Abstract
WRKYs are important regulators in plant development and stress responses. However, knowledge of this superfamily in soybean is limited. In this study, we characterized the drought- and salt-induced gene GmWRKY12 based on RNA-Seq and qRT-PCR. GmWRKY12, which is 714 bp in length, encoded 237 amino acids and grouped into WRKY II. The promoter region of GmWRKY12 included ABER4, MYB, MYC, GT-1, W-box and DPBF cis-elements, which possibly participate in abscisic acid (ABA), drought and salt stress responses. GmWRKY12 was minimally expressed in different tissues under normal conditions but highly expressed under drought and salt treatments. As a nucleus protein, GmWRKY12 was responsive to drought, salt, ABA and salicylic acid (SA) stresses. Using a transgenic hairy root assay, we further characterized the roles of GmWRKY12 in abiotic stress tolerance. Compared with control (Williams 82), overexpression of GmWRKY12 enhanced drought and salt tolerance, increased proline (Pro) content and decreased malondialdehyde (MDA) content under drought and salt treatment in transgenic soybean seedlings. These results may provide a basis to understand the functions of GmWRKY12 in abiotic stress responses in soybean.
Keywords: WRKY, stress responsive mechanism, drought tolerance, salt tolerance, transgenic hairy root assay, soybean
1. Introduction
Drought and salinity are the most important abiotic stress factors affecting plants growth and crop yield. On average, 1/3 of cultivable land suffers drought and salinization, which is equivalent to a loss of about 1,500,000 ha of crop land per year [1]. The damage caused by drought and salt are almost the sum of losses caused by other stress factors. Under limited land and water resources, it is necessary to breed new stress-resistant varieties to increase yield and ensure food security. Cultivation of stress-resistant crop varieties is also an important way to ensure high and stable yield of crops. Transgenic technology has become an important way to learn the function of genes in crops [2,3,4].
Being unable to move, plants encounter numerous biotic and abiotic stresses at different developmental stages which include drought, salinity, temperature changes, nutritional deficiency, pathogen invasion and competition from alien species. To overcome these unfavorable conditions, plants have evolved a complex and efficient signaling network, which can produce a series of responses to external stress signals and induce the expression of stress-related genes to protect the normal activities of the cells [5]. Inducible genes encoding proteins can be divided into three categories based on function: the first is functional genes, which are directly involved in stress response and are located downstream in the signaling network, such as HKT [6,7], SALT [8], NHX [9,10], CAX and CHX [11,12,13]. Another is transcription factors (TFs) that regulate the expression of functional genes in the middle of the signaling network, like DREB [14,15], MYB [16], WRKY [17,18], NAC [19,20], bZIP [21,22] and ERF [23,24]. The last group includes a variety of protein kinases, which conduct stress signals and are located upstream of the signaling network, such as GST [25], LEA [26] and FNS [27].
Among the three classes of stress-related genes, the TFs form a connecting link between the beginning and end of the signaling network; WRKYs are among the largest family of plant TFs. The WRKY domain is about 60 residues in length and is named by a conserved WRKY domain, containing the WRKYGQK heptapeptide at the N-terminus followed by a zinc-finger motif CX4-5CX22-23HXH or CX7CX23HXC [28,29]. Based on the number of WRKY domains and the structure of zinc finger motifs, WRKY TFs are divided into three groups. Group I includes two WRKY domains and either a CX4-5CX22-23HXH or CX7CX23HXC zinc-finger motif. Group II WRKY proteins contain a single WRKY domain and a CX4-5CX22-23HXH zinc-finger motif; due to differences in the primary amino acid sequence, Group II can be divided into five subgroups IIa-IIe [29,30]. Group III WRKY proteins have a single WRKY domain and a CX7CX23HXC zinc-finger motif.
As one of the members of the plant TF family, WRKY is heavily studied. Researchers have determined that WRKY TFs participate in various physiological and developmental processes [29], such as seed development [31], seed dormancy and germination [32], senescence [33], development [34], plant immune response [35], pathogen defense [18,36] and insect resistance [37,38]. Recent studies have revealed that WRKY proteins are involved in the signal transduction of plant hormones, like abscisic acid (ABA) [39,40], jasmonic acid (JA) [41] and gibberellin (GA) [39]. Numerous studies have demonstrated that WRKY TFs respond to abiotic stresses [42,43], such as salt [4], drought [44], cold [45] and heat [46,47,48]. There are 74 WRKY TF members in model plant Arabidopsis [49] and 18 WRKYs have been suggested to be induced by exposure to salt stress; overexpression of WRKY25 or WRKY33 was sufficient to increase Arabidopsis NaCl tolerance [50]. Overexpressing TaWRKY2 and TaWRKY19 exhibited salt and drought tolerance in transgenic Arabidopsis [51]. Moreover, researchers found that OsWRKY11 directly bound to the promoter of a drought-responsive gene, RAB21, as well as enhanced heat and drought tolerance in transgenic rice seedlings [52,53]. Ectopic expression of ZmWRKY33 and ZmWRKY58 in Oryza and Arabidopsis improved drought and salt tolerance, respectively, in transgenic plants [54,55]. In addition, there is extensive cross-talk between responses to biotic/abiotic stresses and exogenous hormones, for example drought and salt stress with the plant hormones. Arabidopsis WRKY46, WRKY54 and WRKY70 are involved in Brassinosteroid-mediated drought response and plant growth [43]. Novel cotton WRKY-genes GhWRKY25 and GhWRKY6-like confer tolerance to abiotic and biotic stresses in transgenic Nicotiana and enhanced salt tolerance by activating the ABA signaling pathway and scavenging reactive oxygen species [56]. SA-inducible poplar PtrWRKY73 is also involved in disease resistance in Arabidopsis [37]. All of these studies illustrated that WRKY TFs play a significant role in plant developmental and physiological processes and abiotic and biotic stresses.
Soybean (Glycine max), is an important global cash crop, accounting for 59 percent of the world’s oilseed production (http://soystats.com). Currently, due to its high protein content it is often treated as an important source of protein for both human consumption and as fodder. The demand for soybean is thus increasing rapidly and improving soybean yield has become a major research goal. Soybean productivity is greatly affected by growing environment, such as climatic and soil conditions (drought, salt, metallic pollution and fungus infection). Therefore, it is vital to cultivate soybean varieties that are resistant to stressors.
Recently, many studies based on biotechnological and RNA-Seq approaches have been conducted on soybean WRKY TFs. Researchers have identified 188 soybean WRKY genes genome-wide and 66 of the genes have been shown to respond rapidly and transiently to the imposition of salt stress [30]. In the latest version of the soybean genome (Wm82.a2v1), 176 GmWRKY proteins were confirmed and the expression of GmWRKY47 and GmWRKY58 decreased upon dehydration, while GmWRKY92, GmWRKY144 and GmWRKY165 increased under salt treatment [57]. GmWRKY13 may function in plant growth and abiotic stress. GmWRKY21 and GmWRKY54 conferred tolerance to cold stress and salt and drought stress, respectively [58]. Here, based on RNA-Seq and several databases and bioinformatics methods, we identified GmWRKY12, which is associated with abiotic stress tolerance by quantitative RT-PCR. Overexpression of GmWRKY12 could improve tolerance of soybean to drought and salt.
2. Results
2.1. Identification of GmWRKYs Up-Regulated under Drought/Salt Treatment
The GmWRKYs are distributed in different tissues or located upstream of soybean genes to bind the W-box consensus (TTGACY) in the promoters of target genes, initiating functions such as plant development, pathogen defense, insect resistance, response to biotic and abiotic stress and participating in signal transduction mediated by plant hormones [59,60]. In order to identify the function of genes or to explore whether GmWRKY mRNA expression goes up under biotic and abiotic stress, we conducted RNA-Seq (Tables S5 and S6). RNA-Seq data were used to screen GmWRKYs that are responsive to drought and salt. There were 105 GmWRKYs upregulated after drought treatment and fifty-three GmWRKYs were selected based on the rule that log2 (GH_treat/CK1_treat) >1 (Table 1). Nine GmWRKYs were selected from salt treatment RNA-Seq data based on the rule that log2 (NaCl_treat/CK2_treat) >1 (Table 2).
Table 1.
Annotation of Glycine max WRKY transcription factors responding to drought stress (up-regulation).
| Gene ID a | Name b | Chr | CDS (bp) | Protein (aa) | Group c |
|---|---|---|---|---|---|
| GLYMA_14G103100 | GmWRKY40 | 14 | 849 | 282 | IIb |
| GLYMA_18G056600 | GmWRKY62 | 18 | 1689 | 542 | IIb |
| GLYMA_17G042300 | GmWRKY6 | 17 | 1173 | 390 | IIe |
| GLYMA_04G054200 | GmWRKY50 | 4 | 486 | 161 | IIe |
| GLYMA_01G222300 | GmWRKY22 | 1 | 738 | 245 | IIc |
| GLYMA_02G293400 | GmWRKY31 | 2 | 1278 | 425 | IIa |
| GLYMA_04G218700 | GmWRKY21 | 4 | 591 | 196 | I |
| GLYMA_06G147100 | GmWRKY51 | 6 | 591 | 196 | III |
| GLYMA_01G224800 | GmWRKY12 | 1 | 714 | 237 | IIc |
| GLYMA_11G163300 | GmWRKY19 | 11 | 1647 | 548 | I |
| GLYMA_06G061900 | GmWRKY17 | 6 | 885 | 294 | IIb |
| GLYMA_10G011300 | GmWRKY54 | 10 | 972 | 323 | IIa |
| GLYMA_04G223300 | GmWRKY58 | 4 | 954 | 317 | III |
| GLYMA_18G213200 | GmWRKY57 | 18 | 900 | 299 | III |
| GLYMA_06G125600 | GmWRKY53 | 6 | 1095 | 364 | IIa |
| GLYMA_19G217800 | GmWRKY23 | 19 | 873 | 290 | IId |
| GLYMA_09G280200 | GmWRKY33 | 9 | 1632 | 543 | I |
| GLYMA_03G002300 | GmWRKY70 | 3 | 747 | 248 | IIc |
| GLYMA_13G310100 | GmWRKY36 | 13 | 1845 | 614 | IIc |
| GLYMA_14G200200 | GmWRKY49 | 14 | 1728 | 575 | IIc |
| GLYMA_16G026400 | GmWRKY60 | 16 | 1122 | 373 | IIc |
| GLYMA_16G0544001 | GmWRKY75 | 16 | 588 | 195 | IIb |
| GLYMA_04G223200 | GmWRKY55 | 4 | 1020 | 339 | IId |
| GLYMA_02G232600 | GmWRKY39 | 2 | 1743 | 580 | III |
| GLYMA_05G0290001 | GmWRKY72 | 5 | 1785 | 594 | I |
| GLYMA_03G220100 | GmWRKY41 | 5 | 762 | 253 | IIe |
| GLYMA_08G021900 | GmWRKY46 | 8 | 1080 | 356 | III |
| GLYMA_15G003300 | GmWRKY27 | 15 | 921 | 306 | IIb |
| GLYMA_17G097900 | GmWRKY61 | 17 | 1803 | 600 | IIc |
| GLYMA_01G128100 | GmWRKY5 | 1 | 1527 | 508 | IId |
| GLYMA_12G212300 | GmWRKY16 | 12 | 792 | 263 | IIc |
| GLYMA_08G082400 | GmWRKY28 | 8 | 881 | 293 | III |
| GLYMA_07G227200 | GmWRKY3 | 7 | 1602 | 533 | IIc |
| GLYMA_03G256700 | GmWRKY43 | 66 | 1089 | 362 | IIe |
| GLYMA_15G168200 | GmWRKY42 | 15 | 882 | 293 | IIb |
| GLYMA_13G289400 | GmWRKY52 | 13 | 798 | 265 | IIc |
| GLYMA_08G011300 | GmWRKY25 | 8 | 444 | 147 | IId |
| GLYMA_09G061900 | GmWRKY47 | 19 | 1573 | 296 | IIc |
| GLYMA_17G222300 | GmWRKY30 | 4 | 555 | 184 | IIa |
| GLYMA_01G053800 | GmWRKY9 | 1 | 1368 | 455 | IIc |
| GLYMA_08G118200 | GmWRKY48 | 7 | 789 | 262 | IIc |
| GLYMA_01G056800 | GmWRKY32 | 1 | 894 | 297 | IId |
| GLYMA_08G218600 | GmWRKY56 | 8 | 942 | 313 | III |
| GLYMA_07G262700 | GmWRKY34 | 7 | 1554 | 517 | IIb |
| GLYMA_03G159700 | GmWRKY15 | 1 | 1017 | 338 | I |
| GLYMA_11G053100 | GmWRKY14 | 11 | 963 | 320 | I |
| GLYMA_05G096500 | GmWRKY11 | 17 | 1050 | 334 | I |
| GLYMA_17G222500 | GmWRKY63 | 17 | 849 | 278 | IIa |
| GLYMA_08G240800 | GmWRKY4 | 2 | 1572 | 523 | I |
| GLYMA_03G176600 | GmWRKY29 | 5 | 1308 | 436 | IIc |
| GLYMA_08G325800 | GmWRKY35 | 8 | 1734 | 577 | IIc |
| GLYMA_10G138300 | GmWRKY1 | 14 | 1449 | 482 | IIb |
| GLYMA_06G077400 | GmWRKY37 | 6 | 903 | 300 | III |
a—The annotated GmWRKYs according to NCBI (https://www.ncbi.nlm.nih.gov/pubmed) and. PlantTFDB (http://planttfdb.cbi.pku.edu.cn/); b—The names of GmWRKYs are given according to SoyDB (http://soykb.org/); c—The grouping is according to [30,61].
Table 2.
Annotation of Glycine max WRKY transcription factors responding to salt stress (up-regulation).
| Gene ID a | Name b | Chr | CDS (pb) | Protein (aa) | Group c |
|---|---|---|---|---|---|
| GLYMA_11G053100 | GmWRKY14 | 9 | 963 | 320 | I |
| GLYMA_08G325800 | GmWRKY35 | 8 | 1734 | 577 | IIc |
| GLYMA_04G218700 | GmWRKY21 | 10 | 591 | 196 | I |
| GLYMA_14G200200 | GmWRKY49 | 18 | 1728 | 575 | IIc |
| GLYMA_07G227200 | GmWRKY3 | 18 | 1602 | 533 | IIc |
| GLYMA_02G115200 | GmWRKY28 | 8 | 881 | 293 | III |
| GLYMA_03G256700 | GmWRKY43 | 16 | 1089 | 362 | III |
| GLYMA_06G320700 | GmWRKY59 | 6 | 2331 | 776 | IIc |
| GLYMA_01G224800 | GmWRKY12 | 7 | 714 | 237 | IIc |
a—The annotated GmWRKYs according to NCBI (https://www.ncbi.nlm.nih.gov/pubmed) and PlantTFDB (http://planttfdb.cbi.pku.edu.cn/). b—The names of GmWRKYs are given according to SoyDB (http://soykb.org/). c—The grouping is according to [30,61].
2.2. Tissue-Specific Expression Patterns of GmWRKYs
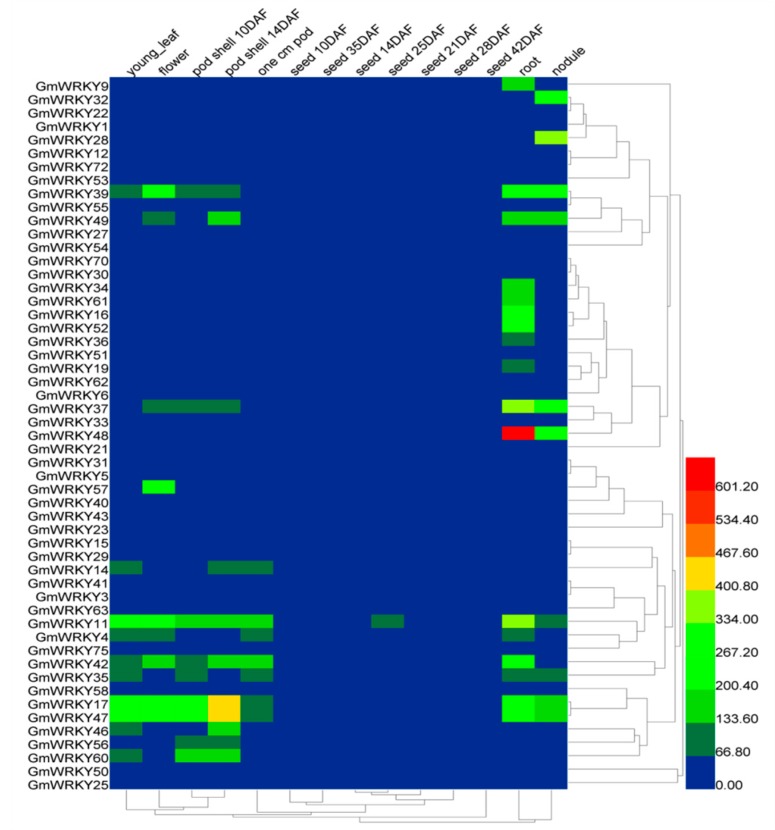
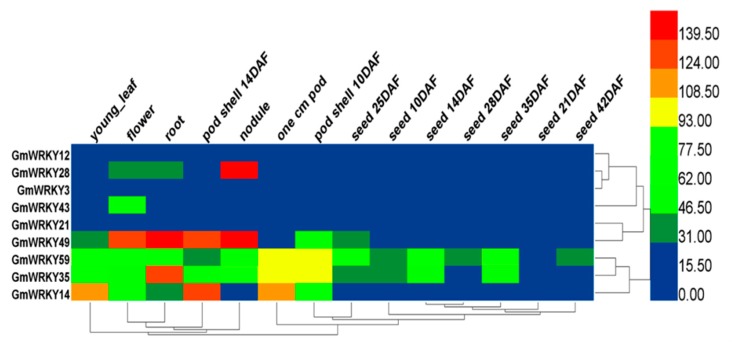
To thoroughly study GmWRKY expression profiles under normal conditions, hierarchical clustering was conducted using expression levels of fifty-three (drought-responsive) and nine (salt-responsive) GmWRKY genes in young leaf, flower, one cm pod, pod shell 10 days after flowering (DAF), pod shell 14 DAF, seed 10 DAF, seed 14 DAF, seed 21 DAF, seed 25 DAF, seed 28 DAF, seed 35 DAF, seed 42 DAF, root and nodule (Figure 1 and Figure 2). Approximately 28% of GmWRKYs from different tissues were expressed at low levels or unexpressed (GmWRKY3, 5, 6, 21, 22, 25, 29, 30, 31, 47, 50, 55, 63, 70 and 72); by contrast, 45% of GmWRKYs were highly expressed in different tissues (GmWRKY4, 9, 11, 14, 16, 17, 19, 28, 32, 34, 35, 36, 37, 39, 41, 42, 46, 48, 49, 52, 56, 57, 60 and 61). Among these GmWRKYs, GmWRKY11 and GmWRKY17 had the highest expression in four different tissues. GmWRKY28, 35, 37, 48 and 57 are highly expressed in nodule, seed 10 DAF, seed 42 DAF, root and flower. Within the nine GmWRKYs related to salt response, GmWRKY3 and GmWRKY21 had low expression and GmWRKY14, 28, 35, 49 and 59 were highly expressed in at least four different tissues. The analysis data are available in Tables S1 and S2.
Figure 1.
Expression pattern of fifty-three GmWRKYs in six different tissues (young leaf, flower, pod shell, seed, root and nodule). The fifty-three GmWRKYs were selected from drought treatment RNA-Seq data based on the rule that log2 (GH_treat/CK1_treat) >1. The tissue expression is from SoyDB (http://soykb.org/). The color legend refers to the different expression level under normal condition. “DAF” in the tissue label indicates days after flowering.
Figure 2.
Expression pattern of nine GmWRKYs in six different tissues (young leaf, flower, pod shell, seed, root and nodule). The nine GmWRKYs were selected from salt treatment RNA-Seq data based on the rule that log2 (NaCl_treat/CK2_treat) >1. The tissue expression is from SoyDB (http://soykb.org/). The color legend refers to the different expression level under normal condition. “DAF” in the tissue label indicates days after flowering.
2.3. GmWRKYs Responsive to Both Drought and Salt Treatments
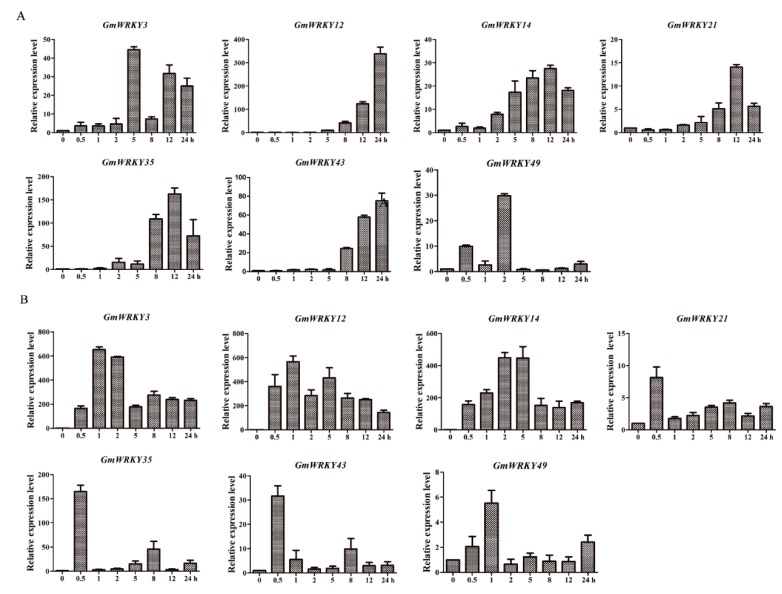
Based on RNA-Seq data and result of Venn method [62], seven GmWRKY genes were found to respond to both drought and salt treatments (GmWRKY3, 12, 14, 21, 35, 43 and 49) (Figure S1A). In order to confirm whether the seven GmWRKY genes are responsive to drought and salt, 10-day-old soybean seedlings were subjected to stress treatments. For drought treatment, soybean seedlings were put on filter paper to stimulate drought and then sampled 0.1 g of leaf on different periods (0, 0.5, 1, 2, 5, 8, 12 and 24 h); for salt treatment, the roots of soybean were soaked in 100 mM NaCl solution then sampled 0.1 g of leaf on different periods (0, 0.5, 1, 2, 5, 8, 12 and 24 h), all samples were submerged immediately in liquid nitrogen and stored at −80 °C for RNA extraction then quantitative real-time PCR (qRT-PCR) was conducted. Results confirmed that the seven GmWRKY genes were responsive to both treatments (Figure 3). Under drought treatment, the expression levels of GmWRKY12 and GmWRKY43 were gradually increased at 0, 0.5, 1, 2, 5, 8, 12 and 24 h. GmWRKY12 was highly expressed after 12 h of drought treatment. While GmWRKY14, GmWRKY21 and GmWRKY35 had a tendency to rise first and then decrease, GmWRKY49 was highly expressed at 2 h. Under drought conditions, the expression profiles of five GmWRKY genes were little changed at 0 to 5 h and then increased significantly at 12 h.
Figure 3.
Quantitative RT-PCR of seven GmWRKYs under drought and salt treatment. (A) qRT-PCR of seven GmWRKYs under drought treatment. (B) qRT-PCR of seven GmWRKYs under salt treatment. The expression level of GmActin as a loading control. The data represent means ± SD of three biological replications.
Under salt treatment, the expression profile increased first and then decreased, meanwhile, there was a notable change at 0 to 0.5 h and GmWRKY3, 12, 14 and 35 were highly expressed. GmWRKY12, which was 714 bp in length, encoded 237 amino acids and had low expression in different tissues under normal conditions but was highly expressed under drought and salt treatments was selected for further investigation (Figure S1B).
2.4. Multiple Sequence Alignment and Phylogenetic Analysis of GmWRKY12
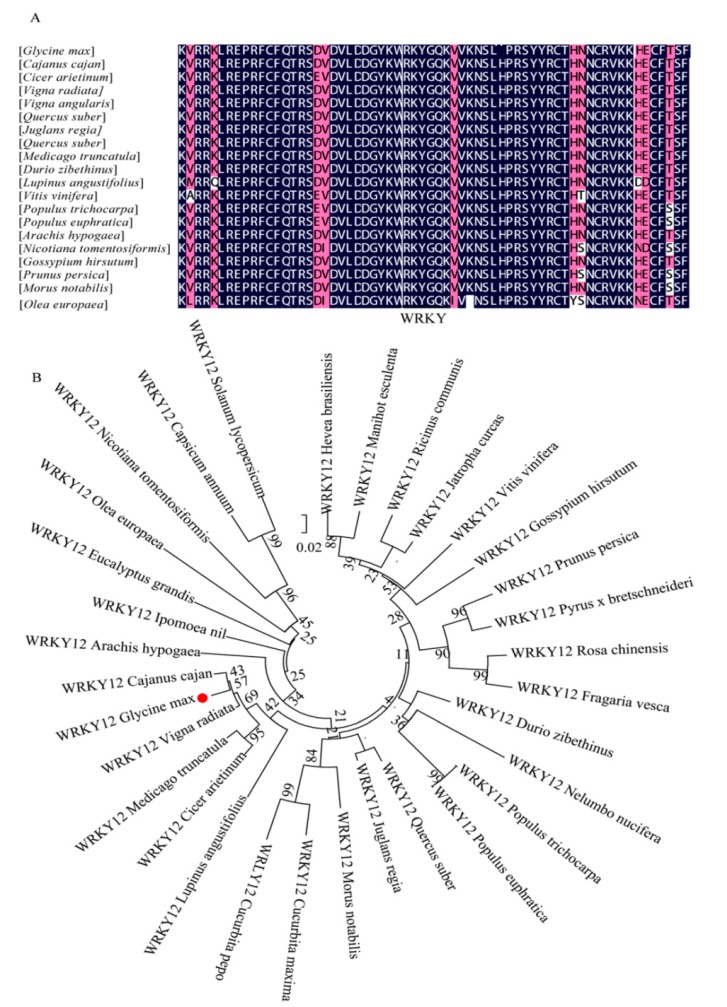
Although WRKYGQK sequence is a conservative motif of WRKY proteins, WRKY variant domains, such as WRKYGEK, WRKYGKK, WQKYGQK, WSKYGQK and WRKYGM have been found in the genomes of Arabidopsis [28], rice [63], grape [64] and tomato [65]. This difference may be a variation of WRKY TFs developed over long-term evolution. The domain of these variations is unique and may represent a new type. Therefore, to identify conservation of GmWRKY12, WRKY12 from 20 different species were selected for multiple sequence alignment (Figure 4A). Results showed that 20 species only harbored one WRKY variant WRKYGQK, with amino acid sequence similarity of 75%, which illustrated that GmWRKY12 was highly conserved. To further evaluate the evolutionary relationship between GmWRKY12 and WRKY12 of 32 different species, a phylogenetic tree was constructed with the neighbor-joining method [66]. Phylogenetic results showed that the relationship between GmWRKY12 and VrWRKY12 (XP_014515898.1) was the closest (Figure 4B).
Figure 4.
Multiple alignment and phylogenetic relationship of GmWRKY12 with different species. (A) Multiple alignment of GmWRKY12 with other WRKY12 proteins from other species. (B) Phylogenetic relationship of GmWRKY12 in different species. The red dot in (B) means GmWRKY12. The number of nodes is the bootstrap value and the number on the branch is the evolutionary distance. Bootstrap replications are 1000.
2.5. Expression Patterns of GmWRKY12 under Different Treatments
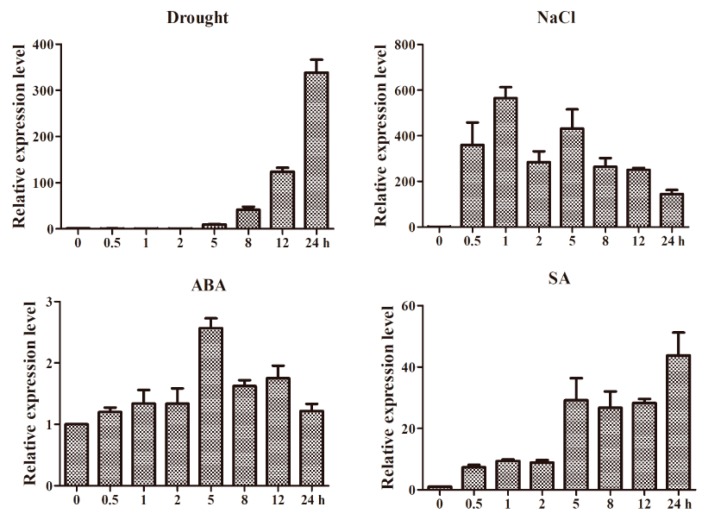
GmWRKY12 was responsive to drought and salt treatments (Figure 3). WRKY proteins are reported to be involved in signal transductions of plant hormones [39]. In order to identify whether GmWRKY12 was responsive to other abiotic stresses, expression patterns were identified using qRT-PCR. Results indicated that GmWRKY12 not only participated in drought and salt response but was also responsive to ABA and SA. Under low concentrations of SA, the expression profile of GmWRKY12 was increased about 50-fold (Figure 5).
Figure 5.
Expression patterns of GmWRKY12 under drought, NaCl, exogenous ABA and SA. The ordinates are the relative expression level (fold) of GmWRKY12 compared to non-stressed control. The horizontal ordinate is treatment time for 0, 0.5, 1, 2, 5, 8, 12 and 24 h. The expression level of GmActin as a loading control. All experiments were repeated three times. Error bars represent standard deviations (SDs). All data represent the means ± SDs of three independent biological replicates.
2.6. Cis-Acting Elements in Promoter
To further understand the regulatory mechanism of GmWRKY12, we isolated its promoter region. Cis-elements correlated to stress were present in the promoter region, including the ABA and wound responsive elements ABER4 and MYC, drought responsive element MYB, salt stress responsive element GT-1 and wound responsive element W-box. In addition, there was another element that participated in heat and GA response in the promoter region of GmWRKY12 (Table 3). This analysis suggested that GmWRKY12 may function in abiotic stress response.
Table 3.
Cis-elements analysis of GmWRKY12 promotor.
| Cis-Elements | Numbers | Target Sequences | Functions |
|---|---|---|---|
| MYC | 32 | CANNTG | ABA and wound responsive element |
| W-box | 21 | TTGAC/TTTGACY/TGACY | SA responsive element |
| ABER4 | 18 | ACGT | ABA responsive element |
| MYB | 14 | C/TAACNA/G | Drought responsive element |
| CCAATB | 10 | CCAAT | Heat-responsive element |
| GT-1 | 7 | GAAAAA | Salt stress responsive element |
| DPBF | 6 | ACACNNG | Dehydration-responsive element |
| GARE | 2 | TAACAAR | GA-responsive element |
“Numbers” corresponds to the number of cis-elements of each type present in the promoter.
2.7. GmWRKY12 was Located in the Nucleus
To investigate GmWRKY12 subcellular localization, GmWRKY12 were fused to the N-terminus of the humanized green fluorescent protein (hGFP) and co-transformed into wheat mesophyll protoplasts with the nucleus marker AT2G03340 (AtWRKY3)-mCherry [67,68]. The 35S::GFP vector was transformed as the control. Fluorescence of GmWRKY12 was specifically detected in the nucleus, whereas GFP fluorescence was distributed throughout the cell (Figure 6).
Figure 6.
Co-localization of GmWRKY12. The recombinant plasmid of GmWRKY12-GFP and At2G03340-mCherry were co-transformed into wheat mesophyll protoplasts under the control of the CaMV 35S promoter. GmWRKY12 was localized in the nucleus of wheat mesophyll protoplasts protoplasts. Results were observed by a confocal laser scanning microscope (LSM700; CarlZeiss, Oberkochen Germany) after incubating in darkness at 22 °C for 18–20 h. Scale bars = 10 μm.
2.8. GmWRKY12 Improved Drought and Salt Tolerance of Soybean
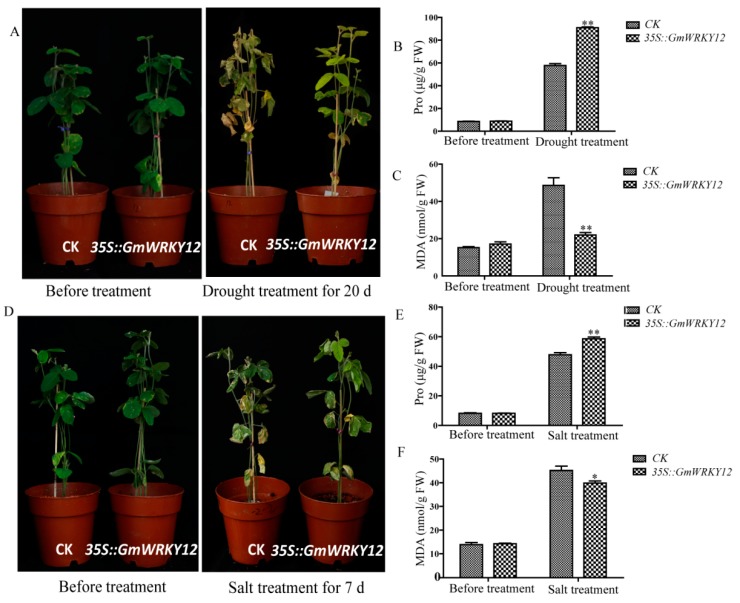
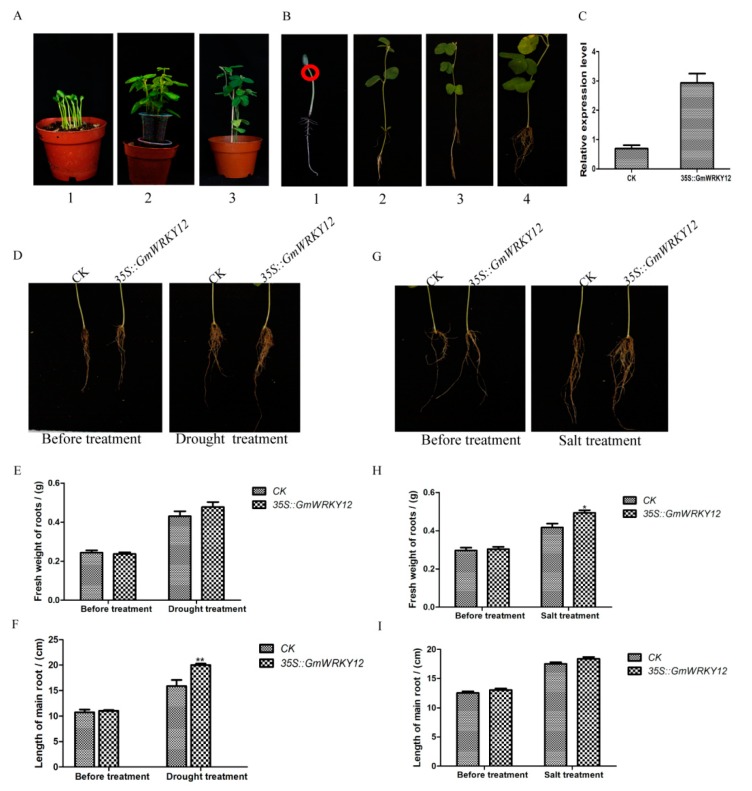
We further used transgenic hairy root assays to investigate the roles of GmWRKY12 in abiotic stress responses. Amplified cDNA sequence of GmWRKY12 was constructed into pCAMBIA3301 to create an overexpression transgenic line and the control was pCAMBIA3301 plant expression vector with CaMV35S promoter. Two constructs were transferred into Agrobacterium rhizogenes strain K599 (NCPPB2659) [69] then transformed into soybean hairy roots as previously described [70,71]. After drought treatment for 20 days, both control and over-expression soybean seedlings had leaf shedding to different degrees, especially the old leaves of the plants (Figure 7A). However, compared with transgenic soybean seedlings, the control seedlings were severely wilted and almost 99% of the leaves had serious dehydration and drying. By contrast, there was slight shedding of the old leaves of transgenic soybean seedlings but the new leaves were still growing vigorously. Results of proline and malondialdehyde (MDA) content determination showed that overexpression of GmWRKY12 increased proline content in transgenic lines, while the MDA content was decreased due to drought stress (Figure 7B,C). Fresh weight and main length of transgenic soybean hair roots under drought treatment were measured (Figure 8E,F), results showed overexpressed GmWRKY12 in soybean roots enhanced drought tolerance of soybean by increasing the length of transgenic hair roots and the number of transgenic hair roots.
Figure 7.
Phenotype identification of GmWRKY12 under drought and salt treatments. (A) Images of drought stress resistance phenotypes of CK and 35S::GmWRKY12 soybean seedlings after drought treatment for 20 days. (B) Proline contents in CK and 35S::GmWRKY12 soybean seedlings under normal growth conditions and drought treatment. (C) MDA contents in in CK and 35S::GmWRKY12 soybean seedlings under normal growth conditions and drought treatment. (D) Images of salt stress resistance phenotypes of CK and 35S::GmWRKY12 soybean seedlings after 200 mM NaCl treatment for 7 days. (E) Proline contents in CK and 35S::GmWRKY12 soybean seedlings under normal growth conditions and salt treatment. (F) MDA contents in CK and 35S::GmWRKY12 soybean seedlings under normal growth conditions and salt treatment. All data represent the means ± SDs of three independent biological replicates. ANOVA tests demonstrated that there were significant differences (* p < 0.05, ** p < 0.01).
Figure 8.
Different growth stage of transgenic soybean seedlings and phenotypes of transgenic soybean hair roots. (A) Images of different growth stage of transgenic soybean seedlings cultivated in flowerpot before any treatment. (A1) Soybean seedlings of 5-days-old without injected A. rhizogenes carrying GmWRKY12. (A2) Soybean seedlings which have injected A. rhizogenes carrying GmWRKY12 for 7 days. (A3) Soybean seedlings which have injected A. rhizogenes carrying GmWRKY12 for 14 days (The original main roots were removed by cutting from 1 cm below the infection site and the hairy roots of the seedlings were cultivated in nutritious soil with full water and grown with 16 h light (100 μM photons m−2·s−1)/8 h dark at 25 °C). (B) Images of different growth stage of signal transgenic soybean seedling before any treatment. (B1) Soybean seedling of 5-days-old without injected A. rhizogenes carrying GmWRKY12 and the red circle shows the inject site of A. rhizogenes. (B2) Soybean seedling which have injected A. rhizogenes carrying GmWRKY12 for 7 days and new hair roots have generated. (B3) Soybean seedling which have injected A. rhizogenes carrying GmWRKY12 for 14 days. (B4) Soybean seedling that have salt treatment for 7days. (C) Relative expression of CK and 35S::GmWRKY12 transgenic soybean hair roots under normal growth conditions. (D) Images of drought stress resistance phenotypes of CK and 35S::GmWRKY12 transgenic soybean hair roots after drought treatment for 20 days. (E) Fresh weight in CK and 35S::GmWRKY12 transgenic soybean hair roots under normal growth conditions and drought treatment. (F) Length in CK and 35S::GmWRKY12 transgenic soybean hair roots under normal growth conditions and drought treatment. (G) Images of salt stress resistance phenotypes of CK and 35S::GmWRKY12 transgenic soybean hair roots after 200 mM NaCl treatment for 7 days. (H) Fresh weight in CK and 35S::GmWRKY12 transgenic soybean hair roots under normal growth conditions and salt treatment. (I) Length in CK and 35S::GmWRKY12 transgenic soybean hair roots under normal growth condition and salt treatment. All data represent the means ± SDs of three independent biological replicates. ANOVA tests demonstrated that there were significant differences (* p < 0.05, ** p < 0.01).
Meanwhile, under NaCl (200 mM) treatment, control and overexpression soybean seedlings had different degrees of leaf shedding (Figure 7D). Compared with the control, transgenic soybean seedlings were slightly wilted and slowly drying out, while the control seedlings were almost dry due to the osmotic stress. Results of Pro and MDA content in transgenic lines (Figure 7E,F) fresh weight and main length of transgenic soybean hair roots (Figure 8H,I) also showed that GmWRKY12 improved salt tolerance of soybean. These results demonstrated that GmWRKY12 confers stress tolerance in transgenic hairy roots.
3. Discussion
The WRKY transcription factor superfamily, as a recently described member of the TF family, has been studied by many researchers due to its numerous and diverse biological functions. Since the first reports of WRKY TFs [72], research conducted in different species [4,52,57,73,74] has shown that WRKY TFs play significant roles in plant development and stress responses. Recently, many studies of GmWRKY TFs have been based on biotechnological and RNA-Seq approaches [30,57]. However, these studies mainly reported genome-wide annotation of the WRKYs and structure analysis of some genes involved in response to abiotic and biotic stresses. Although these genes have been identified through biochemistry and bioinformatics approaches, knowledge about soybean stress tolerance was limited. In this study, based on qRT-PCR and RNA-Seq data, GmWRKY12 was selected for investigation of stress tolerance in soybean (Figure S1).
According to classifications in the WRKY family [18,28,75], WRKY12 belongs to Group IIc and contains a single WRKY domain and a CX4-5CX22-23HXH zinc-finger motif. Recent studies have shown that the WRKYGQK heptapeptide, which can specifically recognize and bind to the W-box consensus sequence (TTGACY) in the promoters of target genes, can be replaced by WRKYGKK, WRKYGEK, WKKYEDK, or WKKYCEDK; variations of the WRKYGQK motif might change the DNA binding specificities to downstream target genes [75]. However, multiple sequence alignment results showed that WRKY12 in different species only harbor the same WRKYGQK heptapeptide, demonstrating that WRKY12 protein is evolutionarily conserved and can recognize and bind to downstream target genes (Figure 4A). The result was consistent with the results observed in other species [54,57,65,76,77]. Structural conservation determines functional specificity: in rice, OsWRKY12 was related to normal plant growth and expression of OsWRKY12 was low at the seedling stage but increased gradually with growth [78]; similar results were found in specific tissues in our study. GmWRKY12 has low expression in young leaf, flower, one cm pod, pod shell 10 DAF, seed 10 DAF, seed 14 DAF, seed 21 DAF, seed 25 DAF, seed 28 DAF, seed 35 DAF, seed 42 DAF and root under normal conditions. At the pod shell 14 DAF and nodule stages, the expression levels gradually increase (Table S2), which may be because genes are differentially expressed at different growth stages, or may perform different activities, such as metabolism, nutrient absorption or material transformation. For example, at the nodule stage, plants are primarily vegetative, while at seed 42 DAF, plants are accumulating nutrients [57]. In addition, WRKY12 was related to plant flowering time: Arabidopsis plants overexpressing MlWRKY12 showed early flowering phenotype [79]. WRKY12 and WRKY13 have opposite effects on flowering time in the action of GA [80]. Overexpression of three Triticum genes, TaWRKY12, TaWRKY18 and TaZFP2 induced the expression of some genes related to Pi absorption and transportation, enhancing the abilities of Pi uptake and Pi use efficiency in plants under low-Pi stress conditions [81]. Thus, GmWRKY12, like other WRKYs, is involved in plant growth and development.
There are many cis-acting elements in the GmWRKY12 promoter region, such as MYC (ABA and wound responsive element), W-box (SA responsive element), ABER4 (ABA responsive element), MYB (drought responsive element), CCAATB (heat-responsive element), GT-1 (salt stress responsive element), DPBF (dehydration-responsive element) and GARE (GA-responsive element) (Table 3). The presence of these elements indicates that GmWRKY12 may take part in various biotic and abiotic responses except for growth and development of plants. Research of tobacco transcription factors NtWRKY12 and TGA2.2 found that NtWRKY12 alone was able to induce PR-1a::GUS expression to high levels, the PR-1a gene was salicylic acid-inducible to activate the expression of SA-inducible genes [82]. SA is an important endogenous molecule that activates plant hypersensitive response and systemic acquired resistance, which are often involved in disease resistance of plants [83]. As the closest orthologue of AtWRKY12, BrWRKY12 from Chinese cabbage conferred enhanced resistance to Pectobacterium carotovorum ssp. carotovorum (Pcc) through transcriptional activation of defense-related genes [84]. Furthermore, LrWRKY12 were responsive to SA and methyl jasmonate (MeJA) treatments and conferred more resistance to B. cinerea than in wild-type plants [85]. These results show that WRKY12 plays an important role in disease defense of plants, mainly because WRKYGQK specifically binds to the W-box to induce expression of downstream target genes.
In addition to the significant roles of WRKY12 identified in development and disease defense of plants, WRKY12 also functions in plant stress responses. Under treatment with NaCl and PEG, the expression level of THWRKY12 in Tamarix tissues was increased, the expression pattern of THWRKY12 after ABA treatment was approximately the same as the expression level changes under NaCl and PEG treatment, showing that the gene may participate in regulating salt and drought tolerance through the signaling pathway regulated by ABA [86]. In our study, GmWRKY12 was first screened following both drought and salt treatment using RNA-Seq. In order to confirm whether it was responsive to salt and drought stress, qRT-PCR was conducted and further showed that GmWRKY12 was highly expressed under drought and salt treatment, which indicated that the gene was related to drought and salt tolerance (Figure 3). Cis-acting elements and expression pattern analysis of GmWRKY12 also showed that it may participate in the ABA signaling pathway (Table 3 and Figure 5). However, compared to the high expression level under drought and salt treatment, on the condition of ABA, GmWRKY12 had low expression. Resistance identification of GmWRKY12 using a soybean hairy root assay further showed that GmWRKY12 may be involved in regulating salt and drought tolerance by promoting the combination of cis-acting elements with drought and salt-related genes, thereby enhancing plant resistance (Figure 7). Similar results were also found in other studies [87,88,89].
4. Materials and Methods
4.1. Identification and Annotation of GmWRKYs Response to Drought/Salt Stress
Identification of the response of GmWRKYs to drought/salt stress was based on RNA-seq data collected from a set of drought and salt stress experiments (Tables S5 and S6). Seeds of Williams 82 were cultivated in a 10 × 10 cm flowerpot (vermiculite: nutritious soil is 1:3), fresh leaf of 10-day-old soybean seedlings were used for RNA-Seq.CK1_treat-Expression represented two independent replicates of plants sampled before any treatment; GH_treat-Expression related to drought treatment for 5 h (put on the filter paper to simulate drought) of soybean plants at room temperature; CK2_treat-Expression without NaCl treatment; and NaCl_treat-Expression salt treatment that soaking soybean roots with 100 mM NaCl solution for 1 h and then sampled for RNA-seq [57,68]. Both log2 (GH_treat/CK1_treat) >1, log2 (NaCl_treat/CK2_treat) >1 and up-regulated were treated as the rule to select GmWRKYs responding to drought/salt stress. Several databases: NCBI (https://www.ncbi.nlm.nih.gov/pubmed), PlantTFDB (http://planttfdb.cbi.pku.edu.cn/), Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) and SoyDB (http://soykb.org/), were used to annotate Gene ID, Name, Chromosomal localization, CDS, Protein and Group.
4.2. Tissue-Specific Expression Patterns of GmWRKYs
Data of six different tissues (young leaf, flower, pod shell, seed, root and nodule) from different growth periods was available from SoyBase (https://www.soybase.org/soyseq/). Heml1.0 software (http://www.patrick-wied.at/static/heatmapjs/) was used to perform hierarchical clustering of fifty-three and nine GmWRKYs under normal conditions. The analysis data are available in Tables S1 and S2.
4.3. RNA Extraction and qRT-PCR
Seeds of Williams 82 was cultivated in a 10 × 10 cm flowerpot (vermiculite: nutritious soil is 1:3), fresh leaf tissue of 10-day-old soybean seedlings were used for RNA extraction of different stress treatment. For drought treatment, soybean seedlings were dried on filter paper then sampled 0.1 g of leaf on different periods (0, 0.5, 1, 2, 5, 8, 12 and 24 h), for salt, ABA and SA treatment, the roots of soybean seedlings were soaked in 100 mM NaCl, 100 μmol·L−1 ABA and 100 μmol·L−1 SA solution, respectively [68]. Then sampled 0.1 g of leaf on different periods (0, 0.5, 1, 2, 5, 8, 12 and 24 h), all samples were submerged immediately in liquid nitrogen and stored at −80 °C for RNA extraction using RNA prep plant kit (TIANGEN, Beijing, China); cDNA was synthesized using a Prime Script First-Strand cDNA Synthesis Kit (TransGen, Beijing, China) following the manufacturer’s instructions. cDNA of treatment for 0 h was used for screen one highly expressed gene from seven GmWRKYs that response to both drought and salt treatment (Figure S1B). qRT-PCR was performed with Super Real PreMix Plus (TransGen, Beijing, China) on an ABI Prism 7500 system (Applied Biosystems, Foster City, CA, USA). Specific primers of GmWRKY3, 12, 14, 21, 28, 35, 43, 49 and soybean actin primers are listed in Table S4. Three biological replicates were used for qRT-PCR analysis. The 2−∆∆Ct method was used for quantification.
4.4. Gene Isolation and Phylogenetic Analysis of GmWRKY12
Venn2.0 (http://bioinfogp.cnb.csic.es/tools/venny/index.html) was used to screen GmWRKYs that respond to both drought and salt treatment, then qRT-PCR was used to find genes highly expressed under stresses. Full-length GmWRKY12 was amplified by PCR with specific primers from soybean cDNA (Williams 82); primers of GmWRKY12 are available in Table S4. PCR products were cloned into pLB vector (TIANGEN, Beijing, China) and sequenced for further study. The amino acid sequence of WRKY12 in different species were searched for in the NCBI database on account of the amino acid similarity between GmWRKY12 and WRKY12 in different species is more than 50%. DNAMAN was applied for multiple sequence alignment on the basis of the amino acid similarity between GmWRKY12 and WRKY12 in different species is more than 60%. Phylogenetic trees were constructed using MEGA 6.0 with the neighbor-joining method [66] and 1000 bootstrap replications. Information of WRKY12 in different species is listed in Table S3.
4.5. Co-Localization of GmWRKY
Seeds of Kenong199 were cultivated in a 10 × 10 cm flowerpot (vermiculite: nutritious soil is 1:3), fresh leaf tissue of 7-day-old wheat seedlings were used for preparation of wheat protoplasts. Amplified cDNA sequence of GmWRKY12 was cloned into the N-terminus hGFP protein driven by the CaMV35S promoter. The cDNA coding sequences of AT2G03340 (AtWRKY3) which located in the nucleus [67] were fused to the N-terminus of the mCherry protein (WRKY25-RFP) under the control of the CaMV 35S promoter [68]. The recombinant plasmid of GmWRKY12-GFP and AtWRKY3-mCherry were co-transformed into wheat mesophyll protoplasts via the PEG4000-mediated method. The 35S::GFP vector was transformed as the control. Fluorescence was observed using a confocal laser scanning microscope (LSM700; CarlZeiss, Oberkochen, Germany) after incubating in darkness at 22 °C for 18–20 h. Primers are available in Table S4.
4.6. Cis-acting Elements in Promoter
The 2.0 kb promoter region upstream of the ATG start codon in the promoter of GmWRKY12 was obtained from soybean genomic DNA in the Ensembl Plants website, cis-acting elements were analyzed by PLACE (http://www.dna.affrc.go.jp/PLACE/).
4.7. A. rhizogenes-mediated Drought and Salt Stress Assays
To generate a transgenic line of soybean the amplified cDNA sequence of GmWRKY12 was constructed into pCAMBIA3301 for an overexpression transgenic line (35S::GmWRLY12) and the control was pCAMBIA3301 plant vector with CaMV35S promoter (CK) and two constructs transferred into A. rhizogenes strain K599 (NCPPB2659) [69]. Primers are available in Table S4. Williams 82 was cultivated in a 10 × 10 cm flowerpot (vermiculite: nutritious soil is 1:3) for stress experiments (Figure 8A1), soybean seeds were grown under a photoperiod of 16 h light (100 μM photons m−2·s−1)/8 h dark at 25 °C. When plants displayed two cotyledons (Figure 8A1), A. rhizogenes strain K599 harboring pCAMBIA3301 (CK) and K599 harboring 35S::GmWRLY12 were injected at the cotyledonary node and/or hypocotyl (Figure 8B1). A plastic cup was used to surround the inoculated soybean seedlings to provide high humidity conditions. After 3 days, nutritious soil was prepared and used to fill the gaps in the plastic cup so that soybean seedlings could grow new roots (Figure 8A2); plants typically need two weeks to generate new roots (2–10 cm) at the infection site (Figure 8B2,B3). The original main roots were removed by cutting from 1 cm below the infection site and the hairy roots of the seedlings were cultivated in nutritious soil with full water and grown with 16 h light (100 μM photons m−2·s−1)/8 h dark at 25 °C for 5 days [70,71]. Each flowerpot cultivated 5 transgenic soybean seedlings and 5 replications of each stress treatment (Figure 8A3). Afterward, the transgenic soybean seedlings were subjected to natural dehydration and 200 mM NaCl for drought and salt stress assays [19,68]. For drought stress assay, both CK and transgenic soybean seedlings were grown without water for 20 days. For salt stress assay, CK and transgenic soybean seedlings were treated with 200 mM NaCl solution for 7 days. There are some Supplement Materials need to prepare for culturing A. rhizogenes strain K599 that harbored (35S::GmWRLY12) and the control (CK)., eg: Solidified LB medium with streptomycin sulfate (100 mg/L) and Kanamycin solution(100 mg/L) (10 g tryptone, 5 g yeast extract, 10 g NaCl, 15 g agar per liter), Liquid LB medium containing streptomycin sulfate (100 mg/L) and Kanamycin solution (100 mg/L) [69].
4.8. Measurements of Proline and MDA Contents
Both proline and MDA content were measured with the Pro and MDA assay kit (Comin, Beijing, China) based on the manufacturer’s protocols; all measurements were from three biological replicates.
4.9. Measurements of Fresh Weight and Main Length
Transgenic soybean hair roots were used to measure the fresh weight and main length. All data represent the means ± SDs of three independent biological replicates.
5. Conclusions
In this study, using RNA-Seq, we identified 62 GmWRKY genes in the soybean genome that were differently expressed in six different tissues under normal condition. Seven GmWRKYs responded to both drought and salt treatment. Based on the qRT-PCR, GmWRKY12, a nucleus protein of 237 amino acids, belonging to WRKY Group II was identified. It was responsive to salt, drought and exogenous hormones ABA and SA. Results of Agrobacterium rhizogenes-mediated hairy roots assay showed that overexpressing GmWRKY12 may improve tolerance to drought and salt in soybean. These results provided new insight into the roles of soybean WRKY genes in abiotic stress responses.
Acknowledgments
We are grateful to Hui Zhang (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for providing the pCAMBIA3301 vector and pUC57-GmU6-sgRNA.
Abbreviations
| ABA | abscisic acid |
| ABRE | ABA-responsive element |
| JA | jasmonic acid |
| SA | salicylic acid |
| MeJA | methyl jasmonate |
| qRT-PCR | quantitative real-time PCR |
| Pro | proline |
| MDA | malondialdehyde |
| DAF | days after flowering |
| GFP | green fluorescent protein |
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/12/4087/s1.
Author Contributions
Z.-S.X. coordinated the project, conceived and designed experiments and edited the manuscript; W.-Y.S. performed the experiments and wrote the first draft of the manuscript; Y.-T.D., J.M. and J.C. conducted the bioinformatic work and performed related experiments; D.-H.M. and L.-G.J. provided analytical tools and analyzed the data; M.C., Y.-B.Z. and X.-H.Z. contributed valuable discussion; and Y.-Z.M. coordinated the project. All authors have read and approved the final manuscript.
Funding
This research was financially supported by the National Key R & D Program of China (2016YFD0102000), the National Transgenic Key Project of the Ministry of Agriculture (2018ZX0800909B and 2016ZX08002-002), the National Natural Science Foundation of China (31871624) and the Major Research Development Program of Shaanxi Province (2018NY-026).
Conflicts of Interest
The authors declare that they have no competing interests.
References
- 1.Peng X., Ma X., Fan W., Man S., Cheng L., Alam I., Lee B.H., Qi D., Shen S., Liu G. Improved drought and salt tolerance of Arabidopsis thaliana by transgenic expression of a novel DREB gene from Leymus chinensis. Plant Cell Rep. 2011;30:1493–1502. doi: 10.1007/s00299-011-1058-2. [DOI] [PubMed] [Google Scholar]
- 2.Hong C., Cheng D., Zhang G., Zhu D., Chen Y., Tan M. The role of ZmWRKY4 in regulating maize antioxidant defense under cadmium stress. Biochem. Bioph. Res. Commun. 2016;482:1504–1510. doi: 10.1016/j.bbrc.2016.12.064. [DOI] [PubMed] [Google Scholar]
- 3.Fu J., Liu Q., Wang C., Liang J., Liu L., Wang Q. ZmWRKY79 positively regulates maize phytoalexin biosynthetic gene expression and is involved in stress response. J. Exp. Bot. 2017;69:497–510. doi: 10.1093/jxb/erx436. [DOI] [PubMed] [Google Scholar]
- 4.Liu Q.L., Xu K.D., Pan Y.Z., Jiang B.B., Liu G.L., Jia Y., Zhang H.Q. Functional Analysis of a Novel Chrysanthemum WRKY Transcription Factor Gene Involved in Salt Tolerance. Plant Mol. Biol. Rep. 2014;32:282–289. doi: 10.1007/s11105-013-0639-3. [DOI] [Google Scholar]
- 5.Zhu J.K. Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 2001;4:401–406. doi: 10.1016/S1369-5266(00)00192-8. [DOI] [PubMed] [Google Scholar]
- 6.Horie T., Hauser F., Schroeder J.I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009;14:660–668. doi: 10.1016/j.tplants.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi R., Liu S., Takano T. Cloning and functional comparison of a high-affinity K+ transporter gene PhaHKT1 of salt-tolerant and salt-sensitive reed plants. J. Exp. Bot. 2007;58:4387–4395. doi: 10.1093/jxb/erm306. [DOI] [PubMed] [Google Scholar]
- 8.Guan R., Qu Y., Guo Y., Yu L., Liu Y., Jiang J., Chen J., Ren Y., Liu G., Tian L. Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 2015;80:937–950. doi: 10.1111/tpj.12695. [DOI] [PubMed] [Google Scholar]
- 9.Yokoi S., Quintero F.J., Cubero B., Ruiz M.T., Bressan R.A., Hasegawa P.M., Pardo J.M. Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 2010;30:529–539. doi: 10.1046/j.1365-313X.2002.01309.x. [DOI] [PubMed] [Google Scholar]
- 10.Rodríguez-Rosales M.P., Gálvez F.J., Huertas R., Aranda M.N., Baghour M., Cagnac O., Venema K. Plant NHX cation/proton antiporters. Plant Signal. Behav. 2009;4:265–276. doi: 10.4161/psb.4.4.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shigaki T., Hirschi K. Characterization of CAX-like genes in plants: Implications for functional diversity. Gene. 2000;257:291–298. doi: 10.1016/S0378-1119(00)00390-5. [DOI] [PubMed] [Google Scholar]
- 12.Shigaki T., Hirschi K.D. Diverse functions and molecular properties emerging for CAX cation/H+ exchangers in plants. Plant Biol. 2006;8:419–429. doi: 10.1055/s-2006-923950. [DOI] [PubMed] [Google Scholar]
- 13.Qi X., Li M.W., Xie M., Liu X., Ni M., Shao G., Song C., Kay-Yuen Y.A., Tao Y., Wong F.L. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 2014;5 doi: 10.1038/ncomms5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J.W., Yang F.P., Chen X.Q., Liang R.Q., Zhang L.Q., Geng D.M., Zhang X.D., Song Y.Z., Zhang G.S. Induced expression of DREB transcriptional factor and study on its physiological effects of drought tolerance in transgenic wheat. Acta Genetica Sinica. 2006;33:468–476. doi: 10.1016/S0379-4172(06)60074-7. [DOI] [PubMed] [Google Scholar]
- 15.Morran S., Eini O., Pyvovarenko T., Parent B., Singh R., Ismagul A., Eliby S., Shirley N., Langridge P., Lopato S. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 2011;9:230–249. doi: 10.1111/j.1467-7652.2010.00547.x. [DOI] [PubMed] [Google Scholar]
- 16.Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 18.Eulgem T., Somssich I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007;10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 19.Hao Y.J., Wei W., Song Q.X., Chen H.W., Zhang Y.Q., Wang F., Zou H.F., Lei G., Tian A.G., Zhang W.K. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011;68:302–313. doi: 10.1111/j.1365-313X.2011.04687.x. [DOI] [PubMed] [Google Scholar]
- 20.Jin H.X., Huang F., Cheng H., Song H.N., Yu D.Y. Overexpression of the GmNAC2 Gene, an NAC Transcription Factor, Reduces Abiotic Stress Tolerance in Tobacco. Plant Mol. Biol. Rep. 2013;31:435–442. doi: 10.1007/s11105-012-0514-7. [DOI] [Google Scholar]
- 21.Guerinot M.L. The ZIP family of metal transporters. BBA Biomembr. 2000;1465:190–198. doi: 10.1016/S0005-2736(00)00138-3. [DOI] [PubMed] [Google Scholar]
- 22.Liao Y., Zou H.F., Wei W., Hao Y.J., Tian A.G., Huang J., Liu Y.F., Zhang J.S., Chen S.Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta. 2008;228:225–240. doi: 10.1007/s00425-008-0731-3. [DOI] [PubMed] [Google Scholar]
- 23.Xu Z.S., Chen M., Ma L.C., Ma Y.Z. Functions and application of the AP2/ERF franscription factor family in crop improvement. Bull Bot. 2011;61:570–585. doi: 10.1111/j.1744-7909.2011.01062.x. [DOI] [PubMed] [Google Scholar]
- 24.Xu Z.S., Chen M., Li L.C., Ma Y.Z. Functions of the ERF transcription factor family in plants. Botany. 2008;86:969–977. doi: 10.1139/B08-041. [DOI] [Google Scholar]
- 25.Fukuda A., Okada Y., Suzui N., Fujiwara T., Yoneyama T., Hayashi H. Cloning and characterization of the gene for a phloem-specific glutathione S -transferase from rice leaves. Physiol. Plant. 2010;120:595–602. doi: 10.1111/j.0031-9317.2004.0253.x. [DOI] [PubMed] [Google Scholar]
- 26.Hundertmark M., Hincha D.K. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9 doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan J., Wang B., Jiang Y., Cheng L., Wu T. GmFNSII-Controlled Soybean Flavone Metabolism Responds to Abiotic Stresses and Regulates Plant Salt Tolerance. Plant Cell Physiol. 2014;55:74–86. doi: 10.1093/pcp/pct159. [DOI] [PubMed] [Google Scholar]
- 28.Fei C., Yue H., Vannozzi A., Wu K., Cai H., Yuan Q. The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 2018;36:1–25. [Google Scholar]
- 29.Rushton P.J., Somssich I.E., Ringler P., Shen Q.J. WRKY transcription factors. Plant Signal. Behav. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Yu Y., Wang N., Hu R., Xiang F. Genome-wide identification of soybean WRKY transcription factors in response to salt stress. Springerplus. 2016;5 doi: 10.1186/s40064-016-2647-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagacã M., Matton D.P. Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta. 2004;219:185–189. doi: 10.1007/s00425-004-1253-2. [DOI] [PubMed] [Google Scholar]
- 32.Zentella R., Zhang Z., Park M., Thomas S., Endo A., Murase K., Fleet C., Jikumaru Y., Nambara E., Kamiya Y. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19:3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silke R., Imre E.S. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002;16:1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson C.S., Kolevski B., Smyth D.R. Transparent TESTA glabra2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14:1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pandey S.P., Somssich I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150:1648–1655. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Birkenbihl R.P., Diezel C., Somssich I.E. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 2012;159:266–285. doi: 10.1104/pp.111.192641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan Y., Jiang Y., Ye S., Karim A., Ling Z., He Y., Yang S., Luo K. PtrWRKY73, a salicylic acid-inducible poplar WRKY transcription factor, is involved in disease resistance in Arabidopsis thaliana. Plant Cell Rep. 2015;34:831–841. doi: 10.1007/s00299-015-1745-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu Y.P., Yu D.Q. Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 2009;65:35–47. doi: 10.1016/j.envexpbot.2008.07.002. [DOI] [Google Scholar]
- 39.Zhang L., Gu L., Ringler P., Smith S., Rushton P.J., Shen Q.J. Three WRKY transcription factors additively repress abscisic acid and gibberellin signaling in aleurone cells. Int. J. Exp. Plant Biol. 2015;236:214–222. doi: 10.1016/j.plantsci.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 40.Zou X., Seemann J.R., Neuman D., Shen Q.J. A WRKY gene from creosote bush encodes an activator of the abscisic acid signaling pathway. J. Biol. Chem. 2004;279:55770–55779. doi: 10.1074/jbc.M408536200. [DOI] [PubMed] [Google Scholar]
- 41.Shimono M., Sugano S., Nakayama A., Jiang C.J., Ono K., Toki S., Takatsuji H. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell. 2007;19:2064–2076. doi: 10.1105/tpc.106.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen L., Song Y., Li S., Zhang L., Zou C., Yu D. The role of WRKY transcription factors in plant abiotic stresses. BBA Gene Regul. Mech. 2012;1819:120–128. doi: 10.1016/j.bbagrm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Chen J., Nolan T., Ye H., Zhang M., Tong H., Xin P., Chu J., Chu C., Li Z., Yin Y. Arabidopsis WRKY46, WRKY54 and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought response. Plant Cell. 2017;29:1425–1439. doi: 10.1105/tpc.17.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erpen L., Devi H.S., Grosser J.W., Dutt M. Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ. 2018;132:1–25. doi: 10.1007/s11240-017-1320-6. [DOI] [Google Scholar]
- 45.Taji T., Ohsumi C., Iuchi S., Seki M., Kasuga M., Kobayashi M., YamaguchiShinozaki K., Shinozaki K. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 2010;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- 46.Wang C.T., Ru J.N., Liu Y.W., Li M., Zhao D., Yang J.F., Fu J.D., Xu Z.S. Maize WRKY Transcription Factor ZmWRKY106 Confers Drought and Heat Tolerance in Transgenic Plants. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19103046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He G.H., Xu J.Y., Wang Y.X., Liu J.M., Li P.S., Ming C., Ma Y.Z., Xu Z.S. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016;16 doi: 10.1186/s12870-016-0806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang C.T., Ru J.N., Liu Y.W., Yang J.F., Li M., Xu Z.S., Fu J.D. The Maize WRKY transcription factor ZmWRKY40 confers drought resistance in transgenic Arabidopsis. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19092580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulker B., Somssich I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004;7:491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 50.Jiang Y., Qiu Y., Hu Y., Yu D. Heterologous expression of AtWRKY57 confers drought tolerance in Oryza sativa. Front. Plant Sci. 2016;7 doi: 10.3389/fpls.2016.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niu C.F., Wei W., Zhou Q.Y., Tian A.G., Hao Y.J., Zhang W.K., Ma B., Lin Q., Zhang Z.B., Zhang J.S. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012;35:1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x. [DOI] [PubMed] [Google Scholar]
- 52.Wu X., Shiroto Y., Kishitani S., Ito Y., Toriyama K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009;28:21–30. doi: 10.1007/s00299-008-0614-x. [DOI] [PubMed] [Google Scholar]
- 53.Lee H., Cha J., Choi C., Choi N., Ji H.S., Park S.R., Lee S., Hwang D.J. Rice WRKY11 plays a role in pathogen defense and drought tolerance. Rice. 2018;11 doi: 10.1186/s12284-018-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cai R., Zhao Y., Wang Y., Lin Y., Peng X., Li Q., Chang Y., Jiang H., Xiang Y., Cheng B. Overexpression of a maize WRKY58 gene enhances drought and salt tolerance in transgenic rice. Plant Cell Tissue Organ. 2014;119:565–577. doi: 10.1007/s11240-014-0556-7. [DOI] [Google Scholar]
- 55.Li H., Gao Y., Xu H., Dai Y., Deng D., Chen J. ZmWRKY33, a WRKY maize transcription factor conferring enhanced salt stress tolerances in Arabidopsis. Plant Growth Regul. 2013;70:207–216. doi: 10.1007/s10725-013-9792-9. [DOI] [Google Scholar]
- 56.Ullah A., Sun H., Hakim, Yang X., Zhang X. A novel cotton WRKY-gene, GhWRKY6-like, improves salt tolerance by activating the ABA signalling pathway and scavenging of reactive oxygen species. Physiol. Plant. 2017;162:439–454. doi: 10.1111/ppl.12651. [DOI] [PubMed] [Google Scholar]
- 57.Song H., Wang P., Hou L., Zhao S., Zhao C., Xia H., Li P., Zhang Y., Bian X., Wang X. Global analysis of WRKY genes and their response to dehydration and salt stress in soybean. Front. Plant Sci. 2016;7 doi: 10.3389/fpls.2016.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Q.Y., Tian A.G., Zou H.F., Xie Z.M., Lei G., Huang J., Wang C.M., Wang H.W., Zhang J.S., Chen S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21 and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2010;6:486–503. doi: 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- 59.Li J., Wang J., Wang N., Guo X., Gao Z. GhWRKY44, a WRKY transcription factor of cotton, mediates defense responses to pathogen infection in transgenic Nicotiana benthamiana. Plant Cell Tissue Organ. 2015;121:127–140. doi: 10.1007/s11240-014-0688-9. [DOI] [Google Scholar]
- 60.Liu X., Song Y., Xing F., Wang N., Wen F., Zhu C. GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana. Protoplasma. 2015;253:1–17. doi: 10.1007/s00709-015-0885-3. [DOI] [PubMed] [Google Scholar]
- 61.Wang Y., Jiang L., Chen J., Tao L., An Y., Cai H. Overexpression of the alfalfa WRKY11 gene enhances salt tolerance in soybean. PLoS ONE. 2018;13:e0192382. doi: 10.1371/journal.pone.0192382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mehdi P., Vijayaraj N., Youping D. GeneVenn—A web application for comparing gene lists using Venn diagrams. Bioinformation. 2007;1:420–422. doi: 10.6026/97320630001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berri S., Abbruscato P., FaivreRampant O., Brasileiro A.C., Fumasoni I., Satoh K., Kikuchi S., Mizzi L., Morandini P., Pè M.E. Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biol. 2009;9:1–22. doi: 10.1186/1471-2229-9-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo C., Guo R., Xu X., Gao M., Li X., Song J., Zheng Y., Wang X. Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 2014;65:1513–1528. doi: 10.1093/jxb/eru007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang S., Gao Y., Liu J., Peng X., Niu X., Fei Z., Cao S., Liu Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genomics. 2012;287:495–513. doi: 10.1007/s00438-012-0696-6. [DOI] [PubMed] [Google Scholar]
- 66.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miller M.J., Barrett-Wilt G.A., Zhihua H., Vierstra R.D. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2011;107:16512–16517. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Du Y.T., Zhao M.J., Wang C.T., Gao Y., Wang Y.X., Liu Y.W., Chen M., Chen J., Zhou Y.B., Xu Z.S. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018;18 doi: 10.1186/s12870-018-1551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kereszt A., Li D., Indrasumunar A., Nguyen C.D., Nontachaiyapoom S., Kinkema M., Gresshoff P.M. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007;2:948–952. doi: 10.1038/nprot.2007.141. [DOI] [PubMed] [Google Scholar]
- 70.Cao D., Hou W.S., Song S.K., Sun H.B., Wu C.X., Gao Y.S., Han T.F. Assessment of conditions affecting Agrobacterium rhizogenes-mediated transformation of soybean. Plant Cell Tissue Organ. 2009;96:45–52. doi: 10.1007/s11240-008-9458-x. [DOI] [Google Scholar]
- 71.Wang F., Chen H.W., Li Q.T., Wei W., Li W., Zhang W.K., Ma B., Bi Y.D., Lai Y.C., Liu X.L. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 2015;83:224–236. doi: 10.1111/tpj.12879. [DOI] [PubMed] [Google Scholar]
- 72.Ishiguro S., Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Genet. Genomics. 1994;244:563–571. doi: 10.1007/BF00282746. [DOI] [PubMed] [Google Scholar]
- 73.Wei K.F., Chen J., Chen Y.F., Wu L.J., Xie D.X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in Maize. DNA Res. 2012;19:153–164. doi: 10.1093/dnares/dsr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu K.L. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005;12:9–26. doi: 10.1093/dnares/12.1.9. [DOI] [PubMed] [Google Scholar]
- 75.Jue D., Sang X., Liu L., Shu B., Wang Y., Liu C., Xie J., Shi S. Identification of WRKY Gene Family from Dimocarpus longan and its expression analysis during flower induction and abiotic stress responses. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19082169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim C.Y., Vo K.T.X., Cong D.N., Jeong D.H., Lee S.K., Kumar M., Kim S.R., Park S.H., Kim J.K., Jeon J.S. Functional analysis of a cold-responsive rice WRKY gene, OsWRKY71. Plant Biotechnol. Rep. 2016;10:13–23. doi: 10.1007/s11816-015-0383-2. [DOI] [Google Scholar]
- 77.Ding M., Chen J., Jiang Y., Lin L., Cao Y., Wang M., Zhang Y., Rong J., Ye W. Genome-wide investigation and transcriptome analysis of the WRKY gene family in Gossypium. Mol. Genet. Genomics. 2015;290:151–171. doi: 10.1007/s00438-014-0904-7. [DOI] [PubMed] [Google Scholar]
- 78.Shi J.N., Li-Yun L.I., Wen-Jing X.U., Guan M.L., Xue-Jiao L.I., Niu D.D., Lan J.P., Dou S.J., Liu L.J., Liu G.Z. Expression analysis of eight WRKY transcription factors in rice leaf growth and disease resistance response. Acta Phytopathol. Sin. 2014;44:54–64. [Google Scholar]
- 79.Yu Y., Hu R., Wang H., Cao Y., He G., Fu C., Zhou G. MlWRKY12, a novel Miscanthus transcription factor, participates in pith secondary cell wall formation and promotes flowering. Int. J. Exp. Plant Biol. 2013;212:1–9. doi: 10.1016/j.plantsci.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 80.Li W., Wang H., Yu D. Arabidopsis WRKY Transcription Factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions. Mol. Plant. 2016;9:1492–1503. doi: 10.1016/j.molp.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 81.Li X.J., Guo C.J., Lu W.J., Duan W.W., Zhao M., Ma C.Y., Gu J.T., Xiao K. expression pattern analysis of Zinc finger protein genes in wheat (Triticum aestivum L.) under phosphorus deprivation. J. Integr. Agric. 2014;13:1621–1633. doi: 10.1016/S2095-3119(13)60739-X. [DOI] [Google Scholar]
- 82.Van Verk M.C., Neeleman L., Bol J.F., Linthorst H.J. Tobacco transcription factor NtWRKY12 interacts with TGA2.2 in vitro and in vivo. Front. Plant Sci. 2011;2:1085–1091. doi: 10.3389/fpls.2011.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thurow C., Schiermeyer A., Krawczyk S., Butterbrodt T., Nickolov K., Gatz C. Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J. 2010;44:100–113. doi: 10.1111/j.1365-313X.2005.02513.x. [DOI] [PubMed] [Google Scholar]
- 84.Kim H.S., Park Y.H., Nam H., Lee Y.M., Song K., Choi C., Ahn I., Park S.R., Lee Y.H., Hwang D.J. Overexpression of the Brassica rapa transcription factor WRKY12 results in reduced soft rot symptoms caused by Pectobacterium carotovorum in Arabidopsis and Chinese cabbage. Plant Biol. 2015;16:973–981. doi: 10.1111/plb.12149. [DOI] [PubMed] [Google Scholar]
- 85.Cui Q., Yan X., Gao X., Zhang D.M., He H.B., Jia G.X. Analysis of WRKY transcription factors and characterization of two Botrytis cinerea-responsive LrWRKY genes from Lilium regale. Plant Physiol. Biochem. 2018:525–536. doi: 10.1016/j.plaphy.2018.04.027. [DOI] [PubMed] [Google Scholar]
- 86.Wang J.X., Tang Y.K., Liu Q., Liang C.Q., Wang Y.C. Cloning and expression analysis of THWRKY12 gene from Tamarix hispida. Chin. J. Agric. Biotechol. 2013;5:55–57. [Google Scholar]
- 87.Baranwal V.K., Negi N., Khurana P. Genome-wide identification and structural, functional and evolutionary analysis of WRKY components of Mulberry. Sci. Rep. 2016;6 doi: 10.1038/srep30794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tao X., Chen C., Li C., Liu J., Liu C., He Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genomics. 2018;19 doi: 10.1186/s12864-018-4880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tan C.K., Carey A.J., Cui X., Webb R.I., Ipe D., Crowley M., Cripps A.W., Benjamin B.W., Jr., Ulett K.B., Schembri M.A. Genome-wide mapping of cystitis due to Streptococcus agalactiae and Escherichia coli in mice identifies a unique bladder transcriptome that signifies pathogen-specific antimicrobial defense against urinary tract infection. Infect. Immun. 2012;80:3145–3160. doi: 10.1128/IAI.00023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.