Abstract
The interaction between plant mitochondria and the nucleus markedly influences stress responses and morphological features, including growth and development. An important example of this interaction is cytoplasmic male sterility (CMS), which results in plants producing non-functional pollen. In current research work, we compared the phenotypic differences in floral buds of different Brassica napus CMS (Polima, Ogura, Nsa) lines with their corresponding maintainer lines. By comparing anther developmental stages between CMS and maintainer lines, we identified that in the Nsa CMS line abnormality occurred at the tetrad stage of pollen development. Phytohormone assays demonstrated that IAA content decreased in sterile lines as compared to maintainer lines, while the total hormone content was increased two-fold in the S2 stage compared with the S1 stage. ABA content was higher in the S1 stage and exhibited a two-fold decreasing trend in S2 stage. Sterile lines however, had increased ABA content at both stages compared with the corresponding maintainer lines. Through transcriptome sequencing, we compared differentially expressed unigenes in sterile and maintainer lines at both (S1 and S2) developmental stages. We also explored the co-expressed genes of the three sterile lines in the two stages and classified these genes by gene function. By analyzing transcriptome data and validating by RT-PCR, it was shown that some transcription factors (TFs) and hormone-related genes were weakly or not expressed in the sterile lines. This research work provides preliminary identification of the pollen abortion stage in Nsa CMS line. Our focus on genes specifically expressed in sterile lines may be useful to understand the regulation of CMS.
Keywords: cytoplasmic male sterility (CMS), phytohormones, differentially expressed genes, pollen development, Brassica napus
1. Introduction
Oilseed rape is one of most important oil crops worldwide, producing food, biofuel, and industrial compounds, including lubricants and surfactants. Hybrid breeding is a key technique to enhance crop production [1,2,3], in which cytoplasmic male sterility (CMS) plays an important role in seed production [4]. CMS is a maternally inherited trait and is beneficial for the production of F1 hybrid seeds by generating infertile pollen without changing vegetative growth and female fertility [5]. CMS systems are not only a useful component for studying pollen development, but also an important way to utilize hybrid vigor [6]. The existence of CMS systems in plants eliminates the laborious and painstaking work of sterilization and manual emasculation in a broad range of crops. CMS can arise spontaneously in breeding lines after wide crosses, interspecific exchange of nuclear or cytoplasmic genomes, and mutagenesis [7]. Initially, it was thought that sterility was caused by mutation within the mitochondrial genome [8], however, further research has revealed that a major cause of CMS is mitochondrial DNA rearrangement, which results in plants unable to generate functional pollen [9]. Mitochondria are important cellular components for energy (ATP, NADH, FADH2)-dependent metabolic pathways, including oxidative phosphorylation, respiratory electron transfer, biosynthesis of amino acids, vitamin cofactors, the Krebs cycle, and programmed cell death [10,11,12]. Therefore, CMS proteins were hypothesized to cause mitochondrial energy deficiency and failure to meet energy requirements during male reproductive development [13].
Currently, 10 types of CMS systems have been reported in Brassica napus, including the natural mutation pol CMS [14] and shan2A CMS [15], and intergeneric hybridization CMS nap CMS [16] and Nsa CMS [17]. Nsa CMS [17], Ogu CMS [18] and tour CMS [19] were generated by protoplast fusion of different species, resulting in a source of genetic variation within the cytoplasmic organelles [20]. Both Pol CMS and Ogu CMS are commonly used as CMS systems for B. napus hybrid breeding. CMS is sensitive to harsh environmental factors, including air temperature and exposure time to sunlight [21,22,23]. However, the Nsa CMS system has demonstrated stable male sterility under different environmental conditions, ensuring seed purity during hybrid seed production.
Preliminary work has demonstrated significant differences in plant endogenous hormones between CMS lines and their maintainer lines in different species [24,25]. In sugar beet, it was found the level of endogenous IAA (indole-3-acetic acid), GA3 (gibberellic acid), and ZR (zeatin-riboside), in relation to ABA (abscisic acid), differed at three developmental stages (vegetative, early flowering, and bud development) [26]. It was also demonstrated that pepper CMS line ‘Bei-A’ and maintainer line ‘Bei-B’ showed significant hormonal differences [27], with a higher IAA and ABA content and lower ZR5 and GA3 content observed within the CMS line [27]. The relationship between phytohormones and CMS has been widely investigated in many species, including B. napus [28,29], flax [30], and rice [31]. It has been shown that phytohormones ABA and IAA may be major contributors for CMS. The concentration of ABA and IAA changes at different stages of bud development between male sterile lines and their maintainer lines [29,30]. These studies collectively provide evidence for the importance of determining the endogenous level of ABA and IAA in CMS and maintainer lines when studying cytoplasmic male sterility.
Most recently, attention has focused on the provision of next-generation sequencing (NGS) technology [32,33,34] and the use of NGS to make studies on expressed genes and genomes in higher plants more feasible [35,36,37]. Currently, RNA-Seq has been used in higher plants with CMS systems in many species, including tomato [37], rice [38], and B. napus [36,39]. A large and growing body of literature has investigated floral buds of CMS and maintainer lines using RNA sequencing and comparative gene expression. In Pol CMS, unigenes related to pollen development were analyzed through transcriptome sequencing [36]. These high-throughput results will be useful for understanding the sterility mechanism of pol CMS in detail. Another transcriptome study of SaNa-1A CMS was also conducted in B. napus [40]. By comparing the sterile line and the maintainer line, many differentially expressed genes (DEGs) involved in metabolic, protein synthesis, and other pathways were identified. These results provide a basis for future research on the CMS mechanism in SaNa-1A. The existence of various CMS lines with different mitochondrial patterns offer new opportunities to explore the genetic regulation of CMS and its associated developmental effects [41].
In the current study, Pol CMS, Ogu CMS, Nsa CMS, and their corresponding maintainer lines (with the same nuclear genome but fertile cytoplasm) were used to carry out transcriptomic and DEG analysis. Simultaneously, we compared the morphological differences in sterile and fertile lines, and analyzed the IAA and ABA contents. We investigated the pollen abortion stage of the Nsa CMS line by semi-thin sectioning. This study confirms the stage of pollen abortion in Nsa CMS, and illustrates the mode of regulation of the different CMS systems during pollen development at the transcriptomic level.
2. Results
2.1. Phenotypic Characterization of CMS Lines and Maintainer Lines
The flower structure of rapeseed includes four sepals, four petals, six stamens (four long and two short), and one pistil from outwards to inwards. When a flower blooms, mature pollen sticks to the pistil. The pistil is almost the same height as the long stamens, allowing pollination to occur easily. In this study, we obtained three CMS systems (Nsa CMS, Pol CMS, and Ogu CMS) with corresponding maintainer lines. All sterile lines and their maintainer line harbor the same nuclear genome but different cytoplasm. We found that all sterile floral petals were visually wrinkled and smaller than fertile flowers in three CMS systems (Figure 1). Degeneration of stamens and shorter stamen length was observed in sterile lines as compared with the normal fertile flowers. Among the three CMS systems, the stamens of the pol CMS sterile line were more seriously degenerated (Figure 1F). However, the pistils of all the sterile floral buds were the same as fertile lines (Figure 1).
Figure 1.
Flower morphology of maintainer and sterile lines of the Pol, Nsa, and Ogu cytoplasmic male sterility (CMS) systems. (A–B) Maintainer line; (C–D) Nsa CMS line; (E–F) Pol CMS line; (G–H) Ogu CMS line; Bar = 0.5 cm.
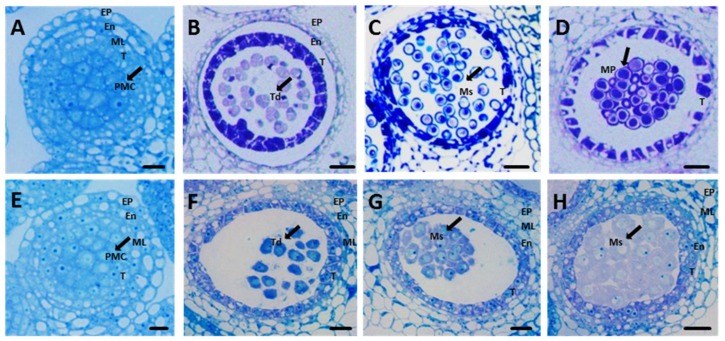
The stage at which pollen abortion occurs within the Nsa CMS system has not been determined clearly. For detailed characterization of the developing pollen, ultrathin specimens were observed under a microscope. By observing a semi-thin section of anthers, we conclude that the abortion period of Nsa CMS occurred during the tetrad period (Figure 2F). After the tetrad stage, Nsa CMS could not produce normal spores at the uni-nuclear stage (Figure 2G). Normal anthers form mature pollen, as shown in Figure 2D. The sterile line did not produce mature pollen but formed a large number of abnormal spores (Figure 2H).
Figure 2.
Comparison of maintainer line “ZS4” (A–D) and sterile line (E–H) anthers of Nsa CMS with toluidine blue staining. Bar = 10 μm, Ep, epidermis; En, endothecium; ML, middle layer; T, tapetum; Ms, microspore; MP, mature pollen; PMC, primary mother cells.
2.2. IAA and ABA Concentration in CMS and Maintainer Lines
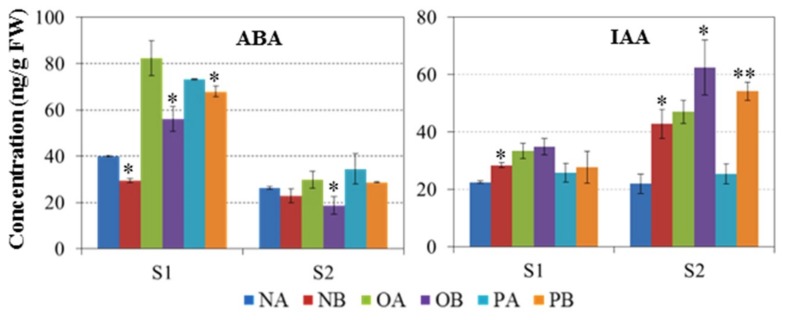
Plant hormones were assessed in CMS and maintainer lines to clarify how plant hormones are altered in the three CMS systems (Figure 3). The ABA and IAA contents in flower buds were detected at S1 (<2.5 mm size of floral buds) and S2 stages (>2.5 mm size of floral buds) in CMS and maintainer lines, respectively. We found that ABA levels were significantly higher in all three CMS lines as compared to maintainer lines at both stages. Conversely, IAA content was significantly lower in the Nsa CMS line than its maintainer line at the S1 stage, while Ogu and pol CMS lines showed no significant difference with their maintainer lines. However, IAA content was significantly lower in all CMS lines as compared to the maintainer lines at the S2 stage. These results indicate that a significantly higher content of endogenous ABA and lower content of IAA may enhance pollen abortion in sterile lines. ABA content showed increasing and IAA decreasing trends at the S1 stage compared to the S2 stage. The ABA content was significantly higher at both stages in CMS lines than in their corresponding maintainer line.
Figure 3.
ABA and IAA contents of developing buds in maintainer and male sterile lines of the Pol, Nsa, and Ogu systems. NA, Nsa sterile line; NB, Nsa maintainer line; OA, Ogu sterile line; OB, Ogu maintainer line; PA, Pol sterile line; PB, Pol maintainer line. Asterisks indicate a significant difference was detected between CMS line and maintainer line in S1 and S2 stage by t-test at *p<0.05, **p<0.01.
2.3. Differentially Expressed Genes in CMS and Maintainer Lines
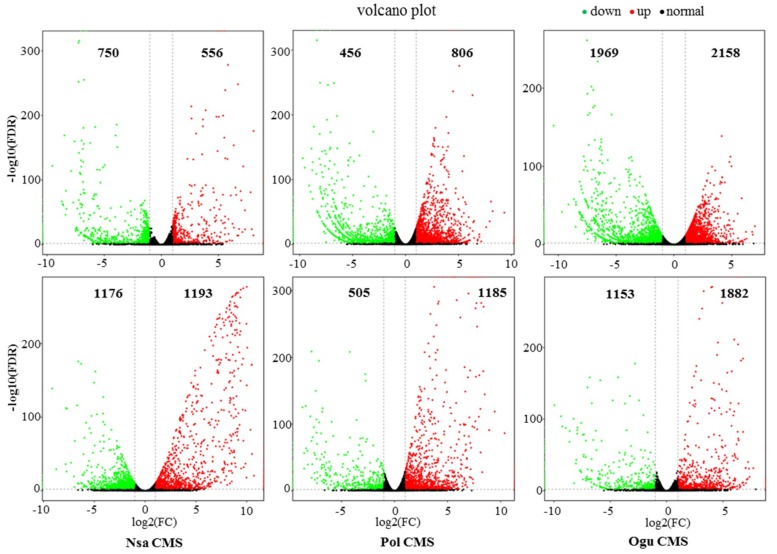
Using high-throughput sequencing, differentially expressed genes were detected in the sterile and corresponding maintainer lines. The flower buds used to determine phytohormone levels were also subjected to transcriptome sequence analysis. Three biological replicates were performed with the reproducibility between replicates being ≥90%. In total, 222.15 Gb of clean data were generated (with all samples Q30 ≥ 90%). Differentially expressed genes (DEGs) were identified in Biocloud (Biomarker Technologies). For each CMS system, DEGs were found between the male sterile line and the corresponding maintainer line. DEGs exhibiting a two-fold change or greater were selected according to the q-values [39]. At the S1 stage, we identified 1306, 1262, and 4127 DEGs in the Nsa, Pol, and Ogu systems, respectively. More DEGs (2369, 1690, and 3035) were discovered at the S2 stage in the three CMS systems. Among the three CMS systems, the largest number of DEGs were observed in the Ogu CMS system at the S1 stage. Among the total 4127 DEGs, 2158 genes were upregulated and 1969 genes were downregulated. The smallest number of DEGs was observed in the Pol CMS system, in which 806 genes were upregulated and 456 genes were downregulated at the S1 stage (Figure 4). Many more upregulated DEGs with high-fold change (>5-fold) were found at the S2 stage compared to the S1 stage in all three systems (Figure 4). Furthermore, only the Ogu CMS system exhibited more DEGs, including upregulated and downregulated genes, in the S1 stage than the S2 stage. More DEGs were observed in the Pol CMS, and especially in the Nsa CMS system at the S2 stage than the S1 stage.
Figure 4.
Differentially expressed unigenes and corresponding genes in the sterile and maintainer lines. The genes were selected with “p ≤ 0.01” and “fold change ≥2”. The X-axis is the log of 2-fold change in expression between the sterile and maintainer lines at two stages. Y-axis shows the statistical significance of the differences with the value of log10 (FDR). The spots in different colors are representing expression of different genes. Black spots represent genes without significant expression. Red spots mean 2-fold upregulated genes from maintainer lines to sterile lines. Green spots represent significantly 2-fold down-expressed genes from maintainer lines to sterile lines.
2.4. Gene Ontology and Classification of Three CMS Lines
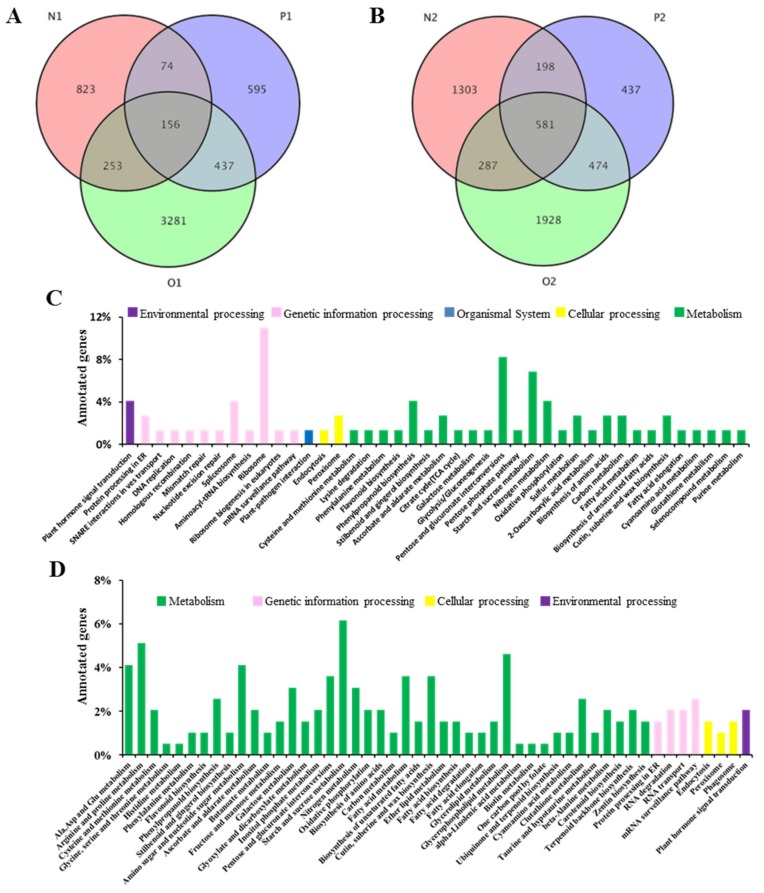
At the S1 stage, we observed that only 156 unigenes were co-differentially expressed in the three CMS lines, compared to 581 unigenes at the S2 stage (Figure 5A,B). KEGG classification and functional enrichment was performed for DEGs at both stages (Figure 5C,D). At the S1 stage, five categories were identified, including environmental information processing, genetic information processing, organismal systems, cellular processes, and metabolism (Figure 5C). At the S2 stage, genes were divided into four categories, including metabolism, genetic information processing, cellular processes, and environmental information processing (Figure 5D). At the S1 stage, in the environmental information processing category, 3% of DEGs were associated with plant hormone signal transduction. Only 1% of the DEGs were relative to plant–pathogen interaction. Within the cellular processes category, the highest number of DEGs was related to the peroxisome. Significantly enriched DEGs were identified as being involved in pentose–glucuronate interconversions and starch–sucrose metabolism among the metabolic components category. At the S2 stage, metabolic components were significantly enriched, including starch–sucrose, arginine–proline, glycerophospholipid, alanine–aspartate–glutamate, and amino–nucleotide sugar metabolism. From these analyses, we can determine starch and sucrose play an important role in metabolism at this stage. Transcriptomic data also revealed that many DEGs were enriched in plant hormonal signal transduction pathways.
Figure 5.
Co-differentially expressed unigenes (A, B) and GO annotations (C, D) of differentially expressed genes (DEGs). Venn diagrams of differentially expressed genes at (A) the S1 stage and (B) the S2 stage in Nsa (N), Ogu (O), and Pol (P) male sterile lines. N1, S1 stage of Nsa CMS system; O1, S1 stage of Ogu CMS system; P1, S1 stage of Pol CMS system; N2, S2 stage of Nsa CMS system; O2, S2 stage of Ogu CMS system; P2, S2 stage of Pol CMS system; (C) and (D), the X-axis indicates the percentage of genes in each categories, and y-axis showed classification of unigenes.
2.5. Verification of DEGs by RT-PCR
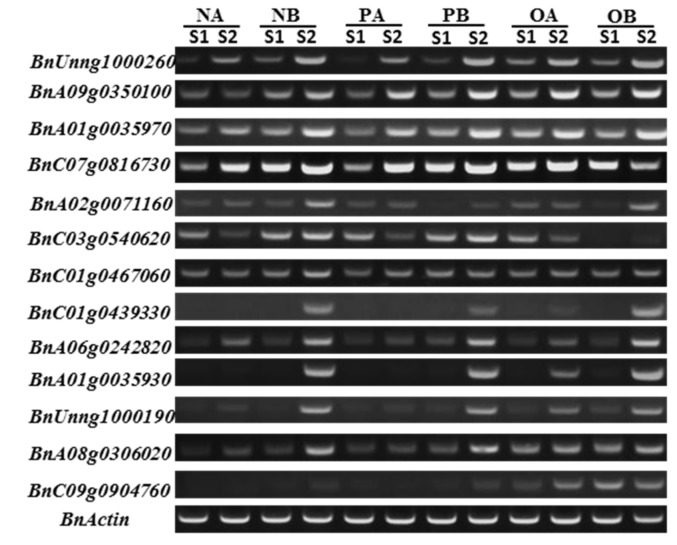
We conducted RT-PCR to validate the results generated by RNA-Seq. To determine whether transcription factors and hormone-related genes were differentially expressed, we quantified expression of these genes in the three CMS lines by semi-quantitative polymerase chain reaction (RT-PCR). Expression of genes encoding transcription factors or involved in ABA or IAA signaling were enriched in maintainer lines compared to male sterile lines (Figure 6). From the RT-PCR results, we found that almost all selected genes were highly expressed in maintainer lines compared to sterile lines. Most of the genes showed higher expression levels at the S2 stage compared to the S1 stage in all CMS systems (Pol, Ogu, and Nsa). This result was consistent with the RNA-Seq results.
Figure 6.
Gene expression difference in three cytoplasmic male sterile materials of S1 and S2 stages. NA, Nsa sterile line; NB, Nsa maintainer line; OA, Ogu sterile line; OB, Ogu maintainer line; PA, Pol sterile line; PB, Pol maintainer line. The BnActin gene was used as the control.
3. Discussion
The widespread existence of CMS in plant species may be related to the potential to promote outcrossing and prevent inbreeding depression. Independent CMS lines differ not only in their sequences and origins [42], but also in their phenotype, including changes in microspore development [43] and breakdown pattern of tapetum structure [44]. Pollen development comprises a series of defined physiological events. A large volume of published studies describe the role of energy (ATP, NADH, FADH2) in the pollen abortion process [45]. Our results also show that many energy production or conversion related genes are differentially expressed between male sterile and maintainer lines. In the semi-thin sections of the Nsa CMS and maintainer lines (Figure 2), we identified differences in the epidermis, endothecium, tapetum, microspore, and mature pollen. Our results revealed that pollen abortion occurred at the tetrad stage in the Nsa CMS line. Following the tetrad stage, Nsa CMS plants could not produce normal spores at the uni-nuclear stage (Figure 2H). Pollen abortion occurred due to the breakdown of tapetum and premature or delayed degeneracy [46,47]. In pol CMS, abortion was started at stage 4 (pollen development period). Anthers of the sterile line could not differentiate sporogenous cells, with the middle layer, endothecium, and tapetum being indistinguishable. The results in sterile anthers filled with numerous, highly vacuolated cells [36]. Due to abnormal development in the early stage, pol CMS lines cannot produce normal tetrads. In Ogu CMS, abortion was also identified to start at the tetrad stage by comparing the cell morphology of three central stages (the tetrad, mid-microspore, vacuolated microspore) of pollen development [44]. It was found that the tapetal cells developed a large vacuole at tetrad stage in the Ogu CMS line. The anther development of sterile line SaNa-1A, a line with CMS derived from somatic hybrids between B. napus and Sinapis alba, is also abnormal from the tetrad stage [40]. The abortion phenotype of Nsa CMS is the same as the SaNa-1 sterile line in all four stages of pollen development. Similar to SaNa-1 CMS, Nsa CMS was derived from somatic hybrids between B. napus and Sinapis arvensis. Together with Ogu CMS, which is derived from intergeneric hybridization between B. napus and Raphanus sativa, all the alloplasmic CMS systems have the same pollen development abortion stage.
Phytohormones (IAA and ABA) are generally known for their specific role in the induction and promotion of DNA synthesis, and play a role in metabolic pathways [48,49]. It was observed that high and exogenous application of ABA induced pollen abortion by specifically suppressing apoplastic sugar transport in pollen [22,50,51]. The ABA content in younger floral buds was higher than that of elder ones in all three CMS systems, whereas the IAA content showed a converse trend. However, similar to previous studies in other species, sterile lines of the three CMS systems showed some differences in IAA and ABA content in both male sterile and their corresponding maintainer lines [52,53].
Transcriptomic analysis detected a total of 5619 DEGs at the S1 stage, with 156 co-differentially expressed in all three CMS lines, and 5208 DEGs at the S2 stage, with 581 co-differentially expressed in all three CMS lines. KEGG analysis divided these co-differentially expressed genes into 42 and 50 categories at the S1 and S2 stages, respectively. At both the S1 and S2 stages, half of the genes were involved in metabolism. Many genes for mitochondrial energy metabolism and pollen development were also differentially expressed in multiple CMS systems [36,54,55]. It has been shown that the presence of infertility genes affects the transcription of genes involved in the energy metabolism of the mitochondria, resulting in impairment of the normal physiological functions of the mitochondria, which leads to infertility [13].
Previous studies identified a link between plant hormones and cytoplasmic male infertility [22,24]. In addition, transcription factors have also been implicated in pollen infertility [56,57]. Therefore, the expression of some transcription factors and hormone-related genes were selected to verify the results generated by RNA-Seq. RT-PCR results indicated that the expression level of selected genes in maintainer lines was significantly higher than that of the male sterile lines. This provides further support that levels of phytohormone precursor genes and some transcription factors may be correlated with cytoplasmic male sterility. Increased IAA content was detected in maintainer lines compared to male sterile lines in all CMS systems. Coincident with this result, we observed IAA signaling-related genes, including two IAA19 genes, were significantly enriched in maintainer lines (Figure 6).
4. Materials and Methods
4.1. Plant Materials
Pol CMS, Ogu CMS, Nsa CMS and their corresponding maintainer lines were used in this study. Materials were cultivated in the field of Oil Crops Research Institute, Chinese Academy of Agricultural Sciences (OCRI-CAAS), Wuhan, China. Anthers at different stages were collected for morphological study, and the abortion stage was studied by semi-thin sectioning of floral buds. Samples of floral buds (<2.5 mm and >2.5 mm) were collected and stored at −70 °C for further RNA-sequencing and hormonal quantification.
4.2. Morphology and Semi-Thin Sections
The floral buds of sterile and fertile lines were examined under the microscope (Olympus: CX31RTSF). At different stages, samples collected from the sterile and fertile line were fixed in FAA solution [38% formaldehyde, 70% ethanol, and 100% acetic acid (1:1:18)]. A vacuum chamber was used to evacuate the air and volatiles from the sample bottles. Fixed floral buds were dehydrated by a graded series of ethanol (70, 85, 95, and 100%) for one hour. Pre-infiltration and penetration by Technovit 7100 resin steps were undertaken to produce semi-thin sections ~3 μm thick. Samples were stained by 1% toluidine blue (Sigma Aldrich, St. Louis, MO, USA) for 3 min, and 5 specimens of each stage were observed to take images under an optical microscope (Olympus: CX31RTSF, Tokyo, Japen).
4.3. Phytohormone (ABA and IAA) Quantification
About 60 floral buds (smaller than 2.5 mm and larger than 2.5 mm) of Pol CMS, Ogu CMS, Nsa CMS, and their corresponding maintainer lines were quantified for ABA and IAA phytohormones. Samples were collected and extracted using methanol compounds [58]. The extraction was carried out by adding 1 mL of MeOH (methyl alcohol) with water (8:2) into each tube containing fresh plant material. Samples were shaken for 30 min before centrifugation at 12000 rpm at 4 °C for 10 min. The supernatant was transferred to a new microcentrifuge tube and dried in a speed vacuum. After drying, 100 µL of MeOH was added to each sample. Each sample was homogenized using a vortex mixer and centrifuged at 12000 RPM at 4 °C for 10 min. Phytohormones within the supernatant were separated by HPLC (Agilent 1200) and analyzed by a hybrid triple quadrupole/linear ion trap mass spectrometry (ABI 4000 Q-Trap, Applied Biosystems, Foster City, CA, USA).
4.4. Illumina Sequencing and Analysis of DEGs
About 60 floral buds were harvested from the plants of each line (Ogu CMS, Nsa CMS, and their corresponding maintainer lines) at the same time. Samples collected from each line were pooled, frozen in liquid nitrogen, and stored at −70 °C for RNA preparation. Total RNA from two stages of floral buds (<2.5 mm and >2.5 mm) of pol CMS, Ogu CMS, Nsa CMS, and their corresponding maintainer lines were extracted by using RNA kits (Tiangen, Beijing, China) in accordance with the manufacturer’s protocol. The integrity of the total RNA was checked by 1% agarose gel electrophoresis. The concentration was detected by Nano-Drop (Thermo Scientific, Madison, WI, USA) and purity of RNA was determined by Agilent 2100 Bio-analyzer (Agilent, Waldbronn, Germany). RNA (10 μL) was sequenced using the Illumina HiSeq 2000 (Illumina, San Diego, CA, USA) and 150 bp of data collected per run. After removing adapters and low-quality data, the resulting clean data was aligned to the B. napus reference genome [59]. Potential duplicate molecules were removed from the aligned BAM/SAM format records. FPKM (fragments per kilobase of exon per million fragments mapped) values were used to analyze gene expression by the software Cufflinks [60]. Three biological replicates were performed for each sample.
4.5. Semi-Quantitative (RT-PCR) Analysis of DEGs
The DEG results were confirmed by RT-PCR using the same RNA samples which were used for RNA library construction. Complementary DNA was generated from the RNA template by using the reverse transcription kit (Vazyme, Nanjing, China). Specific primers for differentially expressed genes were designed to amplify 600–750 bp sequences (Table S1). RT-PCR was carried out by using a program of 95 °C for 5 min (initial hot start), 30 cycles of 95 °C for 30 s, 56 °C for 35 s, and 72 °C for 5 min. Three biological replicates were analyzed for each sample.
5. Conclusions
Considerable effort has been taken to identify the pollen abortion stage in Pol and Ogu CMS lines. Conversely, the pollen abortion stage in the Nsa CMS line had not been determined clearly. From this study, we identified the tetrad stage of Nsa CMS for pollen abortion by using the semi-thin sectioning of floral buds. This information will help support the application of Nsa CMS in plant breeding. Higher content of ABA and lower content of IAA was observed in sterile lines when compared to maintainer lines in all male sterile systems. This result may reveal that ABA and IAA play different roles in fertile pollen development. During the two stages, genes involved in energy production were enriched in maintainer lines in comparison to sterile lines for all CMS systems investigated.
Acknowledgments
We thank Rachel Wells in John Innes Center for proof-reading and English correction.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/12/4022/s1.
Author Contributions
Data curation, M.H.; Formal analysis, B.D.; Funding acquisition, H.C. and Q.H.; Investigation, S.S.; Project administration, D.M. and Q.H.; Resources, H.W. and L.F.; Software, W.W.; Writing—original draft, B.D., Q.U.Z. and H.C.; Writing—review & editing, Q.H. All authors read and approved the final manuscript.
Funding
This research was funded by the National Key Research and Development program of China (No. 2016YFD0101300), National Natural Science Foundation of China (No. 31600226), Fundamental Research Funds for Central Non-profit Scientific Institution (1610172017005), Hubei Provincial Natural Science Foundation of China (2018CFB248), Science and Technology Innovation Project of the Chinese Academy of Agricultural Sciences (Group No. 118).
Conflicts of Interest
The authors declare no conflict of interests.
References
- 1.Wang H.Z. Review and future development of rapeseed industry in China. Chin. J. Oil Crop Sci. 2010;2:300–302. [Google Scholar]
- 2.Fu T.D. Breeding and utilization of rapeseed hybrid. Hubei Sci. Technol. 2000:167–169. [Google Scholar]
- 3.Allender C.J., King G.J. Origins of the amphiploid species Brassica napus L. Investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 2010;10:54. doi: 10.1186/1471-2229-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamagishi H., Bhat S.R. Cytoplasmic male sterility in brassicaceae crops. Breed Sci. 2014;64:38–47. doi: 10.1270/jsbbs.64.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shinada T., Kikuchi Y., Fujimoto R., Kishitani S. An alloplasmic male-sterile line of Brassica oleracea harboring the mitochondria from Diplotaxis muralis expresses a novel chimeric open reading frame, orf72. Plant Cell Physiol. 2006;47:549–553. doi: 10.1093/pcp/pcj014. [DOI] [PubMed] [Google Scholar]
- 6.Shiga T., Baba S. Cytoplasmic male sterility in oil seed rape. Brassica napus L., and its utilization to breeding. Japan J. Breed. 1973;23:187–197. [Google Scholar]
- 7.Hanson M.R., Bentolila S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell. 2004;16:154–169. doi: 10.1105/tpc.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnable P.S., Wise R.P. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 1998;3:175–180. doi: 10.1016/S1360-1385(98)01235-7. [DOI] [Google Scholar]
- 9.Horn R., Guptac K.J., Colombo N. Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion. 2014;19:198–205. doi: 10.1016/j.mito.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Siedow J.N., Umbach A.L. Plant mitochondrial electron transfer and molecular biology. Plant Cell. 1995;7:821–831. doi: 10.1105/tpc.7.7.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Logan D.C. The mitochondrial compartment. J. Exp. Bot. 2006;57:1225–1243. doi: 10.1093/jxb/erj151. [DOI] [PubMed] [Google Scholar]
- 12.Youle R.J., Karbowski M. Mitochondrial fission in apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:657. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- 13.Chen L., Liu Y.G. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 2014;65:579–606. doi: 10.1146/annurev-arplant-050213-040119. [DOI] [PubMed] [Google Scholar]
- 14.Fu T.D. Production and research of rapeseed in the People’s Republic of China. Eucarpia. Crucif. News. 1981;6:6–7. [Google Scholar]
- 15.Shen J.X., Wang H.Z., Fu T.D., Tian B.M. Cytoplasmic male sterility with self-incompatibility, a novel approach to utilizing heterosis in rapeseed (Brassica napus L.) Euphytica. 2008;162:109–115. doi: 10.1007/s10681-007-9606-0. [DOI] [Google Scholar]
- 16.Thompson K.F. Cytoplasmic male-sterility in oil-seed rape. Heredity. 1972;29:253–257. doi: 10.1038/hdy.1972.89. [DOI] [Google Scholar]
- 17.Hu Q., Andersen S., Dixelius C., Hansen L. Production of fertile intergeneric somatic hybrids between brassica napus and sinapis arvensis for the enrichment of the rapeseed gene pool. Plant Cell Rep. 2002;21:147–152. [Google Scholar]
- 18.Ogura H. Studies on the new male-sterility in japanese radish, with special reference to the utilization of this sterility towerds the practical raising of hybrid seeds. Mém. Fac. Agric. Kagoshima Univ. 1967;6:1446–1459. [Google Scholar]
- 19.Rawat D.S., Anand I.J. Male sterility in indian mustard. Indian J. Genet. Plant Breed. 1979;39:412–414. [Google Scholar]
- 20.Kemble R.J., Barsby T.L. Use of protoplast fusion systems to study organelle genetics in a commercially important crop. Biochem. Cell Biol. 1988;66:665–676. doi: 10.1139/o88-076. [DOI] [Google Scholar]
- 21.Burns D.R., Scarth R., McVetty P.B.E. Temperature and genotypic effects on the expression of pol cytoplasmic male sterility in summer rape. Can. J. Plant Sci. 1991;71:655–661. doi: 10.4141/cjps91-097. [DOI] [Google Scholar]
- 22.Zhang J.K., Zong X.F., Yu G.D., Li J.N., Zhang W. Relationship between phytohormones and male sterility in thermo-photo-sensitive genic male sterile (TGMS) wheat. Euphytica. 2006;150:241–248. doi: 10.1007/s10681-006-9114-7. [DOI] [Google Scholar]
- 23.Zhou Y.M., Fu T.D. Genetic improvement of rapeseed in China; Proceedings of the 12th International Rapeseed Congress; Wuhan, China. 26–30 March 2007. [Google Scholar]
- 24.Sawhney V.K., Shukla A. Male sterility in flowering plants: Are plant growth substances involved? Am. J. Bot. 1994;81:1640–1647. doi: 10.1002/j.1537-2197.1994.tb11477.x. [DOI] [Google Scholar]
- 25.Tian C.G., Zhang M.Y., Duan J. Preliminary study on the changes of phytohormones at different development stage in cytoplasmic male sterility line and its maintainer of rape. Sci. Agric. Sin. 1998;31:20–25. [Google Scholar]
- 26.Wang H.Z., Wu Z.D., Han Y. Relationships between endogenous hormone contents and cytoplasmic male sterility in sugarbeet. Sci. Agric. Sin. 2008;41:1134–1141. [Google Scholar]
- 27.Wu Z.M., Hu K.L., Fu J.Q., Qiao A.M. Relationships between cytoplasmic male sterility and endogenous hormone content of pepper bud. J. South China Agric. Univ. 2010;31:1–4. [Google Scholar]
- 28.Dubas E., Janowiak F., Krzewska M., Hura T., Żur I. Endogenous aba concentration and cytoplasmic membrane fluidity in microspores of oilseed rape (Brassica napus L.) genotypes differing in responsiveness to androgenesis induction. Plant Cell Rep. 2013;32:1465–1475. doi: 10.1007/s00299-013-1458-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shukla A., Sawhney V.K. Abscisic acid: One of the factors affecting male sterility in Brassica napus. Physiol. Plant. 2010;91:522–528. doi: 10.1111/j.1399-3054.1994.tb02983.x. [DOI] [Google Scholar]
- 30.Guan T., Dang Z., Zhang J. Studies on the changes of phytohormones during bud development stage in thermo-sensitivity genic male-sterile flax. Chin. J. Oil Crop Sci. 2007;3:248–253. [Google Scholar]
- 31.Huang S., Zhou X. Relationship between rice cytoplasmic male sterility and contents of GA (1+4) and IAA. Acta Agric. Bor-Sin. 1994;9:16–20. [Google Scholar]
- 32.Wang Z., Gerstein M., Snyder M. RNA-seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi Y.X., Liu Y.B., Rong W.H. RNA-Seq and its applications: A new technology for transcriptomics. Hereditas. 2011;33:1191. doi: 10.3724/SP.J.1005.2011.01191. [DOI] [PubMed] [Google Scholar]
- 35.Torti S., Fornara F., Vincent C., Andrés F., Nordström K., Göbel U., Knoll D., Schoof H., Coupland G. Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell. 2012;24:444–462. doi: 10.1105/tpc.111.092791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.An H., Yang Z., Yi B., Wen J., Shen J., Tu J., Ma C., Fu T. Comparative transcript profiling of the fertile and sterile flower buds of pol CMS in B. napus. BMC Genom. 2014;15:258. doi: 10.1186/1471-2164-15-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeong H.J., Kang J.H., Zhao M., Kwon J.K., Choi H.S., Bae J.H., Lee H.A., Joung Y.H., Choi D., Kang B.C. Tomato male sterile 1035 is essential for pollen development and meiosis in anthers. J. Exp. Bot. 2014;65:6693–6709. doi: 10.1093/jxb/eru389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan J., Zhang H., Zheng Y., Ding Y. Comparative expression profiling of mirnas between the cytoplasmic male sterile line meixianga and its maintainer line meixiangb during rice anther development. Planta. 2015;241:109–123. doi: 10.1007/s00425-014-2167-2. [DOI] [PubMed] [Google Scholar]
- 39.Qu C., Fu F., Liu M., Zhao H., Liu C., Li J., Tang Z., Xu X., Qiu X., Wang R., et al. Comparative transcriptome analysis of recessive male sterility (RGMS) in sterile and fertile Brassica napus lines. PLoS ONE. 2015;10:e0144118. doi: 10.1371/journal.pone.0144118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du K., Liu Q., Wu X.Y., Jiang J.J., Wu J., Fang Y.J., Li A.M., Wang Y. Morphological structure and transcriptome comparison of the cytoplasmic male sterility line in Brassica napus (SaNa-1A) derived from somatic hybridization and its maintainer line SaNa-1B. Front. Plant Sci. 2016;7:1313. doi: 10.3389/fpls.2016.01313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leino M., Teixeira R., Landgren M., Glimelius K. Brassica napus lines with rearranged arabidopsis mitochondria display CMS and a range of developmental aberrations. Theor. Appl. Genet. 2003;106:1156–1163. doi: 10.1007/s00122-002-1167-y. [DOI] [PubMed] [Google Scholar]
- 42.Mackenzie S. Male sterility and hybrid seed production. Plant Biotechnol. Agric. 2012:185–194. [Google Scholar]
- 43.Datta R., Chamusco K.C., Chourey P.S. Starch biosynthesis during pollen maturation is associated with altered patterns of gene expression in maize. Plant Physiol. 2002;130:1645–1656. doi: 10.1104/pp.006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.González-Melendi P., Uyttewaal M., Morcillo C.N., Hernández Mora J.R., Fajardo S., Budar F., Lucas M.M. A light and electron microscopy analysis of the events leading to male sterility in Ogu-INRA CMS of rapeseed (Brassica napus) J. Exp. Bot. 2008;59:827–838. doi: 10.1093/jxb/erm365. [DOI] [PubMed] [Google Scholar]
- 45.Yang J.H., Huai Y., Zhang M.F. Mitochondrial atpa gene is altered in a new orf220-type cytoplasmic male-sterile line of stem mustard (Brassica juncea) Mol. Biol. Rep. 2009;36:273–280. doi: 10.1007/s11033-007-9176-1. [DOI] [PubMed] [Google Scholar]
- 46.Luo X.D., Dai L.F., Wang S.B., Wolukau J., Jahn M., Chen J.F. Male gamete development and early tapetal degeneration in cytoplasmic male sterile pepper investigated by meiotic, anatomical and ultrastructural analyses. Plant Breed. 2006;125:395–399. doi: 10.1111/j.1439-0523.2006.01238.x. [DOI] [Google Scholar]
- 47.Li N., Zhang D.S., Liu H.S., Yin C.S., Li X.X., Liang W.Q., Yuan Z., Xu B., Chu H.W., Wang J. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell. 2006;18:2999–3014. doi: 10.1105/tpc.106.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minocha S.C. The role of auxin and abscisic acid in the induction of cell division in jerusalem artichoke tuber tissue cultured in vitro. Z. Pflanzenphysiol. 1979;92:431–441. doi: 10.1016/S0044-328X(79)80188-9. [DOI] [Google Scholar]
- 49.Rook F., Hadingham S.A., Li Y., Bevan M.W. Sugar and aba response pathways and the control of gene expression. Plant Cell Environ. 2006;29:426–434. doi: 10.1111/j.1365-3040.2005.01477.x. [DOI] [PubMed] [Google Scholar]
- 50.Ji X., Dong B., Shiran B., Talbot M.J., Edlington J.E., Hughes T., White R.G., Gubler F., Dolferus R. Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiol. 2011;156:647–662. doi: 10.1104/pp.111.176164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Storme N., Geelen D. The impact of environmental stress on male reproductive development in plants: Biological processes and molecular mechanisms. Plant Cell Environ. 2014;37:1–18. doi: 10.1111/pce.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duca M. Genetic-phytohormonal interactions in male fertility and male sterility phenotype expression in sunflower (Helianthus annuus L.)/interacciones genético-fitohormonales en la expresión fenotípica de la androfertilidad y androesterilidad en girasol (helianthus annuus l.)/interactions génétiques phytohormonales dans l’expression phénotypique d’un male fertile et male sterile du tournesol (helianthus annuus L.) Helia. 2008;31:27–38. [Google Scholar]
- 53.Singh S., Sawhney V. Abscisic acid in a male sterile tomato mutant and its regulation by low temperature. J. Exp. Bot. 1998;49:199–203. doi: 10.1093/jxb/49.319.199. [DOI] [Google Scholar]
- 54.Yang J.H., Zhang M.F., Yu J.Q. Mitochondrial nad2 gene is co-transcripted with CMS-associated orfB gene in cytoplasmic male-sterile stem mustard (Brassica juncea) Mol. Biol. Rep. 2009;36:345–351. doi: 10.1007/s11033-007-9185-0. [DOI] [PubMed] [Google Scholar]
- 55.Heng S., Liu S., Xia C., Tang H., Xie F., Fu T., Wan Z. Morphological and genetic characterization of a new cytoplasmic male sterility system (oxa CMS) in stem mustard (Brassica juncea) Theor. Appl. Genet. 2018;131:59–66. doi: 10.1007/s00122-017-2985-2. [DOI] [PubMed] [Google Scholar]
- 56.Xing M., Sun C., Li H., Hu S., Lei L., Kang J. Integrated analysis of transcriptome and proteome changes related to the Ogura cytoplasmic male sterility in cabbage. PLoS ONE. 2018;13:e0193462. doi: 10.1371/journal.pone.0193462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y., Ding X., Wang X., He T., Zhang H., Yang L., Wang T., Chen L., Gai J., Yang S. Genome-wide comparative analysis of DNA methylation between soybean cytoplasmic male-sterile line NJCMS5A and its maintainer NJCMS5B. BMC Genom. 2017;18:596. doi: 10.1186/s12864-017-3962-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H., Cheng H., Wang W., Liu J., Hao M., Mei D., Zhou R., Fu L., Hu Q. Identification of BnaYUCCA6 as a candidate gene for branch angle in Brassica napus by QTL-seq. Sci. Rep. 2016;6:38493. doi: 10.1038/srep38493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chalhoub B., Denoeud F., Liu S., Parkin I.A., Tang H., Wang X., Chiquet J., Belcram H., Tong C., Samans B., et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 60.Cheng H., Hao M., Wang W., Mei D., Wells R., Liu J., Wang H., Sang S., Tang M., Zhou R., et al. Integrative RNA- and miRNA-Profile Analysis Reveals a Likely Role of BR and Auxin Signaling in Branch Angle Regulation of B. napus. Int. J. Mol. Sci. 2017;18:887. doi: 10.3390/ijms18050887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.