Abstract
Paenibacillus larvae (P. larvae) is a bacterial pathogen causing American foulbrood (AFB), the most serious disease of honeybee larvae. The food of young larvae could play an important role in the resistance of larvae against AFB. It contains antibacterial substances produced by honeybees that may inhibit the propagation of the pathogen in larval midguts. In this study, we identified and investigated the antibacterial effects of one of these substances, trans-10-hydroxy-2-decenoic acid (10-HDA), against P. larvae strains including all Enterobacterial Repetitive Intergenic Consensus (ERIC) genotypes. Its inhibitory activities were studied by determining the minimum inhibitory concentrations (MICs). It was found that 10-HDA efficacy increases substantially with decreasing pH; up to 12-fold differences in efficacy were observed between pH = 5.5 and pH = 7.2. P. larvae strains showed different susceptibility to 10-HDA; up to 2.97-fold differences existed among various strains with environmentally important ERIC I and ERIC II genotypes. Germinating spores of the pathogen were generally more susceptible to 10-HDA than vegetative cells. Our findings suggest that 10-HDA could play significant role in conferring antipathogenic activity to larval food in the midguts of young larvae and contribute to the resistance of individual larvae to P. larvae.
Keywords: honeybee, 10-hydroxy-2-decenoic acid, antibacterial, bacterial spores, royal jelly, worker jelly, larval jelly, ERIC-PCR genotyping, American foulbrood
1. Introduction
American foulbrood (AFB) is a redoubtable infectious disease of honeybee (Apis mellifera) colonies. It is the most destructive brood disease that causes financial losses to beekeepers and farmers worldwide. The causative agent of AFB are spores of the Gram-positive bacterium Paenibacillus larvae that may contaminate larval food. The infectivity of larvae depends on their age (the most susceptible are larvae younger than 36 h) and on the dose of spores in food [1,2]. A precondition for a fatal ailment of larvae seems to be the massive proliferation of vegetative bacterial cells in the larval midgut [3], associated with the production of enzymes and substances that help the bacteria to penetrate through the peritrophic matrix and gut epithelium into the hemocoel [4,5,6]. The subsequent cell proliferation in hemocoel causes the death of the larva, its decomposition and creation of billions of spores that under suitable conditions drive intra-colonial and inter-colonial larval infections that, without the beekeeper’s intervention, can lead to the collapse of the colony [2,7,8].
It has been found that speed of the progression and the severity of AFB in colonies depend on the virulence of P. larvae strains. Strains with the ERIC (Enterobacterial Repetitive Intergenic Consensus) I genotype are less virulent to larvae and kill them mostly after cell capping. The strains possessing the ERIC II, ERIC III and ERIC IV genotypes are highly virulent and kill larvae before cell capping [9,10]. The highly virulent strains paradoxically cause the slower destruction of colonies than less virulent ones. This is associated with the ability of honeybees to detect and remove diseased larvae from the hive (hygienic behaviour), which is more effective when larvae die before cell capping [11,12].
The control of AFB is based on prevention, early detection, disinfection and treatment strategies in the case of infection and the destruction of clinically infected hives by burning [2,8,13,14,15,16]. The prospective way to fight against the disease seems to be the breeding of honeybee lines that are more resistant to AFB. Such lines should possess more effective individual and social defense mechanisms that act in colonies against AFB [2,11,17,18]. It has been shown that the selective breeding of colonies based on the hygienic behavior of bees leads to increased AFB resistance of colonies [15,19,20].
We assume that one possible but poorly explored social protective mechanism of larvae and colonies against AFB could be based on the antimicrobial substances present in larval food and their ability to prevent massive proliferation of bacterial cells in the midguts of individual larvae. It is known that the food of larvae younger than 3 days, i.e., larval jelly (LJ) including royal, worker and drone jelly (RJ, WJ and DJ) [21], contain antimicrobial substances such as peptides royalisin/defensin1 and jelleines [22,23,24], the protein apalbumin2a [25] and some hydroxy or dicarboxylic derivatives of medium-chain fatty acids including trans-10-hydroxy-2-decenoic acid (10-HDA) [26,27,28,29] that produce nurse honeybees. At present, there is a little information about in vitro effects of these substances against P. larvae and no information about their in vivo action in larvae. There are several reports demonstrating the antibacterial effects of WJ [30], RJ [31] and their extracts [32] on P. larvae and a few demonstrating the inhibitory effects of native defensin1 [33,34], recombinant defensin 1 [35] and protein apalbumin 2a [25] on the growth of the pathogen.
Trans-10-hydroxy-2-decenoic acid or (E)-10-hydroxydec-2-enoic acid (IUPAC name) (10-HDA) is the most abundant fatty acid (FA) and a major lipid component of RJ [26,36]. This unique FA shows various biological and pharmacological activities [37,38,39] and is used as a marker of RJ quality and authenticity [40]. Although several studies have demonstrated its antimicrobial activity against various Gram-positive and Gram-negative bacteria (mostly human pathogens) [26,27,28,41], no study has examined its antibacterial properties against P. larvae.
Therefore, we aimed to investigate the antibacterial effect of 10-HDA against P. larvae using reference strains and field isolates having all known ERIC genotypes. Due to the fact that the effect of 10-HDA in honeybee larvae could be affected by different levels of acidity of consumed LJ and by the pH value of the midgut environment, its antibacterial efficacy was studied in media with different pH levels. Furthermore, the susceptibility of vegetative cells and germinating spores of P. larvae to 10-HDA was compared. Based on the obtained findings, we assessed the potential of 10-HDA to inhibit the propagation of P. larvae in larval midguts, either individually or with the help of other factors associated with LJ.
2. Results
2.1. ERIC Genotypes of Bacterial Strains
ERIC genotypes were determined for all P. larvae strains used in this work. Representative results from genotyping of selected strains are shown in supplementary materials in Figure S1. The individual strains and their genotypes are given in Table 1. The characterization confirmed previously specified ERIC I genotypes of reference strain CCUG 28515 and LMG 9820 (identical with DSM 7030) [10]. Another three reference strains, i.e., CCM 4483, CCM 4486 and CCM 39 had the ERIC I, ERIC II and ERIC III genotype, respectively. The reference strain CCM 38 contained two morphologically different strains, one with ERIC III and the other with the ERIC IV genotype. All field isolates of P. larvae had the same ERIC II genotype.
Table 1.
P. larvae strains, their characteristics and susceptibility to 10-HDA.
| Strain | Source | ERIC Genotype | MIC* (μg/μL) |
|---|---|---|---|
| CCUG 28515 | CCUG | I | 1.05 ± 0.16 |
| LMG 9820 | LMG | I | 0.60 ± 0.00 |
| CCM 4483 | CCM | I | 1.20 ± 0.00 |
| CCM 4486 | CCM | II | 1.28 ± 0.15 |
| CCM 38a | CCM | III | 2.80 ± 0.00 |
| CCM 38b | CCM | IV | 2.60 ± 0.00 |
| CCM 39 | CCM | III | 2.60 ± 0.00 |
| 1-99 | diseased brood | II | 1.47 ± 0.14 |
| 2-99 | diseased brood | II | 1.40 ± 0.00 |
| 3-99 | diseased brood | II | 1.02 ± 0.18 |
| 4-99 | diseased brood | II | 1.27 ± 0.14 |
| 1-09 | diseased brood | II | 1.20 ± 0.00 |
| 3-09 | diseased brood | II | 1.78 ± 0.04 |
| 4-09 | diseased brood | II | 1.11 ± 0.09 |
| 5-09 | diseased brood | II | 1.40 ± 0.00 |
| 6-09 | diseased brood | II | 1.58 ± 0.04 |
| 7-09 | diseased brood | II | 1.35 ± 0.09 |
| 1-11 | ground stock | II | 0.98 ± 0.04 |
| 2-11 | ground stock | II | 1.35 ± 0.09 |
| 4-11 | ground stock | II | 1.60 ± 0.00 |
| 7-11 | ground stock | II | 1.33 ± 0.11 |
| 8-11 | diseased brood | II | 1.38 ± 0.04 |
| 11-11 | ground stock | II | 1.50 ± 0.11 |
| 13-11 | ground stock | II | 1.18 ± 0.04 |
| 14-11 | ground stock | II | 1.20 ± 0.00 |
| 17-11 | ground stock | II | 0.93 ± 0.11 |
| 21-11 | ground stock | II | 1.78 ± 0.04 |
| 22-11 | ground stock | II | 1.50 ± 0.11 |
| 23-11 | ground stock | II | 1.38 ± 0.04 |
| 24-11 | ground stock | II | 1.40 ± 0.00 |
| 25-11 | ground stock | II | 1.20 ± 0.00 |
| 26-11 | ground stock | II | 1.43 ± 0.17 |
| 34-11 | ground stock | II | 1.58 ± 0.04 |
| 1-14 | diseased brood | II | 1.38 ± 0.04 |
| 2-14 | diseased brood | II | 1.58 ± 0.04 |
| 3-14 | diseased brood | II | 1.40 ± 0.00 |
| 4-14 | diseased brood | II | 0.60 ± 0.00 |
| 5-14 | diseased brood | II | 1.38 ± 0.04 |
| 6-14 | diseased brood | II | 1.00 ± 0.00 |
| 7-14 | diseased brood | II | 1.40 ± 0.00 |
* Determined at pH = 6.6.
2.2. Effect of pH and Substance PIPES on Growth of P. larvae
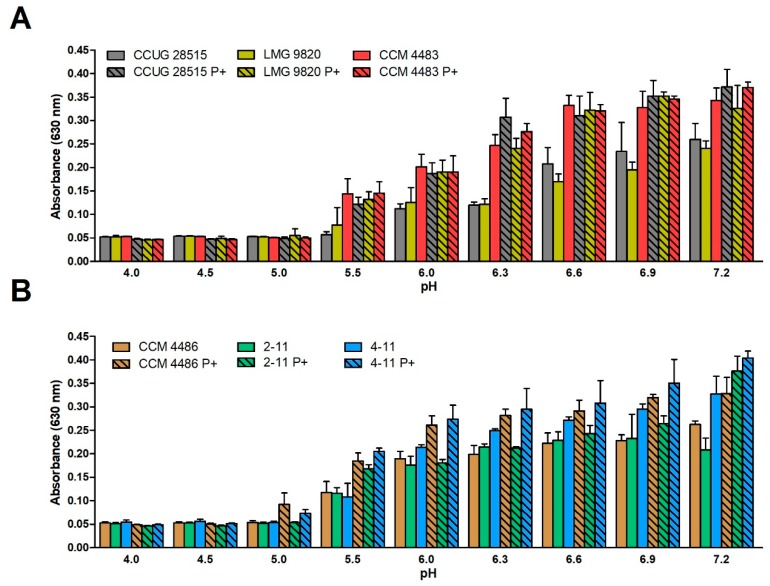
Before studying the inhibitory effect of 10-HDA against P. larvae, we examined suitable conditions for bacterial cultivation. Firstly, we started to cultivate the bacterium in MYPGP medium which is most frequently used for this purpose. The preliminary tests showed that adding 10-HDA (weak acid) to the medium having low buffering capacity caused the not negligible reduction in the pH of the medium dependent on FA concentration. Therefore, MYPGP medium was adjusted with 50 mM PIPES to increase its buffering capacity and the modified medium was designated as MYPGP+P medium. Then, we examined the effect of pH on the growth of six selected strains of P. larvae carrying two environmentally significant ERIC genotypes in both media, MYPGP and MYPGP+P, to determine the effect of PIPES on bacterial growth. The examined pH range included pH values at which this bacterium can occur in its natural environment, beginning from pH = 4 in LJ to pH = 7.2 used ordinarily for laboratory cultivations. The results of these tests are presented in Figure 1.
Figure 1.
Growth of selected Paenibacillus larvae strains representing two environmentally significant Enterobacterial Repetitive Intergenic Consensus (ERIC) genotypes at various pH in MYPGP media. (A) ERIC I genotype, (B) ERIC II genotype. P+ indicates the growth of bacteria in MYPGP+P medium containing the buffering compound PIPES. Data are expressed as mean values with standard deviations.
It was found that the bacterial strains did not grow at pH = 4 or pH = 4.5, and the most of them did not grow at pH = 5. They started to grow at pH = 5.5 and the level of growth rose with increasing pH to pH = 6.9–7.2. There were no substantial growth differences between the strains possessing the ERIC I and ERIC II genotypes. Individual strains showed partial differences in the bacterial mass grown up during the cultivation. The presence of 50 mM PIPES in the medium had a generally positive effect on the bacterial growth of all strains.
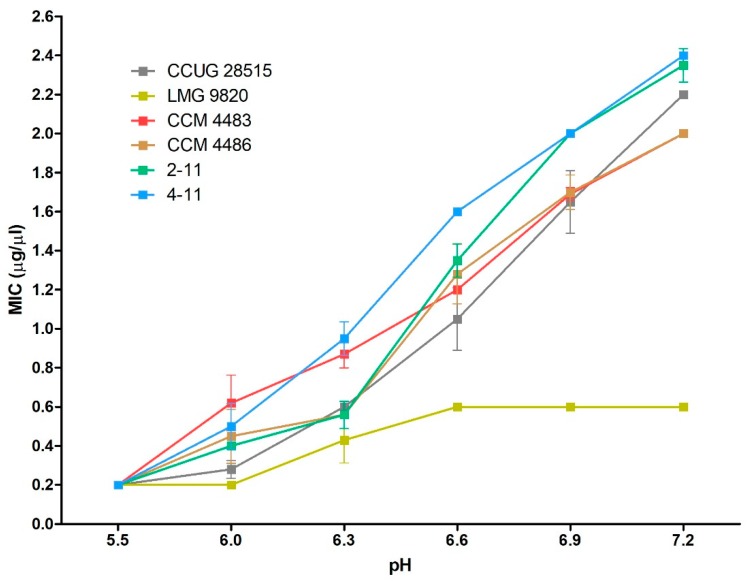
2.3. Efficacy of 10-HDA Against P. larvae at Various pH Levels
The efficacy of 10-HDA against vegetative cells of six bacterial strains (used previously in the growth tests) was estimated by the determination of minimum inhibitory concentrations (MICs) in media with the pH between 5.5 and 7.2. The results are shown in Figure 2. 10-HDA had the strongest inhibitory effect on the growth of P. larvae at pH = 5.5. The inhibition at this pH occurred at the same concentration of 0.2 μg/μL of 10-HDA in all tested strains. With increasing pH, the susceptibility of P. larvae to FA continuously decreased in five strains and reached MIC values greater than 2 μg/μL at pH = 7.2. The strains with the ERIC I and ERIC II genotypes showed a similar dependence of susceptibility on pH, except for one strain with the ERIC I genotype, LMG 9820. This strain did not show pH-dependent susceptibility between pH = 5.5–6 and 6.6–7.2. We assume that this could be connected with the high susceptibility of this strain to 10-HDA.
Figure 2.
Effect of pH on the antibacterial efficacy of 10-HDA in selected P. larvae strains. Minimum inhibitory concentration (MIC) values are graphically depicted to compare pH-dependent susceptibility profiles of individual strains to 10-HDA. Most strains of the ERIC I and ERIC II genotypes showed similar profiles of pH-dependent susceptibility to 10-HDA. Data are expressed as mean values with standard deviations.
2.4. Susceptibility of Culture Strains, Field Isolates, Vegetative Cells and Spores of P. larvae to 10-HDA
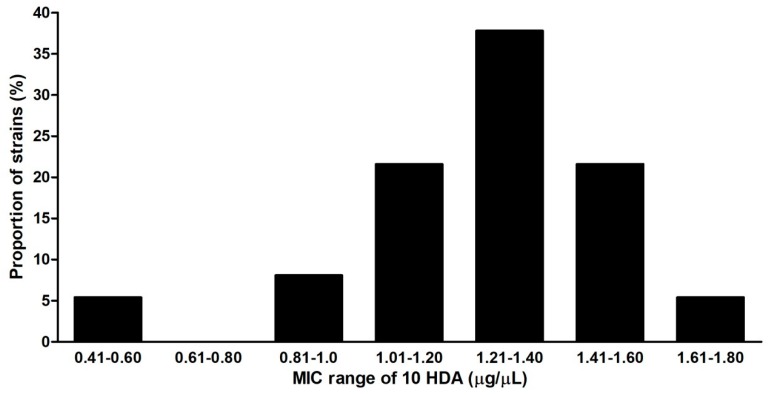
The susceptibility of seven reference strains and 33 Slovak field isolates to 10-HDA was compared by the determination of MICs at pH = 6.6 (assumed to be the highest pH that may occur in the midguts of young larvae). The determined MIC values ranged between 0.6 and 2.8 μg/μL (Table 1). The strains with ERIC I and ERIC II genotypes were more susceptible to 10-HDA (MICs between 0.6–1.2 and 0.6–1.78 μg/μL, respectively) than those with ERIC III and IV genotypes (MICs 2.6 and 2.8 μg/μL). The results showed that there was relatively high variation in the susceptibility of P. larvae strains to 10-HDA considering all ERIC genotypes. The maximal difference in MIC values was 4.67-fold. The strains with ERIC I and ERIC II genotypes showed less variation in susceptibility as the maximal MIC difference was 2.97-fold. Among the tested strains, distinct MICs occurred in various frequencies. The most representative strains were those having MICs between 1.21 and 1.4 μg/μL (Figure 3). The susceptibility of vegetative cells and germinating spores to 10-HDA were compared with strains used in the previous tests. The germinating spores of most strains had MIC values of 10-HDA lower than vegetative cells (Table 2). This result suggests that germinating spores are generally more susceptible to 10-HDA than vegetative cells.
Figure 3.
Frequency of P. larvae strains having different MICs in a group of strains with ERIC I and ERIC II genotypes.
Table 2.
Antibacterial efficacy of 10-HDA on vegetative cells and spores of P. larvae.
| Strain | ERIC Genotype | MIC (μg/μL) * | |
|---|---|---|---|
| Vegetative Cells | Spores | ||
| CCUG 28515 | I | 1.05 ± 0.16 | 1.05 ± 0.16 |
| LMG 9820 | I | 0.60 ± 0.00 | 0.60 ± 0.00 |
| CCM 4483 | I | 1.20 ± 0.00 | 0.98 ± 0.04 |
| CCM 4486 | II | 1.28 ± 0.15 | 0.60 ± 0.00 |
| PL 2-11 | II | 1.35 ± 0.09 | 1.18 ± 0.04 |
| PL 4-11 | II | 1.60 ± 0.00 | 1.20 ± 0.00 |
* MIC determined at pH = 6.6.
3. Discussion
Honeybee colonies differ in their resistance to AFB, in terms of their ability to tolerate certain levels of contamination in bees and food stores with P. larvae spores without clinical signs of disease [2,42,43]. Several mechanisms are assumed to act in larvae and colonies influencing the resistance of colonies against AFB. These mechanisms are: (1) The ability of nurse bees to remove spores from contaminated honey by honey-stopper [44,45], (2) changes in the midgut peritrophic membrane with increasing age of the larva connected with the lower ability of the pathogen to penetrate into the hemocoel [46,47], (3) the presence of some microbial species in the larval midgut that inhibit the propagation of P. larvae [48,49,50], (4) pathogen-induced immune responses including the expression of antimicrobial peptides [51,52,53] and other immune factors [54] in larvae, (5) the presence of unknown non-induced substance(s) in 2- to 4-day-old larvae inhibiting the growth of P. larvae [32,55], (6) the hygienic behavior of honeybees consisting in the detection and removal of diseased larvae from a colony before the pathogen reached the spore stage [11,15,19,56].
Besides these mechanisms, several studies have suggested that resistance of colonies to AFB could also be associated with larval food. Rose and Briggs [30] demonstrated that WJ collected from an AFB resistant line was more effective in inhibiting spore germination and in reducing the numbers of vegetative cells of P. larvae than the WJ from a susceptible line. Hornitzky [31] observed that RJ has bactericidal effect on vegetative cells of the pathogen within 5 min. The authors of these two studies suggested that the antibacterial effects could be mediated by 10-HDA which at the time was the only known antibacterial substance in RJ. Crailsheim and Riessberger-Gallé [32] found that water-ethanol extracts of RJ and WJ inhibited the in vitro growth of P. larvae; the RJ extract showed greater effectiveness than WJ extracts. Furthermore, it was revealed that the LJ peptide defensin1 has in vitro inhibitory activity against P. larvae [33,34,35]. It was documented that the content of the peptide varies in RJ and WJ samples collected from different colonies as well as in RJ samples collected in one colony [57]. Moreover, Bíliková et al. [25] identified the protein apalbumin2a in RJ, which also inhibited the growth of P. larvae in vitro. These findings suggested that defensin1 and apalbumin2a could be additional substances contributing to the antipathogenic activity of larval food and AFB resistance.
In the present work, we showed for the first time that 10-HDA inhibits the growth of vegetative cells and germinated spores of P. larvae. We demonstrated that the antibacterial efficacy of the 10-HDA depends on pH and the susceptibility of individual strains of the pathogen to FA. The strains possessing ERIC I and ERIC II genotypes causing AFB [8,58] seem to have similar susceptibilities to FA. The strains with ERIC III and IV genotypes isolated from colonies in the past and currently available only from culture collections were less susceptible to FA than the ERIC I and ERIC II strains. The molecular reasons for these differences among distinct ERIC genotypes are unknown.
The growth of P. larvae and the efficacy of 10-HDA against the different strains of the bacterium were examined here under conditions that partially resemble those occurring during the development of young larvae. The bacterial incubations were done at 35 °C, which is the common temperature in hives and at pH levels assumed to occur in larval food and the midguts of young larvae (pH = 4–6.6). This enabled us to make some conclusions regarding the action of P. larvae in the natural honeybee system. We did not observe the growth of P. larvae in MYPGP media at pH = 4 or pH = 4.5, and some strains did not grow at pH = 5.0. This suggests that pH itself has bacteriostatic and/or bactericidal effect on the vegetative cells of the bacterium. A lethal effect of saline solution at pH = 4 on vegetative cells within 20 min has been described by Hornitzky [31]. However, our results suggest that the pH range with a detrimental effect on bacteria is higher than pH = 4.
The efficacy of 10-HDA against most of the tested bacterial strains was substantially affected by the pH. The inhibitory potency of FA increased when the pH decreased from 7.2 to 5.5. This suggests that the un-ionised molecules of FA are more antibacterial effective than ionised molecules. This phenomenon seems to be typical for medium-chain FAs because similar pH-dependent potency against some bacteria was observed at caprylic (C8:0), capric (C10:0) and lauric (C12:0) acids [59,60]. The fact that pH has a strong effect on the inhibitory activity of 10-HDA is of great importance due to its possible impact on the ability of this FA to inhibit the growth of pathogen cells in the midguts of young larvae. It is likely that the midgut environment of larvae can have variable pH. The source of pH variations could be genetic factors, the age of the larvae and differences in the acidity of larval food consumed by individual larvae. Chauvin [61] reported that the larval midgut is at pH = 6.8, Bailey and Ball [1] mentioned that the pH in larval intestine is about 6.6 and Wardell [62] stated that the pH is 5–5.5. It seems that the pH in the midgut of young larvae consuming RJ or WJ may have lower values and shows larger variations in values than in older larvae. The reason is that the LJ may show considerable differences in pH and acidity; in RJ, the pH can vary between 3.4 and 4.5 and the acidity (volume of 0.1 N NaOH in mL needed for the adjustment of 1 g of jelly to pH = 7.0) can be between 3.0 and 6.0 [40]. Thus, it is possible consider that 10-HDA at the same concentrations will have greater inhibitory activity in the midguts of young larvae consuming more acidic jellies than in those consuming weakly acidic ones. Differences in inhibitory activities could be as much as 3–8 times between pH = 5.5 and 6.6 according to our results.
Current knowledge about P. larvae pathogenesis [3,6,8] suggests that the inhibition of bacterial growth in the larval midgut could forestall the invasion of bacterial cells into larval hemocoel and enable individual larvae to survive the infection. The determination of the MICs of 10-HDA for 37 epidemiologically significant P. larvae strains having ERIC I and ERIC II genotypes allowed us to think about the possible antipathogenic action of this FA in midguts of individual larvae. The determined MICs (0.6–1.78 μg/μL) can be considered as concentrations of 10-HDA molecules that should inhibit the multiplication of vegetative cells of the pathogen in the midguts of young larvae at pH = 6.6 (pH used for the MIC determination). The question is whether 10-HDA reaches such inhibitory concentrations in the larval midguts. The contents of 10-HDA have been analyzed in numerous RJ samples of various geographic and hive origins and were found to vary between 0.75% and 6.4% [63,64,65,66,67,68,69,70,71,72]. These percentages correspond to 7.5–64 μg of 10-HDA per 1 mg of RJ. From these data, it can be deduced that RJ containing such amounts of 10-HDA could be diluted from 4.2 to 107 times in the larval midguts in order to exert the MIC within ranges 0.6 and 1.78 μg/μL. The given dilution data (its high values) indicate that the MICs may be theoretically reached in larvae consuming food with low amounts of 10-HDA in cases when infections are induced by more susceptible P. larvae strains. Nevertheless, we suppose that the real antipathogenic effect of 10-HDA in the midguts of larvae may be much stronger than indicated dilution data. Several factors can have a positive effect on the inhibitory activities of FA in larvae. Firstly, the inhibitory effect can be multiplied at lower pH assumed to occur in the midguts of young larvae (documented here). Secondly, the inhibitory effect is generally higher against germinating spores (the primary event at pathogen’s infection) than against vegetative cells (also documented here). Thirdly, the inhibitory activity of 10-HDA may be modified by the inhibitory activity of additional proteinous and lipidic antibacterial substances of LJ [33,34,69]. It is likely that the combined action of more inhibitory substances will exhibit much stronger antipathogenic activity in larval midguts than 10-HDA itself. Fourthly, we suppose that the antipathogenic activity of LJ is modified by digestion in larval midguts. We believe that the hydrolytic processes acting during digestion increase the concentrations of free medium-chain FA molecules in the midgut in comparison with their concentrations present in LJ. The free molecules can arise by the hydrolysis of FA monoesters and diesters which together bind molecules of 10-HDA and other abundant medium-chain FAs. The presence of such esters in RJ was revealed by Noda et al. [73]. Thus, the inhibitory activity of larval food processed in midguts might be higher than the inhibitory activity of the consumed natural LJ. The observations of Crailsheim and Riessberger-Gallé [32] support this scenario. They found that the water-ethanol extracts of 2–4 days old larvae were more active than extracts prepared from RJ and WJ samples obtained from the same colonies (extracted LJ samples had the same weights as the larvae). They explained this by the presence of unknown antibacterial substance(s) produced by the larvae. Our scenario represents a possible other explanation or, at least, suggests that hydrolytic processing of the larval foods in extracted larvae contributed to the higher activity of larval extracts. Besides these factors with positive effects on the inhibitory activity of 10-HDA in larval midguts, it is necessary to mention that WJ generally contain lower portions of lipid fraction [74] containing FAs and show lower activities against P. larvae than RJ [32]. However, at present there is little information about variations of 10-HDA contents in WJs within a colony and among colonies [63,75]. Taken together, these findings suggest that 10-HDA participates in determining the anti-P. larvae activity of larval food in the midguts of young larvae. The final inhibitory effects of 10-HDA in individual larvae might depend on amounts of free molecules of 10-HDA occurring during digestion of RJ and WJ in the midgut, on the effects of other factors supporting the inhibitory activity of 10-HDA (pH and the action of other antibacterial substances of LJ) and finally also on the susceptibility of P. larvae strains infecting the larvae to FA.
An interesting finding in this study concerns the ERIC genotype characteristics of the Slovak P. larvae isolates. All 33 isolates collected over a large interval of 15 years had the ERIC II genotype. This result is surprising in the context of studies performed in several other European countries. These studies have shown that the ERIC II genotype occurs together with the ERIC I genotype in all these countries [10,58]. In Austria, a neighboring country to Slovakia, a prevalence of 58% for the ERIC I genotype among isolates has been reported [76]. This finding could reflect the specific evolution of P. larvae in Slovakia in the past and/or it could be associated with Slovak regulations that prohibited the import of foreign honeybee subspecies into the country and this could prevent the dissemination of P. larvae strains with ERIC I genotype in Slovakia. However, more data associated with our findings are needed in order to validate these explanations.
4. Materials and Methods
4.1. Fatty Acid
10-HDA of 98% purity was purchased from AK Scientific, Inc. (Union City, CA, USA). The crystalline powder of 10-HDA was stored at −20 °C. Methanolic stock solutions of 10-HDA were prepared fresh before each experiment.
4.2. P. larvae Strains
Six reference strains of P. larvae (CCUG 28515, LMG 9820, CCM 4483, CCM 4486, CCM 38 and CCM 39) were purchased from different culture collections. The 33 field isolates were obtained from infected material of AFB positive hives (ground stocks or decomposed larval samples) from apiaries located in different regions of Slovakia in 1999, 2009, 2011 and 2014 (Table 1). All isolates were verified to be P. larvae by the specific amplification of 16S rDNA using the specific primers PL1 and PL2 as described by Dobbelaere et al. [14].
4.3. ERIC-PCR Genotyping
The ERIC genotypes of P. larvae strains were determined by specific repetitive element PCR. The PCR reactions (20 μL) contained 2x HotStartTaq Master Mix (Qiagen, Venlo, The Netherlands), 1 μM primers ERIC1R 5′-ATGTAAGCTCCTGGGGATTCAC-3′ and ERIC2 5′-AAGTAAGTGACTGGGGTGAGCG-3′ [77] and 2 μL of DNA solution. The amplification conditions were the same as described Genersch et al. [10].
4.4. Cultivation of P. larvae
Vegetative cells were cultivated in two growth media, each prepared in several variants at different pH levels. One medium was MYPGP [78]; the other designated as MYPGP+P was a modified version of MYPGP containing 50 mM PIPES (Sigma-Aldrich, St. Louis, MI, USA) as a pH buffering compound. The media with different pH values were freshly prepared for each experiment. The bacterial cultivations were performed at 35 °C (the ordinary temperature in bee hives) as follows. Overnight cultures of the examined strains were prepared in standard MYPGP (pH = 7.2) on an orbital shaker and then diluted in media with different pH levels to a final concentration of 1 × 105 cells/mL. Then, 150 μL aliquots of diluted bacterial culture were transferred into the wells of a sterile 96-well polystyrene microplates in triplicate and incubated stationarily for 43 h, whereby the plates were shaken on microplate shaker (Biofil, Merci, Paris, France) at 1200 rpm for 5 min before and after 18 h and 43 h of cultivation. Bacterial growth was determined spectrophotometrically by measuring the absorbance at 630 nm using a plate reader. All cultivations were repeated three times.
4.5. Preparation of Spores
P. larvae cells of selected strains (~1 000 CFU) were cultivated on MYPGP agar plates (pH = 7.2) at 35 °C for 10 days. Grown bacterial mass was removed from the surface of agar plates, homogenised and washed three times by vortexing and centrifugation at 15,000 g for 15 min in 15 mL of sterile deionised water in 15 mL centrifuge tubes (Labcon, Petaluma, CA, USA). Pelleted material was suspended in 2 mL of sterile deionised water and vegetative cells were killed by heating at 65 °C for 15 min. The spores were purified from the obtained suspensions by centrifugation on a Nycodenz (Axis-Shield PoC, Dundee, Scotland) gradient. Briefly, 2 mL of the suspensions were mixed with 11 mL of 30% sterile Nycodenz water solution in 15 mL centrifuge tubes. The mixtures were frozen twice at −20 °C and thawed at laboratory temperature to form gradients and then centrifuged at 15,000 g for 60 min. The content of the tubes was discarded and spores adhering to the tubes, visible as small pellets, were homogenised and washed three times with sterile deionised water as described above. Finally, 2 mL spore stock cultures were prepared in sterile deionised water and stored at 4 °C. The numbers of vital spores (CFU) in stock cultures were determined on standard MYPGP agar plates supplemented with 3 mM uric acid and L-tyrosine [79] after cultivation at 35 °C for 5 days.
4.6. Minimum Inhibitory Concentration Assay
MIC of 10-HDA was determined by the broth microdilution method using a modified MYPGP+P medium at variable pH. Cultures of bacterial vegetative cells or vital spores were prepared in the corresponding media at a concentration of 1 × 105 cells or spores per 1 mL and aliquots of 147 μL were transferred into a sterile 96-well polystyrene microplate. A total of 3 μL from several 10-HDA stock solutions in methanol were added into the wells to prepare cultures with the concentrations of the substance in the range between 0.2 and 3 μg/μL with increments of 0.2 μg/μL. Positive and negative growth controls contained 3 μL of methanol. The microplates with P. larvae vegetative cells were incubated stationarily at 35 °C for 43 h, whereby they were shaken for 5 min on a rotary microplate shaker before and after 18 h and 43 h of cultivation. The spores were stationarily cultivated for 115 h and shaking was performed before and after 72, 96 and 115 h of cultivation. The bacterial growth inhibition was determined by monitoring the absorbance at 630 nm. The MIC was defined as the lowest concentration of 10-HDA inhibiting 100% of bacterial growth. All tests were performed in triplicate and were repeated three times. Finally, the mean of MIC values and standard deviations were determined.
5. Conclusions
This work expands our knowledge about substances contained in LJ that could inhibit the propagation of P. larvae in the midguts of individual larvae. Our results suggest that 10-HDA could play a significant role among these substances. We revealed that the inhibitory effects of 10-HDA against this pathogen are stronger at more acidic pH values thought to occur in the midguts of young larvae. Moreover, we found that environmentally important P. larvae strains (their vegetative form and also germinating spores) exhibited considerable differences in susceptibility to 10-HDA. We believe that these findings support our idea that larval food could play an important role in determining the resistance of individual larvae against P. larvae. We infer that variations in the contents of free molecules of 10-HDA and other active substances in LJ (not yet clearly identified) and in their inhibitory efficacies against different P. larvae strains as well as variations in the acidity and pH of LJ in larval midguts on the level of individual larvae within a colony and in different colonies can contribute to differences in colony resistance to AFB. However, further in vivo research is needed to verify these statements. Future in vivo experiments should evaluate the inhibitory effects of larval foods with different compositions possessing various combinations of the parameters important for anti-P. larvae activity in larval midguts. Finally, we believe that our data might be useful for the prospective breeding of new honeybee lines with improved larvae and colony resistance to AFB.
Supplementary Materials
The following are available online at http://www.mdpi.com/1420-3049/23/12/3236/s1, Figure S1: ERIC-PCR genotyping of selected P. larvae strains.
Author Contributions
Conceptualization, J.K.; Methodology, M.Š, and J.K.; Formal Analysis, M.Š, J.M.; Investigation, M.Š., M.L., L.K.; Resources, A.M., J.K., M.Š; Writing–Original Draft Preparation, J.K., M.Š.; Writing–Review & Editing, J.K., M.Š., J.M., M.L.; Visualization, M.Š., J.M., J.K.; Funding Acquisition, J.K.
Funding
This work was supported by the Scientific grant Agency of the Ministry of Education of the Slovak Republic and the Slovak Academy of Sciences, the project VEGA 2/0175/15.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Not available.
References
- 1.Bailey L., Ball B.V. Honey Bee Pathology. Academic Press; London, UK: 1991. pp. 53–62. [Google Scholar]
- 2.Hansen H., Brødsgaard C.J. American foulbrood: A review of its biology, diagnosis and control. Bee World. 1999;80:5–23. doi: 10.1080/0005772X.1999.11099415. [DOI] [Google Scholar]
- 3.Yue D., Nordhoff M., Wieler L.H., Genersch E. Fluorescence in situ hybridization (FISH) analysis of the interactions between honeybee larvae and Paenibacillus larvae, the causative agent of American foulbrood of honeybees (Apis mellifera) Environ. Microbiol. 2008;10:1612–1620. doi: 10.1111/j.1462-2920.2008.01579.x. [DOI] [PubMed] [Google Scholar]
- 4.Antúnez K., Anido M., Schlapp G., Evans J.D., Zunino P. Characterization of secreted proteases of Paenibacillus larvae, potential virulence factors involved in honeybee larval infection. J. Invertebr. Pathol. 2009;102:129–132. doi: 10.1016/j.jip.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Gonzalez E., Poppinga L., Fünfhaus A., Hertlein G., Hedtke K., Jakubowska A., Genersch E. Paenibacillus larvae chitin-degrading protein PlCBP49 is a key virulence factor in American foulbrood of honey bees. PLoS Path. 2014;10:e1004284. doi: 10.1371/journal.ppat.1004284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebeling J., Knispel H., Hertlein G., Fünfhaus A., Genersch E. Biology of Paenibacillus larvae, a deadly pathogen of honey bee larvae. Appl. Microbiol. Biotechnol. 2016;100:7387–7395. doi: 10.1007/s00253-016-7716-0. [DOI] [PubMed] [Google Scholar]
- 7.Lindström A., Korpela S., Fries I. The distribution of Paenibacillus larvae spores in adult bees and honey and larval mortality, following the addition of American foulbrood disease brood or spore-contaminated honey in honey bee (Apis mellifera) colonies. J. Invertebr. Pathol. 2008;99:82–86. doi: 10.1016/j.jip.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Genersch E. American Foulbrood in honeybees and its causative agent, Paenibacillus larvae. J. Invert. Pathol. 2010;87:87–97. doi: 10.1016/j.jip.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 9.Genersch E., Ashiralieva A., Fries I. Strain- and genotype-specific differences in virulence of Paenibacillus larvae subsp. larvae, the causative agent of American foulbrood disease in honey bees. Appl. Environ. Microbiol. 2005;71:7551–7555. doi: 10.1128/AEM.71.11.7551-7555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genersch E., Forsgren E., Pentikäinen J., Ashiralieva A., Rauch S., Kilwinski J., Fries I. Reclassification of Paenibacillus larvae subsp. pulvifaciens and Paenibacillus larvae subsp. larvae as Paenibacillus larvae without subspecies differentiation. Int. J. Syst. Evol. Microbiol. 2006;56:501–511. doi: 10.1099/ijs.0.63928-0. [DOI] [PubMed] [Google Scholar]
- 11.Spivak M., Gilliam M. Hygienic behaviour of honey bees and its application for control of brood diseases and varroa. Part I. Hygienic behaviour and resistance to American foulbrood. Bee World. 1998;79:124–134. doi: 10.1080/0005772X.1998.11099394. [DOI] [Google Scholar]
- 12.Rauch S., Ashiralieva A., Hedtke K., Genersch E. Negative correlation between individual-insect-level virulence and colony-level virulence of Paenibacillus larvae, the ethiological agent of American foulbrood of honeybees. Appl. Environ. Microbiol. 2009;75:3344–3347. doi: 10.1128/AEM.02839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alippi A.M., Albo G.N., Leniz D., Rivera I., Zanelli M., Roca A.E. Comparative study of tylosin, erythromycin and oxytetracycline to control of American foulbrood in honey bees. J. Apic. Res. 1999;38:149–158. doi: 10.1080/00218839.1999.11101005. [DOI] [Google Scholar]
- 14.Dobbelaere W., de Graaf D.C., Peeters J.E., Jacobs F.J. Development of a fast and reliable diagnostic method for American foulbrood disease (Paenibacillus larvae subsp. larvae) using a 16S rDNA gene based PCR. Apidologie. 2001;32:363–370. doi: 10.1051/apido:2001136. [DOI] [Google Scholar]
- 15.Wilson-Rich N., Spivak M., Fefferman N.H., Starks P.T. Genetic, individual, and group facilitation of disease resistance in insect societies. Annu. Rev. Entomol. 2009;54:405–423. doi: 10.1146/annurev.ento.53.103106.093301. [DOI] [PubMed] [Google Scholar]
- 16.Buczek K. Range of susceptibility of Paenibacillus larvae to antibacterial compounds. Med. Weter. 2011;67:87–90. [Google Scholar]
- 17.Evans J.D., Pettis J.S. Colony-level impacts of immune responsiveness in honey bees, Apis mellifera. Evolution. 2005;59:2270–2274. doi: 10.1111/j.0014-3820.2005.tb00935.x. [DOI] [PubMed] [Google Scholar]
- 18.Evans J.D., Spivak M. Socialized Medicine: Individual and communal disease barriers in honey bees. J. Invertebr. Pathol. 2010;103:62–72. doi: 10.1016/j.jip.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 19.Spivak M., Reuter G.D. Resistance to American foulbrood disease by honey bee colonies Apis mellifera bred for hygienic behavior. Apidologie. 2001;32:555–565. doi: 10.1051/apido:2001103. [DOI] [Google Scholar]
- 20.Pérez-Sato J.A., Châlin N., Martin S.J., Hughes W.O.H., Ratnieks F.L.W. Multi-level selection for hygienic behaviour in honeybee. Heredity. 2009;102:609–615. doi: 10.1038/hdy.2009.20. [DOI] [PubMed] [Google Scholar]
- 21.Brouwers E.V.M., Ebert R., Beetsma J. Behavioural and physiological aspects of nurse bees in relation to the composition of larval food during caste differentiation in the honeybee. J. Apic. Res. 1987;26:11–23. doi: 10.1080/00218839.1987.11100729. [DOI] [Google Scholar]
- 22.Fujiwara S., Imai J., Fujiwara M., Yaeshima T., Kawashima T., Kobayashi K. A potent antibacterial protein in royal jelly. J. Biol. Chem. 1990;265:11333–11337. [PubMed] [Google Scholar]
- 23.Fontana R., Mendes M.A., de Souza B.M., Konno K., César L.M.M., Malaspina O., Palma M.S. Jelleines: A family of antibacterial peptides from the royal jelly of honeybees (Apis mellifera) Peptides. 2004;25:919–928. doi: 10.1016/j.peptides.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 24.Klaudiny J., Albert Š., Bachanová K., Kopernický J., Šimúth J. Two structurally different defensin genes, one of them encoding a novel defensin isoform, are expressed in honeybee Apis mellifera. Insect Biochem. Mol. Biol. 2005;35:11–22. doi: 10.1016/j.ibmb.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Bíliková K., Mirgorodskaya E., Bukovská G., Gobom J., Lehrach H., Šimúth J. Towards functional proteomics of minority component of honeybee royal jelly: The effect of post-translational modifications on the antimicrobial activity of apalbumin2. Proteomics. 2009;9:2131–2138. doi: 10.1002/pmic.200800705. [DOI] [PubMed] [Google Scholar]
- 26.Blum M.S., Novak A.F., Taber S. 10-hydroxy-Δ2-decenoic acid, an antibiotic found in royal jelly. Science. 1959;130:452–453. doi: 10.1126/science.130.3373.452. [DOI] [PubMed] [Google Scholar]
- 27.Yatsunami K., Echigo T. Antibacterial action of royal jelly. Bull. Fac. Agr. Tamagawa Univ. 1985;25:13–22. [Google Scholar]
- 28.Melliou E., Chinou I. Chemistry and bioactivity of royal jelly from Greece. J. Agric. Food Chem. 2005;53:8987–8992. doi: 10.1021/jf051550p. [DOI] [PubMed] [Google Scholar]
- 29.Alreshoodi F., Sultanbawa Y. Antimicrobial activity of royal jelly. Anti-Infective Agents. 2015;13:50–59. doi: 10.2174/2211352513666150318234430. [DOI] [Google Scholar]
- 30.Rose R.I., Briggs J.D. Resistance to American foulbrood in honey bees. Effects of honey bee larval food on the growth and viability of Bacillus larvae. J. Invertebr. Pathol. 1969;13:74–80. doi: 10.1016/0022-2011(69)90240-7. [DOI] [Google Scholar]
- 31.Hornitzky M.A.Z. The pathogenicity of Paenibacillus larvae subsp. larvae spores and vegetative cells to honey bee (Apis mellifera) colonies and their susceptibility to royal jelly. J. Apic. Res. 1998;37:267–271. doi: 10.1080/00218839.1998.11100982. [DOI] [Google Scholar]
- 32.Crailsheim K., Riessberger-Gallé U. Honey bee age-dependent resistance against American foulbrood. Apidologie. 2001;32:91–103. doi: 10.1051/apido:2001114. [DOI] [Google Scholar]
- 33.Bíliková K., Gusui W., Šimúth J. Isolation of a peptide fraction from honeybee royal jelly as a potential antifoulbrood factor. Apidologie. 2001;32:275–283. doi: 10.1051/apido:2001129. [DOI] [Google Scholar]
- 34.Bachanová K., Klaudiny J., Kopernický J., Šimúth J. Identification of honeybee peptide active against Paenibacillus larvae larvae through bacterial growth-inhibition assay on polyacrylamide gel. Apidologie. 2002;33:259–269. doi: 10.1051/apido:2002015. [DOI] [Google Scholar]
- 35.Bíliková K., Huang S.C., Lin I.P., Simuth J., Peng Ch.Ch. Structure and antimicrobial activity relationship of royalisin, an antimicrobial peptide from royal jelly of Apis mellifera. Peptides. 2015;68:190–196. doi: 10.1016/j.peptides.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Lercker G., Capella P., Conte L.S., Ruini F., Giordani G. Components of royal jelly: I. Identification of the organic acids. Lipids. 1981;16:912–919. doi: 10.1007/BF02534997. [DOI] [Google Scholar]
- 37.Sugiyama T., Takahashi K., Mori H. Royal jelly acid, 10-hydroxy-trans-2-decenoic acid, as a modulator of the innate immune responses. Endocr. Metab. Immune Disord. Drug Targets. 2012;12:368–376. doi: 10.2174/187153012803832530. [DOI] [PubMed] [Google Scholar]
- 38.Li X., Huang C., Xue Y. Contribution of lipids in honeybee (Apis mellifera) royal jelly to health. J. Med. Food. 2013;16:96–102. doi: 10.1089/jmf.2012.2425. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y.F., Wang K., Zhang Y.Z., Zheng Y.F., Hu F.L. In vitro anti-inflammatory effects of three fatty acids from royal jelly. Mediators Inflamm. 2016;2016 doi: 10.1155/2016/3583684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabatini A.G., Marcazzan G.L., Caboni M.F., Bogdanov S., de Almeida-Muradian L.B. Quality and standardization of royal jelly. J. ApiProd. Apimed. Sci. 2009;1:16–21. doi: 10.3896/IBRA.4.01.1.04. [DOI] [Google Scholar]
- 41.Garcia M.C., Finola M.S., Marioli J.M. Bioassay direct identification of royal jelly’s active compounds against the growth of bacteria capable of infecting cutaneous wounds. Adv. Microbiol. 2013;3:138–144. doi: 10.4236/aim.2013.32022. [DOI] [Google Scholar]
- 42.Alippi A.M., Reynaldi F.J., López A.C., Aguilar O.M., de Giusti M.R. Molecular epidemiology of Paenibacillus larvae larvae and incidence of American foulbrood in Argentinean honeys from Buenos Aires province. J. Apic. Res. 2004;43:135–143. doi: 10.1080/00218839.2004.11101124. [DOI] [Google Scholar]
- 43.Antunez K., D’Alessandro B., Piccini C., Corbella E., Zunino P. Paenibacillus larvae larvae spores in honey samples from Uruguay; a nationwide survey. J. Invertebr. Pathol. 2004;86:56–58. doi: 10.1016/j.jip.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 44.Sturtevant A.P., Revell I.L. Reduction of Bacillus larvae spores in liquid food of honeybees by action of the honey stopper, and its relation to the development of American foulbrood. J. Econ. Entomol. 1953;46:855–860. doi: 10.1093/jee/46.5.855. [DOI] [Google Scholar]
- 45.Thompson V.C., Rothenbuhler W.C. Resistance to American foulbrood in honey bees. I. Differential protection of larvae by adults of different genetic lines. J. Econ. Entomol. 1957;50:731–737. doi: 10.1093/jee/50.6.731. [DOI] [Google Scholar]
- 46.Bamrick J.F., Rothenbuhler W.C. Resistance to American foulbrood in honey bees. IV. The relationship between larval age at inoculation and mortality in a resistant and in susceptible line. J. Insect Pathol. 1961;3:381–390. [Google Scholar]
- 47.Davidson E.W. Ultrastructure of American foulbrood disease pathogenesis in larvae of the worker honey bee, Apis mellifera. J. Invertebr. Pathol. 1973;21:53–61. doi: 10.1016/0022-2011(73)90113-4. [DOI] [Google Scholar]
- 48.Rinderer T.E., Rothenbuhler W.C., Gochnauer T.A. The influence of pollen on the susceptibility honey bee larvae to Bacillus larvae. J. Invertebr. Pathol. 1974;23:347–350. doi: 10.1016/0022-2011(74)90100-1. [DOI] [PubMed] [Google Scholar]
- 49.Reiche R., Martin K., Mollmann J., Hentschel E. Beitrag zur Klärung der Resistenz der Bienenlarven gegen den Erreger der Amerikanischen Faulbrut Paenibacillus (voher Bacillus) larvae. Apidologie. 1996;27:296–297. [Google Scholar]
- 50.Evans J.D., Armstrong T.N. Inhibition of the American foulbrood bacterium, Paenibacillus larvae, by bacteria isolated form honey bees. J. Apic. Res. 2005;44:168–171. doi: 10.1080/00218839.2005.11101173. [DOI] [Google Scholar]
- 51.Evans J.D. Transcriptional immune responses by honey bee larvae during invasion by the bacterial pathogen, Paenibacillus larvae. J. Invertebr. Pathol. 2004;85:105–111. doi: 10.1016/j.jip.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 52.Evans J.D., Aronstein K., Chen Y.P., Hetru C., Imler J.L., Jiang H., Kanost M., Thompson G.J., Zou Z., Hultmark D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006;15:645–656. doi: 10.1111/j.1365-2583.2006.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Decanini L.I., Collins A.M., Evans J.D. Variation and heritability in immune gene expression by diseased honeybees. J. Hered. 2007;98:195–201. doi: 10.1093/jhered/esm008. [DOI] [PubMed] [Google Scholar]
- 54.Chan Q.W.T., Melathopoulos A.P., Pernal S.F., Foster L.J. The innate immune and systemic response in honey bees to a bacterial pathogen, Paenibacillus larvae. BMC Genomics. 2009;10:387. doi: 10.1186/1471-2164-10-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wedenig M., Riessberger-Gallé U., Crailsheim K. A substance in honey bee larvae inhibits the growth of Paenibacillus larvae larvae. Apidologie. 2003;34:43–51. doi: 10.1051/apido:2002043. [DOI] [Google Scholar]
- 56.Rothenbuhler W.C. Behaviour genetics of nest cleaning in honeybees. I. Responses of four inbred lines to disease-killed brood. Anim. Behav. 1964;12:578–583. doi: 10.1016/0003-3472(64)90082-X. [DOI] [Google Scholar]
- 57.Klaudiny J., Bachanová K., Kohútová L., Dzúrová M., Kopernický J., Majtán J. Expression of larval jelly antimicrobial peptide defensin1 in Apis mellifera colonies. Biologia. 2012;67:200–211. doi: 10.2478/s11756-011-0153-8. [DOI] [Google Scholar]
- 58.Schäfer M.O., Genersch E., Fünfhaus A., Poppinga L., Formella N., Bettin B., Karger A. Rapid identification of differentially virulent genotypes of Paenibacillus larvae, the causative organism of American foulbrood of honey bees, by whole cell MALDI-TOF mass spectrometry. Vet. Microbiol. 2014;170:291–297. doi: 10.1016/j.vetmic.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 59.Sun C.Q., O’Connor Ch.J., Roberton A.M. Antibacterial actions of fatty acids and monoglycerides against Helicobacter pylori. FEMS Immunol. Microbiol. 2003;36:9–17. doi: 10.1016/S0928-8244(03)00008-7. [DOI] [PubMed] [Google Scholar]
- 60.Skřivanová E., Marounek M. Influence of pH on antimicrobial activity of organic acids against rabbit enteropathogenic strain of Escherichia coli. Folia Microbiol. 2007;52:70–72. doi: 10.1007/BF02932141. [DOI] [PubMed] [Google Scholar]
- 61.Chauvin R. Nutrition de I´abeille (Bee nutrition) Ann. Nutr. Aliment. 1962;16:41–63. [PubMed] [Google Scholar]
- 62.Wardell G. Honeybee nutrition and European foulbrood; Proceedings of the International Symposium of European Foulbrood; Quebec, Canada. 18–20 October 1981; pp. 111–126. [Google Scholar]
- 63.Bonvehí J.S., Jordá R.E. Studie über die mikrobiologische Qualität und bakteriostatische Aktivität des Weiselfuttersaft (Gelée Royale): Beeinflussung durch organische Säuren. Deutsche Lebensmittel-Rundschau. 1991;87:256–259. [Google Scholar]
- 64.Bloodworth B.C., Harn C.S., Hock C.T., Boon Y.O. Liquid chromatographic determination of trans-10-hydroxy-2-decenoic acid content of commercial products containing royal jelly. J. AOAC. Int. 1995;78:1019–1023. [PubMed] [Google Scholar]
- 65.Genc M., Aslan A. Determination of trans-10-hydroxy-2-decenoic acid content in pure royal jelly and royal jelly products by column liquid chromatography. J. Chromatogr. A. 1999;839:265–268. doi: 10.1016/S0021-9673(99)00151-X. [DOI] [PubMed] [Google Scholar]
- 66.Ferioli F., Marcazzan G.L., Caboni M.F. Determination of (E)-10-hydroxy-2-decenoic acid content in pure royal jelly: A comparison between a new CZE method and HPLC. J. Sep. Sci. 2007;30:1061–1069. doi: 10.1002/jssc.200600416. [DOI] [PubMed] [Google Scholar]
- 67.Garcia-Amoedo L.H., Almeida-Muradian L.B. Physicochemical composition of pure and adulterated royal jelly. Quim. Nova. 2007;30:257–259. doi: 10.1590/S0100-40422007000200002. [DOI] [Google Scholar]
- 68.Zheng H.Q., Hu F.L., Dietemann V. Changes in composition of royal jelly harvested at different times: Consequences for quality standards. Apidologie. 2010;42:39–47. doi: 10.1051/apido/2010033. [DOI] [Google Scholar]
- 69.Isidorov V.A., Bakier S., Grzech I. Gas chromatographic-mass spectrometric investigation of volatile and extractable compounds of crude royal jelly. J. Chromatogr. B. 2012;885:109–116. doi: 10.1016/j.jchromb.2011.12.025. [DOI] [PubMed] [Google Scholar]
- 70.Wytrychowski M., Chenavas S., Daniele G., Casabianca H., Batteau M., Guibert S., Brion B. Physicochemical characterization of French royal jelly: Comparison with commercial royal jellies and royal jellies produced through artificial bee-feeding. J. Food Compos. Anal. 2013;29:126–133. doi: 10.1016/j.jfca.2012.12.002. [DOI] [Google Scholar]
- 71.Kanelis D., Tananaki C., Liolios V., Dimou M., Goras G., Rodopoulou M.A., Karazafiris E., Thrasyvoulou A. A suggestion for royal jelly specifications. Arh.Hig. Rada.Toksikol. 2015;66 doi: 10.1515/aiht-2015-66-2651. [DOI] [PubMed] [Google Scholar]
- 72.Flanjak I., Jakovljevic M., Kenjeric D., Stokanovic M.C., Primorac L., Rajs B.B. Determination of (2E)-10-hydroxydec-2-enoic acid in Croatian royal jelly by high-performance liquid chromatography. Croat. J. Food Sci. Technol. 2017;9:152–157. doi: 10.17508/CJFST.2017.9.2.10. [DOI] [Google Scholar]
- 73.Noda N., Umebayashi K., Nakatani T., Miyahara K., Ishiyama K. Isolation and characterization of some hydroxy fatty and phosphoric acid esters of 10-hydroxy-2-decenoic acid from the royal jelly of honeybees (Apis mellifera) Lipids. 2005;40:833–838. doi: 10.1007/s11745-005-1445-6. [DOI] [PubMed] [Google Scholar]
- 74.Jung-Hoffmann I. Die Determination von Königin und Arbeiterin der Honigbiene. Zeitschrift für Bienenforschung. 1966;8:296–322. [Google Scholar]
- 75.Wang Y., Ma L., Zhang W., Cui X., Wang H., Xu B. Comparison of the nutrient composition of royal jelly and worker jelly of honey bees (Apis mellifera) Apidologie. 2016;47:48–56. doi: 10.1007/s13592-015-0374-x. [DOI] [Google Scholar]
- 76.Loncaric I., Derakhshifar I., Oberlerchner J.T., Köglberger H., Moosbeckhofer R. Genetic diversity among isolates of Paenibacillus larvae from Austria. J. Invertebr. Pathol. 2009;100:44–46. doi: 10.1016/j.jip.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 77.Versalovic J., Schneider M., De Bruijn F.J., Lupski J.R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 1994;5:25–40. [Google Scholar]
- 78.Dingman D.W., Stahly D.P. Medium promoting sporulation of Bacillus larvae and metabolism of medium components. Appl. Environ. Microbiol. 1983;46:860–869. doi: 10.1128/aem.46.4.860-869.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alvarado I., Phui A., Elekonich M.M., Abel-Santos E. Requirements for in vitro germination of Paenibacillus larvae spores. J. Bacteriol. 2013;195:1005–1011. doi: 10.1128/JB.01958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.