Abstract
Sox2 is a pioneer transcription factor that initiates cell fate reprogramming through locus-specific differential regulation. Mechanistically, it was assumed that Sox2 achieves its regulatory diversity via heterodimerization with partner transcription factors. Here, utilizing single-molecule fluorescence spectroscopy, we show that Sox2 alone can modulate DNA structural landscape in a dosage-dependent manner. We propose that such stoichiometric tuning of regulatory DNAs is crucial to the diverse biological functions of Sox2, and represents a generic mechanism of conferring functional plasticity and multiplicity to transcription factors.
Keywords: transcription factors, DNA-protein interactions, Sox2 sequential DNA loading, smFRET, DNA conformational landscape, sequential DNA bending, transcription factor dosage
1. Introduction
Sox2 regulates a remarkable variety of genes differentially; it activates some and represses others [1,2,3]. This functional diversity is assumed to be mediated by Sox2 heterodimerization with other transcription factors (TFs) such as Oct4, Oct1, Pax6, and Nanog [4,5]. Recent reports, however, suggest that these canonical partners often remain spatiotemporally separated from Sox2 during genome engagement [6,7,8,9,10]. This raises an important question regarding the TF’s mechanism of action as to how Sox2 alone can exert differential loci-specific regulatory effects.
Sox2 is a sequence-specific high-mobility group transcription factor (HMG-TF) [11]. These TFs have conserved DNA binding domains [12,13], also known as HMG box. These DNA binding domains are partly disordered and are assumed to undergo binding-induced functional disorder-to-order transitions [14]. HMG-TFs are known to cooperatively form heterodimers on DNA regulatory elements [13,15,16,17]; each heteromeric TF pair induces characteristic DNA bend and differentially regulates target gene transcription [18,19,20]. Interestingly, a number of recent studies suggested that Sox2 can also function as homodimers [21,22,23]. Whether and how such Sox2 assemblies alter DNA conformations remain largely unknown. Here, we utilize the strengths of single-molecule Förster/fluorescence resonance energy transfer (smFRET) measurements along with ensemble methods to understand the effects of Sox2 binding on regulatory DNA structural landscape in the context of the HMG box (Sox2HMG). Our results suggest that Sox2HMG induces stoichiometry-dependent alternate DNA bends and we propose that the resulting alternate DNA conformations may drive different transcriptional outcomes.
2. Results
2.1. Multiple Sox2HMG Domains Cooperatively Interact with dsDNANANOG
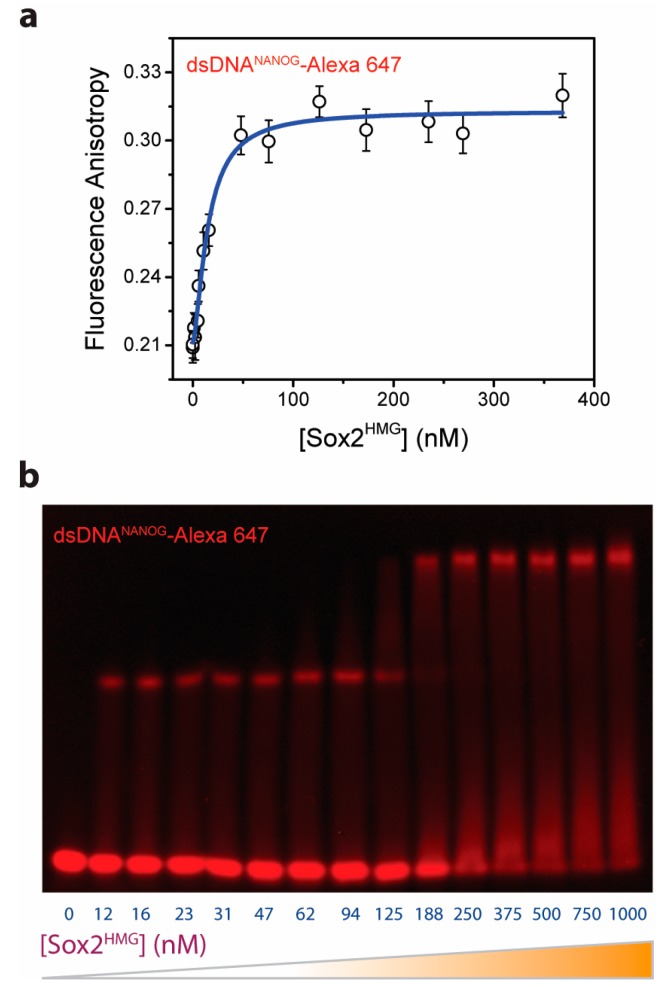
In our initial ensemble experiments, we observe that Sox2HMG cooperatively binds to the NANOG composite promoter (DNANANOG; Figure 1). We utilized fluorescence anisotropy to detect Sox2HMG binding to dsDNANANOG (Supplementary Methods). Anisotropy reports on fluorophore rotational properties, dependent on both probe local and global environment perturbations; fluorescence anisotropy of labeled macromolecule usually increases upon ligand binding. To characterize Sox2HMG-DNA binding, we singly-labeled dsDNANANOG with Alexa Fluor 647 (Supplementary Methods) and monitored changes in DNA fluorescence anisotropy with increasing Sox2HMG concentrations (Figure 1a). Nonlinear least squares (NLS) fitting of the anisotropy data to a Hill equation yields an apparent dissociation constant (KD) of 15.1 (±2.0) nM and Hill coefficient of 1.5 (±0.3). The estimated KD is similar to that previously reported for specific DNA-Sox2 interactions [18]. A Hill coefficient greater than 1 indicates that multiple Sox2 HMG boxes bind to the DNA in a TF concentration-dependent fashion [24]. Anisotropy measurements also indicate that Sox2HMG alone (i.e., the DNA-binding domain in the absence of dsDNANANOG) fails to dimerize/oligomerize (Figure S1). To verify the binding of multiple Sox2 molecules to DNANANOG, we carried out fluorescence electrophoretic mobility shift assay (fEMSA) of DNA with increasing [Sox2HMG]. The fEMSA micrograph shows concentration-dependent appearance of multiple electrophoretic species (Figure 1b). This suggests a multistep Sox2HMG interaction with the NANOG proximal promoter. The non-equilibrium nature of mobility shift assays, however, precludes precise estimation of binding affinities of individual Sox2-DNA assemblies on the basis the fEMSA micrograph [25].
Figure 1.
Sox2 cooperatively binds to the NANOG upstream promoter (DNANANOG). (a) DNA binding of Sox2HMG was probed by monitoring changes in fluorescence anisotropy of Alexa Fluor 647-labeled dsDNA with increasing [Sox2HMG]. The solid line represents nonlinear least squares (NLS) fit of the data to a Hill equation. NLS-derived parameters: KD = 15.1 (±2.0) nM, Hill coefficient = 1.5 (±0.3). (b) Fluorescence electrophoretic mobility assay (fEMSA) of Sox2HMG-DNANANOG binding suggests a multistep Sox2HMG complex formation with dsDNANANOG involving multiple protein molecules that are able to bind the DNA partner. (See also Figure S2.)
2.2. Sox2HMG Induces Sequential dsDNANANOG Bending Transitions
Next, we focused on understanding the mechanism of the TF-DNA complex formation. Although the mobility shift assay clearly demonstrates a multistep higher-order Sox2HMG complex formation with the dsDNA (Figure 1b), our ensemble experiments (i.e., fluorescence anisotropy and fEMSA) were not sensitive enough to determine the stoichiometries of respective TF-DNA complexes. To directly observe Sox2HMG-DNANANOG binding steps, we performed single-molecule fluorescence microscopy experiments that provide key advantages over conventional ensemble methods: (1) individual conformational sub-populations that are averaged out in ensemble measurements can be directly detected; and (2) experiments can be performed with extremely low concentrations of the labeled molecule (typically 50–100 pM). The ability to carry out experiments at low biomolecule concentration provides access and resolution for characterizing individual interaction steps in tightly interacting systems.
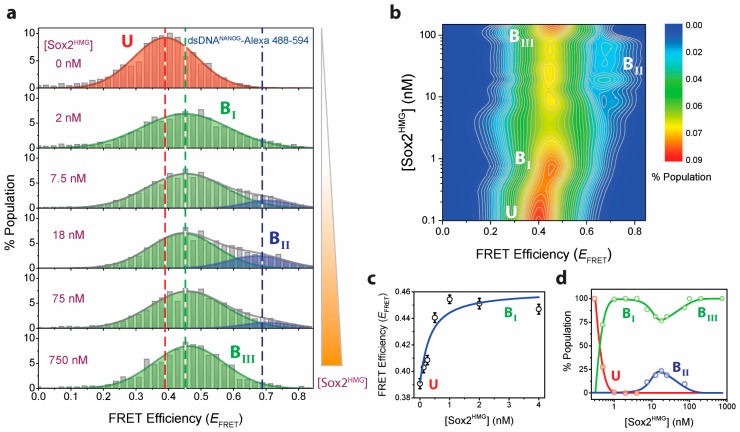
We utilized the distance-dependence of FRET to characterize Sox2HMG-DNANANOG interaction at single-molecule resolution. smFRET is sensitive to distance changes in the 20–70 Å range [26], and provides the necessary spatial resolution to probe changes in dsDNANANOG conformations as induced by TF binding (estimated end-to-end distance of DNANANOG is 57.4 Å, assuming inter-base axial rise of 3.4 Å [27]). For the smFRET experiments, we labeled DNANANOG with Alexa Fluor 488 and 594 donor-accepter dye-pair (Supplementary Methods). Bursts of fluorescence from donor and acceptor dyes were recorded as dual-labeled NANOG promoter DNA passed through the sub-fL observation volume of our custom-built ISS Alba confocal laser microscopy system (described previously [28]). These fluorescence intensities were converted to FRET efficiency (EFRET) histograms, providing a scheme for direct visualization of DNA conformational distributions. Without Sox2HMG, the dual-labeled DNA showed a single-peak in its EFRET histogram with histogram width typical of smFRET studies of freely diffusing dsDNA molecules [29,30] (Figure 2a; top panel). An NLS fit of the histogram to a Gaussian function yielded EFRET value of 0.39 (±0.04). On the basis of this EFRET value, we estimate the apparent distance between the two dyes to be approximately 64.6 Å (assuming a Förster distance of 60 Å between Alexa 488/594 dyes [31]). This is consistent with the estimated end-to-end distance of dsDNANANOG, where the slight increase in the apparent distance (compared to the estimated distance) can be attributed to the linkers present in Alexa dyes.
Figure 2.
smFRET reveals Sox2HMG concentration-dependent multistep bending of DNANANOG. (a) EFRET histograms of DNANANOG with increasing [Sox2HMG]. (b) [Sox2HMG]-EFRET contour map color coded based on fractional occupancy of individual DNA conformations. Corresponding DNA conformations are marked on the contour map. (c) Sox2 binding isotherm of the U ⇋ BI transition as probed by detecting changes in EFRET, linked to dsDNA bending transition. The NLS-derived apparent KD for this binding step is 0.30 (±0.04) nM (binding equation with fixed Hill coefficient of 1). (d) dsDNANANOG conformational distributions as modulated by Sox2HMG concentration, determined from NLS fitting of individual smFRET histograms to Gaussian functions.
Often, histograms of data collected in diffusion-based smFRET experiments show an additional peak at zero EFRET that arise from molecules with active donor(s) and either inactive or absent acceptor [29,30,32,33,34,35]. These zero EFRET peaks tend to significantly overlap with low EFRET peak populations and hamper direct estimation of the position of the non-zero peak(s) [36,37,38,39]. Interestingly, our smFRET histograms lack zero EFRET peaks (Figure 2a). We attribute this to the absence of dual donor-labeled dsDNA molecules as ensured by sequential labeling of individual DNA strands (Supplementary Methods). Therefore, sequential labeling and purification of individual fluorophore-conjugated oligos prior to duplex formation can be utilized to minimize zero peaks.
DNA bending (also known as DNA looping) is critical for many eukaryotic TF function [40,41,42,43,44]. Accordingly, Sox2 was shown to induce binding-mediated FGF (fibroblast growth factor) enhancer bending [18]. We postulate that similar spatially precise bending is induced in Sox2-DNANANOG complexes during gene regulation. To characterize Sox2HMG binding-induced NANOG promoter DNA bending, we carried out isothermal smFRET Sox2HMG titration against approximately 100 pM dual-labeled DNA (Figure 2). Our smFRET experiments provide a direct way to distinguish between subtle conformational changes of DNANANOG induced upon Sox2 binding. In our smFRET experiments, we observed a multistep bending transition in the DNA structural landscape (Figure 2a). Initially, DNANANOG undergoes a cooperative bending to a 0.45 (±0.01) EFRET state that corresponds to 32.1° (±1.4°) apparent bend angle at low Sox2HMG concentrations (≤4 nM) (see Supplementary Methods for the details of FRET-to-apparent-angle conversion). NLS fit of the data yields an estimated KD of 305 (±39) pM (Figure 2c). Such a tight interaction is unlikely to be driven by higher order Sox2HMG assemblies and we therefore postulate that this dsDNA conformation (henceforth referred as BI) is induced by binding to single Sox2HMG molecules.
Our ensemble results suggested that multiple Sox2HMG can form higher order TF-DNA assemblies (Figure 1). To characterize the complex formation, we probed for changes in DNANANOG conformations upon further addition of Sox2 on preformed monomeric Sox2HMG-DNANANOG complexes. With increasing [Sox2HMG], we observe a progressive reduction of the BI population and the emergence of a new population exhibiting higher EFRET (~0.68). This higher EFRET population corresponds to a DNANANOG apparent bend angle of 70° (±2.4°; henceforth referred to as BII DNA conformation). We infer that this DNA conformation is induced by sequential binding of two individual Sox2 TFs on the dsDNA, where binding of each monomer induces an approximate 32° bend at respective binding sites. Our observed apparent bend angle in the ternary complex (two Sox2 monomers and DNA) is similar to the DNA bend angle previously resolved for heterodimeric HMG box TF-DNA complexes [11,17].
Interestingly, an additional transition is visible in our isothermal smFRET titration when additional Sox2HMG is added (i.e., >75 nM [Sox2HMG]). We observe progressive depopulation of the BII bent DNA conformation and coupled emergence of a population at EFRET ~0.44 as [Sox2HMG] increases further (henceforth referred as BIII; Figure 2a). We estimate the apparent bend angle for the BIII population to be 30.4° (±4.5°) from the EFRET data (Supplementary Methods). A longer fEMSA run also indicates higher-order oligomer formation that is consistent with the formation of BIII population (Figure S2). Mechanistically, Sox family TFs induce DNA bends via FM dipeptide intercalation between two Thymine (T) bases at the minor groove interface [45,46]. Within the NANOG composite promoter, three TT pairs are present: two within the two HMG-TF binding sites (Oct/Sox motifs) identified by Rodda et al. [47] and one in between. We hypothesize that the initial two DNA bends are induced by sequential Sox2 binding to the two high-affinity HMG-TF binding motifs, where each binding induces an apparent 32° bend at the sites of interactions (a net 70° DNA apparent bend angle in the ternary complex). As Sox2HMG concentration further increases (>75 nM), an additional TF molecule interacts with the DNA at the remaining TT site and induces similar bend albeit at the opposite DNA face. This results in effective reversal of the second bend as evidenced by the increased inter-dye distance (i.e., reduced EFRET) at higher [Sox2HMG]. The final bend remains relatively unchanged upon further increase in Sox2 (up to 1 µM; Figure 2d). Overall, our smFRET data directly demonstrates multistep sequential DNA bending transitions dependent on Sox2 concentration.
3. Discussion
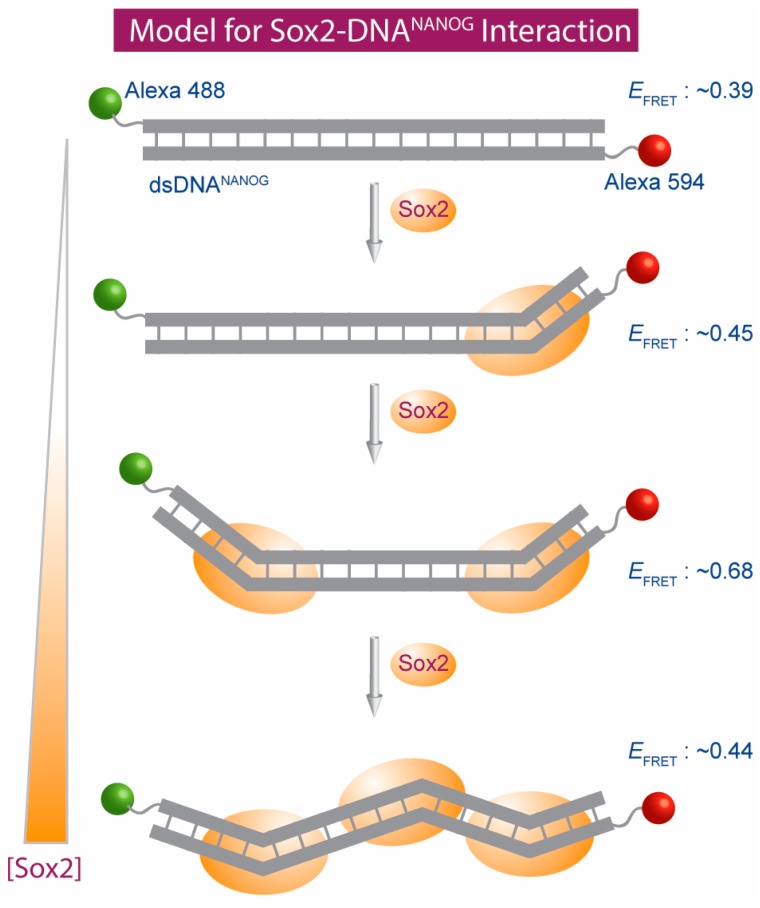
Sox2 is a tightly regulated transcription factor; both significant increases and decreases in Sox2 dosage can be detrimental to its biological function [48,49]. Alterations in Sox2 dosage result in multiple developmental and acquired disorders [50,51,52,53,54]. We show that the Sox2 HMG box can induce concentration-dependent alternate DNA bends (Figure 3). Alternate promoter bends are likely to regulate genes differentially and initiate downstream cascades crucial for Sox2’s diverse functions. Our results provide a mechanism for Sox2’s strict dosage dependence in its function-dysfunction dichotomy [50,55,56,57,58].
Figure 3.
Schematic representation of the Sox2 stoichiometry-dependent dsDNA bending transitions.
In summary, our smFRET experiments clearly demonstrate the role of Sox2 dosage in modulating the conformational landscape of HMG box-binding DNA motifs. Previous studies on Sox family members suggested that heterodimeric homeodomain TFs can induce sequential bending as they interact with their DNA partners [59,60,61,62]. Here, we utilize the strengths of smFRET to demonstrate that a representative sequence-specific HMG-TF alone induces concentration-dependent multistep DNA bending transitions. We envision additional layers of tunability for heteromeric HMG-TFs in respective regulatory complexes where affinities of individual transcription factors for DNAs as well as inter-TF interactions can vary dramatically.
4. Materials and Methods
Experimental details are provided in the Supplementary Materials. Briefly, ensemble fluorescence anisotropy and fluorescence electrophoretic mobility assay (fEMSA) experiments were performed in Buffer E (20 mM Tris, 50 mM NaCl, 0.10 mg/mL BSA, 5% glycerol, 0.1 mM DTT/0.05 mM TCEP, pH 8) with Alexa Fluor-647 labeled dsDNANANOG (Forward: ACTTTTGCATTACAATG; 17 bp). smFRET experiments were performed in the same buffer using a custom-built confocal fluorescence microscopy set up as described previously [28].
Abbreviations
| fEMSA | fluorescence electrophoretic mobility shift assay |
| FGF | fibroblast growth factor |
| HMG | high mobility group |
| NLS | nonlinear least squares |
| smFRET | single-molecule Förster resonance energy transfer |
| TF | transcription factor |
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/12/3865/s1.
Author Contributions
A.C.M.F. and J.C.F. conceived and designed the experiments; M.M.M., P.S.T., K.-J.C., A.C.M.F. and J.C.F. performed the experiments; M.M.M., P.S.T., A.C.M.F. and J.C.F. analyzed the data; M.M.M., A.C.M.F. and J.C.F. wrote the paper.
Funding
This work was supported by laboratory startup funds from the Baylor College of Medicine (A.C.M.F. and J.C.F.). J.C.F. is supported by R01 GM122763 from the NIGMS, NIH.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Chew L.J., Gallo V. The Yin and Yang of Sox proteins: Activation and repression in development and disease. J. Neurosci. Res. 2009;87:3277–3287. doi: 10.1002/jnr.22128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang S., Cui W. Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J. Stem Cells. 2014;6:305–311. doi: 10.4252/wjsc.v6.i3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y.R., Laghari Z.A., Novoa C.A., Hughes J., Webster J.R., Goodwin P.E., Wheatley S.P., Scotting P.J. Sox2 acts as a transcriptional repressor in neural stem cells. BMC Neurosci. 2014;15:95. doi: 10.1186/1471-2202-15-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kondoh H., Kamachi Y. SOX-partner code for cell specification: Regulatory target selection and underlying molecular mechanisms. Int. J. Biochem. Cell Biol. 2010;42:391–399. doi: 10.1016/j.biocel.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Kondoh H., Kamachi Y. Sox2. Academic Press; Boston, MA, USA: 2016. Chapter 8—SOX2–Partner Factor Interactions and Enhancer Regulation; pp. 131–144. [Google Scholar]
- 6.Thomson M., Liu S.J., Zou L.N., Smith Z., Meissner A., Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soufi A., Donahue G., Zaret K.S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J., Zhang Z., Li L., Chen B.C., Revyakin A., Hajj B., Legant W., Dahan M., Lionnet T., Betzig E., Tjian R. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soufi A., Garcia M.F., Jaroszewicz A., Osman N., Pellegrini M., Zaret K.S. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White M.D., Angiolini J.F., Alvarez Y.D., Kaur G., Zhao Z.W., Mocskos E., Bruno L., Bissiere S., Levi V., Plachta N. Long-Lived Binding of Sox2 to DNA Predicts Cell Fate in the Four-Cell Mouse Embryo. Cell. 2016;165:75–87. doi: 10.1016/j.cell.2016.02.032. [DOI] [PubMed] [Google Scholar]
- 11.Hou L., Srivastava Y., Jauch R. Molecular basis for the genome engagement by Sox proteins. Semin. Cell Dev. Biol. 2017;63:2–12. doi: 10.1016/j.semcdb.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Soullier S., Jay P., Poulat F., Vanacker J.M., Berta P., Laudet V. Diversification pattern of the HMG and SOX family members during evolution. J. Mol. Evol. 1999;48:517–527. doi: 10.1007/PL00006495. [DOI] [PubMed] [Google Scholar]
- 13.Malarkey C.S., Churchill M.E. The high mobility group box: The ultimate utility player of a cell. Trends Biochem. Sci. 2012;37:553–562. doi: 10.1016/j.tibs.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiss M.A. Floppy SOX: Mutual induced fit in hmg (high-mobility group) box-DNA recognition. Mol. Endocrinol. 2001;15:353–362. doi: 10.1210/mend.15.3.0617. [DOI] [PubMed] [Google Scholar]
- 15.Schlierf B., Ludwig A., Klenovsek K., Wegner M. Cooperative binding of Sox10 to DNA: Requirements and consequences. Nucleic Acids Res. 2002;30:5509–5516. doi: 10.1093/nar/gkf690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng C.K., Li N.X., Chee S., Prabhakar S., Kolatkar P.R., Jauch R. Deciphering the Sox-Oct partner code by quantitative cooperativity measurements. Nucleic Acids Res. 2012;40:4933–4941. doi: 10.1093/nar/gks153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clore G.M. Chapter 3—Dynamics of SOX2 Interactions with DNA A2—Kondoh, Hisato. In: Lovell-Badge R., editor. Sox2. Academic Press; Boston, MA, USA: 2016. pp. 25–41. [Google Scholar]
- 18.Scaffidi P., Bianchi M.E. Spatially precise DNA bending is an essential activity of the sox2 transcription factor. J. Biol. Chem. 2001;276:47296–47302. doi: 10.1074/jbc.M107619200. [DOI] [PubMed] [Google Scholar]
- 19.Dragan A.I., Read C.M., Makeyeva E.N., Milgotina E.I., Churchill M.E., Crane-Robinson C., Privalov P.L. DNA binding and bending by HMG boxes: Energetic determinants of specificity. J. Mol. Biol. 2004;343:371–393. doi: 10.1016/j.jmb.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 20.Slattery M., Riley T., Liu P., Abe N., Gomez-Alcala P., Dror I., Zhou T., Rohs R., Honig B., Bussemaker H.J., Mann R.S. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell. 2011;147:1270–1282. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J., Pan G., Cui K., Liu Y., Xu S., Pei D. A dominant-negative form of mouse SOX2 induces trophectoderm differentiation and progressive polyploidy in mouse embryonic stem cells. J. Biol. Chem. 2007;282:19481–19492. doi: 10.1074/jbc.M702056200. [DOI] [PubMed] [Google Scholar]
- 22.Cox J.L., Mallanna S.K., Luo X., Rizzino A. Sox2 uses multiple domains to associate with proteins present in Sox2-protein complexes. PLoS ONE. 2010;5:e15486. doi: 10.1371/journal.pone.0015486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia P., Wang S., Ye B., Du Y., Huang G., Zhu P., Fan Z. Sox2 functions as a sequence-specific DNA sensor in neutrophils to initiate innate immunity against microbial infection. Nat. Immunol. 2015;16:366–375. doi: 10.1038/ni.3117. [DOI] [PubMed] [Google Scholar]
- 24.Weiss J.N. The Hill equation revisited: Uses and misuses. FASEB J. 1997;11:835–841. doi: 10.1096/fasebj.11.11.9285481. [DOI] [PubMed] [Google Scholar]
- 25.Hellman L.M., Fried M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreon A.C., Deniz A.A. Protein folding at single-molecule resolution. Biochim. Biophys. Acta. 2011;1814:1021–1029. doi: 10.1016/j.bbapap.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watson J.D., Crick F.H.C. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature. 1953;171:737. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 28.Tsoi P.S., Choi K.J., Leonard P.G., Sizovs A., Moosa M.M., MacKenzie K.R., Ferreon J.C., Ferreon A.C. The N-Terminal Domain of ALS-Linked TDP-43 Assembles without Misfolding. Angew. Chem. Int. Ed. Engl. 2017;56:12590–12593. doi: 10.1002/anie.201706769. [DOI] [PubMed] [Google Scholar]
- 29.Deniz A.A., Dahan M., Grunwell J.R., Ha T., Faulhaber A.E., Chemla D.S., Weiss S., Schultz P.G. Single-pair fluorescence resonance energy transfer on freely diffusing molecules: Observation of Forster distance dependence and subpopulations. Proc. Natl. Acad. Sci. USA. 1999;96:3670–3675. doi: 10.1073/pnas.96.7.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dey S.K., Pettersson J.R., Topacio A.Z., Das S.R., Peteanu L.A. Eliminating Spurious Zero-Efficiency FRET States in Diffusion-Based Single-Molecule Confocal Microscopy. J. Phys. Chem. Lett. 2018;9:2259–2265. doi: 10.1021/acs.jpclett.8b00362. [DOI] [PubMed] [Google Scholar]
- 31.Johnson I.D., Spence M.T.Z. The Molecular Probes Handbook: A Guide to Fluorescent Probes and Labeling Technologies. Molecular Probes; Eugene, OR, USA: 2010. [Google Scholar]
- 32.Pljevaljcic G., Millar D.P., Deniz A.A. Freely diffusing single hairpin ribozymes provide insights into the role of secondary structure and partially folded states in RNA folding. Biophys. J. 2004;87:457–467. doi: 10.1529/biophysj.103.036087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan M.A., Okamoto K., Kahn J.D., English D.S. Single-molecule spectroscopic determination of lac repressor-DNA loop conformation. Biophys. J. 2005;89:2588–2596. doi: 10.1529/biophysj.105.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuler B. Single-molecule FRET of protein structure and dynamics—A primer. J. Nanobiotechnol. 2013;11(Suppl. 1):S2. doi: 10.1186/1477-3155-11-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyagi S., VanDelinder V., Banterle N., Fuertes G., Milles S., Agez M., Lemke E.A. Continuous throughput and long-term observation of single-molecule FRET without immobilization. Nat. Methods. 2014;11:297–300. doi: 10.1038/nmeth.2809. [DOI] [PubMed] [Google Scholar]
- 36.Ferreon A.C., Gambin Y., Lemke E.A., Deniz A.A. Interplay of α-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl. Acad. Sci. USA. 2009;106:5645–5650. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreon A.C., Moran C.R., Ferreon J.C., Deniz A.A. Alteration of the α-synuclein folding landscape by a mutation related to Parkinson’s disease. Angew. Chem. Int. Ed. Engl. 2010;49:3469–3472. doi: 10.1002/anie.201000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gambin Y., VanDelinder V., Ferreon A.C., Lemke E.A., Groisman A., Deniz A.A. Visualizing a one-way protein encounter complex by ultrafast single-molecule mixing. Nat. Methods. 2011;8:239–241. doi: 10.1038/nmeth.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moosa M.M., Ferreon A.C., Deniz A.A. Forced folding of a disordered protein accesses an alternative folding landscape. Chemphyschem. 2015;16:90–94. doi: 10.1002/cphc.201402661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su W., Jackson S., Tjian R., Echols H. DNA looping between sites for transcriptional activation: Self-association of DNA-bound Sp1. Genes Dev. 1991;5:820–826. doi: 10.1101/gad.5.5.820. [DOI] [PubMed] [Google Scholar]
- 41.Lim F.L., Hayes A., West A.G., Pic-Taylor A., Darieva Z., Morgan B.A., Oliver S.G., Sharrocks A.D. Mcm1p-induced DNA bending regulates the formation of ternary transcription factor complexes. Mol. Cell Biol. 2003;23:450–461. doi: 10.1128/MCB.23.2.450-461.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petrascheck M., Escher D., Mahmoudi T., Verrijzer C.P., Schaffner W., Barberis A. DNA looping induced by a transcriptional enhancer in vivo. Nucleic Acids Res. 2005;33:3743–3750. doi: 10.1093/nar/gki689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whittington J.E., Delgadillo R.F., Attebury T.J., Parkhurst L.K., Daugherty M.A., Parkhurst L.J. TATA-binding protein recognition and bending of a consensus promoter are protein species dependent. Biochemistry. 2008;47:7264–7273. doi: 10.1021/bi800139w. [DOI] [PubMed] [Google Scholar]
- 44.Gietl A., Grohmann D. Modern biophysical approaches probe transcription-factor-induced DNA bending and looping. Biochem. Soc. Trans. 2013;41:368–373. doi: 10.1042/BST20120301. [DOI] [PubMed] [Google Scholar]
- 45.Williams D.C., Cai M., Jr., Clore G.M. Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1.Sox2.Hoxb1-DNA ternary transcription factor complex. J. Biol. Chem. 2004;279:1449–1457. doi: 10.1074/jbc.M309790200. [DOI] [PubMed] [Google Scholar]
- 46.Palasingam P., Jauch R., Ng C.K., Kolatkar P.R. The structure of Sox17 bound to DNA reveals a conserved bending topology but selective protein interaction platforms. J. Mol. Biol. 2009;388:619–630. doi: 10.1016/j.jmb.2009.03.055. [DOI] [PubMed] [Google Scholar]
- 47.Rodda D.J., Chew J.L., Lim L.H., Loh Y.H., Wang B., Ng H.H., Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 48.Yamaguchi S., Hirano K., Nagata S., Tada T. Sox2 expression effects on direct reprogramming efficiency as determined by alternative somatic cell fate. Stem Cell Res. 2011;6:177–186. doi: 10.1016/j.scr.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 49.Prakash N. Chapter 4—Posttranscriptional Modulation of Sox2 Activity by miRNAs A2—Kondoh, Hisato. In: Lovell-Badge R., editor. Sox2. Academic Press; Boston, MA, USA: 2016. pp. 43–71. [Google Scholar]
- 50.Bertolini J., Mercurio S., Favaro R., Mariani J., Ottolenghi S., Nicolis S.K. Chapter 11—Sox2-Dependent Regulation of Neural Stem Cells and CNS Development A2—Kondoh, Hisato. In: Lovell-Badge R., editor. Sox2. Academic Press; Boston, MA, USA: 2016. pp. 187–216. [Google Scholar]
- 51.Van Heyningen V. Chapter 13—Congenital Abnormalities and SOX2 Mutations A2—Kondoh, Hisato. In: Lovell-Badge R., editor. Sox2. Academic Press; Boston, MA, USA: 2016. pp. 235–242. [Google Scholar]
- 52.Rizzoti K., Lovell-Badge R. Sox2. Academic Press; Boston, MA, USA: 2016. Chapter 14—Role of SOX2 in the Hypothalamo–Pituitary Axis; pp. 243–262. [Google Scholar]
- 53.Iwafuchi-Doi M., Zaret K.S. Cell fate control by pioneer transcription factors. Development. 2016;143:1833–1837. doi: 10.1242/dev.133900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wuebben E.L., Rizzino A. The dark side of SOX2: Cancer—A comprehensive overview. Oncotarget. 2017;8:44917–44943. doi: 10.18632/oncotarget.16570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarkar A., Hochedlinger K. The sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu K., Lin B., Zhao M., Yang X., Chen M., Gao A., Liu F., Que J., Lan X. The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell Signal. 2013;25:1264–1271. doi: 10.1016/j.cellsig.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kamachi Y., Kondoh H. Sox proteins: Regulators of cell fate specification and differentiation. Development. 2013;140:4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 58.Hagey D.W., Muhr J. Sox2 acts in a dose-dependent fashion to regulate proliferation of cortical progenitors. Cell Rep. 2014;9:1908–1920. doi: 10.1016/j.celrep.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 59.Peirano R.I., Wegner M. The glial transcription factor Sox10 binds to DNA both as monomer and dimer with different functional consequences. Nucleic Acids Res. 2000;28:3047–3055. doi: 10.1093/nar/28.16.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takayama Y., Clore G.M. Impact of protein/protein interactions on global intermolecular translocation rates of the transcription factors Sox2 and Oct1 between DNA cognate sites analyzed by z-exchange NMR spectroscopy. J. Biol. Chem. 2012;287:26962–26970. doi: 10.1074/jbc.M112.382960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morimura H., Tanaka S.I., Ishitobi H., Mikami T., Kamachi Y., Kondoh H., Inouye Y. Nano-analysis of DNA conformation changes induced by transcription factor complex binding using plasmonic nanodimers. ACS Nano. 2013;7:10733–10740. doi: 10.1021/nn403625s. [DOI] [PubMed] [Google Scholar]
- 62.Yamamoto S., De D., Hidaka K., Kim K.K., Endo M., Sugiyama H. Single molecule visualization and characterization of Sox2-Pax6 complex formation on a regulatory DNA element using a DNA origami frame. Nano Lett. 2014;14:2286–2292. doi: 10.1021/nl4044949. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.