | - | [110] |
| Sitosterol cis-ferulate (228) | Bran | - | [98] |
| Bran | Anti-inflammatory activity | [104] | |
| Sitosterol trans-ferulate (229) | Bran | - | [98] |
| ∆7-Sitosterol trans-ferulate (230) | Bran | - | [98] |
| d-Glucopyranosyl-(β1→4)-d-glucopyranosyl-(β1→3′)-β-sitosterol (231) | Bran (Hulls) | - | [111] |
| d-Glucopyranosyl-(β1→3)-d-glucopyranosyl-(β1→3′)-β-sitosterol (232) | Bran (Hulls) | - | [111] |
| d-Glucopyranosyl-(β1→4)-d-glucopyranosyl-(β1→4)-d-glucopyranosyl-(β1→3′)-β-sitosterol (233) | Bran (Hulls) | - | [111] |
| Cellotetraosylsitosterol (234) | Bran | - | [112] |
| Cellopentaosylsitosterol (235) | Bran | - | [112] |
| Stigmastanol cis-ferulate (236) | Bran | Anti-inflammatory activity | [104] |
| Stigmastanol trans-ferulate (237) | Bran | [98] | |
| Bran | Anti-inflammatory activity | [104] | |
| Stigmastanol-3β-p-butanoxy dihydrocoumaroate (238) | Hulls | Weak herbicidal activity | [108] |
| Stigmastanol-3β-p-glyceroxy dihydrocoumaroate (239) | Hulls | - | [108] |
| ∆7-Stigmastenol (240) | Germinating seeds | - | [106] |
| Stigmasterol (241) | Bran | - | [105,107] |
| Seedlings | Drought stress tolerance | [109] | |
| Stigmasterol cis-ferulate (242) | Bran | Anti-inflammatory activity | [104] |
| Stigmasterol trans-ferulate (243) | Bran | - | [98] |
| Bran | Anti-inflammatory activity | [104] |
Figure 7.
Structures of the steroids isolated from rice.
Sterol ferulates are the main components of γ-oryzanol, which is isolated from rice bran. The main sterol ferulates are campesterol trans-ferulate (215) and sitosterol trans-ferulate (229) [102].
Some sterylglycosides (231~235), such as mono-, di-, tri-, tetra-, and pentaglycosylsterols, have been isolated from rice bran. The sugar component is glucose, and the glucose units are linked by β1,4-bonds [111,112].
Stigmastanol-3β-p-butanoxy dihydrocoumaroate (238) and stigmastanol-3β-p-glyceroxy dihydrocoumaroate (239) were isolated from rice hulls. Of them, stigmastanol-3β-p-butanoxy dihydrocoumaroate (238) showed weak growth inhibitory activity toward duckweed (Lemna pausicostata) [108].
2.5. Alkaloids and Their Biological Functions
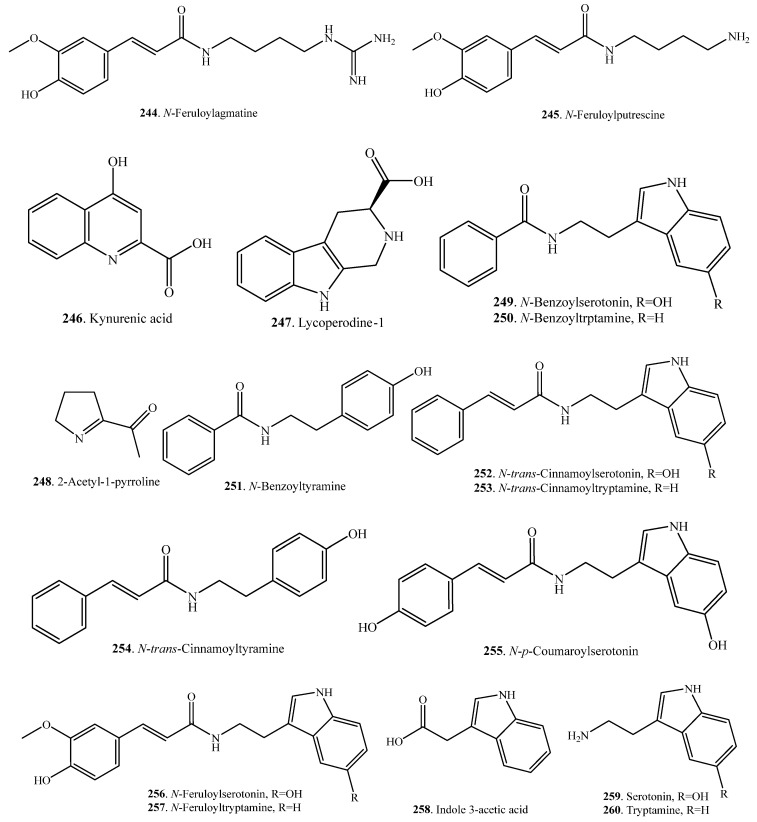
2-Acetyl-1-pyrroline (2AP, 248) is an important nitrogen-containing aroma compound that gives aromatic rice its characteristic flavor [113]. The concentration of 2AP (248) in uncooked Khao Dawk Mali 105 brown rice was quantitatively analyzed by capillary GC and found to be 0.34 μg/g [114]. This compound also occurs naturally in some other plants such as Pandanus amaryllifolius leaves and Vallaris glabra flowers [14]. Proline was proved to be the precursor for the biosynthesis of 2AP (248) in aromatic rice [115].
The main alkaloids in rice are phenylamides containing an indole ring. Rice plants accumulate phenylamides in response to a pathogen attack. If rice leaves are infected with the pathogens Cochliobolus miyabeanus and Xanthomonas oryzae, phenylamides are induced. They include N-feruloylagmatine (FerAgm, 244), N-feruloylputrescine (FerPut, 245), N-benzoylserotonin (BenSer, 249), N-benzoytryptamine (BenTry, 250), N-benzoyltyramine (BenTyr, 251), N-trans-cinnamoylserotonin (CinSer, 252), N-trans-cinnamoyltryptamine (253), N-trans-cinnamoyltyramine (CinTyr, 254), N-p-coumaroylserotonin (CouSer, 255), and N-feruloylserotonin (FerSer, 256). Some of these phenylamides displayed antimicrobial activity against C. miyabeaunus and X. oryzae, indicating that they are phytoalexins [116]. Rice alkaloids and their biological activities are listed in Table 8. Their structures are shown in Figure 8.
Table 8.
Alkaloids and their biological activities.
| Name | Rice Part Used for Isolation | Biological Activity and function | Ref. |
|---|---|---|---|
| N-Feruloylagmatine (244) | Leaves | Antimicrobial activity | [116] |
| N-Feruloylputrescine (245) | Leaves | Antimicrobial activity | [116] |
| Kynurenic acid (246) | Leaves | - | [34] |
| Lycoperodine-1 (247) | Leaves | - | [34] |
| 2-Acetyl-1-pyrroline (248) | Grains | - | [113] |
| N-Benzoylserotonin (249) | Leaves | Antimicrobial activity | [116] |
| N-Benzoyltryptamine (250) | Leaves | Antimicrobial activity | [116] |
| Leaves | Antibacterial activity | [58] | |
| N-Benzoyltyramine (251) | Leaves | Antimicrobial activity | [116] |
| N-trans-Cinnamoylserotonin (252) | Leaves | Antimicrobial activity | [116] |
| N-trans-Cinnamoyltryptamine (253) | Leaves | Antimicrobial activity | [116] |
| Leaves | Antibacterial activity | [58] | |
| N-trans-Cinnamoyltyramine (254) | Whole rice plant | Allelopathic activity; antifungal activity | [117] |
| Leaves | Antibacterial activity | [58] | |
| N-p-Coumaroylserotonin (255) | Leaves | Antimicrobial activity | [116] |
| Leaves | Antibacterial activity | [58] | |
| N-Feruloylserotonin (256) | Leaves | Antimicrobial activity | [116] |
| N-Feruloyltryptamine (257) | Leaves | - | [118] |
| Indole 3-acetic acid (258) | Whole rice plant | Regulation on growth and development | [119] |
| Serotonin = 5-Hydroxytryptamine (259) | Leaves | - | [118] |
| Tryptamine (260) | Leaves | - | [118] |
Figure 8.
Structures of the alkaloids isolated from rice.
2.6. Other Metabolites
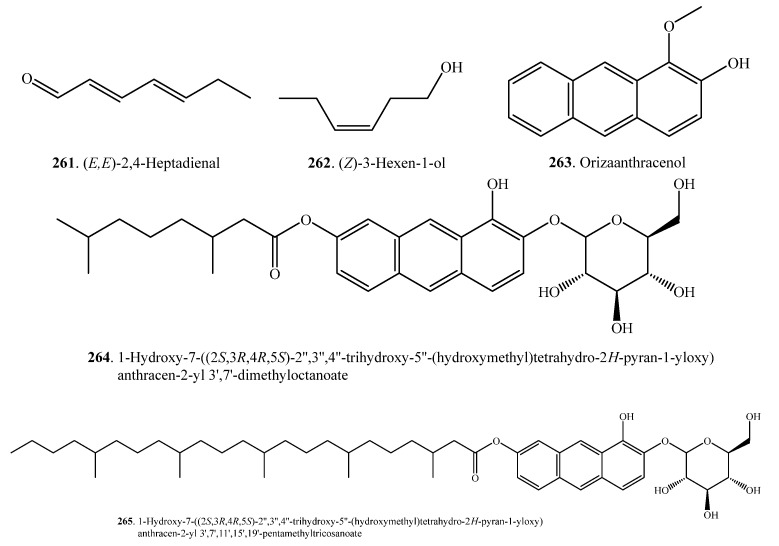
Other secondary metabolites in rice include anthracenes (263~265), tocopherols (269~272), and tocotrienols (273~276). Their names and biological activities are listed in Table 9. Their structures are shown in Figure 9.
Table 9.
Other metabolites and their biological activities.
| Name | Rice Part Used for Isolation | Biological Activity and Function | Ref. |
|---|---|---|---|
| (E,E)-2,4-Heptadienal (261) | Whole phants | Antibacterial and antifungal activities, toxic to rice plants | [120] |
| (Z)-3-Hexen-1-ol (262) | Leaves | - | [66] |
| Orizaanthracenol = 1-Methoxyanthracen-2-ol (263) | Hulls | Strong inhibitory activity in seed germination of radish | [121] |
| 1-Hydroxy-7-((2S,3R,4R,5S)-2″,3″,4″-trihydroxy-5″-(hydroxymethyl)tetrahydro-2H-pyran-1-yloxy)anthracen-2-yl 3′,7′-dimethyloctanoate (264) | Hulls | Weak inhibitory activity in seed germination of radish | [121] |
| 1-Hydroxy-7-((2S,3R,4R,5S)-2″,3″,4″-trihydroxy-5″-(hydroxymethyl)tetrahydro-2H-pyran-1-yloxy)anthracen-2-yl 3′,7′,11′,15′,19′-pentamethyltricosanoate (265) | Hulls | Weak inhibitory activity in seed germination of radish | [121] |
| (5S)-5-(Acetyloxy)-3-(1-methylenthyl)-2-cyclohexen-1-one = 3-Isopropyl-5-acetoxycyclohexene-2-one-1 (266) | Leaves | Allelopathic activity | [55] |
| Seedlings | Allelopathic effects | [40] | |
| cis-12-oxo-Phytodienoic acid (267) | Whole plants | Inducible anti-insect activity | [122] |
| 1-Phenyl-2-hydroxy-3,7-dimethyl-11-aldehydic-tetradecane-2β-d-glucopyranoside (268) | Hulls | Herbicidal activity | [108] |
| α-Tocopherol (269) | Bran | Antioxidative, antihypercholesterolemic, anticancer, neuroprotective activities | [123] |
| β-Tocopherol (270) | Bran | Antioxidative, antihypercholesterolemic, anticancer, neuroprotective activities | [123] |
| γ-Tocopherol (271) | Bran | Antioxidative, antihypercholesterolemic, anticancer, neuroprotective activities | [123] |
| δ-Tocopherol (272) | Bran | Antioxidative, antihypercholesterolemic, anticancer, neuroprotective activities | [123] |
| α-Tocotrienol (273) | Bran | Antioxidative, antihypercholesterolemic, anticancer, neuroprotective activities | [123] |
| β-Tocotrienol (274) | Bran | Antioxidative, antihypercholesterolemic, anticancer, neuroprotective activities | [123] |
| γ-Tocotrienol (275) | Bran | Antioxidative, antihypercholesterolemic, anticancer, neuroprotective activities | [123] |
| δ-Tocotrienol (276) | Bran | Antioxidative, antihypercholesterolemic, anticancer, neuroprotective activities | [123] |
Figure 9.
Structures of the other compounds isolated from rice.
(E,E)-2,4-Heptadienal (261) is a JA-responsive volatile component in rice plants. (E,E)-2,4-Heptadienal (261) has both antibacterial and antifungal activities against Xoo and M. oryzae. In addition, it is also toxic to rice plants. (E,E)-2,4-Heptadienal (261) is essential for rice survival against pathogen attacks [120].
Three anthracene derivatives, i.e., orizaanthracenol (1-methoxyanthracen-2-ol, 263), 1-hydroxy-7-((2S,3R,4R,5S)-2″,3″,4″-trihydroxy-5″-(hydroxymethyl)tetrahydro-2H-pyran-1-yloxy)anthracen-2-yl 3′,7′-dimethyloctanoate (264), and 1-hydroxy-7-((2S,3R,4R,5S)-2″,3″,4″-trihydroxy-5″-(hydroxymethyl)tetrahydro-2H-pyran-1-yloxy)anthracen-2-yl 3′,7′,11′,15′,19′-pentamethyltricosanoate (265), have been isolated from the rice hulls of O. sativa. Among the three compounds, orizaanthracenol (263) exhibited the highest inhibitory activity with respect to the germination of radish (Raphanus sativus) seeds, at 40 μg/mL [121].
(Z)-3-Hexen-1-ol (262) and other volatiles are released from elicitors (CuCl2, JA, UV, Met, and chitosan oligosaccharide)-treated and rice blast fungus-infested rice leaves [66].
(5S)-5-(Acetyloxy)-3-(1-methylenthyl)-2-cyclohexen-1-one (also named 3-isopropyl-5-acetoxycyclohexene-2-one-1 (266) is released from rice seedlings. It inhibited the growth of weeds E. crus-galli and Cyperus difformis [55].
cis-12-oxo-Phytodienoic acid (267) stimulated rice defense response to the brown planthopper (Nilaparvata lungens), a piercing-sucking insect pest of rice. This compound also stimulated the resistance of radish (R. sativus) seedlings to green peach aphid Myzus persicai which indicates the potential application of cis-12-oxo-phytodienoic acid (267) to stimulate plant defense responses to piercing-sucking insect pests in agriculture [122].
3. Biosynthetic Pathways of Rice Secondary Metabolites
In the 1980s, knowledge about the structures of rice secondary metabolites (i.e., phytoalexins) and their biosynthetic pathways accumulated, but no rice secondary metabolite biosynthetic enzyme genes was identified. In 2002, the draft sequences of rice genomes were published for japonica subspecies [15] and indica subspecies [16]. The annotation database platform for the rice genome was also developed and became publicly available (http://www.dna.affrc.go.jp/genome/#ricegenome) [9]. The biosynthetic genes for rice secondary metabolites are usually clustered in the genomes. The elucidation of their biosynthetic pathways is benefitting from the sequence of the rice genome, and many progresses have been achieved.
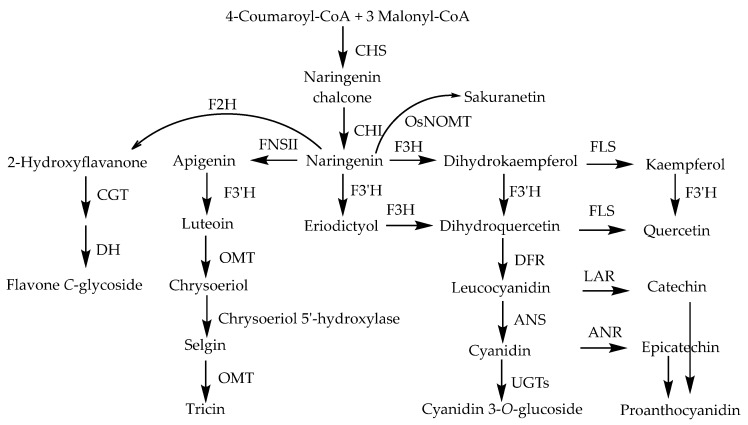
3.1. Biosynthesis of Flavonoids
A rice flavonoid biosynthesis pathway has been suggested by several studies, and the identified genes and enzymes involved in the pathway are shown in Figure 10 [124].
Figure 10.
Proposed biosynthesis pathway of rice flavonoids [124]. Abbreviations: CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; FLS, flavonol synthase; F3′H, flavonoid 3′-hydroxylase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; UGT, UDP-glucosyl transferase; LAR, leucoanthocyanidin reductase; ANR, anthocyanidin reductase; FNSII, flavone synthase II; OMT, O-methyltransferase; F2H, flavanone 2-hydroxylase; CGT, C-glucosyl transferase; and DH, dehydratase; OsNOMT, rice naringenin 7-O-methyltransferase.
The biosynthesis of sakuranetin (81) has been given particular attention as this metabolite is an important phytoalexin in rice. In addition to the phytopathogenic infection induction [125], sakuranetin (81) can be induced by ultraviolet (UV) irradiation [43], treatment with CuCl2 [126], JA [127], methionine [128], the herbicides pretilachlor and butachlor [129], the bacterial phytotoxin coronatine [130], the phytopathogenic stem nematode Ditylenchus angustus [131], and the insect pest white-backed planthopper (Sogatella furcifera) [132].
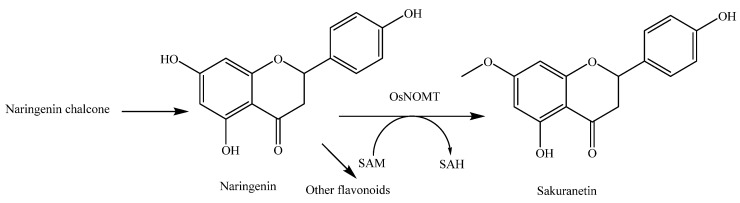
Sakuranetin (81) has been revealed to be biosynthesized from naringenin by S-adenosyl-l-methionine-dependent naringenin 7-O-methyltransferase (NOMT), which is a key enzyme for sakuranetin production. NOMT was successfully purified and identified [42]. As naringenin (79) is a biosynthetic intermediate for a variety of flavonoids, NOMT plays a key role in sakuranetin biosynthesis at a branch point in the common flavonoid biosynthetic pathway (Figure 11). OsMYC2, which is an essential factor for JA-induced sakuranetin production in rice, interacts with MYC2-like proteins that enhance its transactivation ability [133]. The biosynthetic pathway of sakuranetin (81) is shown in Figure 11 [9].
Figure 11.
Biosynthesis pathway of sakuranetin in rice [9]. Abbreviations: SAM, S-adenosyl-l-methionine; SAH, S-adenosyl-l-homocysteine.
3.2. Biosynthesis of Terpenoids
The biosynthesis of rice diterpenoid phytoalexins has been relatively detailed studied. The biosynthesis of the other terpenoids such as abscisic acid (ABA) and GAs in plants was discussed in reviews published elsewhere [134,135]. The production of rice diterpenoid phytoalexins can be induced by a series of stresses. For examples, phytocassanes can be induced by ultraviolet (UV) irradiation [72], and momilactone A (148) can be induced by the bacterial phytotoxins coronatine [130] and methionine [128].
The biosynthetic genes of diterpenoids are organized on the chromosome in functional gene clusters, comprising diterpene cyclase, dehydrogenase, and cytochrome P450 monooxygenase genes. Most of them in the rice genome are present in two gene clusters on chromosomes 2 and 4 (termed the Os02g cluster and the Os04g cluster, respectively) [136,137,138]. Their functions have been studied extensively using in vitro enzyme assay systems. Specifically, P450 genes (CYP71Z6, Z7, CYP76M5, M6, M7, M8) on rice chromosome 2 have multifunctional activities associated with ent-copalyl diphosphate-related diterpene hydrocarbons. Rice diterpenoids mainly contain phytohormones (i.e., gibberellins), phytoallexins (i.e., oryzalexins and phytocassanes), and allelochemicals (i.e., momilactones). They are biosynthesized via the MEP pathway in rice plants and have been well reviewed [139,140,141,142]. Gibberellins are considered phytohormones and are a large family of diterpenoids that possess the tetracyclic ent-gibberellane carbon skeletal structure arranged in either four or five ring systems, where the variable fifth ring is a lactone. Gibberellin biosynthesis and metabolism were well reviewed [143].
Rice diterpenoids are labdane-related. Their biosynthetic pathways in cultivated rice O. sativa are shown in Figure 12 [140,141].
Figure 12.
Biosynthesis pathways of the labdane-related diterpenoids in rice [141].
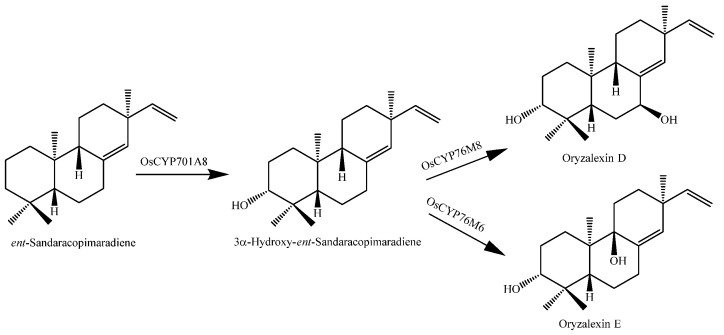
Bioactive rice diterpenoids are commonly elaborated by the addition of at least two spatially separated hydroxyl groups. For example, orzyalexin D (166) is simply 3α,7β-dihydroxylated ent-sandaracopimaradiene, while orzyalexin E (167) is the 3α,9β-dihydroxy derivative. Notably, the production of these phytoalexins appears to proceed via the initial C3α hydroxylation of ent-sandaracopimaradiene catalyzed by OsCYP701A8, while OsCYP76M6 and OsCYP76M8 catalyze the subsequent hydroxylation at C9β or C7β, leading to the production of oryzalexins D (166) and E (167), respectively (Figure 13). These final biosynthetic steps represent the first complete pathways in the production of rice diterpenoid phytoalexins [144].
Figure 13.
Biosynthesis pathways showing ent-sandaracopimaradiene dihydroxylation into oryzalexins D and E [144].
3.3. Biosynthesis of Tocotrienol and Tocopherol
Tocotrienol (T3), an unsaturated form of vitamin E with three double bonds in its isoprenoid side chain, is present in high concentration especially in rice grain [145].
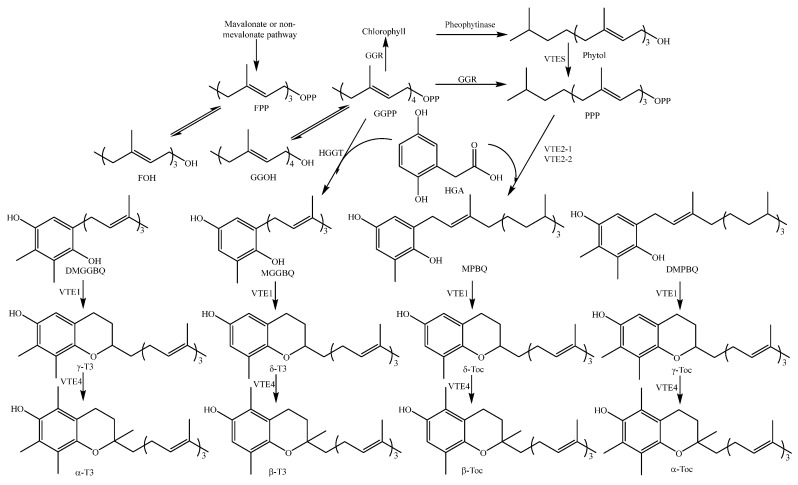
Both tocotrienol and tocopherol (TOC) are biosynthesized through the pathways of mevalonate and shikimate [146]. Their biosynthetic pathways are shown in Figure 14.
Figure 14.
General biosynthetic pathway for vitamin E in plants [146]. Abbreviations: T3, tocotrienol; Toc, tocopherol; FOH, farnesol; GGOH, geranylgeraniol; FPP, farnesyl pyrophosphate; GGPP, geranylgeranyl pyrophosphate; PPP, phytyl pyrophosphate; HGA, homogentisic acid; MGGBQ, 2-methyl-6-geranylgeranylbenzoquinol; DMGGBQ, 2,3-dimethyl-6-geranylgeranylbenzoquinol; MPBQ, 2-methyl-6-phytylbenzoquinol; DMPBQ, 2,3-dimethyl-6-phytylbenzoquinol. The enzymes HGGT, GGR, VTE2-1,2, T3/Toc methyltransferase (VTE3 and VTE4), T3/Toc cyclase (VTE1), pheophytinase, and phytol kinase (VTE5) are believed to be involved in vitamin E biosynthesis.
3.4. Biosynthesis of Alkaloids
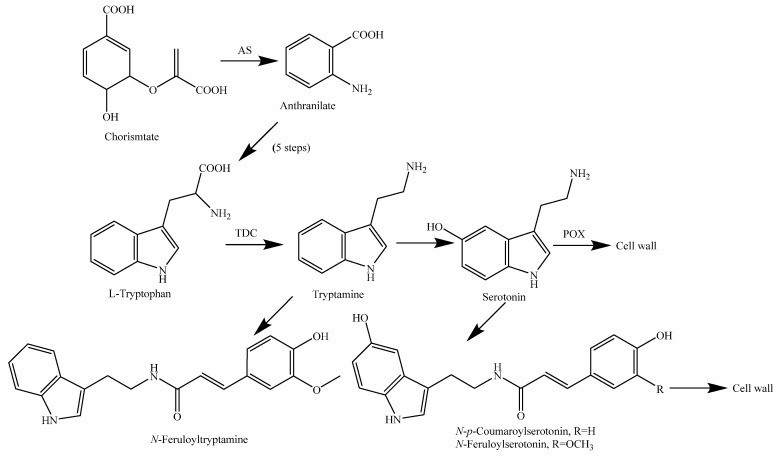
Both 2AP (248) and tryptophan biosynthesis pathways have been studied in detail, as 2AP is an important rice aromatic compound [147], and the tryptophan pathway is involved in rice defense responses against pathogenic infection via serotonin (259) production [148].
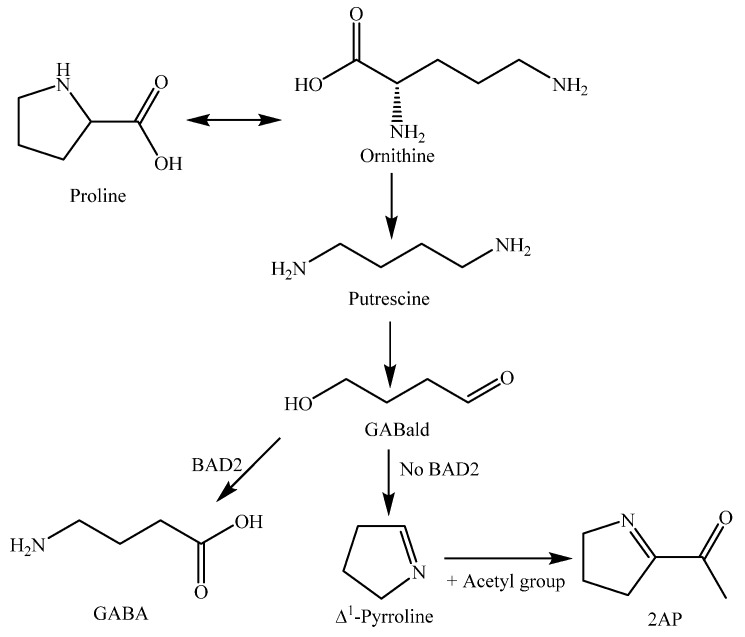
The biosynthesis pathway of 2AP production in rice starts with proline being catabolized via putrescine into γ-guanidinobutyraldehyde (GABald), a substrate of betaine aldehyde dehydrogenase (BAD2). If BAD2 is present and functional, it is able to convert the majority of GABald to γ-aminobutyric acid (GABA), but if BAD2 is absent or non-functional, the majority of GABald is acetylated to form 2AP [149]. The biosynthesis pathway of rice 2AP is shown in Figure 15.
Figure 15.
2-Acetyl-1-pyrroline (2AP) biosynthesis pathways in rice [149]. Abbreviations: GABald, γ-aminobutyraldehyde; BAD2, betaine aldehyde dehydrogenase; GABA, γ-aminobutyric acid.
Tryptophan decarboxylase (TDC) transforms typtophan (Trp) into tryptamine (260), consequently increasing the metabolic flow of tryptophan derivatives into the production of indole-containing metabolites. If the expression cassette containing OsTDC is inserted into an expression plasmid vector containing OASA1D, the overexpression of OASA1D significantly increases Trp levels in rice. The co-expression of OsTDC and OASA1D in rice cells led to almost complete depletion of the Trp pool and the consequent increase in the tryptamine pool. In recent years, the production of indole alkaloids has achieved great success through the metabolic engineering of the tryptophan pathway in rice [150]. The biosynthesis pathways of rice alkaloids are shown in Figure 16.
Figure 16.
Alkaloid biosynthesis pathways in rice [147]. Abbreviations: AS, anthranilate synthase; TDC, tryptophan decarboxylase; POX, peroxidase.
4. Metabolic Regulation of Secondary Meatobolites
The biosynthesis of plant secondary metabolites can be induced and regulated by various biotic and abiotic stresses, including organisms, jasmonic acid, oligosaccharides, and metal ions [9,151]. Among the secondary metabolites, phytoalexins are a kind of inducible antimicrobial metabolites whose biosynthesis is triggered not only by the invasion of pathogens including fungi, bacteria, and viruses, but also by a variety of abiotic elicitors, including phytohormones, oligosaccharides, UV irradiation, heavy metals (i.e., copper chloride), and mechanical stresses [152]. The main achievements regard the regulation of the biosynthesis of rice phytoalexins. The mechanisms of their biosynthetic regulation include signal recognition, signal transduction, gene expression, transcriptional and post-transcriptional pathways, and activation of the key enzymes.
4.1. Metabolic Regulation by Abiotic Stresses
4.1.1. Metabolic Regulation by Phytohormones
Secondary metabolite biosynthesis can be mediated by phytohormones such as ABA), JA, cytokinins (CKs), salicylic acid (SA), ethylene (ET), and their conjugates [17,153].
JA, which is in the form of jasmonates, is a plant hormone which induces the biosynthesis of many secondary metabolites which play roles in plant-environment interactions [154]. JA can induce rice defense responses and plays an important role as a signal transducer for phytoalexin production in stress (e.g., CuCl2, oligosaccharides, phytotoxins)-stimulated rice leaves through gene activation. For example, the endogenous level of JA increased rapidly in CuCl2-stimulated rice leaves, and exogenously applied JA caused a large amount of phytoalexin production in rice leaves [126]. As far as we know, JA can induced the biosynthesis of momilactones A (148) and B (149) as well as of γ-terpinene (117) [62].
The basic leucine zipper transcription factor OsTGAP1 acts as a regulator of the coordinated production of diterpenoid phytoalexins in cultured rice cells. The inductive expression of OstGAP1 under JA treatment was only observed in a root-specific manner, consistent with the JA-inducible expressions of the biosynthetic genes of diterpenoid phytoalexins in roots [155].
In addition, the amino acid conjugates of jasmonic acid N-[(−)-jasmonoyl]-S-isoleucine (JA-Ile) and N-[(−)-jasmonoyl]-S-phenylalanine were found to elicit the production of sakuranetin (81) in rice leaves. The elicitation was considered to arise from the induction of naringenin 7-O-methyltransferase, a key enzyme in sakuranetin biosynthesis [156]. Very interestingly, JA-Ile was not required for diterpenoid production in blast pathogen-infected or CuCl2-treated rice leaves [157].
The treatment with natural and synthetic CKs induced the production of diterpenoid phytoalexins in rice leaves and suspension-cultured cells [158]. However, CK treatment inhibited JA-inducible sakuranetin (81) production in rice leaves [127]. On the other hand, exogenous root applications of SA promoted the accumulation of oryzalexins and momilactone A (148) in the leaves [159]. A synergistic crosstalk of CK and SA signaling was also reported, showing that 0.1 mM CKs with benzothiadiazole (BTH), a plant activator that enhances SA signaling pathway, induced a several-fold enhancement of momilactone and phytocassane biosynthetic genes [160]. In addition, it was reported that the treatment of wounded rice leaves with methionine, the precursor of ET, induced the accumulation of sakuranetin (81) and momilactone A (148). Tiron, a free radical scavenger, counteracted the induction of both sakuranetin (81) and momilactone A (148) production in methionine-treated leaves, indicating that active oxygen species might be important in methionine-induced production of phytoalexins. However, ET treatment of wounded leaves induced the production of sakuranetin but not of momilactone A (148), suggesting that the induction of diterpenoid phytoalexin production by methionine was not regulated by ET alone [128]. In the susceptible rice cultivar Dorella, the bakanae pathogen (Fusarium fujikuroi) induced the production of gibberellin and abscisic acid and inhibited jasmonic acid production, and phytoalexin content in rice was very low [153].
In addition, ethylene-inducing xylanase from Thichoderma viride was a potent elicitor of immune responses in a variety of plant species, such as tobacco (Nicotiana tabacum), tomato (Solanum lycopersicum), and rice. This enzyme induced the expression of defense-related genes involved in the biosynthesis of phytoalexins (i.e., momilactones and phytocassanes) by a cation channel OsTPC1 in suspension-cultured rice cells [161].
4.1.2. Metabolic Regulation by Oligosaccharides
Oligosaccharides derived from fungal and plant cell wall polysaccharides are a class of well characterized elicitors that can induce not only accumulation of secondary metabolites but also MAPK activation, ROS generation, and defense-related enzyme activities at a very low concentration, triggering plant innate immunity [162,163,164,165,166,167,168]. Studies were performed on the structure–activity relationships of the oligosaccharides, the characterization of the corresponding receptors, and the analysis of signal transduction cascades and elicitor-responsive genes. Early studies focused on the inducible effects of oligosaccharides on rice phytoalexin production. Their elicitation mechanisms have been partially revealed in recent years.
A mixture of chitin fragments obtained from the cell walls of the rice pathogen Fusarium moniliforme through either the action of constitutive rice chitinases or partial acid hydrolysis was active to induce diterpenoid phytoalexin biosynthesis in rice cell cultures in suspension [169]. N-Acetylchitooligosaccharides larger than hexaose induced the formation of momilactones A (148) and B (149) as well as oryzalexins A (163), B (164), and D (166) at concentrations of 10−9~10−6 M [170].
β-glucan fragments (oligosaccharides) from the cell walls of the rice blast fungus M. oryzae had the ability to elicit phytoalexin (i.e., momilactone A) biosynthesis in suspension-cultured rice cells. The potent elicitor glucopentaose, namely, tetraglucosyl glucitol, was purified from the digestion of the glucan by an endo-β-(1→3)-glucanase. Interestingly, the obtained tetraglucosyl glucitol from M. oryzae did not induce phytoalexin biosynthesis in the soybean cotyledon cells, indicating differences in the recognition of gluco-oligosaccharide elicitor signals in these two plants [171]. In addition, two purified oligosaccharide elicitors, N-acetylchitohepatose and tetraglucosyl glucitol, derived from M. oryzae cell walls, synergistically activated the biosynthesis of phytoalexin in suspension-cultured rice cells. Inhibition experiments for the binding of the radio labeled N-acetylchitooligosaccharide elicitor to the plasma membrane from rice cells indicated that the two elicitors are recognized by different receptors [172]. Rice cells recognize oligosaccharides for defense singnaling mainly through plasma membrane receptors [173,174].
Diterpenoid phytoalexins are synthesized through the plastidic MEP pathway in rice. OsTGAP1, a basic leucine zipper transcription factor, which is induced by the fungal chitin oligosaccharide elicitor, was identified as a key regulator of the coordinated expression of the clustered biosynthetic genes for diterpenoid phytoalexin production in rice [138]. The overexpression of the bZIP transcription factor OsbZIP79 resulted in the suppression of the chitin oligosaccharide-inducible expression of diterpenoid phytoalexin biosynthetic genes, and thus caused a decrease in the accumulation of diterpenoid phytoalexin in rice cells. OsbZIP79 is considered a negative regulator of rice diterpenoid phytoalexin production [175].
4.1.3. Metabolic Regulation by Cerebrosides
Cerebrosides are categorized as glycosphingolipids. They are important components of a wide variety of tissues and organs in biological systems [176]. Cerebrosides were also found to occur in various fungi, such as Cercospora solani-melogenae, Cochiliobolus miyabeansus, Fusarium oxysporum, Mycosperella pinodes, Rhizoctonia sp., and Trichoderma viride, as the elicitors that can activate plant defense systems. They showed no antifungal activity against pathogens in vitro, showed phytoalexin-inducing activity when applied to plants by spray treatment, and also induced the expression of pathogenesis-related (PR) proteins in rice leaves [177].
Both cerebrosides A and C from the rice blast pathogen M. oryzae elicited hypersensitive cell death and phytoalexin (such as momilactone A and phytocassanes A and B) accumulation in rice plants [178].
The ceramides prepared from the cerebrosides by removal of glucose also showed elicitor activity even at lower concentations compared to the cerebrosides. In field experiments, the cerebroside elicitors effectively protected rice plants against the rice blast fungus. Cerebroside elicitors protected rice plants from other diseases as well and functioned as general elicitors in a wide variety of rice-pathogen interactions [179]. Further studies showed that cerebrosides are non-race-specific elicitors. Treatment of lettuce (Lactuca sativa), tomato (Lycopersicon esculentum), melon (Cucumis melo), and sweet potato (Ipomoea batatas) with cerebroside B resulted in resistance to infection of the fungal pathogens [180].
4.1.4. Metabolic Regulation by Cholic Acid
Cholic acid (CA), a steroid elicitor of rice defense responses, was isolated from human feces. When rice leaves were treated with CA, defense responses were induced, with the accumulation of antimicrobial compounds, hypersensitive cell death, and pathogenesis-related (PR) protein synthesis. The induced antimicrobial compounds were identified as phytocassanes. The structure–activity relationship analysis showed that the hydroxyl groups at C-7 and C-12 and the carboxyl group at C-24 of cholic acid contributed to the elicitor activity [181]. In contrast to the other elicitors (i.e., fungal chitin oligosaccharide elicitor), cholic acid specifically triggered the accumulation of phytocassanes but not of momilactones, suggesting specificity in pathway regulation [180]. Further investigation of the effects of CA on the expression of diterpene cyclase genes showed that CA induced the transcription of the genes OsCPS2 (OsCyc2) and OsKSL7 (OsDTC1) involved in phytocassane biosynthesis. OsCPS2 was particularly strongly induced, suggesting that it is one of the main mechanisms by which CA induces high levels of phytocassanes [182].
4.1.5. Metabolic Regulation by Heavy Metal Ions
The role of heavy metal ions as antifungal agents may consists in part in inducing defense-response genes and in part in inhibiting the pathogens. Among the metal ions, copper ions (Cu2+) were the most effective to induce defense-related genes involved in phytoalexin biosynthesis [183]. Heavy metal ions are abiotic elicitors. Some metal ions can affect the production of plant secondary metabolites including phytoalexins [184]. Typical examples included tanshinone accumulation stimulated by metal ions Co2+, Ag+, and Cd2+ in Salvia miltiorrhiza cell cultures [185], andrographolide production elicitated by Cd2+, Ag+, Cu2+, and Hg2+ in Andrographis paniculata cell cultures [186], resveratrol production enhanced by Co2+, Ag+ and Cd2+ in Vitis vinifera cell cultures [187], and phaseollin production enhanced in Colombian bean (Phaseolus vulgaris) seedlings treated with CuCl2 [188].
The induction of phytoalexins by heavy metal ions in rice leaves was studied by punching detached leaves with a glass capillary tube and applying droplets of a heavy metal salt solution into the holes. Application of 1 mM copper chloride (CuCl2) induced the accumulation of oryzalexins A (163), B (164), C (165), and D (166) and of momilactones A (148) and B (149) in the tissues around the holes and in the droplets. Among the momilactones, momilactone A (148) showed a marked induction. Among the oryzalexins, oryzalexin B (164) was induced to the greatest extent. The accumulation of momilactone A (148) was first noted 12 hr after the application of copper ions, reaching a maximum after 72 h. As these phytoalexins accumulated, brown spots appeared in areas surrounding the punctured holes. Iron and mercury ions made up approximately 37% and 20% of the elicitor activity of copper ion (Cu2+), respectively. Manganese (Mn2+) and cobalt (Co2+) ions hardly showed any elicitor activity [189]. Further study showed that the endogenous level of JA increased rapidly in CuCl2-treated rice leaves. If rice leaves were treated with JA biosynthesis inhibitors, the production of phytoalexin elicited by CuCl2 decreased. JA is thus suggested to play an important role as a signaling molecule in CuCl2-elicited rice phytoalexin biosynthesis [126].
4.1.6. Metabolic Regulation by Ultraviolet Irradiation
UV irraditon is very convenient for inducing the production of rice phytoalexins, because it is easy to irradiate many rice leaves with ultraviolet light [9].
An accumulation of oryzalexins A (163), B (164), C (165), and D (166), and of momilactones A (148) and B (149) accompanied by the appearance of brown spots on the leaf surface was observed in ultraviolet-irradiated detached rice leaves. Momilactone A (148) was detected in abundance, and among the oryzalexins, oryzalexin D (166) was a major substance. The content of these diterpenoid phytoalexins in rice leaves was dependent on leaf aging, the accumulation of these phytoalexins in the uppermost leaves being much lower than that in the aged leaves (lower leaves), and brown spots scarcely ever appeared on the surface of the uppermost leaves [190].
UV irradiation increased not only the concentration of momilactone B (149) in rice seedlings but also the secretion of momilactone B (149) into rice rhizosphere. As momilactone B (149) acts as an antimicrobial and allelopathic agent, the secretion of momilactone B into the rhizosphere may provide a competitive advantage for rice root establishment through the local suppression of soil microorganisms and the inhibition of the growth of competing plant species [191].
UV irradiation also stimulated flavonoid and phenylamide biosynthetic pathways in rice leaves. Five phytoalexins, i.e., sakuranetin (81), N-benzoyltryptamine (250), N-trans-cinnamoyltryptamine (253), N-trans-cinnamoyltyramine (254), and N-p-coumaroylserotonin (255) were isolated from UV-treated rice leaves [58]. In response to UV treatment, sakuranetin accumulated in rice leaves may serve as an antioxidant against UV-induced oxidative stress [42].
4.1.7. Metabolic Regulation by Other Abiotic Stresses
Rice plants were treated with the fungicide 2,2-dichloro-3,3-dimethyl cyclopropanecarboxylic acid (WL28325). The biosynthesis of two phytoalexins, i.e., momilactones A (148) and B (149), was markedly enhanced. This suggested that the fungicide WL28325 has a systemic ability to activate host resistance against rice blast pathogens [192].
Pretilachlor and butachlor, two chloroacetamide herbicides that promote cell death, induced the accumulation of the phytoalexins momilactone A (148) and sakuranetin (81) in rice leaves. The accumulation of these phytoalexins was related to the herbicide concentration and the period of exposure and was followed by the appearance of necrotic lesions on the rice leaves [129].
Carbon dioxide (CO2) has the ability to regulate the biosynthesis of rice phenolics. With elevated CO2 concentrations, the total phenolic content as well as the DPPH radical scavenging capacity decreased, which indicated that these decreases may be meaningful in the preventive ability of rice against free radical-mediated degenerative diseases [193].
4.2. Metabolic Regulation by Biotic Stresses
4.2.1. Metabolic Regulation by Bacteria
The biosynthesis of secondary metabolites can be regulated by either pathogenic and non-pathogenic bacteria or their extracts and components.
If rice leaves were infected with the bacterial pathogen Xanthomonas oryzae, phenylamides were induced. They were identified as N-feruloylagmatine (244), N-feruloylputrescine (245), N-benzoylserotonin (249), N-benzoytryptamine (250), N-benzoyltyramine (251), N-trans-cinnamoylserotonin (252), N-trans-cinnamoyltyramine (254), N-p-coumaroylserotonin (255), and N-feruloylserotonin (256) [116].
The bacterial phytotoxin coronatine was isolated from a Pseudomonas syringae pv. atropurpurea culture broth as a chlorosis-inducing compound in the leaves of Italian ryegrass (Lolium multiflorum). The structure of coronatine, an amide of coronafacic acid and coronamic acid, was somewhat related to that of JA. This phytotoxin could induce the accumulation of sakuranetin (81) and momilactone A (148) in rice leaves. Coronatine-inducible sakuranetin production is under the control of kinetin and ascorbic acid, as observed with JA. The similarity of the structures and elicitation manner of coronatine and JA suggests that they have a similar action mechanism leading to rice phytoalexin production [129].
4.2.2. Metabolic Regulation by Fungi
The biosynthesis of rice secondary metabolites can be also regulated by fungi or their extracts and components. Fungal infection often results in the accumulation of phytoalexins in rice plants [194].
In the rice resistant cultivar Selenio, the presence of the pathogen F. fujikuroi induced a high production of sakuranetin (81), and symptoms of bakanae were not observed. On the contrary, in the susceptible genotype Dorella, the pathogen induced the production of gibberellin and abscisic acid and inhibited jasmonic acid production, and sakuranetin (81) content was very low [153]. Infection with the rice blast pathogen M. oryzae induced the production of momilactones, with greater accumulation of momilactones A (148) and B (149) [195], and also induced sakuranetin (81) production [125]. Young rice leaves in a resistant rice line exhibited a hypersensitive reaction (HR) within three days after the inoculation of a spore suspension of the blast pathogen M. oryzae, and an increase of sakuranetin was detected three days post-inoculation (dpi), increasing to four-fold at 4 dpi. In the susceptible line, increased sakuranetin was detected at 4 dpi but not at 3 dpi, by which time a large fungal mycelia had accumulated without HR. The decrease and detoxification of sakuranetin (81) were detected in both solid and liquid mycelium cultures of the rice blast pathogen [125].
Rice leaves accumulated serotonin (259) in response to infection by Bipolaris oryzae. If serotonin (259) was added to the culture media, it was converted into 5-hydroxyindole-3-acetic acid (5HIAA), which may be a detoxification process in the interaction between B. oryzae and rice [196].
When rice leaves were infected with Cochliobolus miyabeanus, phenylamides were also induced. They included N-feruloylagmatine (244), N-feruloylputrescine (245), N-benzoylserotonin (249), N-benzoytryptamine (250), N-benzoyltyramine (251), N-trans-cinnamoylserotonin (252), N-trans-cinnamoyltyramine (254), N-p-coumaroylserotonin (255), and N-feruloylserotonin (256) [116].
4.2.3. Metabolic Regulation by Insect Pests
The attack by herbivorous insects is one of the major biological stresses that rice plants have to cope with. The secondary metabolites that are derived from the tryptophan pathway have been shown to play defensive roles against insects in rice plants. The biosynthesis of four tryptophan-derived metabolites was induced by the feeding on rice leaves of the rice striped stem-borer (Chilo suppressalis). The amounts of N-p-coumaroylserotonin (255), N-feruloyltryptamine (257), serotonin (259), and tryptamine (260) in the larvae-fed leaves were 12-, 3.5-, 33-, and 140-fold larger than in the control leaves 48 h after the start of feeding [118].
The biosynthesis of serotonin (259) was suppressed by insect infestation in rice, which demonstrates that the regulation of serotonin biosynthesis plays an important role in the defense from insects. In rice, the cytochrome P450 gene CYP71A1 encodes tryptamine 5-hydroxylase, which catalyses the conversion of tryptamine to serotonin. In susceptible wild-type rice, rice brown planthopper (N. lugens) feeding induces the biosynthesis of serotonin, whereas, in mutants with an inactivated CYP71A1 gene, no serotonin is produced, and the plants are more insect-resistant [197].
When rice leaves were infested by the white-backed planthopper (Sogatella furcifera), accumulation of salicylic acid, jasmonic acid, and phytoalexins such as momilactone A (148) and sakuranetin (81) was observed. It is possible that S. furcifera released some elicitor compounds, which might be produced in its salivary glands, into the rice plants during feeding. Next, the defense signal systems, SA- and JA-mediated pathways, were activated by the elicitor. Finally, phytoalexins are induced in rice as antimicrobial compounds mainly through the activation of the JA-mediated pathway [132].
4.2.4. Metabolic Regulation by Nematodes
The elicitation of phytoalexin synthesis was observed in rice in respononse to infection by nematodes. Both chlorogenic acid (11) and sakuranetin (81) were induced in the incompatible varieties of rice after infection by the stem nematode Ditylenchus angustus, and no change occurred in the susceptible varieties of rice [131]. In addition, the systemic suppression of metabolism in the shoot, including the isoprenoid and shikimate pathways, was observed upon rice infection by the root nematode Hirschmanniella oryzae [198].
4.2.5. Metabolic Regulation by Viruses
The rice dwarf virus (RDV) P2 protein interacts with ent-kaurene oxidases, which play a key role in the biosynthesis of the growth hormones gibberellins in rice plants. This leads to reduced biosynthesis of gibberellins and to rice dwarf symptoms. In addition, the interaction between P2 protein and rice ent-kaurene oxidase-like proteins may decrease phytoalexin biosynthesis and make plants more competent for virus replication [199].
4.2.6. Metabolic Regulation by Other Plants
Rice allelopathic activity increased in the presence of seedlings and root exudates of barnyard grass (E. crus-galli). This increase was not due to nutrient competition between the two plant species. Levels of momilactone B (149), both endogenous concentration in rice seedlings and secretion rate, were also increased by the presence of the seedlings and root exudates of barnyard grass. Probably, the active components from the root exudates triggered the production and secretion of momilactone B (149) [200]. Similarly, the production of the sorghum allelochemical sorgoleone was also induced by root extracts of the agriculturally relevant weed velvetleaf [201]. Accordingly, allelopathy potentially acts as an inducible defense mechanism mediated by the recognition of root exudate components specific to other plant species found in the relevant ecosystem [8]. The elicited compounds from rice root exudates need to be identified.
5. Conclusions and Future Perspectives
This review focuses on the elucidation of the structures, biological functions, biosynthesis, and metabolic regulation of rice secondary metabolites carried out during the past 50 years. Some metabolites (i.e., diterpenoid phytoalexins) and their metabolic pathways are unique to rice [141]. Some minor or new rice metabolites should be identified by using new techniques, such as chemoinformatics [202], metabolomics [7,18,34], and compound prediction based on biosynthetic gene clusters [9]. Furthermore, the biological activities of many isolated metabolites (i.e., phenolic acids, flavonoids, and terpenoids) need to be systematically evaluated.
The physiological functions of some rice secondary metabolites remain unknown. Commonly, the development of null mutants and transgenic over-expression lines is enabling the critical examination of biological functions. Phytoalexin-related gene over-expression generally results in increased resistance against pathogens in genetically modified plants [203,204].
The biosynthetic regulation of rice secondary metabolites is very complicated. Concentrated efforts have revealed the relevant factors and signaling pathways that are involved in the regulation of phytoalexin production in rice. It is still unknown, however, how transcription factors regulate phytoalexin biosynthetic genes in concert. Further research investigating the molecular mechanisms of the transcriptional regulation of phytoalexin biosynthetic genes and revealing how upstream signals activate each transcription factor in the signaling cascade is essential. Rice metabolic regulation is certain to far exceed the complexity of the biosynthetic pathways [11,17,205].
In addition, exogenous genes such as stilbene synthase (STS) gene were successfully transferrred from other plant species to rice to improve its resistance against rice blast disease [206]. The importatance of some secondary metabolites, such as 2-acetyl-1-pyrroline (248) [14], phytoalexins [204], and momilactones [207], has been emphasized in agriculture and in the medicinal and food industries. These will be important fields of application for rice secondary metabolites.
Acknowledgments
We acknowledge some authors of cited papers for providing their figures, and Fengke Lin for his assistance in collecting literature.
Author Contributions
W.W. and Y.L. performed bibliographic research, drafted, and corrected the manuscript. P.D. and S.Z. retrieved the literature, participated in the discussions, and supported manuscript corrections. D.L. reviewed the manuscript and helped to revise it. L.Z. conceived the idea, designed the review structure, supervised manuscript drafting, and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was co-financed by grants from the National Key R&D Program of China (2017YFD0201105) and the National Natural Science Foundation of China (31471729).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Zhou Z., Robards K., Helliwell S., Blanchard C. Composition and functional properties of rice. Int. J. Food Sci. Tech. 2002;37:849–868. doi: 10.1046/j.1365-2621.2002.00625.x. [DOI] [Google Scholar]
- 2.Deng G.-F., Xu X.-R., Zhang Y., Li D., Gan R.-Y., Li H.-B. Phenolic compounds and bioactivities of pigmented rice. Crit. Rev. Food Sci. Nutr. 2013;53:296–306. doi: 10.1080/10408398.2010.529624. [DOI] [PubMed] [Google Scholar]
- 3.Izawa T., Shimamoto K. Becoming a model plant: The importance of rice to plant science. Trends Plant Sci. 1996;1:95–99. doi: 10.1016/S1360-1385(96)80041-0. [DOI] [Google Scholar]
- 4.Friedman M. Rice brans, rice bran oils, and rice hulls: Composition, food and industrial uses, and bioactivies in humans, animals, and cells. J. Agric. Food Chem. 2013;61:10626–10641. doi: 10.1021/jf403635v. [DOI] [PubMed] [Google Scholar]
- 5.Afrin S., Gasparrini M., Forbes-Hernandez T.Y., Reboredo-Rodriguez P., Mezzetti B., Varela-Lopez A., Giampieri F., Battino M. Promising health benefits of the strawberry: A focus on clinical studies. J. Agric. Food Chem. 2016;64:4435–4449. doi: 10.1021/acs.jafc.6b00857. [DOI] [PubMed] [Google Scholar]
- 6.Nowak D., Goslinski M., Wojtowicz E., Przygonski K. Antioxidant properties and phenolic compounds of vitamin C-rich juices. J. Food Sci. 2018;83:2237–2246. doi: 10.1111/1750-3841.14284. [DOI] [PubMed] [Google Scholar]
- 7.Chen H., Stout M., Qian Q., Chen F. Genetic, molecular and genomic basis of rice defense against insects. Crit. Rev. Plant Sci. 2012;31:74–91. doi: 10.1080/07352689.2011.616052. [DOI] [Google Scholar]
- 8.Kato-Noguchi H., Peters R.J. The role of momilactones in rice allelopathy. J. Chem. Ecol. 2013;39:175–185. doi: 10.1007/s10886-013-0236-9. [DOI] [PubMed] [Google Scholar]
- 9.Yamane H. Biosynthesis of phytoalexins and regulatory mechanisms of it in rice. Biosci. Biotechnol. Biochem. 2013;77:1141–1148. doi: 10.1271/bbb.130109. [DOI] [PubMed] [Google Scholar]
- 10.Goufo P., Trindade H. Rice antioxidants: Phenolic acids, flavonoids, antocyanins, proanthocyanidins, tocopherols, tocotrienols, γ-oryzanol, and phytic acid. Food Sci. Nutr. 2014;2:75–104. doi: 10.1002/fsn3.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmelz E.A., Huffaker A., Sims J.W., Christensen S.A., Lu X., Okada K., Peters R.J. Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J. 2014;79:659–678. doi: 10.1111/tpj.12436. [DOI] [PubMed] [Google Scholar]
- 12.Cho M.-H., Lee S.-W. Phenolic phytoalexins in rice: Biological functions and biosynthesis. Int. J. Mol. Sci. 2015;16:29120–29133. doi: 10.3390/ijms161226152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samyor D., Das A.B., Deka S.C. Pigmented rice a potential source of bioactive compounds: A review. Int. J. Food Sci. Technol. 2017;52:1073–1081. doi: 10.1111/ijfs.13378. [DOI] [Google Scholar]
- 14.Routray W., Rayaguru K. 2-Acetyl-1-pyrroline: A key aroma component of aromatic rice and other food products. Food Rev. Int. 2018;34:539–565. doi: 10.1080/87559129.2017.1347672. [DOI] [Google Scholar]
- 15.Goff S., Stephen A., Riche D., Lan T.-H., Presting G., Wang R., Dunn M., Glazebrook J., Sessioins A., Oeller P., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–101. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 16.Yu J., Ju S., Wang J., Wong G.K.-S., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., et al. A draft sequence of the rice genome (Oryza sativa L. spp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 17.Miyamoto K., Shimizu T., Okada K. Transcriptional regulation of the biosynthesis of phytoalexin: A lesson from specialized metabolites in rice. Plant Biotechnol. 2014;31:377–388. doi: 10.5511/plantbiotechnology.14.0730a. [DOI] [Google Scholar]
- 18.Kusano M., Yang Z., Okazaki Y., Nakabayashi R., Fukushima A., Saito K. Using metabolomics approaches to explore chemical diversity in rice. Mol. Plant. 2015;8:58–67. doi: 10.1016/j.molp.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Acosta-Estrada B.A., Gutierrez-Uribe J.A., Serna-Saldivar S.O. Bound phenolics in foods, a review. Food Chem. 2013;152:46–55. doi: 10.1016/j.foodchem.2013.11.093. [DOI] [PubMed] [Google Scholar]
- 20.Ti H., Li Q., Zhang R., Zhang M., Deng Y., Wei Z., Chi J., Zhang Y. Free and bound phenolic profiles and antioxidant activity of milled fractions of different indica rice varieties cultivated in Southern China. Food Chem. 2014;159:166–174. doi: 10.1016/j.foodchem.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 21.Pang Y., Ahmed S., Xu Y., Beta T., Zhu Z., Shao Y., Bao J. Bound phenolic compounds and antioxidant properties of whole grain and bran of white, red and black rice. Food Chem. 2018;240:212–221. doi: 10.1016/j.foodchem.2017.07.095. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H., Shao Y., Bao J., Beta T. Phenolic compounds and antioxidant properties of breeding lines between the white and black rice. Food Chem. 2015;172:630–639. doi: 10.1016/j.foodchem.2014.09.118. [DOI] [PubMed] [Google Scholar]
- 23.Zaupa M., Calani L., Rio D.D., Brighenti F., Pellegrini N. Characterization of total antioxidant capacity and (poly)phenolic compounds of differently pigmented rice varieties and their changes during domestic cooking. Food Chem. 2015;187:338–347. doi: 10.1016/j.foodchem.2015.04.055. [DOI] [PubMed] [Google Scholar]
- 24.Ding C., Liu Q., Li P., Pei Y., Tao T., Wang Y., Yan W., Yang G., Shao X. Distribution and quantitative analysis of phenolic compounds in fractions of Japonica and Indica rice. Food Chem. 2019;274:384–391. doi: 10.1016/j.foodchem.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Jun H.-I., Shin J.-W., Song G.-S., Kim Y.-S. Isolation and identification of phenolic antioxidants in black rice bran. J. Food Sci. 2015;89:C262–C268. doi: 10.1111/1750-3841.12754. [DOI] [PubMed] [Google Scholar]
- 26.Adom K.K., Liu R.H. Antioxidant activity of grains. J. Agric. Food Chem. 2002;50:6182–6187. doi: 10.1021/jf0205099. [DOI] [PubMed] [Google Scholar]
- 27.Shen Y., Jin L., Xiao P., Lu Y., Bao J.S. Total phenolics, flavonoids, antioxidant capacity in rice grain and their relations to grain color, size and weight. J. Cereal Sci. 2009;49:106–111. doi: 10.1016/j.jcs.2008.07.010. [DOI] [Google Scholar]
- 28.Seal A.N., Pratley J.E., Haig T., An M. Identification and quantitation of compounds in a series of allelopathic and non-allelopathic rice root exudates. J. Chem. Ecol. 2004;30:1647–1662. doi: 10.1023/B:JOEC.0000042074.96036.14. [DOI] [PubMed] [Google Scholar]
- 29.Olofsdotter M., Rebulanan M., Madrid A., Wang D., Navarez D., Olk D.C. Why phenolic acids are unlikely primary allelochemicals in rice. J. Chem. Ecol. 2002;28:229–242. doi: 10.1023/A:1013531306670. [DOI] [PubMed] [Google Scholar]
- 30.Seal A.N., Haig T., Pratley J.E. Evaluation of putative allelochemicals in rice root exudates for their role in the suppression of arrowhead root growth. J. Chem. Ecol. 2004;30:1663–1678. doi: 10.1023/B:JOEC.0000042075.96379.71. [DOI] [PubMed] [Google Scholar]
- 31.Wang W., Guo J., Zhang J., Peng J., Liu T., Xin Z. Isolation, identification and antioxidant activity of bound phenolic compounds present in rice bran. Food Chem. 2015;171:40–49. doi: 10.1016/j.foodchem.2014.08.095. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka J., Nakanishi T., Shimoda H., Nakamura S., Tsuruma K., Shimazawa M., Matsuda H., Yoshikawa M., Hara H. Purple rice extract and its constituents suppress endoplasmic reticulum stress-induced retinal damage in vitro and in vivo. Life Sci. 2013;92:17–25. doi: 10.1016/j.lfs.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 33.Zeng Z., Hu X., McClements D.J., Luo S., Liu C. Hydrothermal stability of phenolic extracts of brown rice. Food Chem. 2019;271:114–121. doi: 10.1016/j.foodchem.2018.07.180. [DOI] [PubMed] [Google Scholar]
- 34.Yang Z., Nakabayashi R., Okazaki Y., Mori T., Takamatsu S., Kitanaka S., Kikuchi J., Saito K. Toward better annotation in plant metabolomics: Isolation and structure elucidation of 36 specialized metabolites from Oryza sativa (rice) by using MS/MS and NMR analyses. Metabolomics. 2014;10:543–555. doi: 10.1007/s11306-013-0619-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Besson E., Dellamonica G., Chopin J., Markham K.R., Kim M., Koh H.-S., Fukami H. C-Glycosylflavones from Oryza sativa. Phytochemistry. 1985;24:1061–1064. doi: 10.1016/S0031-9422(00)83183-0. [DOI] [Google Scholar]
- 36.Grayer R.J., Harborne J.B., Kimmins F.M., Stevenson P.C., Wijayagunasekera H.N.P. Phenolics in rice phloem sap as sucking deterrents to the brown planthopper, Nilaparvata lugens. Acta Hort. 1994;381:691–694. doi: 10.17660/ActaHortic.1994.381.100. [DOI] [Google Scholar]
- 37.Stevenson P.C., Kimmins F.M., Grayer R.J., Raveendranath S. Schaftosides from rice phloem as feeding inhibitors and resistance factors to brown planthoppers, Nilaparvata lungens. Entomol. Exp. Appl. 1996;80:246–249. doi: 10.1111/j.1570-7458.1996.tb00928.x. [DOI] [Google Scholar]
- 38.Mohanlal S., Maney S.K., Santhoshkumar T.R., Jayalekshmy A. Tricin 4′-O-(erythro-β-guaiacylglyceryl) ether and tricin 4′-O-(threo-β-guaiacylglyceryl) ether isolated from Njavara (Oryza sativa L. var. Njavara), induce apoptosis in multiple tumor cells by mitochondrial pathway. J. Nat. Med. 2013;67:528–533. doi: 10.1007/s11418-012-0710-7. [DOI] [PubMed] [Google Scholar]
- 39.Cho J.-G., Song N.-Y., Nam T.-G., Shrestha S., Park H.-J., Lyu H.-N., Kim D.-O., Lee G., Woo Y.-M., Jeong T.-S., et al. Flavonoids from the grains of C1/R-S transgenic rice, the transgenic Oryza sativa spp. japonica, and their radical scavenging activities. J. Agric. Food Chem. 2013;61:10354–10359. doi: 10.1021/jf403072c. [DOI] [PubMed] [Google Scholar]
- 40.Kong C.H., Li H.B., Hu F., Xu X.H., Wang P. Allelochemicals released by rice roots and residues in soil. Plant Soil. 2006;288:47–56. doi: 10.1007/s11104-006-9033-3. [DOI] [Google Scholar]
- 41.Kong C.H., Zhao H., Xu X.H., Wang P., Gu Y. Activity and allelopathy of soil of flavone O-glycosides from rice. J. Agric. Food Chem. 2007;55:6007–6012. doi: 10.1021/jf0703912. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu T., Lin F., Hasegawa M., Okada K., Nojiri H., Yamane H. Purification and identification of naringenin 7-O-methyltransferase, a key enzyme in biosynthesis of flavonoid phytoalexin sakuranetin in rice. J. Biol. Chem. 2012;287:19315–19325. doi: 10.1074/jbc.M112.351270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kodama O., Miyakawa J., Akatsuka T., Kiyosawa S. Sakuranetin, a flavanone phytoalexin from ultraviolet-irradiated rice leaves. Phytochemistry. 1992;31:3807–3809. doi: 10.1016/S0031-9422(00)97532-0. [DOI] [Google Scholar]
- 44.Katsumata S., Hmamna K., Horie K., Toshima H., Hasegawa M. Identification of sternbin and naringenin as detoxified metabolites from the rice flavanone phytoalexin sakuranetin by Pyricularia oryzae. Chem. Biodivers. 2017;14:e1600240. doi: 10.1002/cbdv.201600240. [DOI] [PubMed] [Google Scholar]
- 45.Katsumata S., Toshima H., Hasegawa M. Xylosylated detoxification of the rice flavonoid phytoalexin sakuranetin by the rice sheath blight fungus Rhizoctonia solani. Molecules. 2018;23:276. doi: 10.3390/molecules23020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saito T., Abe D., Sekiya K. Sakuranetin induces adipopenesis of 3T3-L1 cells through enhanced expression of PPARγ2. Biochem. Biophys. Res. Commun. 2008;372:835–839. doi: 10.1016/j.bbrc.2008.05.146. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X.F., Hung T.M., Phuong P.T., Ngoc T.M., Min B.-S., Song K.-S., Seong Y.H., Bai K.H. Anti-inflammatory activity of flavonoids from Populus davidiana. Arch. Pharm. Res. 2006;29:1102–1108. doi: 10.1007/BF02969299. [DOI] [PubMed] [Google Scholar]
- 48.Miyazawa M., Kinoshita H., Okuno Y. Antimutagenic activity of sakuranetin from Prunus jamasakura. J. Food Sci. 2003;68:52–56. doi: 10.1111/j.1365-2621.2003.tb14113.x. [DOI] [Google Scholar]
- 49.Zhang L., Kong Y., Wu D., Zhang H., Wu J., Chen J., Ding J., Hu L., Jiang H., Shen X. Three flavonoids targeting the β-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: Crystal structure characterization with enzymatic inhibition assay. Protein Sci. 2008;17:1971–1978. doi: 10.1110/ps.036186.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grecco S.S., Reimao J.Q., Tempone A.G., Sartorelli P., Cunha R.L., Romoff P., Ferreira M.J.P., Favero O.A., Lago J.H.G. In vitro antileishmanial and antitrypanosomal activities of flavanones from Baccharis retusa DC (Asteraceae) Exp. Parasitol. 2012;130:141–145. doi: 10.1016/j.exppara.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 51.Drira R., Sakamoto K. Sakuranetin induces melanogenesis in B16BL6 melanoma cells through inhibition of ERK and PI3K/AKT signaling pathways. Phytother. Res. 2016;30:997–1002. doi: 10.1002/ptr.5606. [DOI] [PubMed] [Google Scholar]
- 52.Hao J., Zhu H., Zhang Z., Yang S., Li H. Identification of anthocyanins in black rice (Oryza sativa L.) by UPLC/Q-TOF-MS and their in vitro and in vivo antioxidant activities. J. Cereal Sci. 2015;64:92–99. doi: 10.1016/j.jcs.2015.05.003. [DOI] [Google Scholar]
- 53.Yang Z., Nakabayashi R., Mori T., Takamatsu S., Kitanaka S., Saito K. Metabolome analysis of Oryza sativa (rice) using liquid chromatography-mass spectrometry for characterizing organ specificity of flavonoids with anti-inflammatory and anti-oxidant activity. Chem. Pharm. Bull. 2016;64:952–956. doi: 10.1248/cpb.c16-00180. [DOI] [PubMed] [Google Scholar]
- 54.Kong C., Xu X., Zhou B., Hu F., Zhang C., Zhang M. Two compounds from allelopathic rice accession and their inhibitory activity on weeds and fungal pathogens. Phytochemistry. 2004;65:1123–1128. doi: 10.1016/j.phytochem.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 55.Kong C., Liang W., Xu X., Hu F., Wang P., Jiang Y. Release and activity of allelochemicals from allelopathic rice seedlings. J. Agric. Food Chem. 2004;52:2861–2865. doi: 10.1021/jf035467i. [DOI] [PubMed] [Google Scholar]
- 56.Ajitha M.J., Mohanlal S., Suresh C.H., Jayalekshmy A. DPPH radical scavenging activity of tricin and its conjugates isolated from “Njavara” rice bran: A density functional theory study. J. Agric. Food Chem. 2012;60:3693–3699. doi: 10.1021/jf204826e. [DOI] [PubMed] [Google Scholar]
- 57.Irakli M.N., Samanidou V.F., Biliaderis C.G., Papadoyannis I.N. Simultaneous determination of phenolic acids and flavonoids in rice using solid-phase extraction and RP-HPLC with photodiode array detection. J. Sep. Sci. 2012;35:1603–1611. doi: 10.1002/jssc.201200140. [DOI] [PubMed] [Google Scholar]
- 58.Park H.L., Yoo Y., Hahn T.-R., Bhoo S.-H., Lee S.-W., Cho M.-H. Antimicrobial activity of UV-induced phenylamides from rice leaves. Molecules. 2014;19:18139–18151. doi: 10.3390/molecules191118139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loypimai P., Moongngarm A., Chottanom P. Thermal and pH degradation kinetics of anthocyanins in natural food colorant prepared from black rice bran. J. Food Sci. Technol. 2016;53:461–470. doi: 10.1007/s13197-015-2002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamura S., Yan K., Shimoda H., Murakami N. Anthocyanins from Oryza sativa L. subsp. indica. Biochem. Syst. Ecol. 2010;38:438–440. doi: 10.1016/j.bse.2010.01.017. [DOI] [Google Scholar]
- 61.Hou Z., Qin P., Zhang Y., Cui S., Ren G. Identification of anthocyanins isolated from black rice (Oryza sativa L.) and their degradation kinetics. Food Res. Int. 2013;50:691–697. doi: 10.1016/j.foodres.2011.07.037. [DOI] [Google Scholar]
- 62.Yoshitomi K., Taniguchi S., Tanaka K., Uji Y., Kazuya A., Gomi K. Rice terpene synthase 24 (PsTPS24) encodes a jamonate-responsive monoterpene synthase that produces an antibacterial γ-terpinene against rice pathogen. J. Plant Physiol. 2016;191:120–126. doi: 10.1016/j.jplph.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 63.Lee G.W., Chung M.-S., Kang M., Chung B.Y., Lee S. Direct suppression of a rice bacterial blight (Xanthomonas oryzae pv. oryzae) by monoterpene (S)-limonene. Protoplasma. 2016;253:683–690. doi: 10.1007/s00709-015-0904-4. [DOI] [PubMed] [Google Scholar]
- 64.Taniguchi S., Hosokawa-Shinonaga Y., Tamaoki D., Yamada S., Akimitsu K., Gomi K. Jasmonate induction of the monoterpene linalool confers resistance to rice bacterial blight and its biosynthesis is regulated by JAZ protein in rice. Plant Cell Environ. 2014;37:451–461. doi: 10.1111/pce.12169. [DOI] [PubMed] [Google Scholar]
- 65.Chumpolsri W., Wijit N., Boontakham P., Nimmanpipug P., Sookwong P., Luangkamin S., Wongpornchai S. Variation of terpenoid flavor odorants in bran of some black and white rice varieties analyzed by GC×GC-MS. J. Food Nutr. Res. 2015;3:114–120. doi: 10.12691/jfnr-3-2-7. [DOI] [Google Scholar]
- 66.Obara N., Hasegawa M., Kodama O. Induced volatiles in elicitor-treated and rice blast fungus-inoculated rice leaves. Biosci. Biotechnol. Biochem. 2002;66:2549–2559. doi: 10.1271/bbb.66.2549. [DOI] [PubMed] [Google Scholar]
- 67.Pichersky E., Gershenzon J. The formation and function of plant volatiles: Perfumes for pollinator attraction and defense. Curr. Opin. Plant Biol. 2002;5:237–243. doi: 10.1016/S1369-5266(02)00251-0. [DOI] [PubMed] [Google Scholar]
- 68.Kiryu M., Hamanaka M., Yoshitomi K., Mochizuki S., Akimitsu K., Gomi K. Rice terpene synthase 18 (OsTPS18) encodes a sesquitperpene synthase that produces and antibacterial (E)-nerolidol against a bacterial pathogen of rice. J. Gen. Plant Pathol. 2018;84:221–229. doi: 10.1007/s10327-018-0774-7. [DOI] [Google Scholar]
- 69.Changan S.S., Ali K., Kumar V., Garg N.K., Tyagi A. Abscisic acid biosynthesis under water stress: Anomalous behavior of the 9-cis-epoxycarotenoid dioxygenase1 (NCED1) gene in rice. Biol. Plantarum. 2018;62:663–670. doi: 10.1007/s10535-018-0807-2. [DOI] [Google Scholar]
- 70.Kurogochi S., Murofushi N., Ota Y., Takahashi N. Identification of gibberellins in the rice plant and quantitative changes of gibberellin A19 throught its life cycle. Planta. 1979;146:185–191. doi: 10.1007/BF00388230. [DOI] [PubMed] [Google Scholar]
- 71.Cartwright D.W., Lancake P., Pryce R.J., Leworthy D.P., Ride J.P. Isolation and characterization of two phytoalexins from rice as momilactones A and B. Phytochemistry. 1981;20:535–537. doi: 10.1016/S0031-9422(00)84189-8. [DOI] [Google Scholar]
- 72.Horie K., Inoue Y., Sakai M., Yao Q., Tanimoto Y., Koga J., Toshima H., Hasegawa M. Identification of UV-induced diterpenes including a new diterpene phytoalexin, phytocassane F, from rice leaves by complementary GC/MS and LC/MS approaches. J. Agric. Food Chem. 2015;63:4050–4059. doi: 10.1021/acs.jafc.5b00785. [DOI] [PubMed] [Google Scholar]
- 73.Akatsuka T., Kadama O., Kato H., Kono Y., Takeuchi S. 3-Hydroxy-7-oxo-sandaraco-pimaradiene (oryzalexin A), a new phytoalexin isolated from rice blast leaves. Agric. Biol. Chem. 1983;47:445–447. doi: 10.1080/00021369.1983.10865658. [DOI] [Google Scholar]
- 74.Kono Y., Takeuchi S., Kodama O., Akatsuka T. Absolute configuration of oryzalexin A and structures of its related phytoalexins isolated from rice blast leaves infected with Pyricularia oryzae. Agric. Biol. Chem. 1984;48:253–255. doi: 10.1271/bbb1961.48.253. [DOI] [Google Scholar]
- 75.Akatsuka T., Kodama O., Sekido H., Kono Y., Takeuchi S. Novel phytoalexins (oryzlexins A, B and C) isolated from rice blast leaves infected with Pyricularia oryzae. Part I: Isolation, characterization and biological activities of oryzalexins. Agric. Biol. Chem. 1985;49:1689–1694. [Google Scholar]
- 76.Kono Y., Takeuchi S., Kodama O., Sekido H., Akatsuka T. Novel phytoalexins (oryzalexins A, B and C) isolated from rice blast leaves infected with Pyricularia oryzae. Part II: Structural studies of oryzalexins. Agric. Biol. Chem. 1985;49:1695–1701. doi: 10.1271/bbb1961.49.1695. [DOI] [Google Scholar]
- 77.Sekido H., Endo T., Suga R., Kodama O., Akatsuka T., Kono Y., Takeuchi S. Oryzalexin D (3,7-dihydroxy-(+)-sandaracopimaradiene), a new phytoalexin isolated from blast-infected rice leaves. Nippon Noyaku Gakkaishi. 1986;11:369–372. [Google Scholar]
- 78.Kato H., Kodama O., Akatrsuka T. Oryzalexin E, a diterpene phytoalexin from UV-irradiated rice leaves. Phytochemistry. 1993;33:79–81. doi: 10.1016/0031-9422(93)85399-C. [DOI] [Google Scholar]
- 79.Kato H., Kodama O., Akatsuka T. Oryzalexin F, a diterpene phytoalexin from UV-irradiated rice leaves. Phytochemistry. 1994;36:299–301. doi: 10.1016/S0031-9422(00)97064-X. [DOI] [Google Scholar]
- 80.Koga J., Shimura M., Oshima K., Ogawa N., Yamauchi T., Ogasawara N. Phytocassanes A, B, C and D, novel diterpene phytoalexins from rice, Oryza sativa L. Tetrahedron. 1995;51:7907–7918. doi: 10.1016/0040-4020(95)00423-6. [DOI] [Google Scholar]
- 81.Koga J., Ogawa N., Yamauchi T., Kikuchi M., Ogasawara N., Shimura M. Functional moiety for the antifungal activity of phytocassane E, a diterpene phytoalexin from rice. Phytochemistry. 1997;44:249–253. doi: 10.1016/S0031-9422(96)00534-1. [DOI] [Google Scholar]
- 82.Yajima A., Mori K. Diterpenoid total synthesis, XXXII synthesis and absolute configuration of (-)-phytocassane D, a diterpene phytoalexin isolated from the rice plant, Oryza sativa. Eur. J. Org. Chem. 2000;2000:4079–4091. doi: 10.1002/1099-0690(200012)2000:24<4079::AID-EJOC4079>3.0.CO;2-R. [DOI] [Google Scholar]
- 83.Horie K., Sakai K., Okugi M., Toshima H., Hasegawa M. Ultraviolet-induced amides and casbene diterpenoids from rice leaves. Phytochm. Lett. 2016;15:57–62. doi: 10.1016/j.phytol.2015.11.009. [DOI] [Google Scholar]
- 84.Inoue Y., Sakai M., Yao Q., Tanimoto Y., Toshima H., Hasegawa M. Identification of a novel casbane-type diterpene phytoalexin, ent-10-oxodepressin, from rice leaves. Biosci. Biotechnol. Biochem. 2013;77:760–765. doi: 10.1271/bbb.120891. [DOI] [PubMed] [Google Scholar]
- 85.Watanabe M., Kono Y., Uzawa J., Teraoka T., Hosokawa D., Suzuki Y., Sakurai A., Teraguchi M. Structures of oryzalic acid B and three related compounds, a group of novel antibacterial diterpenes, isolated from leaves of a bacterial leaf blight-resistant cultivar of rice. Biosci. Biotechnol. Biochem. 1992;56:113–117. doi: 10.1271/bbb.56.113. [DOI] [Google Scholar]
- 86.Kono Y., Kojima A., Nagai R., Watanabe M., Onizawa T., Teraoka T., Watanab M., Koshino H., Uzawa J., Suzuki Y., et al. Antibacterial diterpenenes and their fatty acid conjugates from rice leaves. Phytochemistry. 2004;65:1291–1298. doi: 10.1016/j.phytochem.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 87.Watanabe M., Sakai Y., Teraoka T., Abe H., Kono Y., Uzawa J., Kobayashi K., Suzuki Y., Sakurai A. Novel C19-kaurane type of diterpene (Oryzalide A), a nve antimicrobial compound isolated from healthy leaves of a bacterial leaf blight-resistant cultivar of rice plant. Agric. Biol. Chem. 1990;54:1103–1105. doi: 10.1080/00021369.1990.10870045. [DOI] [Google Scholar]
- 88.Kono Y., Uzawa J., Kobayashi K., Suzuki Y., Uramoto M., Sakurai A., Watanabe M., Teraoka T., Hosokawa D., Watanabe M., et al. Structures of oryzalides A and B, and oryzalic acid A, a group of novel antimicrobial diterpenes, isolated from healthy leaves of a bacterial leaf blight-resistant cultivar of rice plant. Agric. Biol. Chem. 1991;55:803–811. doi: 10.1080/00021369.1991.10870634. [DOI] [Google Scholar]
- 89.Watanabe M., Kono Y., Esumi Y., Teraoka T., Hosokawa D., Suzuki Y., Sakurai A., Watanabe M. Studies on a quantitative analysis of oryzalides and oryzalic acids in rice plants by GC-SIM. Biosci. Biotechnol. Biochem. 1996;60:1460–1463. doi: 10.1271/bbb.60.1460. [DOI] [Google Scholar]
- 90.Kato T., Kabuto C., Sasaki N., Tsunagawa M., Aizawa H., Fujita K., Kato Y., Kitahara Y. Momilactones, growth inhibitors from rice, Oryza sativa L. Tetrahedron Lett. 1973;39:3861–3864. doi: 10.1016/S0040-4039(01)87058-1. [DOI] [Google Scholar]
- 91.Kato-Noguchi H., Ota K., Ino T. Release of momilactone A and B from rice plants into the rhizosphere and its bioactivities. Allelopathy J. 2008;22:321–328. [Google Scholar]
- 92.Kato-Noguchi H., Ino T. Concentration and release level of momilactone B in the seedlings of eight rice cultivars. J. Plant Physiol. 2005;162:965–969. doi: 10.1016/j.jplph.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 93.Chung I.M., Kim J.T., Kim S.-H. Evaluation of allelopathic potential and quantification of momilactone A, B from rice hull extracts and assessment of inhibitory bioactivity on paddy field weeds. J. Agric. Food Chem. 2006;54:2527–2536. doi: 10.1021/jf052796x. [DOI] [PubMed] [Google Scholar]
- 94.Tsunakawa M., Ohba A., Sasaki N., Kabuto C., Kato T., Kitahara Y., Takahashi N. Momilactone C, a minor constituent of growth inhibitors in rice husk. Chem. Lett. 1976;5:1157–1158. doi: 10.1246/cl.1976.1157. [DOI] [Google Scholar]
- 95.Cho J.-G., Cha B.-J., Lee S.-M., Shrestha S., Jeong R.-H., Lee D.S., Kim Y.-C., Lee D.-G., Kang H.-C., Kim J., et al. Diterpenes from the roots of Oryza sativa L. and their inhibitory activity on NO production in LPS-stimulated RAW264.7 macrophages. Chem. Biodivers. 2015;12:1356–1364. doi: 10.1002/cbdv.201400239. [DOI] [PubMed] [Google Scholar]
- 96.Kodama O., Li W.X., Tamogami S., Akatsuka T. Oryzalexin S, a novel stemarane-type diterpene rice phytoalexin. Biosci. Biotechnol. Biochem. 1992;56:1002–1003. doi: 10.1271/bbb.56.1002. [DOI] [PubMed] [Google Scholar]
- 97.Tamogami S., Mitani M., Kodama O., Akatsuka T. Oryzalexin S structure: A new stemarane-type rice plant phytoalexin and its biogenesis. Tetrahedron. 1993;49:2025–2032. doi: 10.1016/S0040-4020(01)86302-X. [DOI] [Google Scholar]
- 98.Fang N., Yu S., Badger T.M. Characterization of triterpene alcohol and sterol ferulates in rice bran using LC-MS/MS. J. Agric. Food Chem. 2003;51:3260–3267. doi: 10.1021/jf021162c. [DOI] [PubMed] [Google Scholar]
- 99.Luo H.-F., Li Q., Yu S., Badger T.M., Fang N. Cytotoxic hydroxylated triterpene alcohol ferulates from rice bran. J. Nat. Prod. 2005;68:94–97. doi: 10.1021/np049889d. [DOI] [PubMed] [Google Scholar]
- 100.Kuljanabhagavad T., Wink M. Biological activities and chemistry of saponins from Chenopodium quinoa Willd. Phytochem. Rev. 2009;8:473–490. doi: 10.1007/s11101-009-9121-0. [DOI] [Google Scholar]
- 101.Lerma-Garcia M.J., Herrero-Martinez J.M., Simo-Alfonso E.F., Mendoca C.R.B., Ramis-Ramos G. Composition, industrial processing and applications of rice bran γ-oryzanol. Food Chem. 2009;115:389–404. doi: 10.1016/j.foodchem.2009.01.063. [DOI] [Google Scholar]
- 102.Minatel I.O., Francisqueti F.V., Correa C.R., Lima G.P.P. Antioxidant activity of γ-oryzanol: A complex network of interactions. Int. J. Mol. Sci. 2016;17:1107. doi: 10.3390/ijms17081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Okahara F., Suzuki J., Hashizume K., Osaki N., Shimotoyodome A. Triterpene alcohols and sterols from rice bran reduce postprandial hyperglycemia in rodents and humans. Mol. Nutr. Food Res. 2016;60:1521–1531. doi: 10.1002/mnfr.201500897. [DOI] [PubMed] [Google Scholar]
- 104.Akihisa T., Yasukawa K., Yamaura M., Ukiya M., Kimura Y., Shimizu N., Arai K. Triterpene alcohol and sterol ferulates from rice bran and their anti-inflammatory effects. J. Agric. Food Chem. 2000;48:2313–2319. doi: 10.1021/jf000135o. [DOI] [PubMed] [Google Scholar]
- 105.Verardo V., Gomez-Caravaca A.M., Marconi E., Segura-Carretero A., Garrido-Frenich A., Fernandez-Gutierrez A. Determination of lipophilic and hydrophilic bioactive compounds in raw and parboiled rice bran. RSC Adv. 2016;6:50786. doi: 10.1039/C6RA04836F. [DOI] [Google Scholar]
- 106.Shu X.-L., Frank T., Shu Q.-Y., Engel K.-H. Metabolite profiling of germinating rice seeds. J. Agric. Food Chem. 2008;56:11612–11620. doi: 10.1021/jf802671p. [DOI] [PubMed] [Google Scholar]
- 107.Suttiarporn P., Chumpolsri W., Mahatheeranont S., Luangkamin S., Teepsawang S., Leardkamokkarn V. Structures of phytosterols and triterpenoids with potential anti-cancer activity in bran of black non-glutinous rice. Nutrients. 2015;7:1672–1687. doi: 10.3390/nu7031672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chung I.M., Ali M., Ahmad A., Lim J.D., Yu C.Y., Kim J.S. Chemical constituents of rice (Oryza sativa) hulls and their herbicidal activity against duckweed (Lemna paucicostata Hegelm 381) Phytochem. Anal. 2006;17:36–45. doi: 10.1002/pca.879. [DOI] [PubMed] [Google Scholar]
- 109.Kumar M.S.S., Ali K., Dahuja A., Tyagi A. Role of phytosterols in drought stress tolerance in rice. Plant Physiol. Biochem. 2015;96:83–89. doi: 10.1016/j.plaphy.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 110.Jung Y.-J., Park J.-H., Shrestha S., Song M.-C., Cho S., Lee C.-H., Han D., Baek N.-I. Phytosterols from the rice (Oryza sativa) bran. J. Appl. Biol. Chem. 2014;57:175–178. doi: 10.3839/jabc.2014.028. [DOI] [Google Scholar]
- 111.Ohnishi M., Fujino Y. Novel glycolipids; cellobiosylsterol and cellotriosylsterol in rice bran. Agric. Biol. Chem. 1978;42:2423–2425. [Google Scholar]
- 112.Ohnishi M., Fujino Y. Structural study on new sterylglycosides in rice bran: Cellotetraosylsitosterol and cellopentaosylsitosterol. Agric. Biol. Chem. 1980;44:333–338. [Google Scholar]
- 113.Buttery R.G., Ling L.C., Juliano B.O., Turnbaugh J.G. Cooked rice aroma and 2-acetyl-1-pyrroline. J. Agric. Food Chem. 1983;31:823–826. doi: 10.1021/jf00118a036. [DOI] [Google Scholar]
- 114.Mahatheeranont S., Keawsa-ard S., Dumri K. Quantification of the rice aroma compound, 2-acetyl-1-pyrroline, in uncooked Khao Dauk Mali 105 brown rice. J. Agric. Food Chem. 2001;49:773–779. doi: 10.1021/jf000885y. [DOI] [PubMed] [Google Scholar]
- 115.Kaikavoosi K., Kad T.D., Zanan R.L., Nadaf A.B. 2-Acetyl-1-pyrroline augmentation in scented indica rice (Oryza sativa L.) varieties through ∆1-pyrroline-5-carboxylate synthetase (P5CS) gene transformation. Appl. Biochem. Biotechnol. 2015;177:1466–1479. doi: 10.1007/s12010-015-1827-4. [DOI] [PubMed] [Google Scholar]
- 116.Morimoto N., Ueno K., Teraishi M., Okumoto Y., Mori N., Ishihara A. Induced phenylamide accumulation in response to pathogen infection and hormone treatment in rice (Oryza sativa) Biosci. Biotechnol. Biochem. 2018;82:407–416. doi: 10.1080/09168451.2018.1429889. [DOI] [PubMed] [Google Scholar]
- 117.Thi H.L., Lin C.-H., Smeda R.J., Leigh N.D., Wycoff W.G., Fritschi F.B. Isolation and identification of an allelopathic phenylethylamine in rice. Phytochemistry. 2014;108:109–121. doi: 10.1016/j.phytochem.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 118.Ishihara A., Hashimoto Y., Miyagawa H., Wakasa K. Induction of serotonin accumulation by feeding of rice striped stem borer in rice leaves. Plant Signal. Behav. 2008;3:714–716. doi: 10.4161/psb.3.9.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yamamoto Y., Kamiya N., Morinaka Y., Matsuoka M., Sazuka T. Auxin biosynthesis by YUCCA genes in rice. Plant Physiol. 2007;143:1362–1371. doi: 10.1104/pp.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tanaka K., Taniguchi S., Tamaoki D., Yoshitomi K., Akimitsu K., Gomi K. Multiple roles of plant volatiles in jasmonate-induced defense response in rice. Plant Signal. Behav. 2014;9:e29247. doi: 10.4161/psb.29247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chung I.-M., Lim Y.-H., Ali M., Sultana S., Ahmad A. Novel anthracene derivatives isolated from rice hulls of Oryza sativa and their growth inhibitory activity of radish seed. Bull. Korean Chem. Soc. 2006;27:995–1000. doi: 10.1002/chin.200702211. [DOI] [Google Scholar]
- 122.Guo H.-M., Li H.-C., Zhou S.-R., Xue H.-W., Miao X.-X. cis-12-Oxo-phytodiennoic acid stimulates rice defense response to a piercing-sucking insect. Mol. Plant. 2014;7:1683–1692. doi: 10.1093/mp/ssu098. [DOI] [PubMed] [Google Scholar]
- 123.Sookwong P., Murata K., Nakagawa K., Shibata A., Kimura T., Yamaguchi M., Kojima Y., Miyazawa T. Cross-fertilization for enhancing tocotrienol biosynthesis in rice plants and QTL analysis of their F2 progenies. J. Agric. Food Chem. 2009;57:4620–4625. doi: 10.1021/jf900394t. [DOI] [PubMed] [Google Scholar]
- 124.Park S., Choi M.J., Lee J.Y., Kim J.K., Ha S.-H., Lim S.-H. Molecular and biochemical analysis of two rice flavonoid 3′-hydroxylase to evaluate their roles in flavonoid biosynthesis in rice grain. Int. J. Mol. Sci. 2016;17:1549. doi: 10.3390/ijms17091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hasegawa M., Mitsuhara I., Seo S., Okada K., Yamane H., Iwai T., Ohashi Y. Analysis on blast fungus-responsive characters of a flavonoid phytoalexin sakuranetin; accumulation in infected rice leaves, antifungal activity and detoxification by fungus. Molecules. 2014;19:11404–11418. doi: 10.3390/molecules190811404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rakwal R., Tamogami S., Kodama O. Role of jasmonic acid as a signaling molecule in copper chloride-elicited rice phytoalexin production. Biosci. Biotechnol. Biochem. 1996;60:1046–1048. doi: 10.1271/bbb.60.1046. [DOI] [Google Scholar]
- 127.Tamogami S., Rakwal R., Kodama O. Phytoalexin production elicited by exogenously applied jasmonic acid in rice leaves (Oryza sativa L.) is under the control of cytokinins and ascorbic acid. FEBS Lett. 1997;412:61–64. doi: 10.1016/S0014-5793(97)00743-6. [DOI] [PubMed] [Google Scholar]
- 128.Nakazato Y., Tamogami S., Kawai H., Hasegawa M., Kodama O. Methionine-induced phytoalexin production in rice leaves. Biosci. Biotechnol. Biochem. 2000;64:577–583. doi: 10.1271/bbb.64.577. [DOI] [PubMed] [Google Scholar]
- 129.Tamogami S., Shigeru K., Osamu H., Hirose K., Akatsuka T. Pretilachlor [2-chloro-N-(2,6-diethylphenyl)-N-(2-propoxyethyl) acetamide]- and butachlor [N-(butoxymethl)-2-chloro-N-(2,6-diethylphenyl) acetamide]-induced accumulation of phytoalexin in rice (Oryza sativa) plants. J. Agric. Food Chem. 1995;43:1695–1697. doi: 10.1021/jf00054a052. [DOI] [Google Scholar]
- 130.Tamogami S., Kodama O. Coronatine elicits phytoalexin production in rice leaves (Oryza sativa L.) in the same manner as jasmonic acid. Phytochemistry. 2000;54:689–694. doi: 10.1016/S0031-9422(00)00190-4. [DOI] [PubMed] [Google Scholar]
- 131.Plowright R.A., Grayer R.J., Gill J.R., Rahman M.L., Harborne J.B. The induction of phenolic compounds in rice after infection by the stem nematode Ditylenchus angustus. Nematologica. 1996;42:564–578. [Google Scholar]
- 132.Kanno H., Hasegawa M., Kodama O. Accumulation of salicylic acid, jasmonic acid and phytoalexins in rice, Oryza sativa, infested by the white-backed planthopper, Sogatella furcifera (Hemiptera: Delphacidae) Appl. Entomol. Zool. 2012;47:27–34. doi: 10.1007/s13355-011-0085-3. [DOI] [Google Scholar]
- 133.Ogawa S., Miyamoto K., Nemoto K., Sawasaki T., Yamane H., Nojiri H., Okada K. OsMYC2, an essential factor for JA-inductive sakuranetin production in rice, interacts with MYC2-like proteins that enhance its transactivation ability. Sci. Rep. 2017;7:40175. doi: 10.1038/srep40175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Oritani T., Kiyota H. Biosynthesis and metabolism of abscisic acid and related compounds. Nat. Prod. Rep. 2003;20:414–425. doi: 10.1039/b109859b. [DOI] [PubMed] [Google Scholar]
- 135.Heden P., Thomas S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012;444:11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- 136.Peters R.J. Unconvering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry. 2006;67:2307–2317. doi: 10.1016/j.phytochem.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 137.Toyomasu T. Recent advances regarding diterpene cyclase genes in higher plants and fungi. Biosci. Biotechnol. Biochem. 2008;72:1168–1175. doi: 10.1271/bbb.80044. [DOI] [PubMed] [Google Scholar]
- 138.Okada K. The biosynthesis of isoprenoids and the mechanisms regulating it in plants. Biosci. Biotechnol. Biochem. 2011;75:1219–1225. doi: 10.1271/bbb.110228. [DOI] [PubMed] [Google Scholar]
- 139.Okada A., Shimizu T., Okada K., Kuzuyama T., Koga J., Shibuya N., Nojiri H., Yamane H. Elicitor induced activation of the methylerythritol phosphate pathway toward phytoalexins biosynthesis in rice. Plant Mol. Biol. 2007;65:177–187. doi: 10.1007/s11103-007-9207-2. [DOI] [PubMed] [Google Scholar]
- 140.Miyamoto K., Fujita M., Shenton M.F., Shenton M.R., Akashi S., Sugawara C., Sakai A., Horie K., Hasegawa M., Kawide H., et al. Evolutionary trajectory of phytoalexin biosynthetic gene clusters in rice. Plant J. 2016;87:293–304. doi: 10.1111/tpj.13200. [DOI] [PubMed] [Google Scholar]
- 141.Toyomasu T., Goda C., Sakai A., Miyamoto K., Shenton M.R., Tomiyama S., Mitsuhashi W., Yamane H., Kurata N., Okada K. Characterization of diterpene synthase genes in the wild rice species Oryza brachyatha provides evolutionary insight into rice phytoalexin biosynthesis. Biochem. Biophys. Res. Commun. 2018;503:1221–1227. doi: 10.1016/j.bbrc.2018.07.028. [DOI] [PubMed] [Google Scholar]
- 142.Ye Z., Yamazaki K., Minoda H., Miyamoto K., Miyazaki S., Kawaide H., Yajima A., Nojiri H., Yamane H., Okada K. In planta functions of cytochrome P450 monooxygenase genes in the phytocassane biosynthetic gene cluster on rice chromosome 2. Biosci. Biotechnol. Biochem. 2018;82:1021–1030. doi: 10.1080/09168451.2017.1398067. [DOI] [PubMed] [Google Scholar]
- 143.Salazar-Cerezo S., Martinez-Montiel N., Garcia-Sanchez J., Perez-y-Terron R., Martinez-Contreras R.D. Gibberellin biosynthesis and metabolism: A convergent route for plants fungi and bacteria. Microbiol. Res. 2018;208:85–98. doi: 10.1016/j.micres.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 144.Wu Y., Wang Q., Hillwig M.L., Peters R.J. Picking sides: Distinct roles for CYP76M6 and CYP76M8 in rice oryzalexin biosynthesis. Biochem. J. 2013;454:209–216. doi: 10.1042/BJ20130574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rohrer C.A., Siebenmorgen T.J. Nutraceutical concentrations within the bran of various rice kernel thickness fractions. Biosyst. Eng. 2004;88:453–460. doi: 10.1016/j.biosystemseng.2004.04.009. [DOI] [Google Scholar]
- 146.Matsuzuka K., Kimura E., Nakagawa K., Murata K., Kimura T., Miyazawa T. Investigation of tocotrienol biosynthesis in rice (Oryza sativa L.) Food Chem. 2013;140:91–98. doi: 10.1016/j.foodchem.2013.02.058. [DOI] [PubMed] [Google Scholar]
- 147.Wakte K., Zanan R., Hinge V., Khandagale K., Nadaf A., Henry R. Thirty-three years of 2-acetyl-1-pyrroline, a principal basmati aroma compound in scented rice (Oryza sativa L.): A status review. J. Sci. Food Agric. 2017;97:384–395. doi: 10.1002/jsfa.7875. [DOI] [PubMed] [Google Scholar]
- 148.Ishihara A., Hashimoto Y., Tanaka C., Dubouzet J.G., Nakao T., Matsuda F., Nishioka T., Miyagawa H., Wakasa K. The tryptophan pathway is involved in the desense responses of rice against pathogenic infection via serotonin production. Plant J. 2008;54:481–495. doi: 10.1111/j.1365-313X.2008.03441.x. [DOI] [PubMed] [Google Scholar]
- 149.Bradlbury L.M.T., Gillies S.A., Brushett D.J., Waters D.L.E., Henry R.J. Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice. Plant Mol. Biol. 2008;68:439–449. doi: 10.1007/s11103-008-9381-x. [DOI] [PubMed] [Google Scholar]
- 150.Dubouzet J.G., Matsuda F., Ishihara A., Miyagawa H., Wakasa K. Production of indole alkaloids by metabolic engineering of the tryptophan pathway in rice. Plant Biotechnol. J. 2013;11:1103–1111. doi: 10.1111/pbi.12105. [DOI] [PubMed] [Google Scholar]
- 151.Murthy H.N., Lee E.-J., Paek K.-Y. Production of secondary metabolites from cell and organ cultures: Strategies and approaches for biomass improvement and metabolite accumulation. Plant Cell Tiss. Organ Cult. 2014;118:1–16. doi: 10.1007/s11240-014-0467-7. [DOI] [Google Scholar]
- 152.Ahuja I., Kissen R., Bones A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012;17:73–90. doi: 10.1016/j.tplants.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 153.Siciliano I., Carneiro G.A., Spadaro D., Garibaldi A., Gullino M.L. Jamonic acid, abscisic acid, and salicylic acid are involved in the phytoalexin responses of rice to Fusarium fujikuroi, a high gibberellin producer pathogen. J. Agric. Food Chem. 2015;63:8134–8142. doi: 10.1021/acs.jafc.5b03018. [DOI] [PubMed] [Google Scholar]
- 154.Zhou M., Memelink J. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 2016;34:441–449. doi: 10.1016/j.biotechadv.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 155.Yoshida Y., Miyamoto K., Yamane H., Nishizawa Y., Minami E., Nojiri H., Okada K. OsTGAP1 is responsible for JA-inducible diterpenoid phytoalexin biosynthesis in rice roots with biological impacts on allelopathic interaction. Physiol. Plantarum. 2017;161:532–544. doi: 10.1111/ppl.12638. [DOI] [PubMed] [Google Scholar]
- 156.Tamogami S., Rakwal R., Kodama O. Phytoalexin production by amino acid conjugates of jasmonic acid through induction of naringenin-7-O-methyltransferase, a key enzyme on phytoalexin biosynthesis in rice (Oryza sativa L.) FEBS Lett. 1997;401:239–242. doi: 10.1016/S0014-5793(96)01482-2. [DOI] [PubMed] [Google Scholar]
- 157.Miyamoto K., Enda I., Okada T., Sato Y., Watanabe K., Sakazawa T., Yumoto E., Shibata K., Asahina M., Lino M., et al. Jasmonoyl-L-isoleucine is required for the production of a flavonoid phytoalexin but not diterpenoid phytoalexins in ultraviolet-irradiated rice leaves. Biosci. Biotechnol. Biochem. 2016;80:1934–1938. doi: 10.1080/09168451.2016.1189319. [DOI] [PubMed] [Google Scholar]
- 158.Ko K.W., Okada K., Koga J., Jikumaru Y., Nojiri H., Ymane H. Effects of cytokinin on production of diterpenoid phytoalexins in rice. J. Pestic. Sci. 2010;35:412–418. doi: 10.1584/jpestics.G09-63. [DOI] [Google Scholar]
- 159.Daw B.D., Zhang L.H., Wang Z.Z. Salicylic acid enhances antifungal resistance to Magnaporthe grisea in rice plants. Aust. Plant Pathol. 2008;37:637–644. doi: 10.1071/AP08054. [DOI] [Google Scholar]
- 160.Akagi A., Fukushima S., Okada K., Jiang C.J., Yoshida R., Nakayama A., Shimono M., Sugano S., Yamane H., Takatsuji H. WRKY45-dependent priming of diterpeoid phytoalexin biosynthesis in rice and the role of cytokinin in triggering the reaction. Plant Mol. Biol. 2014;86:171–183. doi: 10.1007/s11103-014-0221-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hamada H., Kurusu T., Nokajima H., Kiyoduka M., Yano K., Kuchitsu K. Regulation of xylanase elicitor-induced expression of defense-related genes involved in phytoalexin biosynthesis by a cation channel OsTPC1 in suspension-cultured rice cells. Plant Biotechnol. 2014;31:329–334. doi: 10.5511/plantbiotechnology.14.0805b. [DOI] [Google Scholar]
- 162.Zhou L., Zheng G., Wang S., Gan F. Effects of oligosaccharins on callus growth and saponin content of Panax notoginseng. Cell Res. 1992;2:83–87. doi: 10.1038/cr.1992.8. [DOI] [Google Scholar]
- 163.Shibuya N., Minami E. Oligosaccharide signaling for defence responses in plant. Physiol. Mol. Plant Pathol. 2001;59:223–233. doi: 10.1006/pmpp.2001.0364. [DOI] [Google Scholar]
- 164.Zhou L., Cao X., Zhang R., Peng Y., Zhao S., Wu J. Stimulation of saponin production in Panax ginseng hairy roots by two oligosaccharides from Paris polyphylla var. yunnanensis. Biotechnol. Lett. 2007;29:631–634. doi: 10.1007/s10529-006-9273-6. [DOI] [PubMed] [Google Scholar]
- 165.Zhang R., Li P., Xu L., Chen Y., Sui P., Zhou L., Li J. Enhancement of diosgenin production in Dioscorea zingiberensis cell culture by oligosaccharide elicitor from its endophytic fungus Fusarium oxysporum Dzf17. Nat. Prod. Commun. 2009;4:1459–1462. [PubMed] [Google Scholar]
- 166.Li P., Mao Z., Lou J., Li Y., Mou Y., Lu S., Peng Y., Zhou L. Enhancement of diosgenin production in Dioscorea zingiberensis cell cultures by oligosaccharides from its endophytic fungus Fusarium oxysporum Dzf17. Molecules. 2011;16:10631–10644. doi: 10.3390/molecules161210631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li Y., Shan T., Mou Y., Li P., Zhao J., Zhao W., Peng Y., Zhou L., Ding C. Enhancement of palmarumycin C12 and C13 production in liquid culture of endophytic fungus Berkleasmium sp. Dzf12 by oligosaccharides from its host plant Dioscorea zingiberensis. Molecules. 2012;17:3761–3773. doi: 10.3390/molecules17043761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Li P., Luo H., Meng J., Sun W., Wang X., Lu S., Peng Y., Zhou L. Effects of oligosaccharides from endophytic Fusarium oxysporum Dzf17 on activities of defense-related enzymes in Diosocrea zingiberensis suspension cell and seedling cultures. Electron. J. Biotechnol. 2014;17:156–161. doi: 10.1016/j.ejbt.2014.04.012. [DOI] [Google Scholar]
- 169.Ren Y.-Y., West C.A. Elicitation of diterpene biosynthesis in rice (Oryza sativa L.) by chitin. Plant Physiol. 1992;99:1169–1178. doi: 10.1104/pp.99.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yamada A., Shibuya N., Kodama O., Akatsuka T. Induction of phytoalexins formation in suspension-cultured rice cells by N-acetylchitooligosaccharides. Biosci. Biotechnol. Biochem. 1993;57:405–409. doi: 10.1271/bbb.57.405. [DOI] [Google Scholar]
- 171.Yamaguchi T., Yamada A., Hong N., Ogawa T., Ishii T., Shibuya N. Differences in the recognition of glucan elicitor signals between rice and soybean: β-glucan fragments from the rice blast disease fungus Pyricularia oryzae that elicit phytoalexin biosynthesis in suspension-cultrued rice cells. Plant Cell. 2000;12:817–826. doi: 10.1105/tpc.12.5.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yamaguchi T., Maehara Y., Kodama O., Okada M., Matsumura M., Shibuya N. Two purified oligosaccharide elicitors, N-acetylchitohepatose and tetraglucosyl glucitol, derived from Magnaporthe grisea cell walls, synergistically activate biosynthesis of phytoalexin in suspension-cultured rice cells. J. Plant Physiol. 2002;159:1147–1149. doi: 10.1078/0176-1617-00877. [DOI] [Google Scholar]
- 173.Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., Shibuya N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA. 2006;103:11086–11091. doi: 10.1073/pnas.0508882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kouzai Y., Kaku H., Shibuya N., Minami E., Nishizawa Y. Expression of the chimeric receptor between the chitin elicitor receptor CEBiP and the receptor-like protein kinase Pi-d2 leads to enhanced responses to the chitin elicitor and disease reistance against Magnaporthe oryzae in rice. Plant Mol. Biol. 2013;81:287–295. doi: 10.1007/s11103-012-9998-7. [DOI] [PubMed] [Google Scholar]
- 175.Miyamoto K., Nishizawa Y., Minami E., Nojiri H., Yamane H., Okada K. Overexpression of the bZIP transcription factor OsbZIP79 suppresses the production of diterpenoid phytoalexin in rice cells. J. Plant Physiol. 2015;173:19–27. doi: 10.1016/j.jplph.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 176.Tan R.X., Chen J.H. The cerebrosides. Nat. Prod. Rep. 2003;20:509–534. doi: 10.1039/b307243f. [DOI] [PubMed] [Google Scholar]
- 177.Umemura K., Ogawa N., Koga J., Iwata M., Usami H. Elicitor activity of cerebroside, a sphingolipid elicitor, in cell suspension cultures of rice. Plant Cell Physiol. 2002;43:778–784. doi: 10.1093/pcp/pcf090. [DOI] [PubMed] [Google Scholar]
- 178.Koga J., Yamauchi T., Shimura M., Ogawa N., Oshima K., Umemura K., Kikuchi M., Ogasawara N. Cerebrosides A and C, sphingolipid elicitors of hypersensitive cell death and phytoalexin accumulation in rice plants. J. Biol. Chem. 1998;273:31985–31991. doi: 10.1074/jbc.273.48.31985. [DOI] [PubMed] [Google Scholar]
- 179.Umemura K., Ogawa N., Yamauchi T., Iwata M., Shimura M., Koga J. Cerebroside elicitors found in diverse phytopathogens activate defense responses in rice plants. Plant Cell Physiol. 2000;41:676–683. doi: 10.1093/pcp/41.6.676. [DOI] [PubMed] [Google Scholar]
- 180.Umemura K., Tanino S., Nagatsuka T., Koga J., Iwata M., Nagashima K., Amemiya Y. Cerebroside elicitor confers resistance to Fusarium disease in various plant species. Phytopathology. 2004;94:813–818. doi: 10.1094/PHYTO.2004.94.8.813. [DOI] [PubMed] [Google Scholar]
- 181.Koga J., Kubota H., Gomi S., Umemura K., Ohnishi M., Kono T. Cholic acid, a bile acid elicitor of hypersensitive cell death, pathgenesis-related protein synthesis, and phytoalexin accumulation in rice. Plant Physiol. 2006;140:1475–1483. doi: 10.1104/pp.105.070334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shimizu T., Jikumaru Y., Okada A., Okada K., Koga J., Umemura K., Minami E., Shibuya N., Hasegawa M., Kodama O., et al. Effects of a bile acid elicitor, cholic acid, on the biosynthesis of diterpenoid phytoalexins in suspension-cultured rice cells. Phytochemistry. 2008;69:973–981. doi: 10.1016/j.phytochem.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 183.Sudo E., Itouga M., Yoshida-Hatanaka K., Ono Y., Sakakibara H. Gene expression and sensitivity in response to copper stress in rice leaves. J. Exp. Bot. 2008;59:3465–3474. doi: 10.1093/jxb/ern196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zhou L.G., Wu J.Y. Development and application of medicinal plant tissue cultures for production of drugs and herbal medicinals in China. Nat. Prod. Rep. 2006;23:789–810. doi: 10.1039/b610767b. [DOI] [PubMed] [Google Scholar]
- 185.Zhao J.-L., Zhou L.-G., Wu J.-Y. Effects of biotic and abiotic elicitors on cell growth and tanshinone accumulation in Salvia miltiorrhiza cell cultures. Appl. Microbiol. Biotechnol. 2010;87:137–144. doi: 10.1007/s00253-010-2443-4. [DOI] [PubMed] [Google Scholar]
- 186.Gandi S., Rao K., Chodisetti B., Giri A. Elicitation of andrographolide in the suspension cultures of Andrographis paniculata. Appl. Biochem. Biotechnol. 2012;168:1729–1738. doi: 10.1007/s12010-012-9892-4. [DOI] [PubMed] [Google Scholar]
- 187.Cai Z., Kastell A., Speiser C., Smetanska I. Enhanced resveratrol production in Vitis vinifera cell suspension cultures by heavy metals without loss of cell viability. Appl. Biochem. Biotechnol. 2013;171:330–340. doi: 10.1007/s12010-013-0354-4. [DOI] [PubMed] [Google Scholar]
- 188.Durango D., Quinones W., Torres F., Rosero Y., Gil J., Echeverri F. Phytoalexin accumulation in Colombian bean varieties and aminosugars as elicitors. Molecules. 2002;7:817–832. doi: 10.3390/71100817. [DOI] [Google Scholar]
- 189.Kodama O., Yamada A., Yamamoto A., Takemoto T., Akatsuka T. Induction of phytoalexins with heavy metal ions in rice leaves. J. Pestc. Sci. 1988;13:615–617. doi: 10.1584/jpestics.13.615. [DOI] [Google Scholar]
- 190.Kodama O., Suzuki T., Miyakawa J., Akatsuka T. Ultraviolet-induced accumulation of phytoalexins in rice leaves. Agric. Biol. Chem. 1988;52:2469–2473. [Google Scholar]
- 191.Kato-Noguchi H., Kujime H., Ino T. UV-induced momilactone B accumulation in rice rhizosphere. J. Plant Physiol. 2007;164:1548–1551. doi: 10.1016/j.jplph.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 192.Cartwright D.W., Langcake P., Ride J.P. Phytoalexin production in rice and its enhancement by a dichlorocyclopropane fungicide. Physiol. Plant Pathol. 1980;17:259–267. doi: 10.1016/S0048-4059(80)80019-1. [DOI] [Google Scholar]
- 193.Goufo P., Pereira J., Figureiredo N., Oliveira M.B.P.P., Carranca C., Rosa E.A.S., Trindade H. Effect of elevated carbon dioxide (CO2) on phenolic acids, flavonoids, tocopherols, tocotrienols, γ-oryzanol and antioxidant capacities of rice (Oryza sativa L.) J. Cereal Sci. 2014;59:15–24. doi: 10.1016/j.jcs.2013.10.013. [DOI] [Google Scholar]
- 194.Ejike C.E.C.C., Gong M., Udenigwe C.C. Phytoalexins from the Poaceae: biosynthesis, function and prospects in food preservation. Food Res. Int. 2013;52:167–177. doi: 10.1016/j.foodres.2013.03.012. [DOI] [Google Scholar]
- 195.Hasegawa M., Mitsuhara I., Seo S., Imai T., Koga J., Okada K., Yamane H., Ohashi Y. Phytoalexin accumulation in the interaction between rice and the blast fungus. Mol. Plant-Micro. Intrac. 2010;23:1000–1011. doi: 10.1094/MPMI-23-8-1000. [DOI] [PubMed] [Google Scholar]
- 196.Ishihara A., Fukami A., Matsuda Y., Nakajima H., Miyagawa H. Accumulation of indole-3-acetic acid in rice sl mutant leaves infected with Bipolaris oryzae. J. Phytopathol. 2016;164:509–519. doi: 10.1111/jph.12476. [DOI] [Google Scholar]
- 197.Lu H., Luo T., Fu H., Wang L., Tan Y., Huang J., Wang Q., Ye G., Gatehouse A.M.R., Lou Y., et al. Resistance of rice to insect pests mediated by suppression of serotonin biosynthesis. Nat. Plants. 2018;4:338–344. doi: 10.1038/s41477-018-0152-7. [DOI] [PubMed] [Google Scholar]
- 198.Kyndt T., Denil S., Bauters L., Van Criekinge W., De Meyer T. Systemic suppression of the shoot metabolism upon rice root nematode infection. PLoS ONE. 2014;9:e106858. doi: 10.1371/journal.pone.0106858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zhu S., Gao F., Cao X., Chen M., Ye G., Wei C., Li Y. The rice dwarf virus P2 protein interacts with ent-kaurene oxidases in vivo, leading to reduced biosynthesis of gibberellins and rice dwarf symptoms. Plant Physiol. 2005;139:1935–1945. doi: 10.1104/pp.105.072306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kato-Noguchi H. Bamyard grass-induced rice allelopathy and momilactone B. J. Plant Physiol. 2011;168:1016–1020. doi: 10.1016/j.jplph.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 201.Dayan F.E. Factors modulating the levels of the allelochemical sorgolene in Sorghum bicolor. Planta. 2006;224:339–346. doi: 10.1007/s00425-005-0217-5. [DOI] [PubMed] [Google Scholar]
- 202.Boiteau R.M., Hoyt D.W., Nicora C.D., Kinmonth-Schultz H.A., Ward J.K., Bingol K. Structure elucidation of unknown metabolites in metabolomics by combined NMR and MS/MS prediction. Metabolites. 2018;8:8. doi: 10.3390/metabo8010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Delaunois B., Cordelier S., Conreux A., Clement C., Jeandet P. Molecular engineering of resveratrol in plants. Plant Biotechnol. J. 2009;7:2–12. doi: 10.1111/j.1467-7652.2008.00377.x. [DOI] [PubMed] [Google Scholar]
- 204.Jeandet P., Courot E., Clement C., Ricord S., Crouzet J., Aziz A., Cordelier S. Molecular engineering of phytoalexins in plants: Benefits and limitations for food and agriculture. J. Agric. Food Chem. 2017;65:2643–2644. doi: 10.1021/acs.jafc.7b00936. [DOI] [PubMed] [Google Scholar]
- 205.Park H.L., Lee S.-W., Jung K.-H., Hahn T.-R., Cho M.-H. Transcriptomic analysis of UV-treated rice leaves revrals UV-induced phytoalexin biosynthesis pathways and their regulatory networks in rice. Phytochemistry. 2013;96:57–71. doi: 10.1016/j.phytochem.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 206.Stark-Lorenzen P., Nelke B., Hanbler G., Muhlbach H.P., Thomzik J.E. Transfer of a grapevine stilbene synthase gene to rice (Oryza sativa L.) Plant Cell Rep. 1997;16:668–673. doi: 10.1007/s002990050299. [DOI] [PubMed] [Google Scholar]
- 207.Zhao M., Cheng J., Guo B., Duan J., Che C.-T. Momilactone and related diterpenoids as potential agricultural chemicals. J. Agric. Food Chem. 2018;66:7859–7872. doi: 10.1021/acs.jafc.8b02602. [DOI] [PMC free article] [PubMed] [Google Scholar]