Abstract
In plants, the HAK (high-affinity K+)/KUP (K+ uptake)/KT (K+ transporter) family represents a large group of potassium transporters that play important roles in plant growth and environmental adaptation. Although HAK/KUP/KT genes have been extensively investigated in many plant species, they remain uncharacterized in wheat, especially those involved in the response to environmental stresses. In this study, 56 wheat HAK/KUP/KT (hereafter called TaHAKs) genes were identified by a genome-wide search using recently released wheat genomic data. Phylogenetic analysis grouped these genes into four clusters (Ι, II, III, IV), containing 22, 19, 7 and 8 genes, respectively. Chromosomal distribution, gene structure, and conserved motif analyses of the 56 TaHAK genes were subsequently performed. In silico RNA-seq data analysis revealed that TaHAKs from clusters II and III are constitutively expressed in various wheat tissues, while most genes from clusters I and IV have very low expression levels in the examined tissues at different developmental stages. qRT-PCR analysis showed that expression levels of TaHAK genes in wheat seedlings were significantly up- or downregulated when seedlings were exposed to K+ deficiency, high salinity, or dehydration. Furthermore, we functionally characterized TaHAK1b-2BL and showed that it facilitates K+ transport in yeast. Collectively, these results provide valuable information for further functional studies of TaHAKs, and contribute to a better understanding of the molecular basis of wheat development and stress tolerance.
Keywords: abiotic stress, gene family, HAK/KUP/KT, potassium deficiency, wheat (Triticum aestivum L.)
1. Introduction
Potassium is an essential plant macronutrient, which comprises up to 10% of plants dry weight [1]. The decline of potassium amount below 10 g kg−1 dry weight can cause severe growth defects in many plants [2]. Potassium is involved in a variety of biochemical processes, including protein synthesis, carbohydrate metabolism, and enzyme activation, as well as in a number of physiological processes, such as stomatal regulation and photosynthesis [3]. In addition, potassium can enhance the tolerance of crop plants to various biotic (e.g., diseases and pests) and abiotic (e.g., salt, drought, and chilling) stresses [3,4,5].
Potassium uptake by plant roots and its translocation inside plants are undertaken by numerous K+ channels and transporters [6,7]. K+ channels generally function under high external potassium concentrations (>0.5 mM) and are recognized as a low affinity transport system. In contrast, K+ transporters, which were initially identified as a high affinity system, are capable of functioning at low external concentrations of K+ (<0.2 mM) [8]. However, the dividing line between these two types of systems blurs as the functions of an increasing number of K+ channels and transporters were revealed [9,10]. In plants, potassium transporters can be grouped into one of four families: KT (K+ transporter)/HAK (high-affinity K+)/KUP (K+ uptake), Trk (Transport of K+)/HKT (high-affinity K+ transporters), KEA (K+ efflux anti-porter), and CHX (cation/hydrogen exchanger) [11,12]. Early reports on bacterial KUP [13], fungal HAK [14], and Arabidopsis KT [15] transporters used a different set of acronyms, leading to the appearance of composite names HAK/KUP/KT or KT/HAK/KUP or KT/KUP/HAK [16,17,18,19]. These composite names are now widely used to refer to the whole family of K+ transporters in plants. The HAK/KUP/KT family, which is the largest potassium transporter family, has been divided into four groups known as clusters I, II, III, and IV [11,20].
Plants contain multiple HAK/KUP/KT transporters; they play diverse roles in K+ uptake and transport as well as in regulation of plant growth and development, salt tolerance, and osmotic potential regulation [17]. The availability of K+ transport mutants in both Arabidopsis and rice has led to a better understanding of their roles in K+ uptake and transport. AtHAK5 and AtAKT1 are the two major contributors to K+ uptake in Arabidopsis [21]. Under normal K+ conditions, the AtHAK5 transcripts were detected in roots, but not in shoots. Under conditions of K+ deficiency, the AtHAK5 transcripts were present in both roots and shoots, although in roots the levels of expression were higher [22]. In addition, there is evidence that AtKUP7 is also involved in K+ uptake and may partially mediate xylem K+ release [23]. In rice, functions of AtHAK5 and AtAKT1 are fulfilled by the rice homologs of these genes, OsHAK1 and OsAKT1. It was found that potassium concentrations in the xylem sap, as well as net potassium export rates in OsHAK1 mutants, are markedly lower than in wild type control plants, particularly in plants pre-treated with low concentrations of potassium (0.1 mM) [24]. Another K+ transporter, OsHAK5, partially contributes to high-affinity K+ uptake, but at higher K+ concentrations than OsHAK1. OsHAK5 is strongly expressed in the xylem parenchyma and phloem of root vascular tissues. Its expression is particularly apparent under potassium-deficient conditions, suggesting that OsHAK5 may be involved in potassium distribution between roots and shoots [25]. Another family member, OsHAK21, has the K+ transporter activity, but is not directly involved in K+ uptake. OsHAK21 may play a role in long-distance K+ distribution between roots and shoots, because its expression was detected mainly in the root vascular system of stele [26].
It was demonstrated that HAK/KUP/KT transporters protect plants against salt stress. For instance, OsHAK1 is essential for maintaining potassium-mediated growth of rice plants, and confers salt tolerance across low-to-high potassium concentrations. The OsHAK1 knockout mutants have stunted root and shoot growth compared to wild type rice plants. The OsHAK1 mutants grown in a 0.1 mM K+ solution produced 50–55% of the root and 45–50% of the shoot dry weight biomasses of wild type plants. Similarly, when grown in a 1 mM K+ solution, those were approximately 70–75% and 60–65%, respectively, of wild types. OsHAK1 overexpression in rice increased K+ uptake as well as the ratio of K+/Na+ [24]. It is known that an increase of K+/Na+ ratio in plants leads to the enhancement of salt tolerance. Overexpression of OsHAK5 increased the K+/Na+ ratio in shoots and subsequently improved plant salt tolerance, as it was indicated by the better shoot growth of transgenic plants than that of wild type plants. Comparatively, OsHAK5 knockout mutants decreased K+/Na+ ratio in shoots, which resulted in increased sensitivity of transgenic plants to salt stress [25]. OsHAK21 was also suggested to have an essential role in the salt stress response. Expression levels of OsHAK21 considerably increased under conditions of high salinity; particularly, after 4 h of salt stress the transcription level of OsHAK21 in roots increased 600-fold. OsHAK21 mutants accumulated less K+ and significantly more Na+ in both shoots and roots under salt stress than wild type control plants. They also had a remarkably lower K+ net uptake rate and higher Na+ uptake rate compared to wild type plants. This highlights the importance of OsHAK21 in maintaining plant K+ homeostasis under conditions of salt stress [26].
Furthermore, HAK/KUP/KT transporters also play a role in drought tolerance. Overexpression of OsHAK1 in rice enhanced drought tolerance at both vegetative and reproductive stages. OsHAK1 overexpression seedlings (Ox) had decreased levels of lipid peroxidation, increased proline accumulation, and improved activities of antioxidant enzymes compared to control seedlings. Under drought conditions, OsHAK1-Ox plants produced 35% more grain yield than wild type plants [27].
Taken together, the above body of research justify the potential value of HAK/KUP/KT family members in crop breading for improved tolerance to high salinity and drought (occurring in arid and semi-arid areas). Although wheat is one of the most important cereal crops worldwide, information about structures and functions of wheat HAK/KUP/KT genes is scarce. In this study, 56 wheat HAK/KUP/KT genes were identified by a genome-wide search using recently released wheat genome data. We thoroughly analyzed the phylogeny, presence of conserved motifs in proteins, gene structures, locations of genes on chromosomes, and expression patterns of HAK/KUP/KT genes in various wheat tissues and under several environmental stresses. In addition, the TaHAK1b-2BL gene was functionally characterized and it was demonstrated that it facilitates K+ transport in yeast.
2. Results
2.1. Identification of the TaHAKs in Wheat
A hidden Markov model (HMM) search for proteins containing the K+ transporter domain (Pfam accession no. PF02705) was performed using the latest wheat genome database [28]. A total of 56 TaHAKs nucleotide sequences were identified, which encoded 25 family members (Table 1). These 25 family members were named according to their corresponding homologs from rice based on a phylogenetic relationship analysis. Letters a, b, or c were added when more than one wheat family member was clustered together with the product of the same rice gene, e.g., TaHAK1a, TaHAK1b, TaHAK1c. Homoeologs within each of the 25 members were further distinguished by the addition of chromosomal arm symbols to the gene/protein names, such as 1AL, 3BS, or 5DS.
Table 1.
TaHAK genes identified in wheat. Twenty-five TaHAK family members are indicated by white or light grey blocks. TMS, transmembrane segments; S.L., subcellular location; PM, plasma membrane.
| Gene Name | Ensembl ID | Amino Acid Length | TMS | Intron No. | Exon No. | S.L. | Gene Location (bp) | |
|---|---|---|---|---|---|---|---|---|
| Start | End | |||||||
| TaHAK1a-4BL | TRIAE_CS42_4BL_TGACv1_321685_AA1064100.1 | 784 | 11 | 7 | 8 | PM | 600657723 | 600654569 |
| TaHAK1a-4DL | TRIAE_CS42_4DL_TGACv1_343636_AA1137540.1 | 778 | 11 | 8 | 9 | PM | 476306655 | 476302887 |
| TaHAK1b-2BL | TRIAE_CS42_2BL_TGACv1_129472_AA0385230.2 | 768 | 11 | 8 | 9 | PM | 436243088 | 436237762 |
| TaHAK1b-2DL | TRIAE_CS42_2DL_TGACv1_158352_AA0516430.1 | 776 | 11 | 8 | 9 | PM | 367507729 | 367502225 |
| TaHAK1b-2A | TRIAE_CS42_U_TGACv1_641460_AA2095620.1 | 772 | 11 | 8 | 9 | PM | 497657505 | 497652193 |
| TaHAK1c-2BL | TRIAE_CS42_2BL_TGACv1_129437_AA0383630.1 | 774 | 10 | 8 | 9 | PM | 436956667 | 436962029 |
| TaHAK2-3AL | TRIAE_CS42_3AL_TGACv1_194420_AA0632700.2 | 784 | 13 | 8 | 9 | PM | 695496006 | 695500670 |
| TaHAK2-3DL | TRIAE_CS42_3DL_TGACv1_250243_AA0864920.1 | 788 | 13 | 9 | 10 | PM | 559072825 | 559076602 |
| TaHAK3-1BL | TRIAE_CS42_1BL_TGACv1_033126_AA0137190.1 | 818 | 11 | 6 | 7 | PM | 559097952 | 559092195 |
| TaHAK3-1DL | TRIAE_CS42_1DL_TGACv1_062044_AA0207990.1 | 817 | 11 | 6 | 7 | PM | 414287464 | 414282613 |
| TaHAK4-7AS | TRIAE_CS42_7AS_TGACv1_570480_AA1836390.3 | 686 | 11 | 4 | 5 | PM | 36648278 | 36651604 |
| TaHAK4-7DS | TRIAE_CS42_7DS_TGACv1_622074_AA2032320.3 | 725 | 11 | 4 | 5 | PM | 36992201 | 36998396 |
| TaHAK5-3AL | TRIAE_CS42_3AL_TGACv1_193587_AA0613950.2 | 788 | 11 | 9 | 10 | PM | 686834648 | 686837566 |
| TaHAK5-3B | TRIAE_CS42_3B_TGACv1_222217_AA0760010.1 | 782 | 12 | 8 | 9 | PM | 729204607 | 729207978 |
| TaHAK5-3DL | TRIAE_CS42_3DL_TGACv1_249200_AA0841080.1 | 764 | 11 | 9 | 10 | PM | 549623301 | 549628488 |
| TaHAK6-3AL | TRIAE_CS42_3AL_TGACv1_195362_AA0648570.1 | 732 | 11 | 5 | 6 | PM | 689132246 | 689135499 |
| TaHAK6-3D | TRIAE_CS42_U_TGACv1_646122_AA2146080.1 | 732 | 10 | 5 | 6 | PM | 552878679 | 552881787 |
| TaHAK7-2AS | TRIAE_CS42_2AS_TGACv1_114941_AA0370050.1 | 778 | 13 | 8 | 9 | PM | 66798807 | 66805468 |
| TaHAK7-2DS | TRIAE_CS42_2DS_TGACv1_179048_AA0603870.1 | 778 | 13 | 8 | 9 | PM | 67562036 | 67555857 |
| TaHAK9-2AS | TRIAE_CS42_2AS_TGACv1_112740_AA0344400.1 | 782 | 11 | 6 | 7 | PM | 59558656 | 59554035 |
| TaHAK9-2DS | TRIAE_CS42_2DS_TGACv1_177265_AA0571140.1 | 780 | 11 | 6 | 7 | PM | 58776915 | 58781534 |
| TaHAK10-7AL | TRIAE_CS42_7AL_TGACv1_556774_AA1770580.1 | 829 | 11 | 8 | 9 | PM | 571701040 | 571706004 |
| TaHAK10-7BL | TRIAE_CS42_7BL_TGACv1_578444_AA1895080.2 | 825 | 11 | 8 | 9 | PM | 532275080 | 532279899 |
| TaHAK10-7DL | TRIAE_CS42_7DL_TGACv1_604392_AA1997720.1 | 827 | 11 | 8 | 9 | PM | 504174129 | 504179023 |
| TaHAK11-2DL | TRIAE_CS42_2DL_TGACv1_159022_AA0530870.1 | 792 | 14 | 7 | 8 | PM | 533132266 | 533137640 |
| TaHAK12-1AS | TRIAE_CS42_1AS_TGACv1_020203_AA0075600.3 | 790 | 14 | 8 | 9 | PM | 29186381 | 29192163 |
| TaHAK13-7AL | TRIAE_CS42_7AL_TGACv1_557049_AA1775770.1 | 803 | 11 | 8 | 9 | PM | 665751743 | 665743564 |
| TaHAK13-7BL | TRIAE_CS42_7BL_TGACv1_577019_AA1862890.1 | 750 | 11 | 9 | 10 | PM | 637233552 | 637228550 |
| TaHAK13-7DL | TRIAE_CS42_7DL_TGACv1_603280_AA1979940.1 | 803 | 11 | 8 | 9 | PM | 575510919 | 575502903 |
| TaHAK16-6AL | TRIAE_CS42_6AL_TGACv1_471561_AA1510890.1 | 806 | 11 | 9 | 10 | PM | 526114436 | 526120099 |
| TaHAK16-6BL | TRIAE_CS42_6BL_TGACv1_502060_AA1622780.1 | 802 | 11 | 9 | 10 | PM | 574459052 | 574465508 |
| TaHAK16-6DL | TRIAE_CS42_6DL_TGACv1_527466_AA1704460.1 | 805 | 11 | 9 | 10 | PM | 384217110 | 384223492 |
| TaHAK17a-5AL | TRIAE_CS42_5AL_TGACv1_374155_AA1191940.2 | 719 | 12 | 7 | 8 | PM | 448614349 | 448619914 |
| TaHAK17a-5BL | TRIAE_CS42_5BL_TGACv1_405525_AA1329450.1 | 719 | 12 | 7 | 8 | PM | 408955185 | 408959389 |
| TaHAK17a-5DL | TRIAE_CS42_5DL_TGACv1_433406_AA1412260.1 | 719 | 12 | 7 | 8 | PM | 348479742 | 348483109 |
| TaHAK17b-5DL | TRIAE_CS42_5DL_TGACv1_434380_AA1434940.1 | 712 | 11 | 7 | 8 | PM | 348346780 | 348352011 |
| TaHAK18-5BL | TRIAE_CS42_5BL_TGACv1_407556_AA1357920.1 | 785 | 14 | 8 | 9 | PM | 546832103 | 546826324 |
| TaHAK18-5DL | TRIAE_CS42_5DL_TGACv1_433145_AA1403590.1 | 785 | 14 | 8 | 9 | PM | 447886169 | 447880484 |
| TaHAK19a-6AS | TRIAE_CS42_6AS_TGACv1_488672_AA1576040.1 | 735 | 11 | 5 | 6 | PM | 33582679 | 33586043 |
| TaHAK19a-6BS | TRIAE_CS42_6BS_TGACv1_514904_AA1665660.1 | 737 | 11 | 5 | 6 | PM | 61222797 | 61226427 |
| TaHAK19a-6D | TRIAE_CS42_U_TGACv1_644308_AA2138720.1 | 734 | 11 | 7 | 8 | PM | 28943744 | 28939705 |
| TaHAK19b-6BS | TRIAE_CS42_6BS_TGACv1_513620_AA1645910.1 | 736 | 11 | 5 | 6 | PM | 60865245 | 60860866 |
| TaHAK19b-6D | TRIAE_CS42_U_TGACv1_644372_AA2139050.1 | 738 | 11 | 6 | 7 | PM | 28998084 | 29001654 |
| TaHAK19c-3B | TRIAE_CS42_3B_TGACv1_221313_AA0736890.1 | 745 | 10 | 6 | 7 | PM | 765005247 | 765001714 |
| TaHAK19c-3DL | TRIAE_CS42_3DL_TGACv1_251389_AA0881060.1 | 744 | 10 | 6 | 7 | PM | 574916450 | 574919986 |
| TaHAK22-2AL | TRIAE_CS42_2AL_TGACv1_096842_AA0321670.1 | 875 | 12 | 2 | 3 | PM | 392724188 | 392729472 |
| TaHAK22-2BL | TRIAE_CS42_2BL_TGACv1_133206_AA0441840.1 | 873 | 12 | 2 | 3 | PM | 394364701 | 394359570 |
| TaHAK22-2DL | TRIAE_CS42_2DL_TGACv1_158627_AA0523560.1 | 878 | 12 | 2 | 3 | PM | 325365084 | 325359910 |
| TaHAK23-5AL | TRIAE_CS42_5AL_TGACv1_377379_AA1246300.1 | 814 | 10 | 9 | 10 | PM | 403355442 | 403350814 |
| TaHAK23-5BL | TRIAE_CS42_5BL_TGACv1_406332_AA1344100.3 | 916 | 12 | 8 | 9 | PM | 355690579 | 355685671 |
| TaHAK23-5DL | TRIAE_CS42_5DL_TGACv1_435669_AA1453280.1 | 898 | 12 | 8 | 9 | PM | 309789789 | 309784581 |
| TaHAK24-7AS | TRIAE_CS42_7AS_TGACv1_569122_AA1807860.1 | 772 | 11 | 8 | 9 | PM | 172470095 | 172464552 |
| TaHAK24-7BS | TRIAE_CS42_7BS_TGACv1_593324_AA1950450.2 | 771 | 13 | 8 | 9 | PM | 135583478 | 135578011 |
| TaHAK24-7DS | TRIAE_CS42_7DS_TGACv1_621622_AA2021390.1 | 771 | 11 | 8 | 9 | PM | 169916315 | 169910689 |
| TaHAK25-6AL | TRIAE_CS42_6AL_TGACv1_471048_AA1501690.1 | 769 | 12 | 8 | 9 | PM | 523950482 | 523944232 |
| TaHAK25-6BL | TRIAE_CS42_6BL_TGACv1_501501_AA1618220.2 | 768 | 13 | 8 | 9 | PM | 570980883 | 570976570 |
As shown in Table 1, ten TaHAKs had three homoeologs distributed across A, B, and D genomes, including TaHAK1b, -5, -10, -13, -16, -17a, -19a, -22, -23, and -24. Single gene sequence were found for four family members (TaHAK1c, -11, -12, and -17b), while the remaining 11 genes possessed two homoeologous sequences each. The length of the putative TaHAK proteins ranged from 686 (TaHAK4-7AS) to 916 (TaHAK23-5BL) amino acids. The number of transmembrane segments (TMS) ranged from 10 to 14, with the most common being 11–12 TMS (73.2%). All examined TaHAKs were predicted to be localized to the plasma membrane.
2.2. Phylogenetic Analysis of TaHAK Proteins
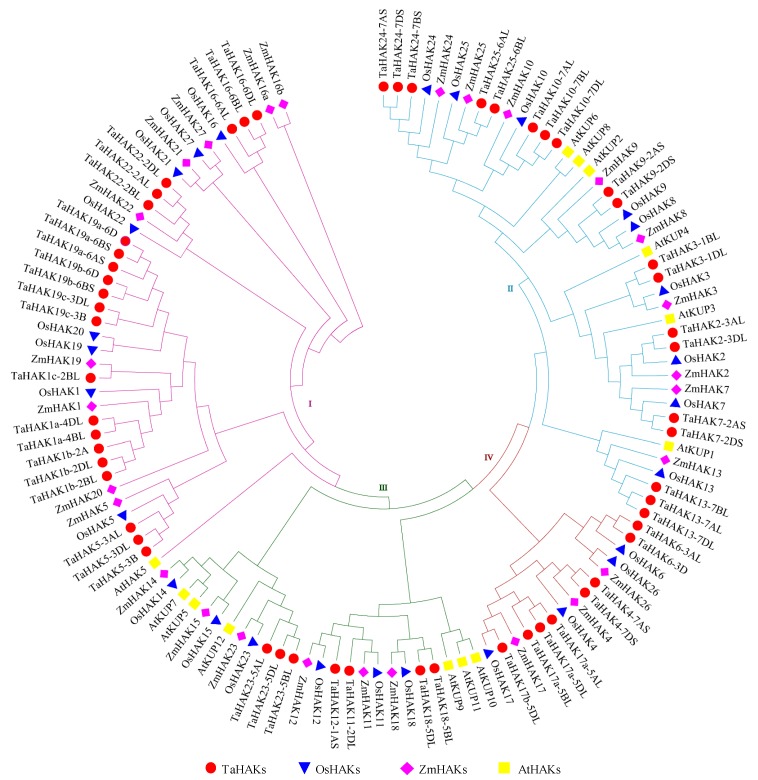
To understand the phylogenetic relationship of the KUP/HAK/KT family proteins, a phylogenetic tree was constructed based on the alignment of the full-length protein sequences of 56 wheat TaHAKs, 27 rice OsHAKs, 27 maize ZmHAKs, and 13 Arabidopsis AtKUP/HAK/KTs (Figure 1). According to the classification criteria used for Arabidopsis and rice [11,22], the wheat HAK proteins were categorized into four clusters. Clusters I, II, III, and IV contained 22, 19, 7, and 8 TaHAK proteins, respectively. Clusters I and II were the most abundant in wheat, maize, and rice, comprising 73%, 67%, and 63% proteins, respectively, while clusters II and III were the most abundant in Arabidopsis, comprising 92% of all AtKUP/HAK/KTs.
Figure 1.
Phylogenetic tree of KUP/HAK/KT family proteins among wheat, rice, maize, and Arabidopsis. The proteins belonging to each of four species are represented by different shapes and colors. The KUP/HAK/KT family proteins were divided into four clusters (I, II, III, IV) and indicated with different colors of lines. The protein loci of rice, maize, and Arabidopsis KUP/HAK/KT family proteins are listed in Table S1. Ta, Triticum aestivum; Os, Oryza sativa; Zm, Zea mays; At, Arabidopsis thaliana.
2.3. Genomic Distribution of TaHAKs
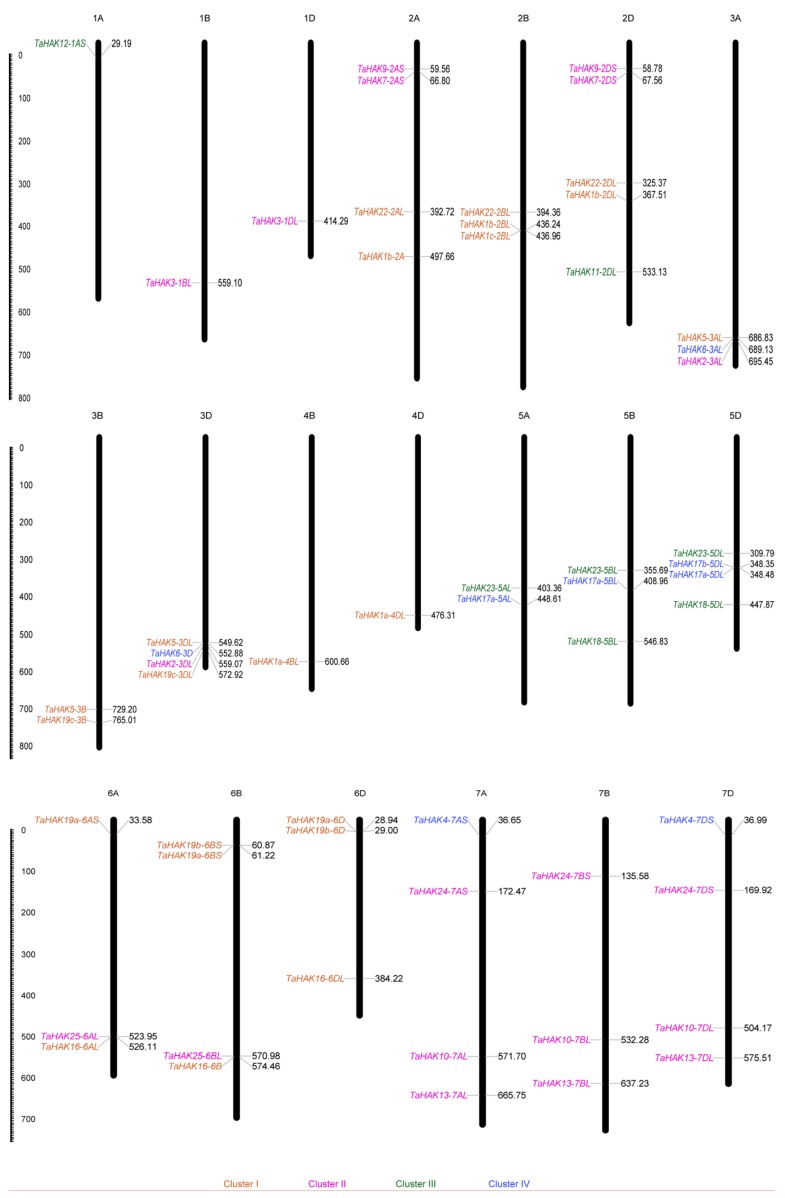
TaHAK genes were mapped to 20 of 21 wheat chromosomes. The physical locations of TaHAKs are listed in Table 1. Fifty-six wheat TaHAK sequences were approximately evenly distributed among A (17), B (17), and D (22) subgenomes. This was in accordance with the observation that most TaHAKs have three homoeologous sequences located on three subgenomes. Nevertheless, TaHAKs were non-randomly distributed among different chromosomal groups. The chromosomal groups one and four contained 3 (5.4%) and 2 (3.6%) sequences, respectively. The remaining 51 sequences were more evenly distributed across chromosomal groups two, three, five, six, and seven, ranging from 9 to 12 genes per group (Figure 2).
Figure 2.
Chromosomal distribution of TaHAK genes. The names of chromosomes are shown above each chromosome. The gene names are indicated on the left side and the starting locations are on the right of chromosomes. The TaHAKs in each cluster are specified by the same color. The lengths of chromosomes are shown in Mb (Millions of bases).
2.4. Gene Structure and Conserved Motif Analyses of TaHAKs
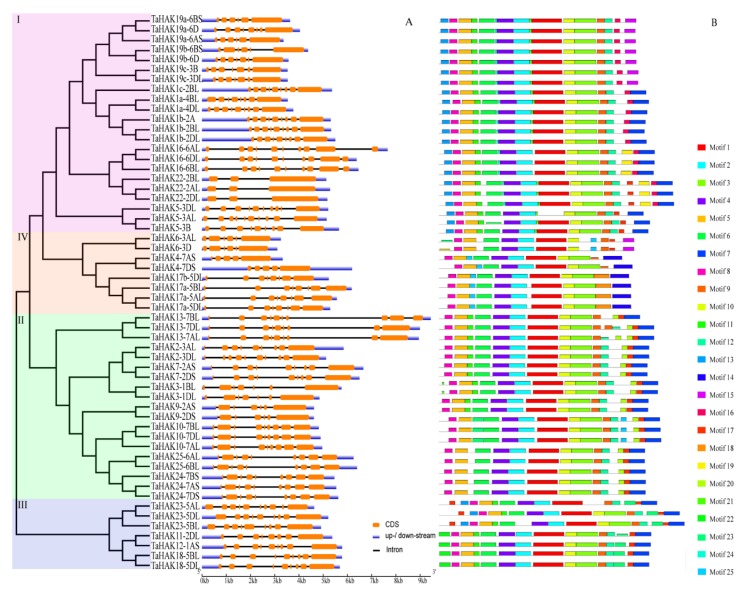
Based on our phylogenetic analyses, the TaHAK family of proteins was divided into four sub-families (Figure 1 and Figure 3). TaHAKs gene structure was revealed using the alignment between CDS and respective genomic sequence by the GSDS [29] online tool (Figure 3A). The examined genes contained 3 to 10 exons and 2 to 9 introns (Figure 3A and Table 1). TaHAK22 had 3 exons and 2 introns, which was the smallest exon/intron number among all identified TaHAK genes. A total of 24 (42.9%) TaHAK genes had 9 exons, which was the largest number of exons in examined TaHAK genes, followed by 8 (14.3%), 7 (12.5%), 7 (12.5%), 5 (8.9%), 3 (5.4%), and 2 (3.6%) gene sequences, possessing 10, 8, 7, 6, 3, and 5 exons, respectively. Exon/intron numbers also varied inside the same sub-family (cluster I: exon: 3 to 10/intron: 2 to 9, cluster II: exon: 7 to 10/intron 6 to 9, cluster III: exon: 8 to 10/intron: 7 to 9, cluster IV: exon: 5 to 8/intron: 4 to 7), and among the homoeologous genes of the same family member. For example, TaHAK5-3B contained 9 exons/8 introns, whereas both TaHAK5-3AL and TaHAK5-3DL had 10 exons/9 introns. These results are similar to the previously reported results for rice and maize HAK genes [11,30].
Figure 3.
Gene structures and conserved protein motifs of TaHAKs. (A) Gene structures. TaHAK genes are displayed in order based on a phylogenetic analysis of their protein products. Introns, exons and noncoding regions are represented with black lines, orange boxes and blue boxes, respectively. (B) Conserved protein motifs. Twenty-five motifs identified in TaHAK proteins marked by different colors.
Protein motifs were analyzed using the online program MEME. In total, 25 conserved motifs were identified in putative TaHAK proteins and designated motifs 1-25 (Figure 3B). More detailed information regarding all conserved motifs is shown in Table S2. As revealed by our NCBI CD (Conserved Domain) search, most conserved motifs were found within the sequence of the K+-superfamily domain (Table S3). This result is not surprising, since the 534 amino acid-long “K_trans” domain (PF02705) accounted for a major part of the TaHAK proteins, ranging from 686 to 916 amino acids. Generally, the motifs were almost evenly distributed, and a similar number of motifs was present in TaHAK proteins from each of the four clusters. Motifs 1, 2, 3, 4, 5, 6, 8, 9 and 10 were conserved in all four TaHAK clusters, with a few exceptions where a particular motif was missing in one gene (Figure 3B and Table S4).
We also identified motifs unique to each cluster. They are motifs 13, 16, 21, and 24 (only appeared in cluster I, with few exceptions); motif 20 (only present in cluster II); motif 22 (only found in cluster III); and motifs 14, 18, and 25 (unique to cluster IV, with the exception of motif 25, which was also present in TaHAK10 (cluster II)). In addition, motifs 7 and 12 were present in clusters I, II, and III, but absent in cluster IV. The only exceptions were that motif 7 was missing in TaHAK19s (cluster I) and motif 12 was missing in TaHAK13-7BL (cluster II).
2.5. Expression Analysis of TaHAK Genes in Various Wheat Tissues
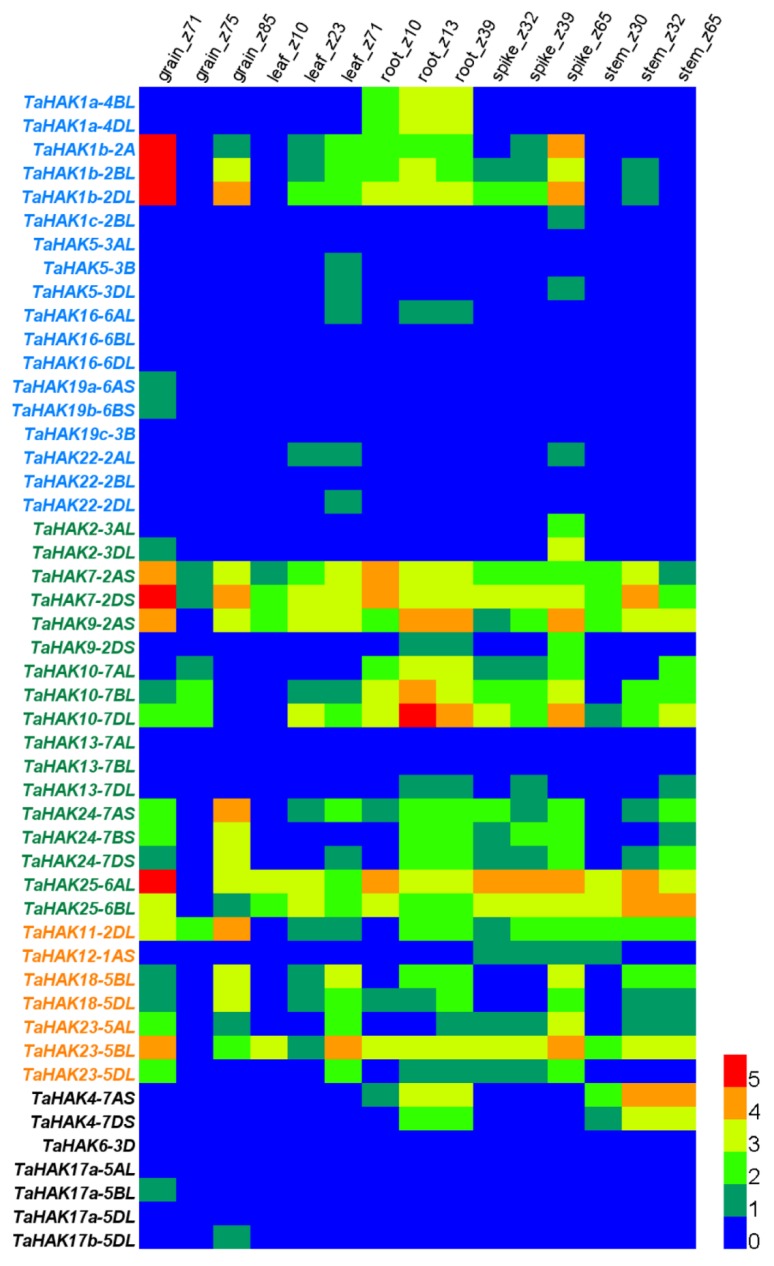
Publicly available RNA-seq databases were used to analyze the expression patterns of TaHAK genes in wheat roots, leaves, stems, spikes, and grains. Of the examined 56 genes, data on 49 genes were obtained from RNA-seq databases, while information about the other seven genes (TaHAK3-1BL and -1DL, TaHAK6-3AL, TaAHK19a-6D and -6BS, TaHAK19b-6D, and TaHAK19c-3DL) was missing (Figure 4 and Table S5). Most genes in cluster I had low expression levels in almost all tissues, with exception of TaHAK1a and TaHAK1b. TaHAK1a-4BL and -4DL were strongly expressed in roots, and TaHAK1b-2A, -2BL, and -2DL were strongly expressed in roots, spikes, and grains, especially in the grain Z71 (2 DAA) stage. For clusters II and III, most genes displayed strong constitutive expression in various tissues, including TaHAK7, TaHAK9-2AS, TaHAK10, TaHAK24, TaHAK25, TaHAK11, TaHAK18, and TaHAK23. In contrast, TaHAK2, TaHAK13, and TaHAK12-1AS had very low expression levels in most wheat tissues at all examined developmental stages. Most genes from cluster IV had very low expression levels in all tissues, except TaHAK4-7AS and TaHAK4-7DS, both of which displayed relatively high expression levels in roots and stems.
Figure 4.
Heat map showing the expression of wheat TaHAK genes in various wheat tissues at different developmental stages. RNA-seq data for bread wheat cultivar Chinese spring were obtained from dataset “developmental timecourse in five tissues” presented in WheatExp database [31] Number 0 to 5 represent the range of expression levels (from the lowest to the highest) of the examined genes.
2.6. Expression Analysis of TaHAK Genes under Various Stresses
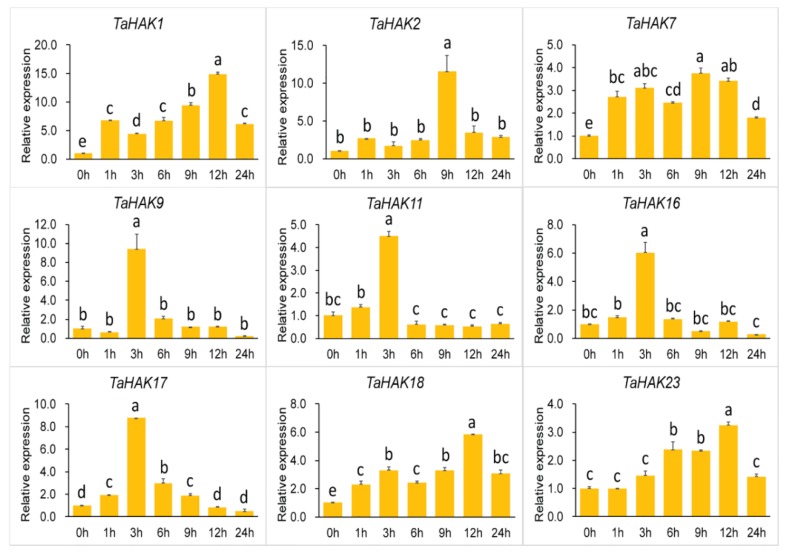
To gain insight into the transcriptional responses of TaHAK genes to environmental stresses, the gene expression levels of nine selected TaHAK genes were assessed using qRT-PCR after exposure of wheat plants to potassium deficiency, high salinity, and drought. As shown in Figure 5, all nine TaHAK genes were upregulated after exposure to potassium deficiency. However, the expression patterns of these genes were different. Basically, expression patterns of all examined genes in plants exposed to the potassium deficiency stress could be divided into two types. The first type is characterized by rapid and continuous upregulation, including TaHAK1, TaHAK7, TaHAK18, and TaHAK23. These genes, with the exception of TaHAK23, were upregulated after 1 h of exposure to stress. The response of TaHAK23 to stress was slower, its upregulation became obvious after 6 h of stress. The second type of expression pattern represented the transient upregulation of genes, which included strong upregulation of genes for a short time after exposure to the potassium deficiency stress, followed by a rapid downregulation to the control level, and the low level of gene expression were unchanged for the remaining time of stress. Such type of expression pattern was observed for TaHAK2, TaHAK9, TaHAK11, TaHAK16, and TaHAK17, most of which showed the highest level of expression after 3 h of stress. The only exception is TaHAK2, expression of which reached the maximum after 9 h of exposure to low potassium stress.
Figure 5.
Expression of TaHAK genes in response to potassium deficiency stress (0.1 mM K+). Expression of TaHAK genes was determined by qRT-PCR using total RNA isolated from wheat roots at different time points (0, 1, 3, 6, 9, 12 and 24 h) of the potassium deficiency stress. One-way ANOVA with Duncan’s multiple range test was conducted using SPSS version 20.0. Different letters on top of error bars indicate significant differences at p = 0.05 level. Error bars indicate the standard error (SE) of three replicates.
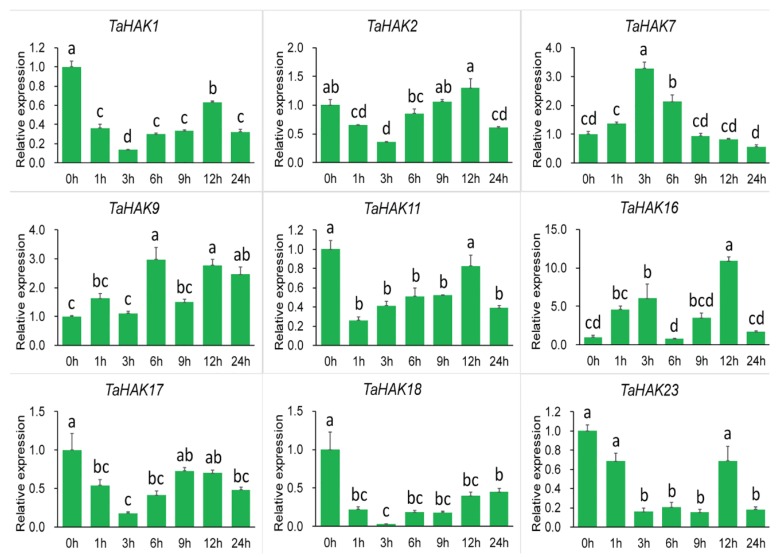
Under salt stress, the expression levels of TaHAK1, TaHAK11, TaHAK17, TaHAK18, and TaHAK23 genes were downregulated. In contrast, under the same stress conditions, expression levels of TaHAK7, TaHAK9, and TaHAK16 were markedly increased (Figure 6).
Figure 6.
Expression of TaHAK genes in response to salt stress (200 mM NaCl). Expression of TaHAK genes was determined by qRT-PCR using total RNA isolated from wheat roots at different time points (0, 1, 3, 6, 9, 12 and 24 h) of salt stress. One-way ANOVA with Duncan’s multiple range test was conducted using SPSS version 20.0. Different letters on top of error bars indicate significant differences at p = 0.05 level. Error bars indicate the SE of three replicates.
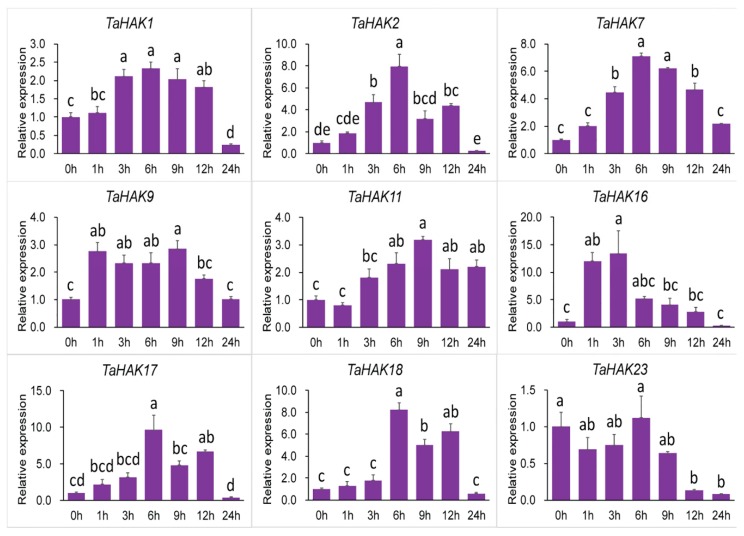
Under dehydration stress simulated by 20% PEG6000, all TaHAK genes were upregulated, albeit to different levels (Figure 7). TaHAK2, TaHAK7, TaHAK16, TaHAK17, and TaHAK18 were all significantly upregulated. In particular, TaHAK16 responded rapidly to dehydration, and its expression reached maximum after 1 h of stress. Four other genes, TaHAK2, TaHAK7, TaHAK17, and TaHAK18, reached the maximum expression level after 6 h exposure to dehydration. Comparatively, TaHAK1, TaHAK9, and TaHAK11 were moderately upregulated. The expression level of TaHAK23 remained near the same during the first 9 h of stress and then started to decrease.
Figure 7.
Expression of TaHAK genes in response to dehydration simulated by 20% PEG6000. Expression of TaHAK genes was determined by qRT-PCR using total RNA isolated from wheat roots at different time points (0, 1, 3, 6, 9, 12 and 24 h) of dehydration. One-way ANOVA with Duncan’s multiple range test was conducted using SPSS version 20.0. Different letters on top of error bars indicate significant differences at p = 0.05 level. Error bars indicate the SE of three replicates.
2.7. TaHAK1b-2BL Functions in Potassium Uptake and Salt Tolerance in Yeast
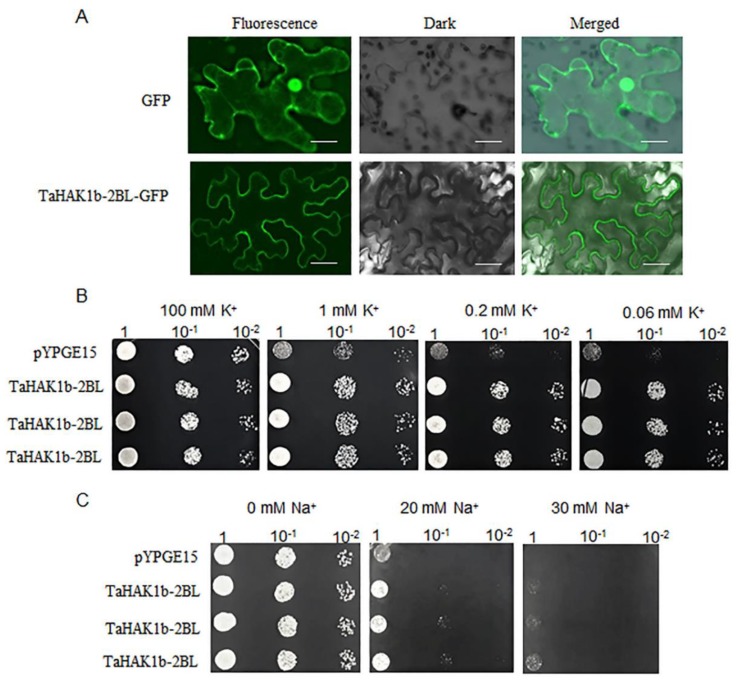
Most of the reported HAK/KUP/KTs are localized in the plasma membrane [17]. Here, we examined the subcellular location of TaHAK1b-2BL, one of TaHAK1 transporters homologous to OsHAK1, which has been previously shown to play a pivotal role in maintaining potassium-mediated growth of rice plants at different salt concentrations. To do this, we generated a construct expressing a fusion protein of TaHAK1b-2BL and green fluorescent protein (GFP). The resulting construct and the control vector (expressing GFP) were introduced into epidermal cells of Nicotiana benthamiana leaves. As shown in Figure 8A, the green fluorescence of the GFP control vector was observed in the nucleus, cytosol, and plasma membrane. In contrast, the TaHAK1b-2BL-GFP fusion protein was localized solely in the plasma membrane.
Figure 8.
Subcellular location of TaHAK1b-2BL and its roles in potassium uptake and salt tolerance. (A). Subcellular location of TaHAK1b-2BL. Scale bars = 20 µm. (B). Functional complementary assay of TaHAK1b-2BL in a high-affinity potassium uptake deficient yeast mutant. Yeasts transformed with the empty vector (pYPGE15) were used as a negative control. The tested potassium concentrations were indicated above each panel. Serial dilutions (10−1) of yeast culture were plated. (C). Salt tolerance assay of TaHAK1b-2BL. The tested sodium concentrations were indicated above each panel. Serial dilutions (10−1) of yeast culture were plated.
Previous studies revealed that OsHAK1 could complement the growth defects of a high-affinity potassium uptake deficient yeast mutant. Furthermore, it was shown that the yeast expressing OsHAK1 could better tolerate high salinity than the yeast harboring an empty vector [24]. In this work, we investigated whether TaHAK1b-2BL has a function in potassium uptake using the yeast strain CY162, which is defective in high-affinity potassium uptake. We found that both the yeast transformed with an empty control vector and the yeast transformed with the TaHAK1b-2BL construct grew well at 100 mM K+ (Figure 8B). However, the yeast expressing TaHAK1b-2BL grew better at K+ concentrations between 0.06 and 1 mM K+ than the control yeast containing an empty vector. In addition, we used the salt-sensitive yeast mutant AXT3K to determine whether the yeast expressing TaHAK1b-2BL could tolerate salt stress. As shown in Figure 8C, TaHAK1b-2BL-containing yeast and yeast without TaHAK1b-2BL demonstrated a similar growth status at 0 mM Na+. However, the yeast containing TaHAK1b-2BL grew better at 20 mM Na+ than the yeast without TaHAK1b-2BL. Notably, only the yeast containing TaHAK1b-2BL could grow at 30 mM Na+.
3. Discussion
Thus far, the HAK/KUP/KT family has been reported in a number of plant species, including Arabidopsis [22,32], rice [11,33], maize [30], peach [34], and pear [16]. The gene and protein structures and characteristic features, expression patterns and functions of some HAK/KUP/KT genes have already been characterized. However, a genome-wide analysis of the HAK family genes have not yet been performed in wheat (Triticum aestivum L.). Recently released wheat genome data allowed us to identify 56 HAK genes (including homoeologs) of 25 family members from wheat (Table 1).
In plants, members of the HAK gene family display a low level of conservation of their exon/introns structures. The number of exons in wheat TaHAK genes ranged from 3 to 10 (Figure 3A and Table 1), which is very similar to that of rice (2 to 10) [11] and maize (3 to 10) [30]. In wheat, 7 out of the 21 TaHAK family members with two or three homoeologs contained different exon numbers. For instance, of the three homoeologues of TaHAK5, TaHAK5-3AL, and -3DL had 10 exons each, whereas TaHAK5-3B had nine exons. This difference in the exon numbers among homoeologs was also found in other families of wheat genes, such as TaJAZs [35] and MADS-box gene families [36]. This suggests that exon acquisition or loss might have occurred in these gene family during wheat evolution, leading to the various structures of homoeologous genes [35]. It was also found that the exon numbers of orthologous HAK genes from different species was rather conserved. For example, TaHAK22 contained three exons, which was the smallest exon number across all of the TaHAK genes. Similarly, ZmHAK22 and OsHAK22 genes contained three and two exons, respectively [11,30]. This conservation is in agreement with the fact that homologs in different species normally share similar functions.
The TaHAK genes are long and encode for proteins ranging from 686 to 916 amino acids. Of these, 534 amino acids constitute the characteristic “K_trans” domain (PF02705). Although the 25 identified motifs were conserved and evenly present in all TaHAK proteins, several motifs specific to a particular cluster of TaHAKs were found (Figure 3B and Table S4). These specific motifs are likely to contribute to the divergence of wheat TaHAKs. Another characteristic feature of HAK transporters is the presence of the consensus motif GVVYGDLGTSPLY [37]. This motif was a part of the motif 8, one of the 25 conserved protein motifs identified in this study (Table S2).
Spatiotemporal gene expression patterns likely reflect their potential functions. To gain a better insight into the functions of TaHAK genes, we performed gene expression analysis for TaHAK genes in a variety of wheat tissues using publicly available RNA-seq data. We found that most genes from clusters I and IV had low expression levels across all five tested tissues. In contrast, most genes from clusters II and III were strongly expressed in all tested tissues (Figure 4). These results are in agreement with previous reports on rice and Arabidopsis HAK genes. In rice, five genes belonging to clusters II and III—OsHAK2, OsHAK10, OsHAK15, OsHAK23, and OsHAK25—were expressed in all tissues across all three genotypes [11]. In Arabidopsis, many AtKT/KUP/HAKs were expressed in roots, leaves, siliques, and flowers [32]. In fact, 12 out of 13 AtKT/KUP/HAK genes belonged to groups II and III. Therefore, the gene expression patterns of AtKT/KUP/HAKs were also in accordance with our results that most TaHAK genes from clusters II and III are constitutively expressed in all wheat tissues.
The expression of genes from clusters II and III across a variety of tissues explains the observation that cluster II and III KT/KUP/HAKs have a substantial functional diversity [38]. In addition to mediating high- and low-affinity K+ uptake, some transporters from clusters II and III are involved in plant growth and development. For instance, AtKUP4/TRH1 from Arabidopsis has a role in the growth of root hair tips [39] and AtKT2/KUP2 affects shoot cell expansion [40]. Transporters from clusters II and III may also participate in responses to high salinity, since AtHAK2, AtHAK6, and AtHAK11 expression levels have been shown to be altered by salt stress [41]. Notably, TaHAK1a-4BL and -4DL were strongly expressed in roots, while TaHAK1b-2A, -2BL, and -2DL were strongly expressed in roots, spikes, and grains (Figure 4). Both TaHAK1a and TaHAK1b were clustered into cluster I together with OsHAK1, which was reported to be expressed in whole rice plants, but its strongest expression was observed in roots [20]. Further detailed studies revealed that OsHAK1 was abundantly expressed in roots, especially at root tips. In contrast, expression levels of this gene in shoots were low, with the exception of meristematic tissues, where expression of OsHAK1 was rather strong [24]. The expression pattern of OsHAK1 allows it to play a major role in K+ uptake in rice [24].
Transcriptional regulation of K+ transporter genes represents a major mechanism in plant responses to low-K+ stress [7]. Generally, K+ transporter genes are upregulated under conditions of low-K+ stress [4]. Previous studies demonstrated that expression of OsHAK1 and OsHAK5 genes is induced by potassium deficiency and, consequently, K+ uptake in rice roots is enhanced [24,25]. In Arabidopsis, AtHAK5 is the most prominent gene and the only gene in the HAK/KUP/KT family induced by low-K+ stress [32,42]. In this study, we found that all nine tested TaHAK genes were upregulated under conditions of potassium deficiency, albeit to different extents (Figure 5). Our results are in good agreement with the previous report on expression of rice OsHAK genes. Seventeen OsHAKs, with the exception of two undetected genes, were all upregulated by potassium starvation [43]. Interestingly, TaHAK2, TaHAK9, TaHAK11, TaHAK16, and TaHAK17 were upregulated after a short period of potassium deficiency treatment and then rapidly downregulated (transient activation), suggesting that they may be involved in low potassium responses in wheat. TaHAK1, TaHAK7, TaHAK18, and TaHAK23 exhibited a continuous upregulation, suggesting that they might function in maintaining plants normal growth under potassium deficiency and mediate potassium ion absorption. Under salt stress, the transcripts of three TaHAK genes (TaHAK7, TaHAK9 and TaHAK16) were clearly increased (Figure 6). Maintaining K+ uptake at high Na+ is essential for K+/Na+ homeostasis and salt tolerance [44,45]. Thus, these three genes likely play important roles in mediating K+ and Na+ homeostasis in wheat under salt stress. Under drought stress, eight of the examined TaHAK genes showed increased expression, and only TaHAK23 was downregulated (Figure 7). Information about expression of HAK genes in response to drought is very limited. A previous study revealed that OsHAK1 expression levels were increased in roots and shoots in response to water deficit. Transgenic rice seedlings overexpressing OsHAK1 exhibited higher tolerance to drought stress than control plants [27]. Osakabe et al. [46] found that three close homologs, KUP6, KUP8, and KUP2, may act as key contributors to osmotic adjustment in Arabidopsis by maintaining K+ homeostasis during cell growth and under drought.
Chen et al. [24] demonstrated that OsHAK1 expression could complement the growth defect of the potassium uptake deficient yeast mutant R5421 under both low (0.05 and 0.1 mM) and high (1mM) external K+ conditions. They also found that the yeast transformed with OsHAK1 could tolerate salt stress much better than control yeast transformed with empty vector in the media containing up to 300 mM NaCl [24]. In this study, we demonstrated that TaHAK1b-2BL, a homolog of OsHAK1, functions in K+ uptake at K+ concentrations between 0.06 and 1 mM. Moreover, the salt sensitive yeast mutant AXT3K expressing TaHAK1b-2BL could tolerate salt stress better than the control yeast at 0 to 30 mM NaCl. AXT3K is very sensitive to salt stress due to the dysfunction of its major Na+ transporters in the plasma and endosomal membranes [47]. This is likely to be the main reason why the yeast mutant expressing TaHAK1b-2BL had stress tolerance only at relatively low (0 to 30 mM) NaCl concentrations. Similarly, Xu et al. showed that the AXT3K expressing TaNHX2 had better salt stress tolerance than control yeast at 0 to 60 mM NaCl [47].
4. Materials and Methods
4.1. Identification of TaHAK Genes in Wheat
Using the wheat genome data embedded in Ensembl plants database (Triticum aestivum TGACv1) [28], we conducted a hidden Markov model (HMM) search using the HMM profile of the K+ potassium transporter family (Pfam ID: PF02705) as queries. In total 150 sequences of 81 gene IDs were retrieved. The longest transcript sequence of a gene was retained while the other sequences under the same gene ID were removed. Furthermore, gene sequences without either a start or termination codon as well as incompletely sequenced genes were removed. Remaining sequences were analyzed using the Pfam tool with e-value < e−5 [48] and Conserved Domain Database (CDD) provided by the National Center for Biotechnology Information (NCBI) [49], and sequences containing incomplete K+ potassium transporter domains were discarded. At the end, 56 non-redundant sequences were identified as wheat TaHAK genes. Subcellular localization of each TaHAK protein was predicted using WOLF PSORT software [50] and the transmembrane structure was obtained using TMHMM Server 2.0 online tool [51]. Default parameters were used for all the programs unless otherwise stated.
4.2. Phylogenetic Analysis of TaHAKs
The protein sequences for AtKUP/HAK/KTs, OsHAKs, and ZmHAKs were retrieved from the NCBI [52], and these protein loci are listed in Table S1. The full-length proteins of AtHAK/KUP/KTs, OsHAKs, ZmHAKs, and the newly identified TaHAKs were aligned using ClustalW (embedded in MEGA7), and the phylogenetic tree was constructed based on the alignment using MEGA7 [53] with the neighbor-joining method and bootstrap replicates set to 1000.
4.3. Physical Localization of TaHAK Genes
To obtain TaHAKs chromosomal locations, the CDS of each gene was used to blast the URGI wheat genome database [54] with the expect threshold <e−10. The information regarding chromosomal length was obtained from the IWGSC CS RefSeq v.1.0 database [55]. The MapInspect software [56] was used to visualize TaHAK genes chromosomal distributions according to their gene starting positions and chromosomal lengths.
4.4. Analysis of Gene Structures and Protein Motifs
The coding and genome sequences of each TaHAK gene were downloaded from the wheat database in Ensembl Plants [28]. The TaHAKs gene structures were generated using the GSDS online tool [57], and the gene structures were arranged according to the protein positions in the phylogenetic tree constructed by the method of 4.2. The conserved motifs of the deduced TaHAK proteins were identified in the MEME website [58] using the following parameters: the finding motif number was set to 25 and the range of the motif length was set to 5–200 aa.
4.5. Plant Materials, Growth Conditions, and Experimental Treatments
Wheat (Triticum aestivum L. cv. Aikang58) seeds were grown for five days in culture dishes containing water in a greenhouse at 22 °C with a photoperiod of 16 h light/8 h dark. The five-day-old seedlings were transplanted into a nutrient solution (Table S6), which was replaced every three days until the seedlings reached the three-leaf stage. Roots at the three-leaf stage of seedling development were then subjected to K+ deficiency (0.1 mM K+) treatment, salt stress (200 mM NaCl), and dehydration (20% PEG6000). Roots were collected at 0, 1, 3, 6, 9, 12 and 24 h after the application of stress and treatments. All plant samples were collected in three biological replicates and were immediately frozen in liquid nitrogen. All samples were then stored at −80 °C until RNA isolation.
4.6. RNA Isolation and qRT-PCR
Total RNA was extracted using TransZOL (TransGEN Biotech, Beijing, China) according to the manufacturer’s instructions. cDNA was synthesized using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Dalian, China). GoTaq® qPCR Master Mix (Promega, Beijing, China) was used for qPCR amplification in Quantstudio™ 5 (Thermo Fisher, Shanghai, China). The qRT-PCR conditions were as follows: 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s, and 61 °C for 1 min, and a final extension at 72 °C for 5 min. The wheat β-actin gene was used as an internal reference gene for all qRT-PCR and expression levels were evaluated using the 2−∆∆Ct method. Publicly available RNA-seq data of the bread wheat cultivar Chinese Spring was retrieved from the WheatExp [31] website and used to evaluate the expression of wheat HAK genes in different wheat tissues. All qRT-PCR primers are listed in Table S7.
4.7. Subcellular Location of the TaHAK1b-2BL Protein
The TaHAK1b-2BL ORF sequence was amplified and inserted in frame to upstream GFP gene sequence in the 35S-GFP vector using ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). All primers are listed in Table S7. The resulting construct was transformed into Agrobaeterium tumefaeiens strain GV3101, which was then introduced into leaf epidermal cells of Nicotiana benthamiana by agro-infiltration. The transformed plants were allowed to grow for two days at 22 °C with a photoperiod of 16/8 h light/dark before visualization under a confocal laser-scanning microscope (LSM 710, ZEISS, Oberkochen, Germany).
4.8. Functional Complementation Assay of TaHAK1b-2BL in Yeast
The TaHAK1b-2BL coding sequence was amplified using the primers listed in Table S7 and cloned into the pYPGE15 vector, which was linearized by double digestion. The vectors pYPGE15-TaHAK1b-2BL and pYPGE15 were then transformed into the potassium uptake-deficient yeast strain CY162 (MATα, ura3-52, his3Δ200, his4-15, trk1Δ, trk2Δ1::pCK64) [59] and the salt-sensitive yeast mutant AXT3K (Δena1::HIS3::ena4, Δnha1::LEU2, Δnhx1::KanMX4) [60], respectively. The yeast transformants were grown to saturation in YNB medium as previously described [61]. These initial yeast cultures were used for making series of dilutions (10−1) and 5 μL samples of each diluted culture were plated onto AP plates containing 100, 1, 0.2, and 0.06 mM KCl or 20 and 30 mM NaCl, as indicated. The plates were incubated at 30 °C for 5 d before pictures were taken.
5. Conclusions
In this study, we identified 56 wheat TaHAK genes from a genome-wide survey of the recently available wheat genome data. Phylogenetic analysis of gene products resulted in identification of four clusters, containing 22, 19, 7, and 8 proteins, respectively. Protein sequences analysis revealed motifs that are conserved in the proteins originating from all four clusters as well as motifs unique for proteins from particular clusters of TaHAKs. Gene expression analysis using publicly available RNA-seq data revealed that TaHAKs from clusters II and III are constitutively expressed in various wheat tissues, while most genes in clusters I and IV have very low expression levels in all examined tissues at different developmental stages. The qRT-PCR analysis showed that TaHAK genes are differentially up- or downregulated in seedlings subjected to K+ deficiency, salinity, or dehydration. Furthermore, functional characterization of the TaHAK1b-2BL gene demonstrated that it facilitates K+ transport in yeast. Overall, our study provides a systematic description of HAK/KUP/KT potassium transporter genes in wheat. The obtained results deliver valuable information for future functional characterization of TaHAKs using transgenic plants, which we are currently planning in our laboratory.
Acknowledgments
We thank Sheng Luan and Renjie Tang from University of California, Berkeley for kindly offering the yeast strain CY162.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/12/3969/s1.
Author Contributions
Conceptualization, H.B. and H.X.; Methodology, X.L. and H.B.; Software, X.L.; Validation, H.B. and H.X.; Formal Analysis, X.L. and H.B.; Investigation, X.C., X.L., W.M., X.Z., and S.C.; Resources, H.B.; Data Curation, X.L.; Writing-Original Draft Preparation, X.L. and H.B.; Writing-Review & Editing, H.X.; Visualization, X.L.; Supervision, H.B. and H.X.; Project Administration, K.Z. and H.X.; Funding Acquisition, K.Z. and H.X.
Funding
This work was funded by the National Transgenic Research Project (grant no.2016ZX08002003-004), the National Key Research and Development Program of China (grant no. 2017YFD0100706), and the National Key Basic Research Program (973 Program 2014CB138105). These funding bodies neither influenced the selected experiments of this study nor their design.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Leigh R., Jones R.W. A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant-cell. New Phytol. 1984;97:1–13. doi: 10.1111/j.1469-8137.1984.tb04103.x. [DOI] [Google Scholar]
- 2.Wang X., Mohamed I., Ali M., Abbas M.H.H., Shah G.M., Chen F. Potassium distribution in root and non-root zones of two cotton genotypes and its accumulation in their organs as affected by drought and potassium stress conditions. J. Plant Nutr. Soil Sci. 2018 doi: 10.1002/jpln.201800026. [DOI] [Google Scholar]
- 3.Hasanuzzaman M., Bhuyan M., Nahar K., Hossain M., Mahmud J., Hossen M., Masud A., Fujita M. Potassium: A vital regulator of plant responses and tolerance to abiotic stresses. Agronomy. 2018;8:31. doi: 10.3390/agronomy8030031. [DOI] [Google Scholar]
- 4.Wang Y., Wu W.H. Regulation of potassium transport and signaling in plants. Curr. Opin. Plant Biol. 2017;39:123–128. doi: 10.1016/j.pbi.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Wang M., Zheng Q., Shen Q., Guo S. The critical role of potassium in plant stress response. Int. J. Mol. Sci. 2013;14:7370–7390. doi: 10.3390/ijms14047370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Very A.A., Nieves-Cordones M., Daly M., Khan I., Fizames C., Sentenac H. Molecular biology of K+ transport across the plant cell membrane: What do we learn from comparison between plant species? J. Plant Physiol. 2014;171:748–769. doi: 10.1016/j.jplph.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y., Wu W.H. Potassium transport and signaling in higher plants. Annu. Rev. Plant Biol. 2013;64:451–476. doi: 10.1146/annurev-arplant-050312-120153. [DOI] [PubMed] [Google Scholar]
- 8.Epstein E., Rains D., Elzam O. Resolution of dual mechanisms of potassium absorption by barley roots. Proc. Natl. Acad. Sci. USA. 1963;49:684–692. doi: 10.1073/pnas.49.5.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coskun D., Britto D.T., Li M., Oh S., Kronzucker H.J. Capacity and plasticity of potassium channels and high-affinity transporters in roots of barley and Arabidopsis. Plant Physiol. 2013;162:496–511. doi: 10.1104/pp.113.215913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordi S., Josep P. Potassium: A neglected nutrient in global change. Glob. Ecol. Biogeogr. 2015;24:261–275. doi: 10.1111/geb.12259. [DOI] [Google Scholar]
- 11.Gupta M., Qiu X., Wang L., Xie W., Zhang C., Xiong L., Lian X., Zhang Q. KT/HAK/KUP potassium transporters gene family and their whole-life cycle expression profile in rice (Oryza sativa) Mol. Genet. Genom. 2008;280:437–452. doi: 10.1007/s00438-008-0377-7. [DOI] [PubMed] [Google Scholar]
- 12.Maser P., Thomine S., Julian S., Ward J.M., Hirschi K. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schleyer M., Bakker E. Nucleotide sequence and 3’-end deletion studies indicate that the K+-uptake protein KUP from Escherichia coli is composed of a hydrophobic core linked to a large and partially essential hydrophilic C terminus. J. Bacteriol. 1993;175:6925–6931. doi: 10.1128/jb.175.21.6925-6931.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bañuelos M., Klein R., Alexander-Bowman S., Rodríguez-Navarro A. A potassium transporter of the yeast Schwanniomyces occidentalis homologous to the Kup system of Escherichia coli has a high concentrative capacity. EMBO J. 1995;14:3021–3027. doi: 10.1002/j.1460-2075.1995.tb07304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quintero F., Blatt M. A new family of K+ transporters from Arabidopsis that are conserved across phyla. FEBS Lett. 1997;415:206–211. doi: 10.1016/S0014-5793(97)01125-3. [DOI] [PubMed] [Google Scholar]
- 16.Li Y., Peng L., Xie C., Shi X., Dong C., Shen Q., Xu Y. Genome-wide identification, characterization, and expression analyses of the HAK/KUP/KT potassium transporter gene family reveals their involvement in K+ deficient and abiotic stress responses in pear rootstock seedlings. Plant Growth Regul. 2018;85:187–198. doi: 10.1007/s10725-018-0382-8. [DOI] [Google Scholar]
- 17.Li W., Xu G., Alli A., Yu L. Plant HAK/KUP/KT K+ transporters: Function and regulation. Semin. Cell Dev. Biol. 2018;74:133–141. doi: 10.1016/j.semcdb.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Grabov A. Plant KT/KUP/HAK potassium transporters: Single family—Multiple functions. Ann. Bot. 2007;99:1035–1041. doi: 10.1093/aob/mcm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gierth M., Maser P. Potassium transporters in plants—Involvement in K+ acquisition, redistribution and homeostasis. FEBS Lett. 2007;581:2348–2356. doi: 10.1016/j.febslet.2007.03.035. [DOI] [PubMed] [Google Scholar]
- 20.Banuelos M.A., Garciadeblas B., Cubero B., Rodriguez-Navarro A. Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol. 2002;130:784–795. doi: 10.1104/pp.007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nieves-Cordones M., Martinez V., Benito B., Rubio F. Comparison between Arabidopsis and rice for main pathways of K+ and Na+ uptake by roots. Front. Plant Sci. 2016;7:992. doi: 10.3389/fpls.2016.00992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubio F., Esanta-Marita G., Rodrıíguez-Navarro A. Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol. Plant. 2000;109:34–43. doi: 10.1034/j.1399-3054.2000.100106.x. [DOI] [Google Scholar]
- 23.Han M., Wu W., Wu W.-H., Wang Y. Potassium transporter KUP7 is involved in K+ acquisition and translocation in Arabidopsis root under K+ limited conditions. Mol. Plant. 2016;9:437–446. doi: 10.1016/j.molp.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 24.Chen G., Hu Q., Luo L., Yang T., Zhang S., Hu Y., Yu L., Xu G. Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ. 2015;38:2747–2765. doi: 10.1111/pce.12585. [DOI] [PubMed] [Google Scholar]
- 25.Yang T., Zhang S., Hu Y., Wu F., Hu Q., Chen G., Cai J., Wu T., Moran N., Yu L., et al. The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 2014;166:945–959. doi: 10.1104/pp.114.246520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen Y., Shen L., Shen Z., Jing W., Ge H., Zhao J., Zhang W. The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice. Plant Cell Environ. 2015;38:2766–2779. doi: 10.1111/pce.12586. [DOI] [PubMed] [Google Scholar]
- 27.Chen G., Liu C., Gao Z., Zhang Y., Jiang H., Zhu L., Ren D., Yu L., Xu G., Qian Q. OsHAK1, a high-affinity potassium transporter, positively regulates responses to drought stress in rice. Front. Plant Sci. 2017;8:1885. doi: 10.3389/fpls.2017.01885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ensembl Plant Database. [(accessed on 9 July 2017)]; Available online: http://plants.ensembl.org.
- 29.GSDS2.0: Gene Structure Display Server. [(accessed on 10 June 2018)]; Available online: http://gsds.cbi.pku.edu.cn/
- 30.Zhang Z., Zhang J., Chen Y., Li R., Wang H., Wei J. Genome-wide analysis and identification of HAK potassium transporter gene family in maize (Zea mays L.) Mol. Biol. Rep. 2012;39:8465–8473. doi: 10.1007/s11033-012-1700-2. [DOI] [PubMed] [Google Scholar]
- 31.Wheat Exp: An Expression Database for Polyploid Wheat. [(accessed on 7 November 2017)]; Available online: https://wheat.pw.usda.gov/WheatExp/
- 32.Ahn S.J. Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol. 2004;134:1135–1145. doi: 10.1104/pp.103.034660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amrutha R., Sekhar P., Varshney R., Pbk K. Genome-wide analysis and identification of genes related to potassium transpoter families in rice (Oryza sativa L.) Plant Sci. 2007;172:708–721. doi: 10.1016/j.plantsci.2006.11.019. [DOI] [Google Scholar]
- 34.Song Z.Z., Ma R.J., Yu M.L. Genome-wide analysis and identification of KT/HAK/KUP potassium transporter gene family in peach (Prunus persica) Genet. Mol. Res. 2015;14:774–787. doi: 10.4238/2015.January.30.21. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y., Qiao L., Bai J., Wang P., Duan W., Yuan S., Yuan G., Zhang F., Zhang L., Zhao C. Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.) BMC Genom. 2017;18:152. doi: 10.1186/s12864-017-3582-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma J., Yang Y., Luo W., Yang C., Ding P., Liu Y., Qiao L., Chang Z., Geng H., Wang P., et al. Genome-wide identification and analysis of the MADS-box gene family in bread wheat (Triticum aestivum L.) PLoS ONE. 2017;12:e0181443. doi: 10.1371/journal.pone.0181443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodrígueznavarro A. Potassium transport in fungi and plants. BBA Rev. Biomembr. 2000;1469:1–30. doi: 10.1016/S0304-4157(99)00013-1. [DOI] [PubMed] [Google Scholar]
- 38.Yang Z., Gao Q., Sun C., Li W., Gu S., Xu C. Molecular evolution and functional divergence of HAK potassium transporter gene family in rice (Oryza sativa L.) J. Genet. Genom. 2009;36:161–172. doi: 10.1016/S1673-8527(08)60103-4. [DOI] [PubMed] [Google Scholar]
- 39.Rigas S., Debrosses G., Haralampidis K., Vicente-Agullo F., Feldmann K.A., Grabov A., Dolan L., Hatzopoulos P. TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell. 2001;13:139–152. doi: 10.1105/tpc.13.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elumalai R.P. A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell Online. 2002;14:119–131. doi: 10.1105/tpc.010322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maathuis F.J. The role of monovalent cation transporters in plant responses to salinity. J. Exp. Bot. 2006;57:1137–1147. doi: 10.1093/jxb/erj001. [DOI] [PubMed] [Google Scholar]
- 42.Gierth M., Maser P., Schroeder J.I. The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 2005;137:1105–1114. doi: 10.1104/pp.104.057216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okada T., Nakayama H., Shinmyo A., Yoshida K. Expression of OsHAK genes encoding potassium ion transporters in rice. Plant Biotechnol. 2008;25:241–245. doi: 10.5511/plantbiotechnology.25.241. [DOI] [Google Scholar]
- 44.Cuin T.A., Bose J., Stefano G., Jha D., Tester M., Mancuso S., Shabala S. Assessing the role of root plasma membrane and tonoplast Na+/H+ exchangers in salinity tolerance in wheat: In planta quantification methods. Plant Cell Environ. 2011;34:947–961. doi: 10.1111/j.1365-3040.2011.02296.x. [DOI] [PubMed] [Google Scholar]
- 45.Munns R., Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 46.Osakabe Y., Arinaga N., Umezawa T., Katsura S., Nagamachi K., Tanaka H., Ohiraki H., Yamada K., Seo S.U., Abo M., et al. Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell. 2013;25:609–624. doi: 10.1105/tpc.112.105700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Y., Zhou Y., Hong S., Xia Z., Cui D., Guo J., Xu H., Jiang X. Functional characterization of a wheat NHX antiporter gene TaNHX2 that encodes a K+/H+ exchanger. PLoS ONE. 2013;8:e78098. doi: 10.1371/journal.pone.0078098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conserved Domain Database (CDD) in NCBI. [(accessed on 11 June 2018)]; Available online: https://www.ncbi.nlm.nih.gov/cdd.
- 50.WoLF PSORT: Protein Subcellular Localization Prediction. [(accessed on 19 October 2017)]; Available online: https://wolfpsort.hgc.jp/
- 51.TMHMM Server v. 2.0: Prediction of Transmembrane Helices in Proteins. [(accessed on 18 October 2017)]; Available online: http://www.cbs.dtu.dk/services/TMHMM/
- 52.National Center for Biotechnology Information (NCBI) [(accessed on 11 July 2017)]; Available online: https://www.ncbi.nlm.nih.gov/
- 53.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.URGI Wheat Genome Database. [(accessed on 3 November 2017)]; Available online: https://urgi.versailles.inra.fr/blast_iwgsc/blast.php.
- 55.IWGSC CS RefSeq v1.0 Database. [(accessed on 3 November 2017)]; Available online: https://urgi.versailles.inra.fr/jbrowseiwgsc/gmod_jbrowse/?data=myData%2FIWGSC_RefSeq_v1.0.
- 56.MapInspect 1.0: Displays and Analyzes Genetic Linkage Maps. [(accessed on 1 November 2017)]; Available online: https://mapinspect.software.informer.com/
- 57.Hu B., Jin J., Guo A.Y., Zhang H., Luo J., Gao G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.MEME: Multiple Em for Motif Elicitation, Version 4.11.4. [(accessed on 20 June 2018)]; Available online: http://memesuite.org/tools/meme.
- 59.Anderson J., Huprikar S., Kochian L., Lucas W., Gaber R. Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1992;89:3736–3740. doi: 10.1073/pnas.89.9.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quintero F.J., Ohta M., Shi H., Zhu J.K., Pardo J.M. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA. 2002;99:9061–9066. doi: 10.1073/pnas.132092099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou Y., Lai Z., Yin X., Yu S., Xu Y., Wang X., Cong X., Luo Y., Xu H., Jiang X. Hyperactive mutant of a wheat plasma membrane Na+/H+ antiporter improves the growth and salt tolerance of transgenic tobacco. Plant Sci. 2016;253:176–186. doi: 10.1016/j.plantsci.2016.09.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.