Abstract
Group B Streptococcus (GBS) is a neonatal pathogen frequently transmitted from maternal asymptomatic vagino-rectal colonization. Co-colonization with multiple GBS serotypes, which has implications for type-specific vaccination strategies, is difficult to detect with standard microbiologic techniques. We designed a nested real-time PCR assay to detect vaginal co-colonization in samples from a cohort of non-pregnant women (N=433). 6/91 (6.6%) GBS-positive samples harbored 2 GBS serotypes, with over-representation of serotype V among co-colonized samples. Serotype IV GBS was more prevalent (>10%) in this cohort than in previously reported United States studies. Ongoing surveillance of GBS serotype epidemiology and co-colonization is indicated.
Keywords: Streptococcus agalactiae, vaginal colonization, serotype, co-colonization
BACKGROUND
Streptococcus agalactiae (Group B Streptococcus [GBS]) invasive infection is a major cause of infant mortality and morbidity worldwide, including long-term neurodevelopmental sequelae.[1, 2] Maternal colonization is the primary risk factor for early-onset disease (EOD), manifesting within six days of birth with rapid bacterial spread that can lead to pneumonia, sepsis or meningitis. In the United States (US) and several other countries the risk of GBS EOD has been mitigated to a large extent by the introduction of intrapartum antibiotic prophylaxis (IAP). However, even in resource-rich settings, IAP is subject to missed opportunities and also has had a lesser effect on late-onset GBS disease.[3] An alternative method for prevention of GBS invasive disease is through vaccination. Immunization with the capsular polysaccharide (CPS) can induce serotype-specific immunity, and candidate CPS-protein conjugate vaccines are currently undergoing clinical trials.[4, 5]
The polysaccharide capsule of GBS is a major virulence factor for the organism, and to date, ten capsular serotypes have been identified: Ia, Ib and II to IX. CPS serotyping is used to characterize strains and to investigate epidemiological trends of GBS infection and colonization in humans to provide important data for vaccine development and to detect events such as capsular switching and serotype replacement. Serotyping of GBS isolates is most commonly performed by latex agglutination using commercially available kits that are generally reliable and easy to perform. Limitations of this method include the inability to type some strains due to lack of or low expression of CPS, the potential to miss co-colonization with multiple serotypes of GBS if only one or a few GBS colonies are tested for each sample, as well a risk of false-positive cross-reactions of a single strain with multiple serotype latex beads. Serotyping is rarely performed in clinical microbiology laboratories, and even in studies of GBS serotype distribution, testing of multiple colonies to assess potential co-colonization is not commonly done.
Polymerase chain reaction (PCR)-based typing methods to detect genes in the cps locus have also been developed, including multiplex approaches.[6] More recently, whole-genome sequencing has also been used to determine GBS serotypes.[7] We combined sip PCR-based detection of GBS colonization with a nested real-time PCR serotyping strategy in order to assess serotype distribution as well as co-colonization in a cohort of healthy non-pregnant adult women.
METHODS
Samples and DNA extraction
Stored deidentified samples from a previous study [Bacterial Vaginosis–Improved Diagnosis by ELISA and Sequencing (BV-IDEAS)] were used for the current study. Briefly, 5-mL sterile saline vaginal lavage specimens were collected after written informed consent from non-pregnant women aged 18–55 years seeking primary gynecologic care at a site in New York City between July 2010 and June 2012. Rectovaginal swab samples for routine GBS culture were not collected as part of this study. Samples were aliquoted immediately after collection, refrigerated for transport (<6 hours), and stored at −80°C as previously described.[8] For each sample, 200 μL from stored aliquots were added to 10 mL of Lim broth and incubated overnight at 37°C. Bacteria were pelleted by centrifugation and the supernatant discarded. The cell pellet was re-suspended in 2.5 mL of phosphate buffered saline (PBS) with 1.8 mL used for DNA extraction using the AllPrep Bacterial DNA/RNA/Protein Kit (Mo Bio Laboratories, Inc. Carlsbad, California, US) according to the manufacturer’s instructions with minor modifications to enhance lysis including the addition of EDTA to the lysis buffer solution and an additional step of heating samples to 65oC for 15 minutes prior to bead-beating for 20 minutes.
Polymerase chain reactions (PCRs)
Extracted DNA from each sample was used to detect GBS by real-time (RT)-PCR using primers and probe targeting a region of the GBS surface immunogenic protein gene (sip) as previously described.[9] Reaction volumes used were: 10 μL TaqMan® Universal Mastermix II, no UNG (Thermo Fisher Scientific); 0.2 μL of each primer (40 μM) and probe (20 μM) and 9.4 μL of DNA template and water (2 ng/well DNA). All samples with amplification within 40 cycles (CT ≤ 40) were also tested by a nested serotype PCR using previously reported serotype-specific primers and probes[10]. The first stage of the serotyping PCR was performed using newly designed primers for the GBS capsule targeting a region conserved across all 10 serotypes. Forward primer: 5’-AGA TAT TAA GAT TAT TST MMK AAC ACT-3’; reverse primer: 5’-TGT ATA AAT AAA ACC AGA TTG AAG TAA TTT-3’. Reaction volumes used were: 0.25 μL Q5 High-Fidelity DNA Polymerase (New England Biolabs Inc.), 5 μL Q5 Reaction Buffer, 0.5 μL dNTPs (10 mM), 1.25 μL of each primer (10 μM) and 16.75 μL of DNA template and water (2 ng/well DNA). PCR conditions: initial denaturation at 98°C for 30 seconds; then 40 cycles of 98°C for 10 seconds, 52°C for 20 seconds, 72°C for 4 minutes; final extension at 72°C for 2 minutes. Reaction volumes for the second stage PCR using serotype-specific primers and probes were: 10 μL TaqMan Universal Mastermix II, no UNG; 0.2 μL of each primer (50 μM) and probe (20 μM), 2.0 μL DNA template from the first stage reaction and 7.4 μL water. Primer/probe pairs and reaction conditions for sip PCR were as previously reported.[9, 10] Positive (known GBS and serotype-specific samples) and negative (no DNA template) controls were included with each PCR run. Samples with sip CT ≤ 37 and at least one serotype CT ≤ 20 were considered positive for GBS.
Latex Agglutination
We tested samples in which more than one GBS serotype was identified by PCR by testing multiple colonies by latex agglutination (IMMULEX™ STREP-B kit, Statens Serum Institut Diagnostica, Hillerød, Denmark). 10 μL of resuspended GBS culture (was streaked onto multiple plates of GBS chromogenic agar (CHROMagar™ StrepB, Chromagar, Paris, France) and incubated for 24–48 hrs at 37°C. Mauve-colored colonies were picked individually and tested by latex agglutination for the two (or three) predicted serotypes and alternative negative control serotypes.
Statistics
Descriptive analyses were performed using Microsoft Excel (2010), and comparison of proportions was performed using GraphPad Prism (version 7).
RESULTS
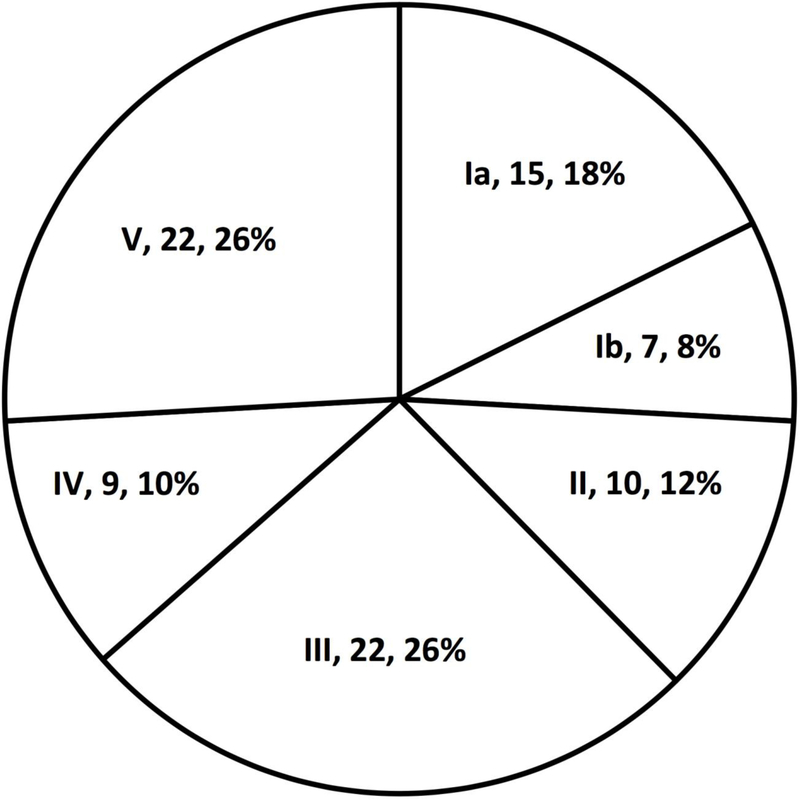
A total of 433 vaginal lavage samples were analyzed, of which 91 (21%) were GBS positive. Of these, 6 (6.6%) samples harbored at least two serotypes of GBS (Table 1). Serotype distribution of samples with a single GBS serotype is provided in Figure 1. Serotype IV was detected in 9/85 monocolonized samples (Fig. 1) and 2/6 samples in which multiple serotypes were detected (Table 1), for an overall rate of 12% of GBS strains. Among co-colonized samples, 1/6 had 3 separate serotypes detected (Ia, Ib, V) and the remainder had 2 serotypes. While all of the serotypes detected in co-colonized samples were also detected in mono-colonized samples, we noted possible over representation of type V among co-colonized samples. We found that 4/6 (66%) of co-colonized samples had serotype V detected, whereas that serotype made up only 22/85 (26%) of GBS in mono-colonized samples (Fisher’s exact test, P=0.05). For 4 out of 6 (66%) co-colonized samples (samples BVI0030, BVI0091, BVI0337 and BVI0341) all predicted serotypes were confirmed to be present by latex agglutination, including the sample with three serotypes. For the other 2 samples only one of the two predicted serotypes could be definitively identified.
Table 1:
Distribution of serotypes in GBS-positive vaginal lavage samples with multiple serotypes detected.
| Sample | Serotypes detected |
|---|---|
| BVI0030 | Ia, IV |
| BVI0058 | Ia, Ib |
| BVI0091 | II, V |
| BVI0236 | IV, V |
| BVI0337 | Ia, Ib, V |
| BVI0341 | III, V |
Figure 1. Serotype distribution of GBS-positive vaginal lavage samples with a single serotype detected.
Total number of samples (N) = 85. Sector labels specify serotype, number of samples (n) and %.
DISCUSSION
This cohort of healthy, non-pregnant women in New York City had a GBS vaginal colonization prevalence of 21%. This colonization rate is similar to those previously reported in samples of both pregnant and non-pregnant women. However, the use of vaginal rather than rectovaginal samples may underestimate the true rate of GBS colonization in this cohort [11]. In addition, the relative sensitivity of lavage compared to swab sampling is unknown.
Serotypes Ia, Ib, II, III and V have previously been reported as the predominant serotypes among both neonatal and adult invasive strains and maternal colonizing strains in the US. In contrast, the current study also identified a significant proportion of serotype IV isolates (12% of overall isolates, including co-colonized samples), consistent with recent reports describing the emergence and clonal expansion of serotype IV in both colonization and invasive disease. Notably, an increase in serotype IV colonization in nonpregnant women in the US [12] and pregnant women in Toronto, Canada [13] has been described over the past decade. Thus, the findings of the current study may reflect a temporal change in epidemiology that occurred around 2010, when the samples for this study were collected. While earlier candidate GBS conjugate vaccines did not contain serotype IV, a hexavalent candidate that includes that serotype is in early clinical trials.[5] Serotype distribution in women and even colonized mothers may correlate imperfectly with serotypes causing invasive neonatal disease, as invasiveness may vary by serotype. Nevertheless, GBS serotype distribution appears to be more diverse than previously reported with significant variation across and within regions, and colonization data from a variety of sources may provide further insight into circulating GBS serotypes in different regions over time. In particular, continued monitoring for the emergence of non-vaccine GBS serotypes is warranted.
The increasing use of molecular methods for detecting GBS serotypes may also add to a shifting paradigm of GBS epidemiology and serotype distribution. Notably, co-colonization with multiple serotypes of GBS may be recognized more frequently than previously reported. The current study identified co-colonization in 6.6% of GBS-positive samples. The serotype distribution among these samples included all of the serotypes detected among the mono-colonized samples; however, we noted an over-representation of serotype V (detected in 4/6 samples) and the suggestion of possible under-representation of serotype III (detected in 1/6 samples). Previous reports using culture and latex agglutination with multiple colony picks have identified co-colonization in 1%[14] to 4%[15] of cultures taken from a single anatomical site, and from 1%[14, 16] to 23%[17] when cultures are performed from multiple sites. Despite a small sample size, the results from our study suggest potential differences in predilection for co-colonization according to serotype.
Accurately identifying co-colonization will potentially be more important if and when a multivalent GBS conjugate vaccine is licensed and becomes widely used. The experience from pneumococcal conjugate vaccines demonstrated rapid serotype replacement with non-vaccine serotypes following the introduction of a heptavalent vaccine. In contrast, serotype replacement has not been a major factor in Haemophilus influenzae disease, despite widespread vaccination against only capsule type b. Lessons learned from these experiences include the importance of accurate and continued surveillance of serotype distribution in the years prior to and following vaccine introduction, as the future impact of GBS vaccination on overall serotype distribution is unknown at this time.
Thus, use of a serotype-specific PCR identifies co-colonization with more than one serotype of GBS in a significant minority of GBS-positive vaginal samples from non-pregnant women, as well as a greater representation of serotype IV among colonizing strains than previously reported from the US. The implications of these findings may become increasingly important in the era of licensed GBS vaccines, and ongoing surveillance is required. Future directions of this work will include use of this technique to study colonization and serotype distribution among different populations, including pregnant women in New York, as well as pregnant and non-pregnant women in different regions.
Acknowledgments
FUNDING
This work was supported by the National Institutes of Health [R56 AI136499 to A.J.R.; K23 HD065844 and R21 AI127957 to T.M.R.] and the Doris Duke Charitable Foundation [DDCF CSDA 2009–039 to A.J.R.] M.G. and E.S. were supported by the Jack Cary Eichenbaum Neonatology Scholars Program at New York University School of Medicine. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding bodies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
PRESENTATION
The results of this study were presented in part at the 1st International Symposium on Streptococcus agalactiae Disease; Cape Town, South Africa; February 20–23, 2018.
CONFLICT OF INTEREST STATEMENT
Potential conflicts: A.J.R. has served as a consultant to Pfizer.
All other authors report no conflicts of interest.
REFERENCES
- [1].Seale AC, Bianchi-Jassir F, Russell NJ, Kohli-Lynch M, Tann CJ, Hall J, et al. Estimates of the Burden of Group B Streptococcal Disease Worldwide for Pregnant Women, Stillbirths, and Children. Clin Infect Dis 2017;65:S200–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kohli-Lynch M, Russell NJ, Seale AC, Dangor Z, Tann CJ, Baker CJ, et al. Neurodevelopmental Impairment in Children After Group B Streptococcal Disease Worldwide: Systematic Review and Meta-analyses. Clin Infect Dis 2017;65:S190–S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Verani JR, McGee L, Schrag SJ. Prevention of perinatal group B streptococcal disease--revised guidelines from CDC, 2010. MMWR Recomm Rep 2010;59:1–36. [PubMed] [Google Scholar]
- [4].Kobayashi M, Vekemans J, Baker CJ, Ratner AJ, Le Doare K, Schrag SJ. Group B Streptococcus vaccine development: present status and future considerations, with emphasis on perspectives for low and middle income countries. F1000Res 2016;5:2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Anderson AL, Y. T, Kanevsky I, I.L. S, D. P, Buurman ET, et al. Updates on the Development of a Multivalent Group B Streptococcal Vaccine. International Society for Vaccines Annual Congress 2017. Abstract O1.3.
- [6].Imperi M, Pataracchia M, Alfarone G, Baldassarri L, Orefici G, Creti R. A multiplex PCR assay for the direct identification of the capsular type (Ia to IX) of Streptococcus agalactiae. J Microbiol Methods 2010;80:212–4. [DOI] [PubMed] [Google Scholar]
- [7].Sheppard AE, Vaughan A, Jones N, Turner P, Turner C, Efstratiou A, et al. Capsular Typing Method for Streptococcus agalactiae Using Whole-Genome Sequence Data. J Clin Microbiol 2016;54:1388–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rosen GH, Randis TM, Desai PV, Sapra KJ, Ma B, Gajer P, et al. Group B Streptococcus and the Vaginal Microbiota. J Infect Dis 2017;216:744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Khatami A, Randis TM, Chamby A, Hooven TA, Gegick M, Suzman E, et al. Improving the Sensitivity of Real-time PCR Detection of Group B Streptococcus Using Consensus Sequence-Derived Oligonucleotides. Open Forum Infect Dis 2018;5:ofy164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Breeding KM, Ragipani B, Lee KD, Malik M, Randis TM, Ratner AJ. Real-time PCR-based serotyping of Streptococcus agalactiae. Sci Rep 2016;6:38523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Badri MS, Zawaneh S, Cruz AC, Mantilla G, Baer H, Spellacy WN, et al. Rectal colonization with group B streptococcus: relation to vaginal colonization of pregnant women. J Infect Dis 1977;135:308–12. [DOI] [PubMed] [Google Scholar]
- [12].Diedrick MJ, Flores AE, Hillier SL, Creti R, Ferrieri P. Clonal analysis of colonizing group B Streptococcus, serotype IV, an emerging pathogen in the United States. J Clin Microbiol 2010;48:3100–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Teatero S, Ferrieri P, Martin I, Demczuk W, McGeer A, Fittipaldi N. Serotype Distribution, Population Structure, and Antimicrobial Resistance of Group B Streptococcus Strains Recovered from Colonized Pregnant Women. J Clin Microbiol 2017;55:412–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Anthony BF, Okada DM, Hobel CJ. Epidemiology of group B Streptococcus: longitudinal observations during pregnancy. J Infect Dis 1978;137:524–30. [DOI] [PubMed] [Google Scholar]
- [15].Ferrieri P, Hillier SL, Krohn MA, Moore D, Paoletti LC, Flores AE. Characterization of vaginal & rectal colonization with multiple serotypes of group B streptococci using multiple colony picks. Indian J Med Res 2004;119 Suppl:208–12. [PubMed] [Google Scholar]
- [16].Whitney CG, Daly S, Limpongsanurak S, Festin MR, Thinn KK, Chipato T, et al. The international infections in pregnancy study: group B streptococcal colonization in pregnant women. J Matern Fetal Neonatal Med 2004;15:267–74. [DOI] [PubMed] [Google Scholar]
- [17].Hoogkamp-Korstanje JA, Gerards LJ, Cats BP. Maternal carriage and neonatal acquisition of group B streptococci. J Infect Dis 1982;145:800–3. [DOI] [PubMed] [Google Scholar]