Abstract
Four new depsidones, mollicellins O–R (compounds 1–4), along with three known compounds 5–7, were isolated from cultures of the fungus Chaetomium sp. Eef-10, an endophyte isolated from Eucalyptus exserta. The structures of the new compounds were elucidated by analysis of the 1D and 2D NMR and HR-ESI-MS spectra. The known compounds were identified by comparison of their spectral data with published values. Compounds 1–7 were evaluated for antibacterial activities against Staphylococcus aureus (sensitive and resistant strains), Escherichia coli, Agrobacterium tumefaciens, Salmonella typhimurium, Pseudomonas lachrymans, Ralstonia solanacearum, Xanthomonas vesicatoria and cytotoxic activities against two human cancer cell lines (HepG2 and Hela). Mollicellin H (6) displayed best antibacterial activity, with IC50 values of 5.14 µg/mL against S. aureus ATCC29213 and 6.21 µg/mL against S. aureus N50, MRSA, respectively. Mollicellin O (1) and mollicellin I (7) also exhibited antibacterial activities against S. aureus ATCC29213 and S. aureus N50. Mollicellin G (5) was active against both two human cancer cell lines, with IC50 values of 19.64 and 13.97 µg/mL while compounds 6 and 7 only showed cytotoxic activity against one cell line. In addition, mollicellin O (1) showed antioxidant activity based on DPPH radical scavenging, with an IC50 value of 71.92 µg/mL.
Keywords: endophytic fungus, secondary metabolites, Chaetomium sp. Eef-10, antibacterial activity, cytotoxic activity, antioxidant activity
1. Introduction
Plant endophytes are regarded as a sort of particular-biotope microorganisms that reside in the tissues of living plants without causing any immediate obvious negative effects in their host plants [1,2]. Metabolites of endophytic fungi such as taxol, sanguinarine and gallic acid are known to possess various rare and novel scaffolds with significant biological activities, including anticancer, antimicrobial and antioxidant activities [3,4,5]. Recently, a great deal of interest has been generated in discovery of remarkable pharmacological agents from endophytic fungi [1]. In the past decades, the Chaetomium genus hase been revealed to be a rich source of natural products, such as alkaloids [6,7,8,9], azaphilones [10,11,12,13,14], chaetoglobosins [15,16,17,18], chaetoindicins [19], diketopiperazines [20,21,22] and terpenoids [23]. Depsidones are a group of fungal metabolites that have attracted the attention of natural product chemists due to their diversified structures and potent biological activities. To date, Chaetomium species have been reported to produce about 14 depsidones, the mollicellins A-N [24,25,26]. Recently, we focused our attention on Chaetomium sp. Eef-10, a fungal strain isolated from the fruits of Eucalyptus exserta, and, as a result, four new (compounds 1–4) and three known depsidones 5–7 were isolated and elucidated (Figure 1). Details of the structure determination and biological activities of these compounds are presented herein.
Figure 1.
Chemical structures of compounds 1–7.
2. Results
2.1. Purification and Characterization
The solid fermentation product of Chaetomium sp. Eef-10 was extracted with methanol (MeOH) and the resulting extracts were partitioned into petroleum ether-, ethyl acetate (EtOAc)- and H2O- soluble fractions. The petroleum ether and EtOAc fractions were further purified by conventional chromatographic techniques to obtain seven compounds 1–7, and their structures were elucidated by 1D and 2D NMR spectroscopy and by comparison with literature data.
2.2. Structure Elucidation of Compounds 1–4
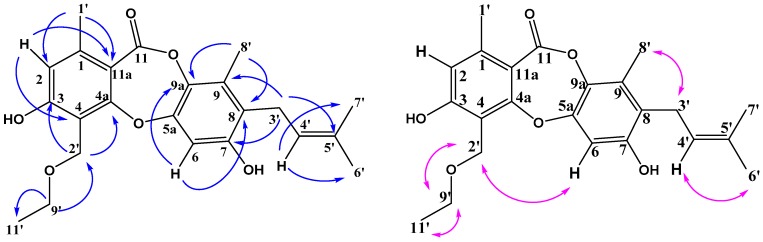
Compound 1 was obtained as a white solid. Its molecular formula was established as C23H26O6, as a prominent pseudomolecular ion peak was observed at m/z 421.1616 [M + Na]+ in the HR-ESI-MS spectrum (Figure S1.8). The IR spectrum exhibited absorption bands at 3308, 1693 cm−1 for hydroxy and lactone carbonyl functional groups (Figure S1.2). The 13C-NMR (Table 1) and HSQC spectra (Figure S1.5) suggested 23 carbon signals for one conjugated ester carbonyl (δC 164.0), eleven quaternary carbons of which five were O-substituted aromatic quaternary carbons (δC 162.45, 161.16, 152.57, 149.90, and 137.00), three methine (δC 123.08, 116.32 and 106.36), two oxymethylene (δC 66.27 and 62.35), one methylene (δC 25.76), and five methyl (δC 25.86, 21.03, 17.94, 15.53 and 12.64) carbons of which four were singlets. According to the above data, two benzene rings and one lactone were also present in the structure. The 1H-NMR spectrum (Table 2) of 1 showed the presence of a hydroxyl proton at δH 9.02 (2H, br s, 3,7-ΟН), two aromatic protons of different pentasubstituted benzenes at δH 6.86 (1H, s, Н-6) and 6.68 (1H, s, Н-2), one double bond proton at δH 5.02 (1H, t, Н-4′), two oxymethylene protons at δH 4.77 (2H, s, H-2′), and a 3-methylbut-2-enyl side chain was revealed by the signals at δH 1.62 (s, H-6′), 1.74 (s, H-7′), 3.33 (d, H-3′), and 5.02 (t, H-4′). In addition, the NOESY spectrum of 1 demonstrated a correlation between H-2′ and H-6 (Figure 2) which indicated that C-8′ was a methyl group. Compared with the NMR data of the known depsidone mollicellin I (7) [25], compound 1 had a similar structure except for one additional ethoxy group (δH 3.60, 2H, q; δH 1.19, 3H, t; 2′-OCH2CH3). The HMBC correlations of δH 3.60 (q, H-9′) with δC 15.55 (C-11′) and 62.35 (C-2′) suggested that the structure of 1 was ethoxylated at C (2′) (Figure 2). Compound 1 was named mollicellin O.
Table 1.
13C-NMR Data (δ, ppm) of compounds 1–7 (1–5, 7 in acetone-d6, 6 in DMSO-d6).
| Position | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| 1 | 144.60 | 144.74 | 150.66 | 154.10 | 153.72 | 151.66 | 144.02 |
| 2 | 116.32 | 116.76 | 116.43 | 118.14 | 117.93 | 117.60 | 116.45 |
| 3 | 161.16 | 161.81 | 161.31 | 165.90 | 166.05 | 164.04 | 161.35 |
| 4 | 114.41 | 114.66 | 114.54 | 111.69 | 111.90 | 112.06 | 116.59 |
| 4a | 162.45 | 162.43 | 162.33 | 165.37 | 166.56 | 164.53 | 161.37 |
| 5a | 149.90 | 153.32 | 144.57 | 151.07 | 143.03 | 148.56 | 149.96 |
| 6 | 106.36 | 101.39 | 108.11 | 107.3 | 130.95 | 105.07 | 106.21 |
| 7 | 152.57 | 157.92 | 150.98 | 151.41 | 143.35 | 152.58 | 152.70 |
| 8 | 125.73 | 127.03 | 118.38 | 119.49 | 123.01 | 125.56 | 125.82 |
| 9 | 129.90 | 128.85 | 129.40 | 127.22 | 105.82 | 129.47 | 129.99 |
| 9a | 137.00 | 138.01 | 137.35 | 137.34 | 154.62 | 135.24 | 137.05 |
| 11 | 164.02 | 163.81 | 163.93 | 162.51 | 162.82 | 162.76 | 164.08 |
| 11a | 114.00 | 113.68 | 113.97 | 113.97 | 114.21 | 112.94 | 113.95 |
| 1′ | 21.03 | 21.03 | 21.01 | 22.07 | 22.27 | 21.80 | 20.92 |
| 2′ | 62.35 | 62.77 | 62.61 | 194.92 | 194.70 | 191.53 | 56.12 |
| 3′ | 25.76 | 72.87 | 30.34 | 119.11 | 25.86 | 25.32 | 25.78 |
| 4′ | 123.08 | 99.27 | 69.64 | 132.46 | 126.01 | 122.39 | 123.15 |
| 5′ | 131.81 | 71.19 | 77.62 | 76.75 | 132.20 | 131.35 | 131.85 |
| 6′ | 25.86 | 26.15 | 25.82 | 27.81 | 25.95 | 25.85 | 25.91 |
| 7′ | 17.94 | 25.50 | 20.43 | 27.81 | 18.06 | 18.21 | 17.97 |
| 8′ | 12.64 | 12.36 | 12.24 | 11.75 | 13.49 | 12.81 | 12.67 |
| 9′ | 66.27 | 66.47 | 66.36 | - | - | - | - |
| 11′ | 15.53 | 15.58 | 15.50 | - | - | - | - |
Table 2.
1H-NMR Data (δ, ppm) of compounds 1–7 (1–5, 7 in acetone-d6, 6 in DMSO-d6).
| Position | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| 2 | 6.68 (s) | 6.71 (s) | 6.69 (s) | 6.80 (s) | 6.67 (s) | 6.80 (s) | 6.64 (s) |
| 6 | 6.86 (s) | 6.72 (s) | 6.71 (s) | 6.85 (s) | - | 6.79 (s) | 6.80 (s) |
| 9 | - | - | - | - | 6.78 (s) | - | - |
| 1′ | 2.36 (s) | 2.37 (s) | 2.37 (s) | 2.51 (s) | 2.49 (s) | 2.40 (s) | 2.36 (s) |
| 2′ | 4.77 (s) | 4.81 (s) | 4.82 (s) | 10.68 (s) | 10.77 (s) | 10.49 (s) | 4.99 (s) |
| 3′a | 3.33 (d, 6.2) | 5.42 (d, 3.8) | 2.86 (dd, 16.8, 5.6) | 6.58 (d,10.2) | 3.37 (d, 6.9) | 3.22 (d, 6.9) | 3.33 (d, 6.9) |
| 3′b | - | - | 2.52 (dd, 16.9, 7.7) | - | - | - | - |
| 4′ | 5.02 (t, 6.2) | 4.29 (d, 3.8) | 3.78 (dd, 7.6, 5.6) | 5.83 (d,10.2) | 5.03 (m) | 4.95 (t, 7.0) | 5.02 (m) |
| 6′ | 1.62 (s) | 1.19 (s) | 1.18 (s) | 1.37 (s) | 1.63 (s) | 1.60 (s) | 1.63 (s) |
| 7′ | 1.74 (s) | 1.25 (s) | 1.30 (s) | 1.37 (s) | 1.75 (s) | 1.70 (s) | 1.75 (s) |
| 8′ | 2.23 (s) | 2.35 (s) | 2.18 (s) | 2.30 (s) | 2.37 (s) | 2.18 (s) | 2.23 (s) |
| 9′ | 3.60 (q, 7.0) | 3.64 (q, 10.2) | 3.63 (q, 7.0) | - | - | - | - |
| 11′ | 1.19 (t, 7.0) | 1.21 (t, 10.2) | 1.21 (t, 7.0) | - | - | - | - |
| OH-3 | 9.02 (br s) | - | - | 12.25 (s) | 12.22 (s) | - | - |
| OH-7 | 9.02 (br s) | - | - | - | - | 9.70 (s) | - |
| OH-8 | - | - | - | - | 8.82 (s) | - | - |
Figure 2.
Key HMBC (H-C) correlations and NOESY (H-H) correlations of mollicellin O (1).
Compound 2 was isolated as a white solid. Its HR-ESI-MS spectrum (Figure S2.8) showed pseudomolecular ion peaks at m/z 431.1696 [M + H]+ and 453.1518 [M + Na]+, consistent with the molecular formula C23H26O8, indicating 11 degrees of unsaturation. The IR spectrum bands at 3363, 1705 cm−1 suggested hydroxy and lactone carbonyl functional groups (Figure S2.2). The UV, 1H- and 13C-NMR spectra of 2 were similar to those of 1, except for the 3-methylbut-2-enyl side chain. The signals of two geminal methyl groups, both at δH 1.19 (s, H-6′), 1.25 (s, H-7′) and two oxymethine protons at δH 4.29 (d, H-4′), 5.42 (d, H-3′) with the same coupling constant of 3.8 Hz were also observed. The HMBC correlations of H-4′ to C-3′ and C-5′; H-6′ to C-4′, C-5′ and C-7′; H-7′ to C-4′, C-5′ and C-6′, together with the degree of unsaturation, suggested that the prenyl group formed a dihydropyran ring with C-8 and C-7. The small value of the coupling constant between H-3 and H-4 (J = 3.8 Hz) indicated that the relative stereochemistry of the two hydroxyl groups at C-3 and C-4 was cis-. HMBC and NOESY data are shown in the Figure 3. On the basis of the above data, the structure of 2 was determined as indicated in Figure 1 and it was named mollicellin P.
Figure 3.
Key HMBC (H-C) correlations and NOESY (H-H) correlations of mollicellin P (2).
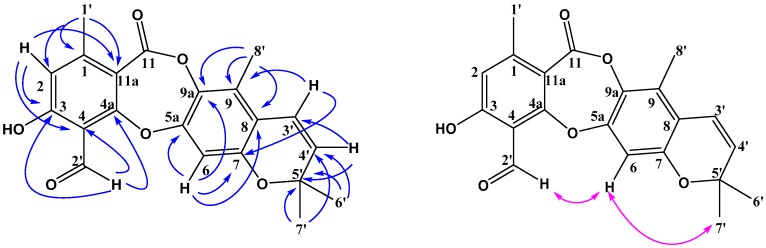
Compound 3 was obtained as a white solid, and its molecular formula C23H26O7 was established by the quasimolecular ion peak at m/z 415.1751 [M + H]+ and 43701565 [M + Na]+ in the HR-ESI-MS (Figure S3.8). The IR bands at 3273 and 1732 cm−1 indicated there were hydroxy and lactone carbonyl groups in compound 3 (Figure S3.2). The UV, 1H- and 13C-NMR spectra of 3 were also similar to those of 1 except for the 3-methylbut-2-enyl side chain. The signals of two geminal methyl groups both appeared at δH 1.18 (s, H-6′), 1.30 (s, H-7′). Only one oxymethine proton at δH 3.78 (t) was seen in the 1H-NMR, and there was one oxygen atom missing in the molecular formula of compound 3 compared with compound 2, HMBC signals of H-4′/C-6′ and C-7′ also suggested there was a hydroxyl group at C-4′ and C-3′ was an unoxygenated carbon. The main HMBC and NOESY data are shown in Figure 4, the structure of 3 is shown in Figure 1 and it was named mollicellin Q.
Figure 4.
Key HMBC (H-C) correlations and NOESY (H-H) correlations of mollicellin Q (3).
Compound 4 was also a white solid, and its molecular formula C21H18O6 was obtained according to the quasimolecular ion peak at m/z 367.1186 [M + H]+ in the HR-ESI-MS (Figure S4.8), indicating 13 degrees of unsaturation. Its IR spectrum (Figure S4.2) exhibited absorption bands at 1726 cm−1 for a lactone carbonyl grous. Compared with 1–3, the 1H-NMR (Table 2) and 13C-NMR spectra (Table 1) indicated that compound 4 contained two aromatic rings, one lactone at δC 164.03, one double bond and one aldehyde group at δC 194.92, plus an additional ring according to the remaining one degree of unsaturation. Comparing the NMR data, the structure of compound 4 may be similar to that of compound 2. The 1H-NMR data showed two aromatic proton singlets at δH 6.86 (H-6) and 6.68 (H-2), as well as two aromatic methyl substituents at δH 2.36 (CH3-1′) and 2.23 (CH3-8′). The HMBC signals (Figure 5 or Figure S4.9) showed correlations of δH 10.68 (H-2′) to δ 165.90 (C-3), δC 111.69 (C-4) and δ 165.37 (C-4a), which suggested the aldehyde group was at C-2′. HMBC correlations of δH 6.58 (H-3′) to δC 151.41 (C-7), 119.49 (C-8), 127.22 (C-9); δH 5.83 (H-4′) to δC 119.49 (C-8), 27.81 (C-6′ and C-7′), together with the NOESY signal of H-6/H-7′, confirmed that the prenyl group formed a pyran ring with C-7 and C-8, so the structure of compound 4 was confirmed and it was named mollicellin R.
Figure 5.
Key HMBC (H-C) correlations and NOESY (H-H) correlations of mollicellin R (4).
The known compounds, mollicellins G–I (compounds 5–7) were identified by comparison of their physical and spectroscopic data with the values reported in the literature. We also provide the correct data and NMR spectra to further confirm the structures.
2.3. Antibacterial Assay
All isolated compounds except compound 2 were evaluated for their antibacterial activities against one Gram-positive bacterium, including drug sensitive and resistant strains and six Gram-negative bacteria. Among them, compounds 1, 6 and 7 showed antibacterial activity against S. aureus ATCC29213 and S. aureus N50 (MRSA) (Table 3). Furthermore, mollicellin H (6) displayed the best antibacterial activity with IC50 values of 5.14 µg/mL (against S. aureus ATCC29213) and 6.21 µg/mL (against S. aureus N50), respectively. However, no compounds showed inhibitory activity against the Gram-negative bacteria.
Table 3.
Antibacterial activities.
| Compound | IC50 (µg/mL) | |
|---|---|---|
| S. aureus ATCC29213 | S. aureus N50 | |
| Mollicellin O (1) | 79.44 | 76.35 |
| Mollicellin H (6) | 5.14 | 6.21 |
| Mollicellin I (7) | 70.14 | 63.15 |
| Streptomycin sulfate | 1.05 | - a |
a “-” indicates inactive (IC50 > 100 µg/mL).
2.4. Cytotoxicity Assay
The cytotoxicities of all the isolated compounds were evaluated against two human cancer cell lines (HepG2 and HeLa) using the MTT assay (Table 4). Mollicellin G (5) showed pronounced cytotoxic activities against the two tested cell lines, with IC50 values of 19.64 µg/mL against HepG2 and 13.97 µg/mL against HeLa, respectively. Mollicellin H (6) showed the best cytotoxic activity against HepG2 with an IC50 value of 6.83 µg/mL, while the IC50 value against the HeLa cell line was greater than 50 µg/mL. In addition, mollicellin I (7) only showed cytotoxicity against HeLa with an IC50 value of 21.35 µg/mL. The other compounds were inactive (IC50 > 50 µg/mL).
Table 4.
Cytotoxic activities.
| Compounds | IC50 (µg/mL) | |
|---|---|---|
| HepG2 | Hela | |
| Mollicellin G (5) | 19.64 | 13.97 |
| Mollicellin H (6) | 6.83 | - a |
| Mollicellin I (7) | - a | 21.35 |
| Camptothecin | 3.6 | 6.3 |
a “-” indicates inactive (IC50 > 50 µg/mL).
2.5. Antioxidant Assay
All the compounds were subjected to screening for possible antioxidant activity by the DPPH free radical scavenging assay (Table 5). Only mollicellin O (1) displayed weak antioxidant activity with an IC50 value of 71.92 µg/mL.
Table 5.
Antioxidant activities.
| Compound | IC50 (µg/mL) |
|---|---|
| Mollicellin O (1) | 71.92 ± 0.09 |
| BHT | 0.15 ± 0.03 |
3. Discussion
Endophytic fungi as important sources of natural active substances have been used to obtain novel bioactive secondary metabolites with potential applications in the agricultural and medical sectors, which can also act as lead targets of pharmaceutical and medicinal potential [27]. As part of our ongoing investigation on bioactive natural products from Eucalyptus-derived fungi, an endophytic fungus Chaetomium sp. Eef-10, which was first isolated from the healthy fruits of Eucalyptus exserta, attracted our attention because the extracts of the fungal culture exhibited significant antibacterial activity against S. aureus. According to a bioassay guided isolation, seven depsidones, including four new ones, were obtained from the extracts of Chaetomium sp. Eef-10. As stated previously, fourteen depsidones had been isolated from different Chaetomium species, and possessed excellent antibacterial activity against Gram-positive bacteria. All compounds in this study can be divided into two major types according to their skeletal structures. Compounds 1, 5, 6 and 7 have three rings, while compounds 2, 3 and 4 have four rings. Compounds 1, 5, 6 and 7 all have a 3-methylbut-2-enyl side chain and displayed stronger antibacterial and cytotoxic activity than the other compounds, which indicated that the side chain was the active group. In this study, mollicellin H (6) displayed best antibacterial activity, with an IC50 value of 5.14 µg/mL against S. aureus ATCC29213. What’s more, mollicellin H (6) also showed strong antibacterial activity against S. aureus N50 (MRSA) and the best cytotoxic activity against HepG2 cancer cell line. This compound could stimulate future potential antimicrobial and cytotoxic agent studies.
It was reported that naturally occurring depsidones possessed remarkable antibacterial, antiproliferative, cytotoxic, antioxidant, radical scavenging, antimalarial, antihypertensive, aromatase and cholinesterase inhibitor, and antifungal bioactivities [28], therefore they have potential beneficial human health effects, which is consistent with the findings of our study. With the rapid development of genetic engineering and metabolic regulation, the search for novel active depsidones through structural modifications could become an efficient approach to new natural medicines.
4. Materials and Methods
4.1. General Experimental Procedures
The UV spectra were scanned on a UV-2600 instrument (Shimadzu, Kyoto, Japan). IR spectra were recorded on a Shimadzu Affinity-1 instrument (Shimadzu, Japan). HRESIMS spectra were obtained on a Q-TOF mass spectrometer from Bruker maXis, with an electrospray ionization (ESI) interface (Bruker, Fremont, CA, USA). Standard 1D and 2D NMR spectra were recorded on a Bruker Avance-600 NMR spectrometer (1H at 600 MHz and 13C at 150 MHz) (Bruker, Fremont, CA, USA). Silica gel (200–300 mesh, Qingdao Marine Chemical Inc., Qingdao, China), and Sephadex LH-20 (GE Healthcare, Uppsala, Sweden) were used for column chromatography. Semi-preparative HPLC separations were carried out on a semi-preparative HPLC instrument equipped with an UC-3281 pump and a UC-3292S UV detector, using a XB-C18 column (250 mm × 10 mm, 10 μm, Welch, Shanghai, China). Precoated silica gel GF-254 plates (Qingdao Marine Chemical Inc., Qingdao, China) were used for analytical TLC. Spots were visualized under UV light (254 or 356 nm) or by spraying with 5% H2SO4 in 95% EtOH followed by heating.
4.2. Fungal Material
The fungus Chaetomium sp. Eef-10 was isolated from healthy seeds of Eucalyptus exserta collected in Guangdong Province in June 2016. The isolate was identified as a species of Chaetomium sp. by analysis of its morphological characteristics and the rDNA gene internal transcribed spacer (ITS) sequence (GenBank accession no. MK120863). The fungus was stored on potato dextrose agar (PDA) slants at 4 °C in College of Forestry and Landscape Architecture, South China Agricultural University, Guangzhou, China.
4.3. Fermentation, Extraction, and Isolation
The endophytic fungus was cultured on potato dextrose agar (PDA) (potato 200 g/L, dextrose 20 g/L, and agar 20 g/L) medium in Petri dishes at 25 °C for 5 days. Then, three agar plugs (0.5 × 0.5 cm) were inoculated in a 500 mL Erlenmeyer flask containing 200 mL of potato dextrose broth (PDB) (potato 200 g/L and dextrose 20 g/L) medium and incubated on a rotary shaker at 150 rpm and 25 °C for 7 days. The obtained liquid seeds were added into the sterilized rice culture medium (6 kg) in the incubators at 28 °C for 60 days before harvest. The leavening was extracted with methanol (MeOH). Then, the MeOH extracts were extracted successively at room temperature with petroleum ether, and ethyl acetate (EtOAc) to give crude extracts of 15.15 g and 51.40 g, respectively. The EtOAc extract was subjected to column chromatography over silica gel (200–300 mesh) eluted with petroleum ether (5 L), CH2Cl2-MeOH (10:1, v/v) (5 L), CH2Cl2-MeOH (5:1, v/v) (5 L), and MeOH (5 L) to give four fractions (Fr. A–D). Fraction B (25.19 g) was subjected to column chromatography over silica gel (200–300 mesh) eluting with a gradient of petroleum ether-CH2Cl2 (100:0–0:100) and CH2Cl2-MeOH (100:0–0:100) to afford eleven subfractions, B1–B11.
Subfraction B3 (1.212 g) was chromatographed over Sephadex LH-20 (eluted with CHCl3-MeOH, 1:1) to give six further subfractions, B3-1 to B3-6, and subfraction B3-4 (75 mg) was further purified by semi-preparative HPLC (MeOH-H2O, 78:22) to afford mollicellin R (4) (7 mg). Subfraction B6 (9.5 g) was divided into two parts, B6-1 to B6-2, by means of recrystallization purification method, and the mollicellin H (6) (3750 mg) was recrystallized from subfraction B6-2. The subfraction B6-1(600 mg) was chromatographed over Sephadex LH-20 (eluted with CHCl3-MeOH, 1:1) to give six further subfractions, B6-1-1 to B6-1-6, and the subfraction B6-1-5 (50 mg) was further purified to yield a colorless needle crystal, mollicellin G (5) (5 mg). Subfraction B7 (1.1 g) was chromatographed over Sephadex LH-20 (eluted with CHCl3-MeOH, 1:1) to give nine further subfractions, B7-1 to B7-9, and subfraction B7-4 (40 mg) was further purified by semi-preparative HPLC (MeOH-H2O, 70:30) to afford mollicellin Q (3) (6.0 mg), and subfraction B7-5 (73 mg) was further purified by semi-preparative HPLC (MeOH-H2O, 65:35) to afford mollicellin P (2) (1.6 mg), and subfraction B7-6 (70 mg) was further purified by semi-preparative HPLC (MeOH-H2O, 75:25) to afford mollicellin O (1) (24 mg). Subfraction B9 (0.58 g) was chromatographed over Sephadex LH-20 (eluted with CHCl3-MeOH, 1:1) to give eight further subfractions, B9-1 to B9-8, and the subfraction B9-5 (50 mg) was further purified to yield mollicellin I (7) (3 mg).
4.4. Compound Identification
Mollicellin O (1): white solid; UV (MeOH) λmax 208.40, 268.60 nm; IR νmax 3308, 2927, 2361, 1694, 1558, 1456, 1435, 1358, 1260, 1211, 1182, 1142, 1098, 1061, 1034, 854, 831, 793, 750 cm−1; HR-ESI-MS m/z 421.1616 [M + Na]+ (calcd for C23H26O6Na, 421.1622), 1H-NMR and 13C-NMR see Table 1 and Table 2.
Mollicellin P (2): white solid; UV (MeOH) λmax 207.40, 267.80 nm; IR νmax 3364, 3242, 2930, 1705, 1611, 1454, 1362, 1267, 1213, 1165, 1136, 1067, 1024, 978, 839, 791, 667 cm−1; HR-ESI-MS m/z 431.1696 [M + H]+ (calcd for C23H27O8, 431.1700), 453.1518 [M + Na]+ (calcd for C23H26O8Na, 453.1520), 1H-NMR and 13C-NMR see Table 1 and Table 2.
Mollicellin Q (3): white solid; UV (MeOH) λmax 206.40, 268.80 nm; IR νmax 3273, 2924, 1732, 1611, 1474, 1456, 1435, 1360, 1331, 1258, 1217, 1161, 1128, 1096, 1070, 1045, 999, 895, 849, 797, 750 cm−1; HR-ESI-MS m/z 415.1751 [M + H]+ (calcd for C23H27O7, 415.1751), 437.1565 [M + Na]+ (calcd for C23H26O7Na, 437.1571), 1H-NMR and 13C-NMR see Table 1 and Table 2.
Mollicellin R (4): white solid; UV (MeOH) λmax 203.00, 229.20, 317.40 nm; IR νmax 2926, 2363, 1726, 1647, 1636, 1576, 1558, 1541, 1474, 1456, 1437, 1389, 1375, 1341, 1300, 1254, 1215, 1202, 1152, 1130, 1111, 1070, 1032, 841, 768 cm−1; HR-ESI-MS m/z 367.1186 [M + H]+ (calcd for C21H19O6, 367.1176), 1H-NMR and 13C-NMR see Table 1 and Table 2.
Mollicellin G (5): colorless needles; UV (MeOH) λmax 205.40, 234.80 nm; IR νmax 3350, 2922, 1699, 1639, 1572, 1504, 1435, 1389, 1344, 1273, 1202, 1153, 1061, 1038, 853, 829, 802, 775 cm−1; HR-ESI-MS m/z 369.1335 [M + H]+ (calcd for C21H21O6, 369.1333), 391.1151 [M + Na]+ (calcd for C21H20O6Na, 391.1152), 1H-NMR and 13C-NMR see Table 1 and Table 2.
Mollicellin H (6): white solid; UV (MeOH) λmax 206.00, 231.40 nm; IR νmax 3368, 2930, 2887, 1694, 1659, 1566, 1435, 1385, 1352, 1271, 1202, 1167, 1072, 829, 781, 750 cm−1; HR-ESI-MS m/z 369.1344 [M + H]+ (calcd for C21H21O6, 369.1333), 1H-NMR and 13C-NMR see Table 1 and Table 2.
Mollicellin I (7): white solid; HR-ESI-MS m/z 371.1495 [M + H]+ (calcd for C21H22O6, 371.1489), 393.1307 [M + Na]+ (calcd for C21H22O6Na, 393.1309), 1H-NMR and 13C-NMR see Table 1 and Table 2.
4.5. Antibacterial Assay
The antibacterial activities were tested against Gram-positive bacteria (S. aureus ATCC29213, S. aureus N50) and Gram-negative bacteria (E. coli ATCC25922, S. typhimurium ATCC14028, A. tumefaciens ATCC11158, P. lachrymans ATCC11921, R. solanacearum ATCC11696, X. vesicatoria ATCC11633). Streotomycin sulfate was used as the positive control. The minimum inhibitory concentrations (MIC) of the compounds and positive control were determined in sterile 96-well plates by the modified broth dilution test [29].
4.6. Cytotoxicity Assay
Cytotoxic activities were tested against two human cancer cell lines (HepG2 and Hela) using the microculture tetrazolium (MTT) assay as described previously [29]. Camptothecin was used as the positive control.
4.7. Antioxidant Activiity Assay
Radical scavenging assay was determined by a microplate spectrophotometric method based on the reduction of a methanol solution of DPPH [30]. Briefly, DPPH solution (80 μL, 0.2 mg/mL) and compounds solution in 30% acetone (20 μL) were added into each well of the microplate and mixed. The mixture was shaken vigorously and left to stand at 37 °C for 30 min in the dark. The absorbance of the solution was then measured at wavelength 515 nm using a microplate spectrophotometer. Inhibition (%) of free radical (DPPH) in percent was determined as [(Acontrol − Asample)/Acontrol] × 100, where A control is the absorbance of the control reaction containing all reagents except the test sample, and A sample is the absorbance of the test compounds. Tests were carried out in triplicate. BHT was used as the positive control. The IC50 value was calculated using linear relation between the compound concentration and probability of the percentage of DPPH inhibition.
Supplementary Materials
The following are available online.
Author Contributions
J.O., Z.M., H.W., J.W. and T.S. performed the research and recorded the spectra. J.O., J.W. and T.S. performed the isolation and identification of the fungal strain. J.O., H.G., Y.X. and T.S. performed the structure elucidation of compounds. Z.C. and J.S. did the antibacterial assay. Y.X. did the cytototic assay. J.O. and Z.M. did the antioxidant assay. X.W., J.W. and T.S. designed the research. J.O., Z.M., H.G. and T.S. completed the manuscript draft. Z.M., X.W., J.W. and T.S. finished the review and editing.
Funding
This research was co-financed by the National Natural Science Foundation of China (NSFC, 31400544), the Province Natural Science Foundation of Guangdong (2017A030313200), and the Distinguish Young Scientist Starting Grant of Guangdong Province (2014KQNCX034, 2017KQNCX016).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are not available from the authors.
References
- 1.Chang H.S., Lin C.H., Chen Y.S., Wang H.C., Chan H.Y., Hsieh S.Y., Wu H.C., Cheng M.J., Yuan G.F., Lin S.Y., et al. Secondary metabolites of the endophytic fungus Lachnum abnorme from Ardisia cornudentata. Int. J. Mol. Sci. 2016;17:1512. doi: 10.3390/ijms17091512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ribeiro A.S., Polonio J.C., Costa A.T., Menicoze dos Santos C., Rhoden S.A., Azevedo J.L., Pamphile J.A. Bioprospection of culturable endophytic fungi associated with the ornamental plant Pachystachys lutea. Curr. Microbiol. 2018;75:588–596. doi: 10.1007/s00284-017-1421-9. [DOI] [PubMed] [Google Scholar]
- 3.Kusari S., Singh S., Jayabaskaran C. Rethinking production of Taxol (paclitaxel) using endophyte biotechnology. Trends Biotechnol. 2014;32:304–311. doi: 10.1016/j.tibtech.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Wang X.J., Min C.L., Ge M., Zuo R.H. An endophytic sanguinarine-producing fungus from Macleaya cordata, Fusarium proliferatum BLH51. Curr. Microbiol. 2014;68:336–341. doi: 10.1007/s00284-013-0482-7. [DOI] [PubMed] [Google Scholar]
- 5.Pan F., Su T.J., Cai S.M., Wu W. Fungal endophyte-derived Fritillaria unibracteata var. wabuensis: Diversity, antioxidant capacities in vitro and relations to phenolic, flavonoid or saponin compounds. Sci. Rep. 2017;7 doi: 10.1038/srep42008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C., Tong Q., Zhu H., Tan D., Zhang J., Xue Y., Yao G., Luo Z., Wang J., Wang Y., et al. Nine new cytochalasan alkaloids from Chaetomium globosum TW1-1 (Ascomycota, Sordariales) Sci. Rep. 2016;6 doi: 10.1038/srep18711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ancheeva E., Mandi A., Kiraly S.B., Kurtan T., Hartmann R., Akone S.H., Weber H., Daletos G., Proksch P. Chaetolines A and B, pyrano[3,2-f]isoquinoline alkaloids from cultivation of Chaetomium sp. in the presence of autoclaved Pseudomonas aeruginosa. J. Nat. Prod. 2018;26:2392–2398. doi: 10.1021/acs.jnatprod.8b00373. [DOI] [PubMed] [Google Scholar]
- 8.Yang M.H., Gu M.L., Han C., Guo X.J., Yin G.P., Yu P., Kong L.Y. Aureochaeglobosins A–C, three [4 + 2] adducts of chaetoglobosin and aureonitol derivatives from Chaetomium globosum. Org. Lett. 2018;20:3345–3348. doi: 10.1021/acs.orglett.8b01243. [DOI] [PubMed] [Google Scholar]
- 9.Yu F.X., Chen Y., Yang Y.H., Li G.H, Zhao P.J. A new epipolythiodioxopiperazine with antibacterial and cytotoxic activities from the endophytic fungus Chaetomium sp. M336. Nat. Prod. Rep. 2018;32:689–694. doi: 10.1080/14786419.2017.1338285. [DOI] [PubMed] [Google Scholar]
- 10.Yamada T., Doi M., Shigeta H., Muroga Y., Hosoe S., Numata A., Tanaka R. Absolute stereostructures of cytotoxic metabolites, chaetomugilins A-C, produced by a Chaetomium species separated from a marine fish. Cheminform. 2008;39:4192–4195. doi: 10.1002/chin.200842211. [DOI] [Google Scholar]
- 11.Ge H.M., Zhang W.Y., Ding G., Saparpakorn P., Song Y.C., Hannongbua S., Tan R.X. Chaetoglobins A and B, two unusual alkaloids from endophytic Chaetomium globosum culture. Chem. Commun. 2009;40:5978–5980. doi: 10.1039/B812144C. [DOI] [PubMed] [Google Scholar]
- 12.Yamada T., Muroga Y., Jinno M., Kajimoto T., Usami Y., Numata A., Tanaka R. New class azaphilone produced by a marine fish-derived Chaetomium globosum. The stereochemistry and biological activities. Bioorg. Med. Chem. 2011;19:4106–4113. doi: 10.1016/j.bmc.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Chen C., Wang J., Zhu H., Wang J., Xue Y., Wei G., Guo Y., Tan D., Zhang J., Yin C., et al. Chaephilones A and B, two new azaphilone derivatives isolated from Chaetomium globosum. Chem. Biodivers. 2016;13:422–426. doi: 10.1002/cbdv.201500117. [DOI] [PubMed] [Google Scholar]
- 14.Wang W., Liao Y., Chen R., Hou Y., Ke W., Zhang B., Gao M., Shao Z., Chen J., Li F. Chlorinated azaphilone pigments with antimicrobial and cytotoxic activities isolated from the deep sea derived fungus Chaetomium sp. NA-S01-R1. Mar. Drugs. 2018;16:61. doi: 10.3390/md16020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sekita S., Yoshihira K., Natori S. Structures of chaetoglobosin A and B, cytotoxic metabolites of Chaetomium globosum. Cheminform. 1973;4:628–631. doi: 10.1002/chin.197337402. [DOI] [Google Scholar]
- 16.Oikawa H., Murakami Y., Ichihara A. New plausible precursors of chaetoglobosin an accumulated by treatment of chaetomium subaffine with cytochrome P-450 inhibitors. Tetrahedron Lett. 1991;32:4533–4536. doi: 10.1016/0040-4039(91)80032-2. [DOI] [Google Scholar]
- 17.Cui C.M., Li X.M., Li C.S., Proksch P., Wang B.G. Cytoglobosins A–G, cytochalasans from a marine-derived endophytic fungus, Chaetomium globosum QEN-14. J. Nat. Prod. 2010;73:729–733. doi: 10.1021/np900569t. [DOI] [PubMed] [Google Scholar]
- 18.Zhao S.S., Zhang Y.Y., Yan W., Cao L.L., Xiao Y., Ye Y.H. Chaetomium globosum CDW7, a potential biological control strain and its antifungal metabolites. FEMS Microbiol. Lett. 2017;364 doi: 10.1093/femsle/fnw287. [DOI] [PubMed] [Google Scholar]
- 19.Li G.Y., Li B.G., Yang T., Liu G.Y, Zhang G.L. Chaetoindicins A–C, three Isoquinoline alkaloids from the fungus Chaetomium indicum. Cheminform. 2006;37:3613–3615. doi: 10.1002/chin.200650209. [DOI] [PubMed] [Google Scholar]
- 20.Kanokmedhakul S., Kanokmedhakul K., Phonkerd N., Soytong K., Kongsaeree P., Suksamrarn A. Antimycobacterial anthraquinone-chromanone compound and diketopiperazine alkaloid from the fungus Chaetomium globosum KMITL-N0802. Planta Med. 2002;68:834–836. doi: 10.1055/s-2002-34415. [DOI] [PubMed] [Google Scholar]
- 21.Wang S., Li X.M., Teuscher F., Diesel A., Ebel R., Proksch P., Wang B.G. Chaetopyranin, a benzaldehyde derivative, and other related metabolites from Chaetomium globosum, an endophytic fungus derived from the marine red alga Polysiphonia urceolata. J. Nat. Prod. 2006;69:1622–1625. doi: 10.1021/np060248n. [DOI] [PubMed] [Google Scholar]
- 22.Wang F., Jiang J., Hu S., Ma H.R., Zhu H.C., Tong Q.Y., Cheng L., Hao X.C., Zhang G., Zhang Y.H. Secondary metabolites from endophytic fungus Chaetomium sp. induce colon cancer cell apoptotic death. Fitoterapia. 2017;121:86–93. doi: 10.1016/j.fitote.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi M., Yoshimura S., Kinoshita T., Abe F., Nitta K., Ezaki M., Sakamoto K., Hashimoto M., Fujie A., Hino M., et al. FR207944, an antifungal antibiotic from Chaetomium sp. No. 217, 2: Isolation and structure elucidation. Biosci. Biotechnol. Biochem. 2005;69:1029–1032. doi: 10.1271/bbb.69.1029. [DOI] [PubMed] [Google Scholar]
- 24.Stark A.A., Kobbe B., Matsuo D., Buchi G., Wogan G.N., Demain A.L. Mollicellins: Mutagenic and antibacterial mycotoxins. Appl. Environ. Microbiol. 1978;36:412–420. doi: 10.1128/aem.36.3.412-420.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G.Y., Li B.G., Yang T., Liu G.Y., Zhang G.L. Secondary metabolites from the fungus Chaetomium brasiliense. Helv. Chim. Acta. 2008;91:124–129. doi: 10.1002/hlca.200890002. [DOI] [Google Scholar]
- 26.Khumkomkhet P., Kanokmedhakul S., Kanokmedhakul K., Hahnvajanawong C., Soytong K. Antimalarial and cytotoxic depsidones from the fungus Chaetomium brasiliense. J. Nat. Prod. 2009;72:1487–1491. doi: 10.1021/np9003189. [DOI] [PubMed] [Google Scholar]
- 27.Bilal S., Ali L., Khan A.L., Shahzad R., Asaf S., Imran M., Kang S.M., Kim S.K., Lee I.J. Endophytic fungus Paecilomyces formosus LHL10 produces sester-terpenoid YW3548 and cyclic peptide that inhibit urease and alpha-glucosidase enzyme activities. Arch. Microbiol. 2018;200:1493–1502. doi: 10.1007/s00203-018-1562-7. [DOI] [PubMed] [Google Scholar]
- 28.Ibrahim S.R.M., Mohame G.A., Al Haidari R.A., El-Kholy A.A., Zayed M.F., Khayat M.T. Biologically active fungal depsidones: Chemistry, biosynthesis, structural characterization, and bioactivities. Fitoterapia. 2018;129:317–365. doi: 10.1016/j.fitote.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 29.Shan T., Tian J., Wang X., Mou Y., Mao Z., Lai D., Dai J., Peng Y., Zhou L., Wang M. Bioactive spirobisnaphthalenes from the endophytic fungus Berkleasmium sp. J. Nat. Prod. 2014;77:2151–2160. doi: 10.1021/np400988a. [DOI] [PubMed] [Google Scholar]
- 30.Wang J., Zhao J., Liu H., Zhou L., Liu Z., Wang J., Han J., Yu Z., Yang F. Chemical analysis and biological activity of the essential oils of two Valerianaceous species from China: Nardostachys chinensis and Valeriana officinalis. Molecules. 2010;15:6411–6422. doi: 10.3390/molecules15096411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.