Abstract
Background
Cellulosic biofuels are sustainable compared to fossil fuels. However, inhibitors, such as acetic acid generated during lignocellulose pretreatment and hydrolysis, would significantly inhibit microbial fermentation efficiency. Microbial mutants able to tolerate high concentration of acetic acid are needed urgently to alleviate this inhibition.
Results
Zymomonas mobilis mutants AQ8-1 and AC8-9 with enhanced tolerance against acetic acid were generated via a multiplex atmospheric and room temperature plasma (mARTP) mutagenesis. The growth and ethanol productivity of AQ8-1 and AC8-9 were both improved in the presence of 5.0–8.0 g/L acetic acid. Ethanol yield reached 84% of theoretical value in the presence of 8.0 g/L acetic acid (~ pH 4.0). Furthermore, a mutant tolerant to pH 3.5, named PH1-29, was generated via the third round of ARTP mutagenesis. PH1-29 showed enhanced growth and ethanol production under both sterilized/unsterilized conditions at pH 4.0 or 3.5. Intracellular NAD levels revealed that mARTP mutants could modulate NADH/NAD+ ratio to respond to acetic acid and low pH stresses. Moreover, genomic re-sequencing revealed that eleven single nucleic variations (SNVs) were likely related to acetic acid and low pH tolerance. Most SNVs were targeted in regions between genes ZMO0952 and ZMO0956, ZMO0152 and ZMO0153, and ZMO0373 and ZMO0374.
Conclusions
The multiplex mutagenesis strategy mARTP was efficient for enhancing the tolerance in Z. mobilis. The ARTP mutants generated in this study could serve as potential cellulosic ethanol producers.
Electronic supplementary material
The online version of this article (10.1186/s13068-018-1348-9) contains supplementary material, which is available to authorized users.
Keywords: Multiplex atmospheric and room temperature plasma (mARTP), Mutagenesis, Inhibitor tolerance, Zymomonas mobilis, Acetic acid, Low pH
Background
Cellulosic biofuels are sustainable alternative compared to fossil fuels. However, inhibitors generated during the pretreatment and hydrolysis of lignocellulosic feedstock, such as acetic acid, furfural, 5′-hydroxymethyl furfural, vanillin, and other organic acids/aldehydes/phenols, can severely inhibit cell growth and microbial fermentation efficiency [1, 2]. Among them, acetic acid is a major inhibitor that is able to destroy membrane integrity and intracellular redox homeostasis, which consequently exert osmotic stress, lower pH value, and reduce carbohydrate metabolism [3]. Series of biological, chemical, and physical methods have been developed to remove or tolerate this inhibitor from cellulosic hydrolysates [4, 5]. Among these methods, creating mutants capable of tolerating acetic acid is efficient for economic production of cellulosic biofuels.
Zymomonas mobilis is an attractive ethanologenic bacterium due to its high ethanol yield and high tolerance to ethanol [6, 7], uncoupling ethanol production with growth [8, 9], and wide environmental adaption under both aerobic and anaerobic conditions [10]. Recently, it has also emerged as a promising chassis for producing other bio-based products [11–13]. However, acetic acid still represents a great challenge to the conversion of lignocellulosic feedstock into bio-based chemicals by Z. mobilis. Rational and/or irrational methods have been widely applied in some biofuel producers, such as S. cerevisiae and Escherichia coli, to enhance tolerance to acetic acid and low pH [14, 15]. Likewise, rational modifications can help relieve acid toxicity to Z. mobilis. For instance, the introduction of an amino acid proton buffing peptide could improve the transient tolerance to low pH and acids in Z. mobilis strain CP4 [16]. The overexpression of Hfq, an RNA chaperon, in Z. mobilis significantly enhanced cellular tolerance against acetate, while its deletion diminished the acetate tolerance of strain AcR [17]. However, rational modification in Z. mobilis is still challenging due to the lack of comprehensive understanding on its functional genomics and regulatory network.
Random mutagenesis has been demonstrated as a powerful alternative to enhance tolerances in Z. mobilis. Recently, a mutant ZMA7-2 that is tolerant to 7 g/L acetic acid was created via adaptive laboratory evolution (ALE) [18]. The strain enabled high ethanol production using unsterilized acidic food waste hydrolysates at low pH values [19]. Alternatively, chemical mutagenesis has been widely applied in Z. mobilis for creating mutants. For example, mutant strain AcR tolerant to acetate [20] and flocculent mutant ZM401 (ATCC31822) with high tolerance to acetate and other inhibitors [21] were obtained by nitrosoguanidine (NTG) mutagenesis.
Atmospheric and room temperature plasma (ARTP) currently emerges as a powerful mutagenesis technique for bio-breeding [22]. It has been widely used in bacteria, fungi, and plants [23] to improve production efficiency [24–27] and/or to enhance robustness [28]. Compared to NTG or ultraviolet mutagenesis, the mutagenesis mediated by ARTP is fast, safe, and efficient with low locus bias [29]. Despite ARTP is powerful, no desirable mutant possibly could possibly be obtained under high selective pressure, such as high acetic acid level or low pH value if only one single round of ARTP mutagenesis was carried out. Recently, multi-round of ARTP mutagenesis has been applied to improve the production efficiency [30], demonstrating that it as an efficient mutagenesis strategy.
In this study, a multi-round ARTP mutagenesis named mARTP is performed with increasing selective pressures to generate mutants tolerant to 8.0 g/L acetic acid or low pH. Genomic mutation sites and NAD levels were analyzed to investigate tolerance mechanisms.
Results and discussion
mARTP mutagenesis in Z. mobilis
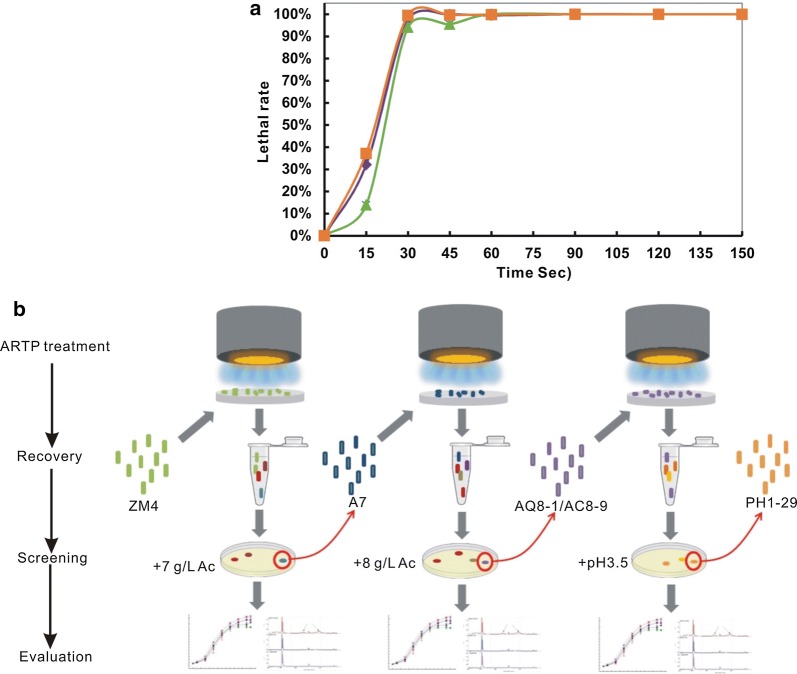
Prior to mutagenesis, the optimal lethal rate of ARTP was determined. Results showed that mutagenesis using a radio-frequency power input at 120 W and flow rate at 10 standard liters per minute (SLM) enabled high lethal rate and stable mutation efficiency in Z. mobilis wild-type strain ZM4, while adjustments to 100 W and 5 or 12 SLM led to unsteady mutagenesis efficiencies (data not shown). As shown in Fig. 1a, Z. mobilis cells were sensitive to ARTP treatment. Treatment by ARTP for 15 s resulted in a cellular death rate between 13.8 and 37.1%, and for over 30 s resulted in no cell survival. Accordingly, Z. mobilis cells were treated by ARTP at 120 W, 10 SLM, and 22 °C for 30 s in this study, leading to a cellular lethal rate of 94.0–99.5%.
Fig. 1.
mARTP mutagenesis in Z. mobilis. a Lethal rate of ARTP mutagenesis. Results were calculated from three independent tests; b flow chart of mARTP mutagenesis. Details on the mutagenesis were described in “Materials and methods” and “Results”
Previously, a Z. mobilis mutant against 7.0 g/L acetic acid, A7-2, was created [18], but mutants against higher concentration of acetic acid are desirable for efficient lignocellulosic ethanol production. Thus, the tolerance was further enhanced via mARTP mutagenesis in this study. After a single round of ARTP, no cells could survive when the ARTP-treated cells were immediately selected in rich medium (RM) containing 8.0 g/L acetic acid (data not shown). Such cell death could be ascribed to the dual toxicity of high acetic and low pH generated by acetic acid, as the pH value of RM containing 8.0 g/L acetic acid was decreased below 4.0 (Additional file 1: Table S1). It is hypothesized that under low pH conditions, the higher acetic acid concentration possibly led to a reduction of transmembrane pH gradient and thus to acidification of intracellular space. To solve this problem, mARTP was carried out with gradually increased selective pressures (Fig. 1b). ZM4 cells treated by the first round of ARTP were screened in RM supplemented with 7.0 g/L acetic acid. A resulting mutant A7, which had fast growth rate in the presence of 7.0 g/L acetic acid, was selected for the second round of ARTP. Consequently, two resulting mutants AQ8-1 and AC8-9 showed dramatically enhanced tolerance to 8.0 g/L acetic acid. The evidence, as shown in Additional file 1: Table S1, that pH value declined in the presence of high concentration of acetic acid suggests that the mutants’ tolerance to low pH was also improved. Hence, the third round of ARTP mutagenesis was performed using mutant AQ8-1 to screen mutants against lower pH value. A resulting mutant PH1-29 against pH 3.5 was finally obtained.
To test genetic stability, mutants were streaked on RM and grown in RM that was supplemented with acetic acid or that adjusted to pH 3.5 or 4.0. Such continuous culturing was repeated at least for three times, and there was no change of tolerance to acetic acid and pH stresses. Moreover, glycerol stocks of the mutants were stored at − 80 °C and kept stable in growth and tolerance capability so far.
By means of the mARTP mutagenesis, Z. mobilis mutants with “designed” tolerance capability could be generated. Thus, the mARTP mutagenesis was demonstrated to be efficient to enhance the tolerance of Z. mobilis.
Improved growth and ethanol production of Z. mobilis against acetic acid stresses
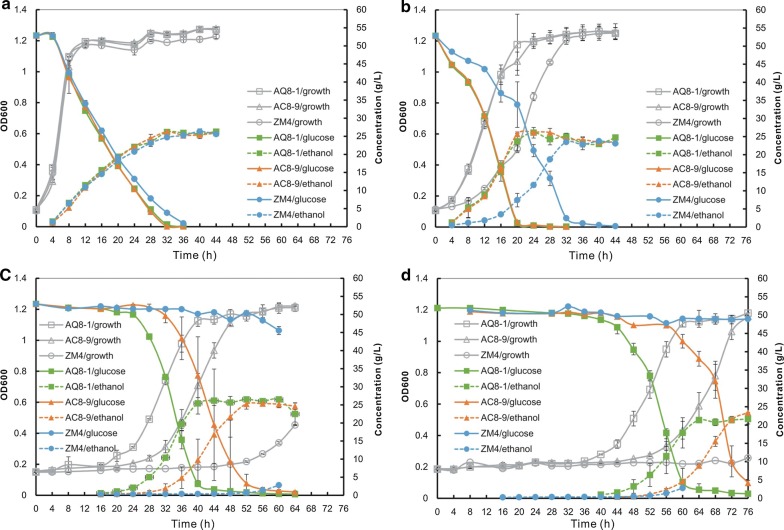
As shown in Fig. 2a, b, the growth and ethanol production of mutants AQ8-1 and AC8-9 were not affected by mild acetic acid concentrations (< 5.0 g/L). In the presence of higher concentration of acetic acid (7.0 or 8.0 g/L), their growth and ethanol production were remarkably improved compared to wild-type strain, though with a longer lag phase (Fig. 2c, d and Table 1). By contrast, wild-type strain ZM4 was dramatically suppressed by over 5.0 g/L acetic acid.
Fig. 2.
Growth and ethanol fermentation of Z. mobilis mutants tolerant to acetic acid. a RM50; b RM50 + 5.0 g/L acetic acid; c RM50 + 7.0 g/L acetic acid, and d RM50 + 8.0 g/L acetic acid. Growth is indicated by OD600 value; glucose indicates the concentration of the sugar remained in cultures; EtOH indicates the concentration of ethanol produced
Table 1.
Ethanol production of Z. mobilis under acetic acid or low pH stresses
| Strain | Fermentation time (h) | Glucose consumed (g/L) | Ethanol | References | ||
|---|---|---|---|---|---|---|
| Titer (g/L) | Yield (g/g glucose consumed) | Productivity (g/L/h) | ||||
| 50 g/L glucose, pH 6.0 | This study | |||||
| AQ8-1 | 28 | 48.06 ± 0.64 | 23.44 ± 1.25 | 0.49 ± 0.03 | 0.84 ± 0.04 | |
| AC8-9 | 28 | 48.65 ± 0.84 | 24.45 ± 1.08 | 0.50 ± 0.02 | 0.87 ± 0.04 | |
| PH1-29 | 24 | 49.32 ± 0.60 | 23.82 ± 0.44 | 0.48 ± 0.00 | 0.99 ± 0.02 | |
| ZM4 | 28 | 45.03 ± 0.28 (< 0.01) |
23.11 ± 0.06 (0.29) |
0.51 ± 0.00 (0.17) |
0.83 ± 0.00 (0.54) |
|
| 50 g/L glucose + 5 g/L acetic acid | ||||||
| AQ8-1 | 20 | 51.77 ± 0.29 | 23.67 ± 2.61 | 0.46 ± 0.05 | 1.18 ± 0.13 | |
| AC8-9 | 20 | 51.42 ± 0.40 | 25.67 ± 0.23 | 0.50 ± 0.01 | 1.28 ± 0.01 | |
| ZM4 | 32 | 50.41 ± 0.54 (0.03) |
23.47 ± 0.45 (0.32) |
0.47 ± 0.00 (0.36) |
0.73 ± 0.01 (< 0.01) |
|
| 50 g/L glucose + 7 g/L acetic acid | ||||||
| AQ8-1 | 40 | 49.74 ± 0.81 | 25.41 ± 0.43 | 0.51 ± 0.01 | 0.64 ± 0.01 | |
| AC8-9 | 56 | 51.24 ± 0.54 | 25.34 ± 0.44 | 0.49 ± 0.00 | 0.45 ± 0.01 | |
| ZM4 | 60 | 7.35 ± 1.10 (< 0.01) |
2.77 ± 0.05 (< 0.01) |
0.38 ± 0.07 (0.01) |
0.05 ± 0.00 (< 0.01) |
|
| 50 g/L glucose + 8 g/L acetic acid | ||||||
| AQ8-1 | 64 | 49.59 ± 0.27 | 20.76 ± 1.16 | 0.42 ± 0.02 | 0.32 ± 0.02 | |
| AC8-9 | 72 | 41.85 ± 4.22 | 21.46 ± 1.11 | 0.43 ± 0.00 | 0.30 ± 0.02 | |
| ZM4 | 76 | 3.00 ± 0.32 (< 0.01) |
0.48 ± 0.03 (< 0.01) |
0.16 ± 0.01 (< 0.01) |
0.01 ± 0.00 (< 0.01) |
|
| 50 g/L glucose, pH 4.0 | ||||||
| PH1-29a | 24 | 49.60 ± 0.45 | 23.84 ± 0.56 | 0.48 ± 0.01 | 0.99 ± 0.02 | |
| PH1-29b | 32 | 50.23 ± 0.01 | 23.09 ± 1.08 | 0.46 ± 0.02 | 0.72 ± 0.03 | |
| ZM4a | 40 | 23.15 ± 1.24 | 11.00 ± 0.38 | 0.48 ± 0.01 | 0.27 ± 0.01 | |
| ZM4b | 40 | 8.51 ± 0.34 (< 0.01) |
3.87 ± 0.06 (< 0.01) |
0.46 ± 0.02 (0.28) |
0.10 ± 0.00 (< 0.01) |
|
| 50 g/L glucose, pH 3.5 | ||||||
| PH1-29a | 52 | 49.89 ± 0.16 | 23.04 ± 2.63 | 0.46 ± 0.05 | 0.44 ± 0.05 | |
| PH1-29b | 88 | 50.05 ± 0.02 | 23.93 ± 0.19 | 0.48 ± 0.00 | 0.27 ± 0.00 | |
| ZM4a | 60 | 3.17 ± 0.48 (< 0.01) |
1.45 ± 0.16 (< 0.01) |
0.46 ± 0.02 (0.73) |
0.02 ± 0.00 (< 0.01) |
|
| 100 g/L glucose 2.1 g/L acetic acid, pH 4.22c | Ma et al. | |||||
| A7-2 | 24 | N/A | 49.05 | 0.49 | 2.04 | |
| 100 g/L glucose 4.2 g/L acetic acid, pH 3.97c | ||||||
| A7-2 | 24 | N/A | 48.69 | 0.49 | 2.03 | |
| 100 g/L glucose 6.3 g/L acetic acid, pH 3.85c | ||||||
| A7-2 | 24 | N/A | 47.53 | 0.48 | 1.98 | |
| 100 g/L glucose 6.3 g/L acetic acid pH 6.0d | Zhao et al. | |||||
| ZM401 | 24 | 99.9 | 48.9 | 0.49 | 2.04 | |
| ZM4 | 24 | 99.9 | 46.0 | 0.46 | 1.92 | |
| 100 g/L glucose 8.4 g/L acetic acid, pH 6.0d | ||||||
| ZM401 | 24 | 99.9 | 48.3 | 0.48 | 2.01 | |
| ZM4 | 24 | 97.6 | 44.0 | 0.45 | 1.83 | |
| 100 g/L glucose 10.5 g/L acetic acid, pH 6.0d | ||||||
| ZM401 | 36 | 97.4 | 46.2 | 0.47 | 1.28 | |
| ZM4 | 48 | 67.5 | 23.9 | 0.35 | 0.50 | |
| 120 g/L glucose 16 g/L sodium acetate, pH 5.0 | Liu et al. | |||||
| ZMA-142e | 56 | 103.9 | 48.0 | 0.46 | 0.86 | |
| ZMA-142f | 36 | 97.2 | 60.8 | 0.63 | 1.69 | |
| ZMA-167e | 64 | 100.7 | 44.3 | 0.44 | 0.69 | |
| ZMA-167f | 38 | 86.6 | 47.4 | 0.55 | 1.25 | |
p values calculated by one-way ANOVA are listed in brackets. Three repeats were performed for each strain, and error bars indicated standard deviation
aSterilized and bunsterilized RM containing 50 g/L glucose
cConcentrations of acetic acid were evaluated based on that original data (0.2%, 0.4%, and 0.6% (v/v) acetic acid was added, respectively)
dAdding 2 N KOH to adjust pH value to 6.0
eSeed culture in RM; fermentation in RM with 195 mM sodium acetate
fSeed culture in RM with 146 mM sodium acetate; fermentation in RM with 195 mM sodium acetate
Although AQ8-1 and AC8-9 were generated in the same mutagenesis process, they still showed difference in acetic acid tolerance. There was no remarkable difference in growth, glucose consumption, and ethanol production between both mutants below 5.0 g/L acetic acid (Fig. 2a, b). However, AQ8-1 grew faster than AC8-9 in the presence of 7.0 g/L or 8.0 g/L acetic acid (Fig. 2c, d), consequently with higher ethanol productivities (Table 1).
In addition, the tolerance of AQ8-1 and AC8-9 against mixtures of acetic acid and furfural, which are produced in cellulosic hydrolysates and are two major inhibitors to Z. mobilis [31, 32], was also investigated. So far, the reported highest concentrations of acetic acid and furfural Z. mobilis could tolerate were 7.0 g/L and 3.0 g/L, respectively [18]. Therefore, the concentration of two inhibitors in mixtures arranged from 1.0 to 7.0 g/L and 1.0 to 3.0 g/L, respectively. The mutants did not show obvious advantages over ZM4 under mixed stresses (Additional file 1: Figure S1), suggesting that the tolerance of both mutants to furfural was not enhanced. In the future, additional work is needed to improve the tolerance of Z. mobilis to multiple stresses via ARTP and/or other techniques.
Improved growth and ethanol production of Z. mobilis at low pH
AQ8-1 and AC8-9 were conferred with enhanced tolerance to low pH as well as acetic acid since the pH value was reduced below 4.0 with the addition of 8 g/L acetic acid (Additional file 1: Table S1). Low pH circumstance can effectively prevent microbial contamination during fermentation, thus can realize the fermentation under non-aseptic conditions [33]. In this study, a mutant pH1-29 against low pH was created using AQ8-1 as the parental strain through mARTP mutagenesis.
The growth of PH1-29 at low pH was enhanced as compared to ZM4 (Fig. 3a). The ethanol production was not inhibited at pH 4.0, while the productivity reduced to 0.5 g/L/h at pH 3.5, nearly by a half of that at pH 6.0. In contrast, ZM4 was severely inhibited at low pH values. It grew poorly at pH 4.0 and hardly survived at pH 3.5 (Fig. 3a), resulting in severe suppression of ethanol production at low pH values (Table 1).
Fig. 3.
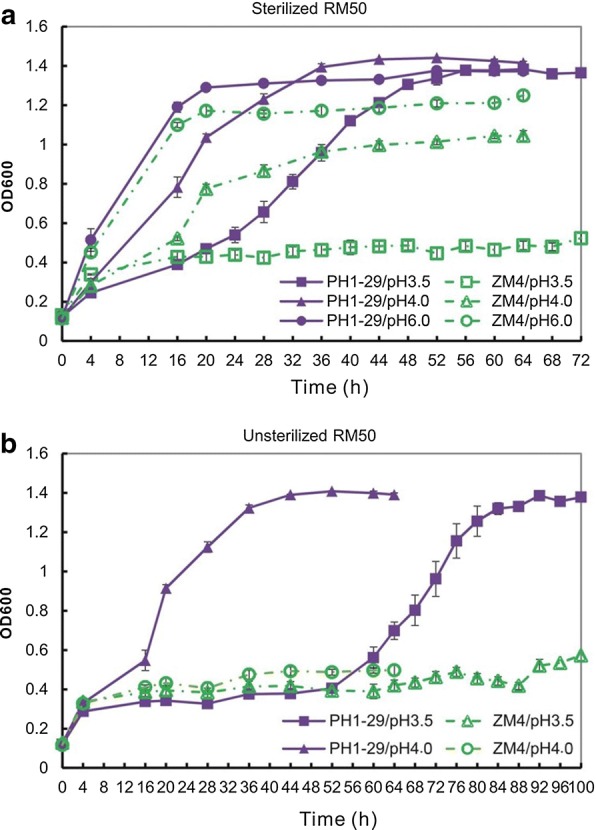
Growth curve of low pH-tolerant mutant PH1-29 in a sterilized and b unsterilized RM
Furthermore, the growth and ethanol production of PH1-29 were also improved under unsterilized conditions. The growth curve of PH1-29 under unsterilized condition showed similar to that under sterilized conditions but with longer lag phase of growth at pH 3.5 (Fig. 3b). Its ethanol yields kept stable under both sterilized and unsterilized cultures (Table 1), but ethanol productivity was declined to 0.3 g/L/h at pH 3.5. These evidences showed that it has stronger tolerance to low pH than the starting strain under non-aseptic conditions.
Comparison of ethanol production under acetic acid stress conditions
Mutants AQ8-1, AC8-9, and PH1-29 showed great potential to produce ethanol under unsterilized and low pH conditions. The ethanol productivities of AQ8-1 and AC8-9 reached at 1.3 g/L/h and 1.4 g/L/h, respectively, in the presence of 5 g/L acetic acid (pH 4.0). Even the addition of 8.0 g/L acetic acid did not dramatically affect their growth and ethanol yields (Fig. 2d), only with the decline of ethanol productivities to 0.4 and 0.3 g/L/h, respectively (Table 1). In comparison, another acetic acid-tolerant strain A7-2 that was mentioned above was able to produce ethanol at the productivity of 2.0 g/L/h in the presence of 2.1 to 6.3 g/L acetic acid with pH values ranging from 4.2 to 3.8 (Table 1). Further optimization of fermentation process will help improve the ethanol productivity of the mutant strains.
A flocculating mutant ZM401 showed strong tolerance to acetic acid. Its ethanol productivity reached at 2.0 g/L/h in the presence of 8.4 g/L acetic acid. Even in the presence of 10.5 g/L acetic acid, its ethanol productivity was far higher than that of AQ8-1 and AC8-9 [34] (Table 1). In that work, pH value was maintained at 6.0. However, the pH value was uncontrolled in this study. As mentioned above, dual effects of acetic ion and low pH resulted from acetic acid could partially explain the low productivity of AQ8-1 and AC8-9 in this study.
The tolerance of AQ8-1 and AC8-9 against sodium acetates was also tested, but there was no advantage over ZM4 (data not shown), suggesting the different effects of acetic acid and acetate salt on cells. In the future, the performance of the mutant generated, AQ8-1, AC8-9, and PH1-29, still remains to improve in the future combining with ALE [35], genome shuffling [36], or metabolic engineering [37, 38].
Mutation analysis
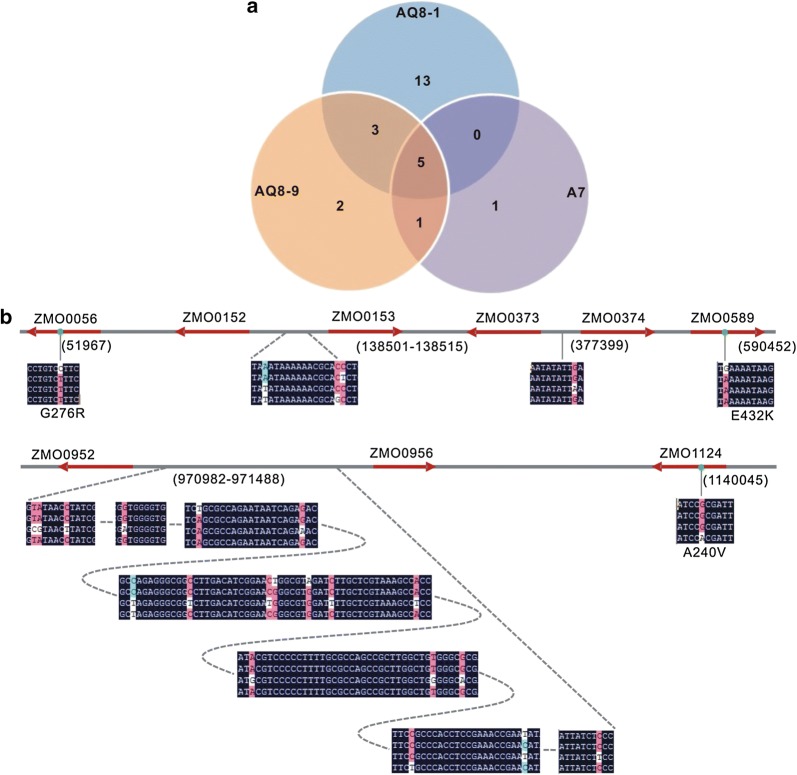
Mutant genomes were sequenced to investigate genetic changes that were caused by ARTP mutagenesis. There were dozens of single-nucleotide variations (SNVs) identified in A7, AQ8-1, AC8-9, and PH1-29, respectively (Fig. 4a, Tables 2, 3). It was found that more SNVs were created with the increase of ARTP treating cycles. Five same SNVs were detected in A7, AQ8-1, and AC8-9, two of which were located in coding sequence (CDS) and three in intergenic regions. SNVs in CDS regions led to two nonsynonymous amino acid (AA) changes in glutamine–fructose-6-phosphate aminotransferase encoded by gene ZMO0056, and RadA encoded by ZMO0589, respectively. Intergenic SNVs detected in all mutants were targeted in regions between gene ZMO0952 and ZMO0956. In addition, more unique SNVs were detected in AQ8-1 than in A7 and AC8-9, being targeted in intergenic regions between ZMO0152 and ZMO0153, ZMO0373 and ZMO0374, and ZMO0952 and ZMO0956 (Fig. 4, Tables 2, 3). Variation loci in mutant PH1-29 were quite similar to that in AQ8-1 except with two distinct SNVs at locus 703683 and 971488. All SNV sites are listed in Tables 2, 3.
Fig. 4.
Variations in the ARTP mutants tolerant to acetic acid. a Venn gram of SNVs in A7, AQ8-1, and AC8-9. b “Hot spots” of mutagenesis loci in the ARTP mutants. Gene names are shown above the lines, arrows indicate transcriptional directions. Variations are highlighted in nucleotide rows which from top to bottom represent ZM4, A7, AQ8-1, and AC8-9, respectively
Table 2.
SNVs by ARTP mutagenesis in CDS
| Locus | Ref | SNV | AA change | AQ 8-1 | AC 8-9 | A7 | PH 1-29 | A7-2 | Gene/product | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 51967 | C | T | G276R | + | + | + | + | + | ZMO0056/glutamine–fructose-6-phosphate aminotransferase |
| 2 | 122147 | G | A | D104N | − | − | − | − | + | ZMO0133/outer membrane-associated acid tolerance proteins |
| 3 | 122153 | C | G | Q106E | − | − | − | − | + | |
| 4 | 122161 | G | C | Syn. | − | − | − | − | + | |
| 5 | 122169 | T | G | F111L | − | − | − | − | + | |
| 6 | 122170 | T | G | F111L | − | − | − | − | + | |
| 7 | 122172 | T | A | F112Y | − | − | − | − | + | |
| 8 | 122179 | A | G | Syn. | − | − | − | − | + | |
| 9 | 122195 | T | A | L120M | − | − | − | − | + | |
| 10 | 122196 | T | A | L120X | − | − | − | − | + | |
| 11 | 122197 | G | T | L120F | − | − | − | − | + | |
| 12 | 122201 | G | T | E122X | − | − | − | − | + | |
| 13 | 122202 | A | C | E122A | − | − | − | − | + | |
| 14 | 122214 | G | A | S126N | − | − | − | − | + | |
| 15 | 122235 | C | A | T133N | − | − | − | − | + | |
| 25 | 590452 | G | A | E432K | + | + | + | + | + | ZMO0589/DNA repair protein RadA |
| 26 | 703683 | A | G | C141R | − | − | − | + | − | ZMO0708/phosphoribosyl-glycinamide formyl transferase |
| 27 | 1140045 | G | A | A240V | − | + | − | − | − | ZMO0843/arginine–tRNA ligase |
Syn.: synonymous, variation in nucleotide led to no amino acid change. +/− indicate the presence/absence of variation in the genome, respectively
Table 3.
SNVs by ARTP mutagenesis in intergenic regions
| Locus | Ref | SNV | AQ8-1 | AC8-9 | A7 | PH 1-29 | A7-2 | Gene/product | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 138501 | A | T | + | + | − | + | + |
ZMO0152/pyruvate kinase ZMO0153/YebC/Pmp R family DNA binding transcriptional factor |
| 2 | 138514 | C | G | − | + | − | − | + | |
| 3 | 138515 | C | T | − | − | + | − | + | |
| 4 | 377399 | G | A | + | − | − | − | − |
ZMO0372/AsnC family transcriptional regulator ZMO0373/hypothetical protein ZMO0374/levansucrase |
| 5 | 970982 | T | C | + | − | − | + | + |
ZMO0952/(tRNA cytidine-2′-O) methyltransferase ZMO0956/ubiquinol–cytochrome C reductase |
| 6 | 970983 | A | G | + | − | − | + | + | |
| 7 | 970988 | C | T | + | − | − | + | + | |
| 8 | 971023 | G | A | + | − | − | + | + | |
| 9 | 971059 | T | A | + | + | + | + | + | |
| 10 | 971077 | G | A | + | − | − | + | + | |
| 11 | 971114 | C | T | + | + | − | + | + | |
| 12 | 971128 | C | T | + | − | − | + | + | |
| 13 | 971129 | T | G | + | + | + | + | + | |
| 14 | 971135 | A | G | + | + | + | + | + | |
| 15 | 971139 | C | T | + | − | − | + | + | |
| 16 | 971154 | A | T | + | − | − | + | + | |
| 17 | 971183 | A | G | + | − | − | + | + | |
| 18 | 971214 | T | G | + | − | − | + | + | |
| 19 | 971219 | G | A | + | − | − | + | + | |
| 20 | 971312 | C | T | + | + | − | + | + | |
| 21 | 971332 | T | C | – | + | + | + | + | |
| 22 | 971488 | C | T | + | − | − | + | + |
+/− indicate the presence/absence of variation in the genome, respectively
Many SNVs were detected in intergenic regions between ZMO0152 and ZMO0153, and between ZMO0952 and ZMO0956 in acetic acid-tolerant mutants A7, AQ8-1, AC8-9, and A7-2 (Fig. 4b). Considering that mutant A7-2 and mutants A7, AQ8-1 and AC8-9 were created by different means, these variation regions are likely to function in the response/tolerance to acetic acid in Z. mobilis. The gene ZMO0152 encodes a pyruvate kinase (PYK), whose upstream region was reported to regulate the acid tolerance. For example, a PYK of Lactobacillus bulgaricus was regulated by the catabolite control protein A by binding to the kinase-encoding gene pyk in the presence of acid stress [39]. The differential proteomic profiling of Streptococcus mutans also revealed that the up-regulated PYK enhanced tolerance against acid stress [40]. Putative A/T-rich binding elements, for example, (5-TGTAAGCCCTAACA-3) upstream the –35 region, were summarized in the above literature. In ARTP mutants created in this study, variations were detected in A/T-rich regions similar to these binding elements (data not shown). Thus, a similar regulation likely exists in these ARTP mutants. Moreover, 18 variations were located in the intergenic region of ZMO0952 and ZMO0956. The former gene encodes a tRNA (cytidine/uridine-2′-O-)-methyltransferase and the latter encodes a Fe–S subunit of ubiquinol–cytochrome c reductase. It was still unknown why so many SNVs were located in the region, and the function of this region in acetic acid tolerance remains to investigate in the future.
ARTP mutagenesis also revealed that several genes likely contributed to acid tolerance/response. The gene ZMO0056 encodes a glutamine–fructose-6-phosphate aminotransferase that was reported to be critical for cells against organic acid stress [41, 42]. Such proteins contain a conserved GlmS superfamily domain that is involved in the cell wall/membrane biosynthesis [43]. Even though there is no direct evidence demonstrating its role in acid tolerance, cell wall/membrane is essential for keeping cellular integrity. The gene ZMO0859 encodes a DNA repair protein RadA, which is essential for the survival and genetic stability when cells suffered from the acid stress [44, 45]. An alanine at position 272 of RadA was replaced with a valine in ARTP mutants as well as A7-2, but the mechanism on how such a mutation can influence gene expression and regulation is yet to be investigated in the future. A unique SNV at locus 377399 is present only in AQ8-1. It is likely to regulate the expression of genes ZMO0372, ZMO0373, and ZMO0374 (Fig. 4 and Tables 2, 3). The gene ZMO0372 encodes a putative leucine responsive regulatory protein/regulator of asparigine synthase C (Lrp/AsnC) transcriptional regulator which is involved in amino acid transport metabolism and cell replication [46]. The gene ZM0374 encodes a levansucrase, a functional protein possibly involved in tolerance to multiple stresses [47]. Even though their functions in tolerating acetic acid and low pH remain unknown, the microarray revealed the down-regulation of ZMO0372 and ZMO0373 under acetate stress in Z. mobilis strain 8b [32]. Therefore, this region likely plays an important role in regulating acetic acid tolerance.
On the other hand, based on variation sites in the mutants generated by different means, we propose that mechanisms of Z. mobilis responding to acetic acid and to acetate salts are different. The same SNVs that were detected in both Z. mobilis acetate-tolerant mutants 8b and AcR were barely detected in acetic acid-tolerating mutants A7-2 [18], A7, AQ8-1 and AC8-9, and vice versa (Tables 2, 3).
Intracellular NAD levels
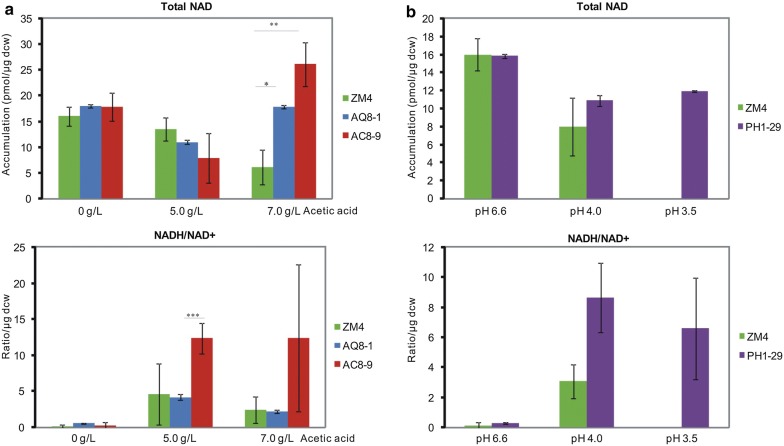
It has been reported that intracellular NADH/NAD+ ratio increased in Z. mobilis as well as S. cerevisiae under stress conditions [48, 49]. Thus, the accumulation of total nicotinamide adenine dinucleotide (NAD) as well as NADH/NAD+ ratio was measured to investigate cellular responses to acetic acid and low pH. Total NAD and NADH/NAD+ remained low in RM without stresses (Fig. 5), which agreed with the reports mentioned above. Cellular total NAD levels of AQ8-1 and AQ8-9 were comparable to that of ZM4 in RM or RM with 5.0 g/L acetic acid, but were significantly higher than ZM4 in the presence of 7.0 g/L acetic acid. However, NADH/NAD+ ratio displayed different trends between strains. The ratio remained low in ZM4 and AQ8-1 while obviously increased in AC8-9 under acetic acid stress conditions (Fig. 5a). Similarly, the accumulation and a high NADH/NAD+ ratio were detected in PH1-29 under low pH stress conditions (Fig. 5b).
Fig. 5.
Accumulation of intracellular NAD levels under a acetic acid conditions, and b low pH conditions. Total NAD comprises NADH and NAD+; NADH/NAD+ = NADH/(total NAD − NADH). Two repeats were performed for each strain, and error bars indicate standard deviation. The change of total NAD or NADH/NAD+ per microgram dry cell weight (DCW) between two time points indicated the accumulation of NAD or NADH/NAD+. DCW was calculated from OD 600 nm (1OD600 = 0.323 gDCW/L). Asterisks above bars indicate t test significance between mutants and ZM4.*p = 0.0415; **p = 0.0384; ***p = 0.0326
Under salinity stress conditions, the increase of NADH/NAD+ was reported in Z. mobilis. The respiratory chain played important roles in maintaining the redox balance by oxidizing NADH to NAD+ [48]. The authors suggested that NADH/NAD+ ratio is likely responsible for a wide stress tolerance. In the present study, many SNVs nearby the gene encoding Fe–S subunit of ubiquinol–cytochrome c reductase, which is an important member in respiratory chain, were detected in mutants (Fig. 4 and Table 3). Therefore, it was speculated that NADH/NAD+ under acetic acid and low pH stress conditions was also regulated by the similar mechanism. This can explain that a relatively low NADH/NAD+ of AQ8-1 was maintained under acetic acid conditions.
In contrast, the ratio in AC8-9 and PH1-29 was relatively high under stress conditions. Although the growth and ethanol productivities of AC8-9 under acetic acid stress conditions were lower than AQ8-1 (Fig. 2 and Table 1). It still enabled relatively high growth and ethanol production under stress conditions. Similar growth and ethanol production were detected in PH1-29 (Fig. 3 and Table 1). Since a high NADH or NADH/NAD+ level is toxic to cells, these results suggested that enhanced ability of maintaining integrity of cellular membrane and resisting high level of protons represents an alternative tolerance mechanism in AC8-9 and PH1-29.
Conclusion
Mutant strains able to tolerate 8.0 g/L acetic acid and pH 4.0 and 3.5 were generated via the mARTP mutagenesis in this study. Therefore, the mARTP mutagenesis is efficient to create Z. mobilis mutants with enhanced tolerance to acetic acid and low pH. These mARTP mutants could modulate NADH/NAD+ ratio to respond to stress. Genome re-sequencing revealed the “hot spots” which are likely involved in tolerance to acetic acid and pH. Mutant strains generated in this study will not only serve as potential bioethanol producers, but also help understand the stress response and regulation in Z. mobilis.
Materials and methods
Cell growth
A single colony of Z. mobilis strain ZM4 was inoculated in 10 mL of RM (20.0 g/L glucose, 10.0 g/L yeast extract, and 2.0 g/L KH2PO4) and grown overnight at 30 °C without shaking. Cell pellets were harvested from 8 mL of RM cultures by centrifuging at 3000 rpm and 4 °C and then inoculated in 80 mL of RM that were supplemented with acetic acid or that at low pH values. Thirty-five or forty-four microliters of concentrated H2SO4 were added in 100 mL of RM to adjust pH values to 4.0 or 3.5. For tolerance tests, glucose concentration in the media was 50.0 g/L (RM50); otherwise, RM was used for the growth of Z. mobilis.
mARTP mutagenesis
ZM4 cells (106 to 108 cells/mL) were centrifuged at 3000 rpm and 4 °C and then suspended in 0.9% NaCl solution. Ten microliters of cell aliquot were spread on a carrier plate and treated for 15 to 150 s in a Type M ARTP Mutagenesis Bio-breeding Machine (Wuxi Tmaxtree Biotechnology Co., Ltd., China). Gas helium (He) of high purity (99.999%) served as working gas at the flow rate of 10–20 SLM. The radio-frequency power input was set at 100 or 120 W and jet temperature was controlled at 22 °C using a water cycler. The treated cells were suspended in 1 mL of RM, followed by spreading on RM supplemented with 7.0 g/L acetic acid. Those resulting colonies able to grow with higher OD600nm values in RM with 7.0 g/L acetic acid were then subjected to a second ARTP mutagenesis. The treated cells were screened in RM containing 8.0 g/L acetic acid. To select low pH-tolerant mutants, a mutant able to tolerate 8.0 g/L acetic acid was subjected to another round of ARTP mutagenesis, followed by screening on RM at pH 3.5. To test genetic stability, the mutants were continuously cultured in RM with acetic acid or RM at pH 4.0 or 3.5 for at least three times, and their glycerol stocks were stored at − 80 °C and their growth and tolerance were checked every 3–4 weeks.
Genomic DNA isolation
Cells from 3 to 5 mL of Z. mobilis overnight cultures were harvested by centrifuging at 13,000 rpm for isolating genomic DNAs (gDNAs) using a Bacterial DNA Kit (Omega Biotek, USA). The concentration of gDNAs was determined using a Qubit 3 Fluorometer (ThermoFisher, USA). The quality of gDNAs was checked via gel electrophoresis (0.7% agarose, 12 V/cm, 60–70 min).
Amplification of 16S rRNA gene by PCR
To ensure that the mutants are affiliated with Z. mobilis, 16S rRNA gene was amplified by polymerase chain reaction (PCR). Fresh cells harvested from 50 μL of overnight cultures were washed and re-suspended in 10 μL of ddH2O. One microliter of the suspension were used for PCR. 16S rRNA genes were amplified using general primer set 15F (AGAGTTTGATCCTGGCTCAG)/1492R (TACGGYTACCTTGTTACGACTT) and KOD-Plus-Neo polymerase (TOYOBO, Japan). PCRs were performed with a program as follows: predenaturing at 95 °C for 5 min, 30 cycles of denaturing at 98 °C for 10 s, annealing at 60 °C for 30 s, and extending at 68 °C for 1 min. Amplicons were sequenced in GENEWIZ Inc. (Suzhou, China).
Genome re-sequencing
Genomic DNAs of the mutants were sequenced to investigate mutations. Briefly, libraries of gDNA were constructed and then re-sequenced using a Illumina HiSeq instrument (Illumina, San Diego, CA, USA). After the removal of adaptors, PCR primers, the content of N bases > 10%, and bases of quality lower than 20, clean data to the reference genome of strain ZM4 (GenBank No. NC_006526.2) were mapped. Annotation for potential SNVs was performed by Annovar (V21 Feb 2013). The sequencing was completed by GenWize Inc. (Suzhou, China).
Analytical methods
Glucose and ethanol were calculated using high-performance liquid chromatography (HPLC; Agilent 1200), according to the description by You et al. [50]. Briefly, an HPX-87H ion exclusion column (Bio-Rad Aminex) was used and 5 mM H2SO4 worked as the mobile phase. The chromatographic system was run at a flow rate of 0.6 mL/min and 35 °C. The injection volume was set to 20 μL.
Cells were harvested for assaying intracellular NAD using a NAD/NADH Assay Kit (ab65348, Abcam Inc.). Briefly, one milliliter of fresh cells was harvested by centrifuging at 4000 rpm and 4 °C for 10 min. After washing with cold phosphate buffer solution (PBS), cells were extracted using NADH/NAD extraction buffer via two freeze/thaw cycles of 20 min at − 80 °C followed by 10 min at room temperature (RT). After a short vortex, supernatants were collected by centrifuging at 4 °C for 5 min. For measuring total NAD, 50 μL of extracted aliquots and 100 μL of reaction mix were added in each well of a 96-well microplate, incubated at RT for 5 min. 10 μL of NADH developer were added in each well and mixed, and OD450nm values were then read using a microplate reader (Spectro MAX190, Molecular Devices, USA). For measuring NADH, extracted aliquots were heated to decompose NAD+ before blending with reaction mix.
Additional file
Additional file 1: Table S1. pH values of RM supplemented with acetic acid or sodium acetate. Figure S1. Tolerance of AQ8-1, AC8-9 and ZM4 to mixture of acetic acid and furfural. “A” indicates acetic acid and “F” indicates furfural. The number after A or F indicates the concentration of corresponding inhibitor. The concentrations of acetic acid ranged from 1.0 to 7.0 g/L, and furfural from 1.0 to 3.0 g/L in the mixture. OD600 nm values were measured at stationery phase.
Authors’ contributions
BW and MH conceived the study, wrote the manuscript, and prepared figures and tables. BW, HQ, YY, GD, CZ, and QZ performed experiments and data analysis, HS performed ANOVA, and t test analysis, BW, SY, FX, FT, MH, and GH discussed and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Dr. Nan Peng at Huazhong Agriculture University for critical comments and reviewing. We also thank Ms. Qinghu Yu for sampling and NADH analysis.
Competing interests
The authors declare that they have no competing interests.
Availability of supporting data
The Z. mobilis AQ8-1, AC8-9, and PH1-29 have been deposited at Guangdong Microbial Culture Center (GDMCC) under the Accession Number GDMCC60258, 60259, and 60260, respectively.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This research was financially supported by the Basic Research Program of CAAS (Grant No. 1610012016020 and 2060302-017-03), the National Natural Science Foundation of China (Grant No. 31570055), and the Elite Program and Basic Research Program of Chinese Academy of Agricultural Sciences to MH.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ARTP
atmospheric and room temperature plasma
- mARTP
multiplex atmospheric and room temperature plasma
- ALE
adaptive laboratory evolution
- NTG
nitrosoguanidine
- RM
rich medium
- OD
optical density
- CDS
coding sequence
- SNV
single-nucleotide variation
- PYK
pyruvate kinase
- G
guanine
- C
cytosine
- AA
amino acid
- PCR
polymerase chain reaction
- HPLC
high-performance liquid chromatography
- RT
room temperature
- gDNA
genomic DNA
- NAD
nicotinamide adenine dinucleotide
- NAD+
oxidative NAD
- NADH
reductive NAD
- SLM
standard liter per minute
- PBS
phosphate buffer solution
- DCW
dry cell weight
Contributor Information
Bo Wu, Email: wubo@caas.cn.
Han Qin, Email: qinhan@caas.cn.
Yiwei Yang, Email: 1036187527@qq.com.
Guowei Duan, Email: duanguowei_1@163.com.
Shihui Yang, Email: Shihui.Yang@hubu.edu.cn.
Fengxue Xin, Email: xinfengxue@njtech.edu.cn.
Chunyan Zhao, Email: 776567991@qq.com.
Huanhuan Shao, Email: 347776721@qq.com.
Yanwei Wang, Email: wangyanwei@caas.cn.
Furong Tan, Email: tanfurong@caas.cn.
Guoquan Hu, Email: huguoquan@caas.cn.
Mingxiong He, Phone: +86-28-85242281, Email: hemingxiong@caas.cn.
References
- 1.Jeon YJ, Xun Z, Rogers PL. Comparative evaluations of cellulosic raw materials for second generation bioethanol production. Lett Appl Microbiol. 2010;51:518–524. doi: 10.1111/j.1472-765X.2010.02923.x. [DOI] [PubMed] [Google Scholar]
- 2.Grzenia DL, Wickramasinghe SR, Schell DJ. Fermentation of reactive-membrane-extracted and ammonium-hydroxide-conditioned dilute-acid-pretreated corn stover. Appl Biochem Biotechnol. 2012;166:470–478. doi: 10.1007/s12010-011-9442-5. [DOI] [PubMed] [Google Scholar]
- 3.Lawford HG, Rousseau JD. Cellulosic fuel ethanol: alternative fermentation process designs with wild-type and recombinant Zymomonas mobilis. Appl Biochem Biotechnol. 2003;105–108:457–469. doi: 10.1385/ABAB:106:1-3:457. [DOI] [PubMed] [Google Scholar]
- 4.Palmqvist E, Hahn-Hägerdal B. Fermentation of lignocellulosic hydrolysates. I: inhibition and detoxification. Bioresour Technol. 2000;74:17–24. doi: 10.1016/S0960-8524(99)00160-1. [DOI] [Google Scholar]
- 5.Mussatto SI, Roberto IC. Alternatives for detoxification of diluted-acid lignocellulosic hydrolyzates for use in fermentative processes: a review. Bioresour Technol. 2004;93:1–10. doi: 10.1016/j.biortech.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Bai FW, Anderson WA, Moo-Young M. Ethanol fermentation technologies from sugar and starch feedstocks. Zymomonas mobilis: a bacterium for ethanol production. Biotechnol Adv. 2008;26:89–105. doi: 10.1016/j.biotechadv.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Sprenger GA. Carbohydrate metabolism in Zymomonas mobilis: a catabolic highway with some scenic routes. FEMS Microbiol Lett. 1996;145:301–307. doi: 10.1111/j.1574-6968.1996.tb08593.x. [DOI] [Google Scholar]
- 8.Kalnenieks U. Physiology of Zymomonas mobilis: some unanswered questions. Adv Microb Physiol. 2006;51:73–117. doi: 10.1016/S0065-2911(06)51002-1. [DOI] [PubMed] [Google Scholar]
- 9.Doelle HW, Kirk L, Crittenden R, Toh H, Doelle MB. Zymomonas mobilis—science and industrial application. Crit Rev Biotechnol. 1993;13:57–98. doi: 10.3109/07388559309069198. [DOI] [PubMed] [Google Scholar]
- 10.Yang S, Tschaplinski TJ, Engle NL, Carroll SL, Martin SL, Davison BH, Palumbo AV, Rodriguez M, Jr, Brown SD. Transcriptomic and metabolomic profiling of Zymomonas mobilis during aerobic and anaerobic fermentations. BMC Genom. 2009;10:34. doi: 10.1186/1471-2164-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He M-X, Wu B, Qin H, Ruan Z-Y, Tan F-R, Wang JL, Shui ZX, Dai LC, Zhu QL, Pan K, Tang XY, Wang W-G, Hu Q-C. Zymomonas mobilis: a novel platform for future biorefineries. Biotechnol Biofuels. 2014;7:101. doi: 10.1186/1754-6834-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang S, Fei Q, Zhang Y, Contreras LM, Utturkar SM, Brown SD, Himmel ME, Zhang M. Zymomonas mobilis as a model system for production of biofuels and biochemicals. Microb Biotechnol. 2016;9:699–717. doi: 10.1111/1751-7915.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, He Q, Yang Y, Wang J, Haning K, Hu Y, Wu B, He M, Zhang Y, Bao J, Contreras LM, Yang S. Advances and prospects in metabolic engineering of Zymomonas mobilis. Metab Eng. 2018. (in press). [DOI] [PubMed]
- 14.Ko JK, Lee SM. Advances in cellulosic conversion to fuels: engineering yeasts for cellulosic bioethanol and biodiesel production. Curr Opin Biotechnol. 2018;50:72–80. doi: 10.1016/j.copbio.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Mills TY, Sandoval NR, Gill RT. Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnol Biofuels. 2009;2:26. doi: 10.1186/1754-6834-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumler DJ, Hung KF, Bose JL, Vykhodets BM, Cheng CM, Jeong K-C, Kaspar CW. Ehancement of acid tolerance in Zymomonas mobilis by a proton-buffering peptide. Appl Biochem Biotechnol. 2006;134:15–26. doi: 10.1385/ABAB:134:1:15. [DOI] [PubMed] [Google Scholar]
- 17.Yang S, Pelletier DA, Lu TY, Brown SD. The Zymomonas mobilis regulator hfq contributes to tolerance against multiple lignocellulosic pretreatment inhibitors. BMC Microbiol. 2010;10:135. doi: 10.1186/1471-2180-10-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shui ZX, Qin H, Wu B, Ruan ZY, Wang LS, Tan FR, Wang JL, Tang XY, Dai LC, Hu GQ, He MX. Adaptive laboratory evolution of ethanologenic Zymomonas mobilis strain tolerant to furfural and acetic acid inhibitors. Appl Microbiol Biotechnol. 2015;99:5739–5748. doi: 10.1007/s00253-015-6616-z. [DOI] [PubMed] [Google Scholar]
- 19.Ma K, Ruan Z, Shui Z, Wang Y, Hu G, He M. Open fermentative production of fuel ethanol from food waste by an acid-tolerant mutant strain of Zymomonas mobilis. Bioresour Technol. 2016;203:295–302. doi: 10.1016/j.biortech.2015.12.054. [DOI] [PubMed] [Google Scholar]
- 20.Joachinsthal E, Haggett KD, Jang J-H, Rogers PL. A mutant of Zymomonas mobilis ZM4 capable of ethanol production from glucose in the presence of high acetate concentrations. Biotechnol Lett. 1998;20:137–142. doi: 10.1023/A:1005320306410. [DOI] [Google Scholar]
- 21.Lee JH, Skotnicki ML, Rogers PL. Kinetic studies on a flocculent strain of Zymomonas mobilis. Biotechnol Lett. 1982;4:615–620. doi: 10.1007/BF00127795. [DOI] [Google Scholar]
- 22.Wu Y-N, Xing X-H, Zhang C, Lin H-P, Wang L-Y. Recent progress on atmospheric and room temperature plasma (ARTP) biobreeding technology, instrumentation and its industrialization. Biotechnol Econ (Sheng Wu Chan Ye Ji Shu) 2017;1:37–45. [Google Scholar]
- 23.Zhang X, Zhang XF, Li HP, Wang LY, Zhang C, Xing XH, Bao CY. Atmospheric and room temperature plasma (ARTP) as a new powerful mutagenesis tool. Appl Microbiol Biotechnol. 2014;98:5387–5396. doi: 10.1007/s00253-014-5755-y. [DOI] [PubMed] [Google Scholar]
- 24.Ma Y, Shen W, Chen X, Liu L, Zhou Z, Xu F, Yang H. Significantly enhancing recombinant alkaline amylase production in Bacillus subtilis by integration of a novel mutagenesis-screening strategy with systems-level fermentation optimization. J Biol Eng. 2016;10:13. doi: 10.1186/s13036-016-0035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong TT, Gong JS, Gu BC, Zhang Q, Li H, Lu ZM, Lu ML, Shi JS, Xu ZH. Significantly enhanced substrate tolerance of Pseudomonas putida nitrilase via atmospheric and room temperature plasma and cell immobilization. Bioresour Technol. 2017;244:1104–1110. doi: 10.1016/j.biortech.2017.08.039. [DOI] [PubMed] [Google Scholar]
- 26.Zheng B, Ma X, Wang N, Ding T, Guo L, Zhang X, Yang Y, Li C, Huo YX. Utilization of rare codon-rich markers for screening amino acid overproducers. Nat Commun. 2018;9:3616. doi: 10.1038/s41467-018-05830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin J, Wang Y, Yao M, Gu X, Li B, Liu H, Ding M, Xiao W, Yuan Y. Astaxanthin overproduction in yeast by strain engineering and new gene target uncovering. Biotechnol Biofuels. 2018;11:230. doi: 10.1186/s13068-018-1227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi F, Kitahara Y, Wang Z, Zhao X, Du W, Liu D. Novel mutant strains of Rhodosporidium toruloides by plasma mutagenesis approach and their tolerance for inhibitors in lignocellulosic hydrolyzate. J Chem Technol Biotechnol. 2014;89:735–742. doi: 10.1002/jctb.4180. [DOI] [Google Scholar]
- 29.Zhang X, Zhang C, Zhou QQ, Zhang XF, Wang LY, Chang HB, Li HP, Oda Y, Xing XH. Quantitative evaluation of DNA damage and mutation rate by atmospheric and room-temperature plasma (ARTP) and conventional mutagenesis. Appl Microbiol Biotechnol. 2015;99:5639–5646. doi: 10.1007/s00253-015-6678-y. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Y, Shang YP, Li H, Zhang C, Pan J, Bai YP, Li CX, Xu JH. Enhancing transglutaminase production of Streptomyces mobaraensis by iterative mutagenesis breeding with atmospheric and room-temperature plasma (ARTP) Bioresour Bioprocess. 2017;4:37. doi: 10.1186/s40643-017-0168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He MX, Wu B, Shui ZX, Hu QC, Wang WG, Tan FR, Tang XY, Zhu QL, Pan K, Li Q, Su XH. Transcriptome profiling of Zymomonas mobilis under furfural stress. Appl Microbiol Biotechnol. 2012;95:189–199. doi: 10.1007/s00253-012-4155-4. [DOI] [PubMed] [Google Scholar]
- 32.Yang S, Franden MA, Brown SD, Chou Y-C, Pienkos PT, Zhang M. Insights into acetate toxicity in Zymomonas mobilis 8b using different substrates. Biotechnol Biofues. 2014;7:140. doi: 10.1186/s13068-014-0140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narayanan V, Sanchez INV, van Niel EWJ, Gorwa-Grauslund MF. Adaptation to low pH and lignocellulosic inhibitors resulting in ethanolic fermentation and growth of Saccharomyces cerevisiae. AMB Express. 2016;6:59. doi: 10.1186/s13568-016-0234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao N, Bai Y, Liu CG, Zhao XQ, Xu JF, Bai FW. Flocculating Zymomonas mobilis is a promising host to be engineered for fuel ethanol production from lignocellulosic biomass. Biotechnol J. 2014;9:362–371. doi: 10.1002/biot.201300367. [DOI] [PubMed] [Google Scholar]
- 35.Ma J-F, Wu M-K, Zhang C-Q, He A-Y, Kong X-P, Li G-L, Wei C, Jiang M. Coupled ARTP and ALE strategy to improve anaerobic cell growth and succinic acid production by Escherichia coli. J Chem Technol Biotechnol. 2016;91:711–717. doi: 10.1002/jctb.4633. [DOI] [Google Scholar]
- 36.Gu C, Wang G, Mai S, Wu P, Wu J, Wang G, Liu H, Zhang J. ARTP mutation and genome shuffling of ABE fermentation symbiotic system for improvement of butanol production. Appl Microbiol Biotechnol. 2017;101:2189–2199. doi: 10.1007/s00253-017-8093-z. [DOI] [PubMed] [Google Scholar]
- 37.Yang S, Land ML, Klingeman DM, Pelletier DA, Lu TY, Martin SL, Guo HB, Smith JC, Brown SD. Paradigm for industrial strain improvement identifies sodium acetate tolerance loci in Zymomonas mobilis and Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2010;107:10395–10400. doi: 10.1073/pnas.0914506107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu YF, Hsieh CW, Chang YS, Wung BS. Effect of acetic acid on ethanol production by Zymomonas mobilis mutant strains through continuous adaptation. BMC Biotechnol. 2017;17:63. doi: 10.1186/s12896-017-0385-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhai Z, An H, Wang G, Luo Y, Hao Y. Functional role of pyruvate kinase from Lactobacillus bulgaricus in acid tolerance and identification of its transcription factor by bacterial one-hybrid. Sci Rep. 2015;5:17024. doi: 10.1038/srep17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Len AC, Harty DW, Jacques NA. Proteome analysis of Streptococcus mutans metabolic phenotype during acid tolerance. Microbiology. 2004;150:1353–1366. doi: 10.1099/mic.0.26888-0. [DOI] [PubMed] [Google Scholar]
- 41.Feehily C, O’Byrne CP, Karatzas KA. Functional gamma-aminobutyrate shunt in Listeria monocytogenes: role in acid tolerance and succinate biosynthesis. Appl Environ Microbiol. 2013;79:74–80. doi: 10.1128/AEM.02184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santiago B, MacGilvray M, Faustoferri RC, Quivey RG., Jr The branched-chain amino acid aminotransferase encoded by ilvE is involved in acid tolerance in Streptococcus mutans. J Bacteriol. 2012;194:2010–2019. doi: 10.1128/JB.06737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernández-Herrero LA, Badet-Denisot M-A, Badet B, Berenguer J. glmS of Thermus thermophilus HB8: an essential gene for cell-wall synthesis identified immediately upstream of the S-layer gene. Mol Microbiol. 1995;17:1–12. doi: 10.1111/j.1365-2958.1995.mmi_17010001.x. [DOI] [PubMed] [Google Scholar]
- 44.Foster JW. Low pH adaptation and the acid tolerance response of Salmonella typhimurium. Crit Rev Microbiol. 1995;21:215–237. doi: 10.3109/10408419509113541. [DOI] [PubMed] [Google Scholar]
- 45.Jeong KC, Hung KF, Baumler DJ, Byrd JJ, Kaspar CW. Acid stress damage of DNA is prevented by Dps binding in Escherichia coli O157:H7. BMC Microbiol. 2008;8:181. doi: 10.1186/1471-2180-8-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yokoyama K, Ishijima SA, Clowney L, Koike H, Aramaki H, Tanaka C, Makino K, Suzuki M. Feast/famine regulatory proteins (FFRPs): Escherichia coli Lrp, AsnC and related archaeal transcription factors. FEMS Microbiol Rev. 2006;30:89–108. doi: 10.1111/j.1574-6976.2005.00005.x. [DOI] [PubMed] [Google Scholar]
- 47.Velazquez-Hernandez ML, Baizabal-Aguirre VM, Cruz-Vazquez F, Trejo-Contreras MJ, Fuentes-Ramirez LE, Bravo-Patino A, Cajero-Juarez M, Chavez-Moctezuma MP, Valdez-Alarcon JJ. Gluconacetobacter diazotrophicus levansucrase is involved in tolerance to NaCl, sucrose and desiccation, and in biofilm formation. Arch Microbiol. 2011;193:137–149. doi: 10.1007/s00203-010-0651-z. [DOI] [PubMed] [Google Scholar]
- 48.Hayashi T, Kato T, Watakabe S, Song W, Aikawa S, Furukawa K. The respiratory chain provides salt stress tolerance by maintaining a low NADH/NAD+ ratio in Zymomonas mobilis. Microbiology. 2015;161:2384–2394. doi: 10.1099/mic.0.000195. [DOI] [PubMed] [Google Scholar]
- 49.Shi X, Zou Y, Chen Y, Zheng C, Ying H. Overexpression of a water-forming NADH oxidase improves the metabolism and stress tolerance of Saccharomyces cerevisiae in aerobic fermentation. Front Microbiol. 2016;7:1427. doi: 10.3389/fmicb.2016.01427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.You Y, Wu B, Yang YW, Wang YW, Liu S, Zhu QL, Qin H, Tan FR, Ruan ZY, Ma KD, Dai LC, Zhang M, Hu GQ, He MX. Replacing process water and nitrogen sources with biogas slurry during cellulosic ethanol production. Biotechnol Biofuels. 2017;10:236. doi: 10.1186/s13068-017-0921-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. pH values of RM supplemented with acetic acid or sodium acetate. Figure S1. Tolerance of AQ8-1, AC8-9 and ZM4 to mixture of acetic acid and furfural. “A” indicates acetic acid and “F” indicates furfural. The number after A or F indicates the concentration of corresponding inhibitor. The concentrations of acetic acid ranged from 1.0 to 7.0 g/L, and furfural from 1.0 to 3.0 g/L in the mixture. OD600 nm values were measured at stationery phase.