Abstract
The two non-visual subtypes, arrestin-2 and arrestin-3, are ubiquitously expressed and bind hundreds of G protein-coupled receptors. In addition, these arrestins also interact with dozens of non-receptor signaling proteins, including c-Src, ERK and JNK, that regulate cell death and survival. Arrestin-3 facilitates the activation of JNK family kinases, which are important players in the regulation of apoptosis. Here we show that arrestin-3 is specifically cleaved at Asp366, Asp405 and Asp406 by caspases during the apoptotic cell death. This results in the generation of one main cleavage product, arrestin-3-(1–366). The formation of this fragment occurs in a dosedependent manner with the increase of fraction of apoptotic cells upon etoposide treatment. In contrast to a caspase-resistant mutant (D366/405/406E) the arrestin-3-(1–366) fragment reduces the apoptosis of etoposide-treated cells. We found that caspase cleavage did not affect the binding of the arrestin-3 to JNK3, but prevented facilitation of its activation, in contrast to the caspaseresistant mutant, which facilitated JNK activation similar to WT arrestin-3, likely due to decreased binding of the upstream kinases ASK1 and MKK4/7. The data suggest that caspasegenerated arrestin-3-(1–366) prevents the signaling in the ASK1-MKK4/7-JNK1/2/3 cascade and protects cells, thereby suppressing apoptosis.
Keywords: arrestin-3, caspase cleavage, MAP kinases, JNK, cell death: apoptosis
1. Introduction
The programmed cell death, or apoptosis, is required to remove damaged cells. It also plays an essential role in the development of multicellular organisms [1]. The removal of damaged cells prevents the development of diseases, such as cancer, neurodegenerative disorders, or immune diseases. Apoptosis can be activated by cellular stress or death receptors. In both cases caspases (aspartate-specific proteases) are the key players involved [2]. Caspase 2, 3, 7, 8, 9 and 10 are apoptotic caspases. The initiator caspases (2, 8, 9 and 10) cleave the inactive forms of effector caspases (3, 6 and 7). These activated effector caspases cleave numerous downstream proteins, triggering the apoptosis pathway and destroying normal cellular functions [3].
Arrestins are well known as ubiquitous regulators of cell signaling, both G protein-coupled receptor (GPCR)-dependent and -independent [4, 5]. Some binding partners of arrestins, such as protein kinases c-Src (proto-oncogene tyrosine-specific protein kinase), ERK (Extracellular Signal-Regulated Kinase) and JNK (c-Jun N-terminal Kinase), are intimately involved in the regulation of cell proliferation, survival, and death. Recently arrestin-21 and arrestin-3 (a.k.a. β-arrestin1 and 2, respectively) have been reported to have a pro- and anti-apoptotic effects. Both non-visual arrestins were shown to block apoptosis [6], as well as to promote cell death via Wnt/β-catenin pathway [7], and β-amyloid-induced death of SH-SY5Y cells [8]. Arrestin-3 facilitated cell death via NF-κB pathway [9], or via Akt inhibition [10, 11] At the same time, arrestin-3 was found to facilitate cardiomyocyte survival after infarction, whereas arrestin-2 in the same situation was cytotoxic [12]. In contrast, arrestin-2 was found to inhibit intestinal stem cell apoptosis [13] and brain cell death after ischemia [14]. Thus, it appears that the effect of either non-visual arrestin on cell death and survival depends on cell type, in line with the accumulating evidence that arrestins are remarkably multi-functional signaling proteins affecting numerous pathways (reviewed in [5, 15]). To rationalize conflicting evidence, the exact mechanisms of their action need to be elucidated. A recent study showed that caspase-3 cleaves arrestin-2 generating a (1–380) fragment, which translocates to the mitochondria and increases cytochrome c release in cooperation with another product of caspase activity, tBid [16]. Cytochrome c release facilitates further caspase activation. Thus, arrestin-2 cleavage by caspases appears to function as part of a positive feedback loop in apoptosis [17].
Here we show that during apoptosis caspases 7 and 9 cleave arrestin-3, producing the arrestin3-(1–366) fragment. This cleavage occurs in response to different apoptosis stimuli in several cell types. In contrast to the cytoplasmic arrestin-3, the fragment is primarily localized to the nucleus. This arrestin-3-(1–366) fragment binds JNK3 like wild type (WT) arrestin-3, but it has lost the ability to facilitate JNK3 activation, likely due to reduced binding of the upstream kinases ASK1 and MKK4/7. This fragment showed a strong cytoprotective effect, in contrast to caspaseresistant arrestin-3-(D366/405/406E) (arrestin-3-(3xE)), which promoted the activation of JNK3, and dose-dependently facilitated cell death. Thus, in contrast to arrestin-2, arrestin-3 cleavage by caspases plays a role in suppression of cell death.
2. Materials and methods
2. 1. Reagents and antibodies
Mouse monoclonal pan-arrestin F4C1 antibody (epitope residues 43–47, DGVVLVD) from Dr. LA Donoso (Wills Eye Institute, Philadelphia, PA, USA) [18]; antibodies against caspase-3 and COX IV were from Cell Signaling Technology (Beverly, MA, USA); caspase-8 mouse monoclonal antibody (1G12) was from Enzo Life Sciences (Plymouth Meeting, PA, USA); antibody for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and rabbit caspase-8 antibody were from Millipore (Billerica, MA, USA); antibody against GFP and cytochrome C were from BD Bioscience (Palo Alto, CA, USA); tissue culture media and reagents were from Invitrogen (Carlsbad, CA, USA); recombinant caspase-1 to −10 were from Millipore or BioMol (Plymouth Meeting, PA, USA); caspase inhibitor Z-VAD was from BioMol; restriction enzymes were from New England Biolabs (Ipswich, MA, USA), kits for DNA preps were from Zymo Research (Irvine, CA).
2.2. Cell culture
COS7 cells (a gift of Dr. S. S. Zinkel, Vanderbilt University, Nashville, TN, USA); arrestin-2 knockout (KO) mouse embryonic fibroblasts (MEFs) (a gift of Dr. R.J. Lefkowitz, Duke University, NC, USA [19]); and Rat-1 cells were cultured in DMEM (supplemented with 10% FBS and 1% penicillin/streptomycin) with 5% CO2 and 37°C. Cells were transfected using lipofectamine 2000 (3:1, lipid:DNA ratio). The amount of DNA used was equalized with empty vector (pcDNA3). For apoptosis induction the indicated amount of TNFα with cycloheximide (CHX) or etoposide was used, as described [16], for specified times at 37°C and 5% CO2. As a control no etoposide or TNFα was added. The nuclear fractionation was performed using the CelLytic NuClear Extraction Kit from Sigma-Aldrich (St. Louis, MO, USA).
2.3. Protein preparation and Western blotting
Cells were disrupted in lysis buffer (Ambion, Brand Island, NY, USA) and incubated for 5 min at 95°C. Proteins were isolated using methanol precipitation (1:9, cell lysate:methanol). Precipitate was pelleted via centrifugation (10,000 x g, 10 min at room temperature). Pellet was washed with 1 ml of 90% methanol, dried and resuspended in SDS-sample buffer. The proteins were separated by SDS-PAGE, where equal amounts of proteins were used, then transferred onto Immobilon-P membrane (Millipore, Bedford, MA, USA). The membrane was blocked with 5% non-fat dry milk in TBS supplemented with 0.1% of Tween-20. Following the incubation with the corresponding primary (overnight at 4oC) and peroxidase-conjugated secondary (1 h, room temperature) antibodies protein bands were visualized with SuperSignal enhanced chemilumescence reagent (Pierce, Rockford, IL, USA). X-ray film (Fujifilm, Stamford, CT, USA) was used for detection.
2.4. Cleavage of arrestin-3 by caspases in vitro
75 ng of recombinant arrestin-3 purified, as described [20], and 1 unit of indicated caspase (Millipore) were incubated 3 h at 37°C in 50 μl caspase cleavage buffer (50 mM HEPES, pH 7.4, 100 mM NaCl, 0.1% CHAPS, 1mM EDTA, 10 mM DTT). The high salt buffer had additionally 1 M ammonium citrate for caspases-2, −8, −9 and −10, as described [16]. The reaction was stopped with SDS-PAGE sample buffer and incubation for 5 min at 95°C. The products were analyzed by Western blot with the monoclonal anti-arrestin antibody F4C1 [18].
2.5. Nuclear fractionation
To determine the localization of the arrestin-3 cleavage fragment a nuclear fractionation assay was performed. Therefore, the Rat1–1 cells were transfected with certain constructs. After 24h of protein expression cells were treated with 100 μM etoposide for 18h. For the nuclear fractionation the CelLytic™ NuCLEAR™ Extraction Kit from Sigma-Aldrich (St. Louis, MO, USA) was used.
2.6. Expression and purification of WT and mutant arrestin-3
WT arrestin-3, arrestin-3-(1–366) and caspase-resistant triple mutant D366E, D405E, D406E (arrestin-3-(3xE)) were expressed and purified as described [20].
2.7. Arrestin interaction with the kinases of JNK signaling cascade.
In vitro arrestin-downstream effector binding was assayed by pull-down, as described [21, 22].
2.8. Arrestin-3-mediated JNK3 activation assay in cells.
Phosphatase inhibitors (50 mM NaF and 10 mM Na3VO4) were added to the medium of COS7 cells 15 min before lysis at 37°C. Then the cells were washed with ice-cold PBS and lysed using SDS-sample buffer supplemented with 10 mM NaF, 100 μm Na3VO4, 2 mM EDTA, 2 mM EGTA, and 1 mM PMSF. The suspension was incubated at 95°C for 5 min., and centrifuged at 10,000 × g for 10 min. Supernatant was used for SDS-PAGE and Western blot. Mouse monoclonal antibodies against FLAG (Sigma), HA (Sigma), GFP (Clontech), and phospho-JNK (Cell Signaling Technology Inc.) were used at 1:1,000 or 1:2,000 dilution.
2.9. In-cell apoptosis assay
COS7 cells were transfected with arrestin-3-(3xE) or arrestin-3-(1–366) fragment. After 24 h cells were treated without or with 80 μM etoposide for 24 h. The cell nuclei were stained with Hoechst 33258. Images were collected using Nikon TE2000-E automated microscope controlled by Nikon NIS-Elements software. Cells with condensed chromatin or fragmented nuclei were considered apoptotic. Approximately 400 cells were scored per data point using Image J software.
2.10. In vitro receptor binding assay was performed, as described [23].
Rhodopsin for this assay was purified in native disc membranes from cow retinas, phosphorylated, and regenerated with 11-cis-retinal (a gift of NIH NEI and Dr. R.K. Crouch, Medical University of South Carolina) after phosphorylation, as described [24]. Indicated arrestins were subcloned into pGEM2 vector for in vitro transcription [25]. Arrestins were prepared in radiolabeld form using purified mRNAs by in vitro translation, as described [26]. To measure receptor binding, 1 nM indicated arrestin (50 fmol) was incubated with 0.3 μg of light activated phosphorylated (P-Rh*) or unphosphorylated (Rh*), or dark inactive phosphorylated (P-Rh) or unphosphorylated (Rh) rhodopsin (11 pmol, yielding final concentration of 0.22 μM) in 50 concentration of 0.22 μM) in 50 l of 50 mM Tris-HCL, pH 7.4, 100 mM potassium acetate, 1 mM EDTA, 1 mM DTT for 5 min at 37oC under room light (Rh* and P-Rh*) or in the dark in light-proof box (P-Rh and Rh). Samples were cooled on ice, then bound and free arrestin was separated in the dark at 4oC by gel-filtration on 2-ml Sepharose 2B-CL column. Arrestin eluting with rhodopsin-containing membranes was quantified by liquid scintillation counting. Non-specific “binding”, determined in samples where rhodopsin was omitted, was subtracted.
2.11. Immunocytochemistry and microscopy.
COS7 cells plated on 4-chambered glass slides (Fisher) were fixed 48 hours post-transfection in 100% methanol at −20°C for 5 min. The cells were rehydrated with PBS and blocked with 3% bovine serum albumin in PBS for 1 h at room temperature. GFP-JNK3 was visualized by its own fluorescence, whereas arrestin-3 species were visualized with rabbit pan-arrestin polyclonal antibodies [20] (24 h at 4oC) followed by five washes with PBS and by Alexa 593 anti-rabbit secondary antibody (1 h at room temperature). After five additional washes with PBS, the slides were mounted with DAPI-containing Vectashield (Vector laboratories) and imaged on Olympus FV-1000 confocal microscope with 40x oil objective. At least 30 cells (expressing both arrestin-3 construct and GFP-JNK3) per condition were scored for GFP-JNK3 and arrestin distribution (C>N means higher concentration in the cytoplasm than in the nucleus). The experiment was performed three times.
2.12. Statistical analysis.
ANOVA statistics was used to analyze the biochemical data with Protein (Arr3, Arr3–3xE, Arr3(1–366)) as main factor followed by Bonferroni post hoc test with correction for multiple comparisons. The cell death data were analyzed by two-way ANCOVA with Protein as a factor and DNA concentration as a co-variate. The ANOVA analysis was supplemented by comparisons using Student’s t-test where appropriate. StatView software (SAS Institute) was used for the statistical analysis. The value of p<0.05 was considered significant.
3. Results
3.1. Arrestin-3 cleavage by caspases in vitro and in cells
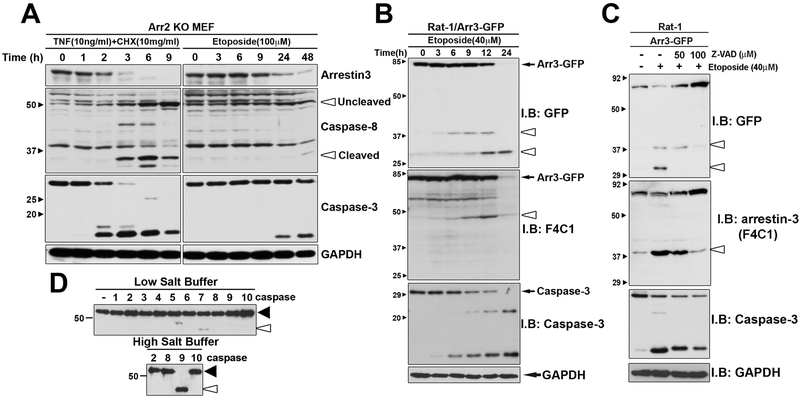
Arrestin-2 and arrestin-3 are ubiquitously expressed in mammals [4, 5, 27]. To exclude interference from arrestin-2, arrestin-2 knockout (Arr2KO) MEFs were used [19]. To induce apoptosis, the cells were treated with the tumor necrosis factor α (TNFα) (in the presence of cycloheximide (CHX) to inhibit protein synthesis) or topoisomerase inhibitor etoposide for different times. TNFα activates tumor necrosis factor receptor 1 (TNFR1) and therefore induces apoptosis via extrinsic pathway [28]. Etoposide treatment causes DNA damage, which induces apoptosis via intrinsic pathway [16]. We observed progressive decrease of full-length arrestin-3 over time with both apoptosis inducers, with TNFα being more efficient (Fig. 1A). The stimulated TNFR1 activates caspase 8 directly, which is likely why it acts faster than etoposide, which induces the activation of multiple caspases indirectly. An increase of activated caspases 3 and 8 is clearly visible in TNFα, and at later time points, in etoposide treated cells (Fig. 1A).
Fig. 1. Arrestin-3 is cleaved by caspases during apoptosis initiated via TNFα or Etoposide.
(A) Arr2 KO MEFs were treated with 10 ng/ml TNFα along with 10 μg/ml CHX or 100 μM etoposide for indicated time. Cells were lysed and the cleavage (seen here as the loss of full-length arrestin-3) was detected by Western blotting with F4C1 mouse monoclonal anti-arrestin antibody. In parallel, Western blotting for caspases 3 and 8 was performed to indicate the progression of apoptosis. Small black arrowheads show protein standards in kDa. The positions of uncleaved (inactive) and cleaved (active) caspase-8 are indicated by open arrowheads. GAPDH was used as a loading control. Representative data from four experiments are shown. (B) Rat-1 cells were transfected with arrestin-3-GFP and treated with 40 μM etoposide for 12 hours. Black arrow, full length arrestin-3-GFP; arrestin-3 cleavage fragments are indicated by open arrowheads. Arrestin-3 cleavage parallels caspase-3 activation. (C) Arrestin-3-GFP transfected Rat-1 cells were treated for 2 hours with etoposide in the absence (−) of presence of caspase inhibitor Z-VAD at 50 μM or 100 μM. Arrestin-3 cleavage fragments are indicated by open arrowheads. Representative blots (out of three repeats) are shown. (D) In-vitro cleavage of arrestin-3. 75 ng of purified arrestin-3 was incubated with different caspases for 3 h at 37°C. The reaction was performed in 50 μl of caspase cleavage buffer (low salt buffer) and in the same buffer with the addition of 1.0 M ammonium citrate (high salt buffer). Caspases were purchased from Millipore, one unit per reaction was used. Cleavage products were analyzed using Western blotting with the pan-arrestin F4C primary antibody. Arrestin-3 without the addition of caspases was used as a control. Black arrowheads point to full-length arrestin-3; open arrowheads – to the cleavage product. The experiment was performed three times with identical results.
Identification of caspase cleavage sites in arrestin-3 (Fig. 1B) was performed using arrestin-3 construct with fused C-terminal GFP in Rat-1 cells. These cells were chosen because they undergo “pure” caspase-driven apoptosis, without interference from necrosis and action of other proteases. After etoposide treatment two cleavage fragments were visible (around 30 and 40 kDa). This indicates the existence of at least two caspase cleavage sites (D/V-XX-D) within arrestin-3, which must be accessible in folded arrestin-3 [29]. To further characterize arrestin-3 cleavage products, arrestin-3-GFP constructs were visualized using F4C1 and GFP antibody (Fig. 1B). Two species of the C-terminal cleavage products and one N-terminal product were detectable. In all cases the amount of the cleavage product was progressively increased with etoposide treatment time.
We found that the amount of arrestin-3 cleavage products decreased with increasing concentration of pan-caspase inhibitor Z-VAD (Fig. 1C), indicating that arrestin-3 was cleaved by caspases, rather than other proteases in the cell. To determine which caspases cleave arrestin-3, we performed an in vitro assay using purified arrestin-3 and individual caspases (Fig. 1D). Because initiator caspases tend to be more active in high salt, which promotes their dimerization [30], the in vitro cleavage was tested in two different conditions, at low and high salt. Purified arrestin-3 was cleaved by caspase 9 at high salt and less efficiently by caspase 7 at low salt (Fig. 1D). The in vitro cleavage yielded fragments of approximately the same size as in-cell experiments (Fig. 1). Thus, caspases 7 and 9 apparently cleave arrestin-3 in apoptotic cells.
3.2. Identification of the aspartic acid residues targeted by caspases in arrestin-3
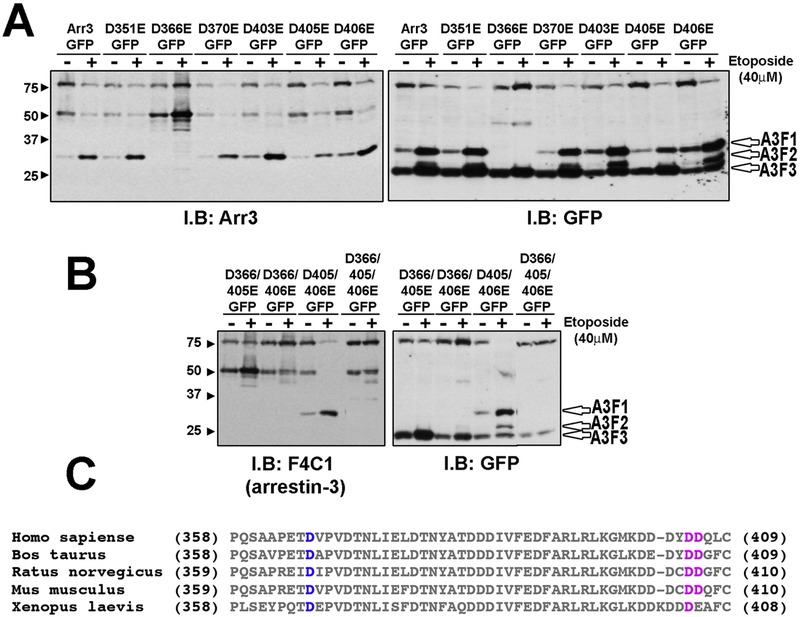
To identify the exact cleavage sites, we generated different mutants with conservative Asp to Glu substitutions in putative caspase cleavage sites, shown in Fig. 2. Rat-1 cells, which undergo apoptosis without the involvement of other mechanisms of cell death, and therefore serve as a useful model [16], were treated with or without etoposide and the cleavage products were visualized using both anti-arrestin F4C1 and anti-GFP antibodies. Several single mutants showed a significant reduction in the amount of cleavage products, such as D405E and D406E. In case of D366E we observed a complete loss of the cleavage products A3F1 and A3F2 (Fig. 2A). The triple mutant D366/405/406E (3xE) was completely resistant to caspase cleavage in etoposidetreated cells (Fig. 2B). These data identify aspartate residues 366, 405, and 406 as caspase cleavage sites. All three sites are in the elements that are not visible in the crystal structure of arrestin-3 in its basal [29] or active [31] conformation, which indicates that these elements are likely very flexible and therefore accessible to proteases. Homologous elements of the visual arrestin-1 in complex with rhodopsin [32, 33] are also not visible in the crystal structure, suggesting that the accessibility of these sites to caspases would not be affected by receptor binding.
Fig. 2. Identification of cleavage sites within arrestin-3.
(A) Rat-1 cells were transfected with indicated arrestin-3-GFP mutants. Apoptosis was induced by 18 h treatment with 40 μM etoposide. The cell lysate was analyzed using arrestin-3 or GFP antibodies. GFP antibody detected three different cleavage products, A3F1, A3F2 and A3F3 (open arrowheads). Arrestin3 antibody detected only the A3F1 product. Mutant D336E showed a loss of cleavage products A3F1 and A3F2. (B) Double and triple mutants were treated similarly. Cleavage of double mutant D405/406E yielded A3F1 band. Arrestin-3-D366/405/406E (3xE) showed a significant loss of caspase cleavage. Representative blots from three experiments that yielded virtually identical results are shown in A and B. (C) Sequence alignment of arrestin-3 C-terminal regions. Conserved D366 is highlighted in blue. D406/D406 (highlighted in pink) are conserved in mammals but only one of these two residues is conserved in Xenopus.
Sequence alignment of the C-termini of cloned vertebrate arrestin-3 species revealed conserved Asp366 and Asp405: Asp366 and Asp405 in human, mouse and rat arrestin-3 and homologous Asp366 and Asp406 in X. laevis (Fig. 2C). The third cleavage site (corresponding to Asp406 in bovine sequence) is conserved in mammals, but not in X. laevis. Thus, arrestin-3 may be cleaved at homologous aspartates during apoptosis in other vertebrate species.
3.3. Receptor binding of full-length and caspase-cleaved arrestins
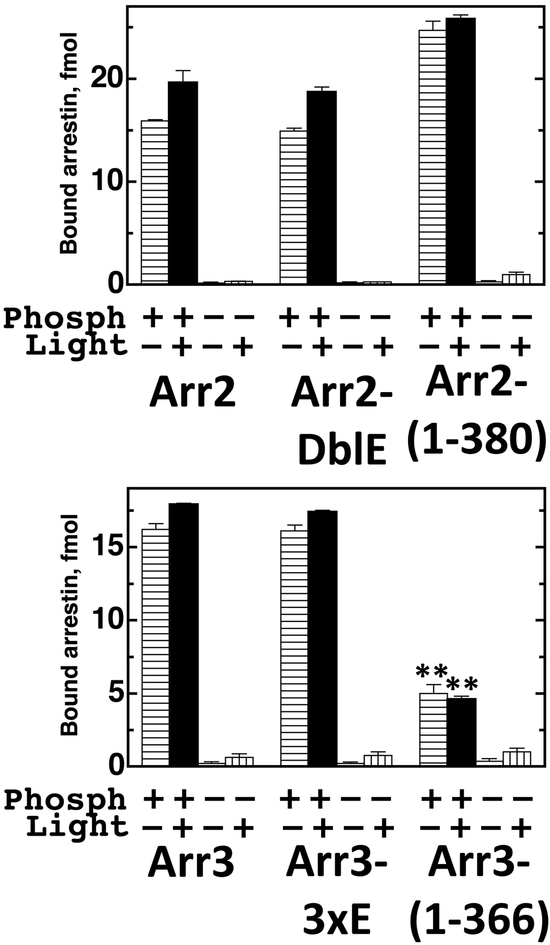
We compared wild type arrestin-3, its caspase-resistant mutant (3xE), and caspase-generated fragment arrestin-3-(1–366), as well as arrestin-2, its caspase-resistant form (DblE) and caspasegenerated fragment arrestin-2-(1–380) for their ability to bind all four functional forms of rhodopsin (phosphorylated inactive, phosphorylated light-activated, unphosphorylated inactive, and unphosphorylated light-activated). To this end, we generated arrestins in radiolabeled form by cell-free translation [26], incubated them with the same amount of all forms of rhodopsin, separated bound arrestins from free by gel-filtration, and quantified binding (Fig. 3). The data show that caspase-resistant forms of both arrestin-2 and arrestin-3 fully retain their ability to bind rhodopsin (Fig. 3). Interestingly, while caspase-generated arrestin-2-(1–380) demonstrates an increase in receptor binding characteristic for enhanced arrestin mutants with short C-terminal deletions [34, 35], caspase-generated arrestin-3-(1–366) shows reduced receptor binding, reminiscent of visual arrestin-1-(1–365) with a similar larger C-terminal deletion [23, 36]. Arrestin-3-(1–366) still shows the preference for phosphorylated rhodopsin with minimal discrimination between active and inactive form characteristic for both non-visual arrestins (Fig. 3) [26].
Fig. 3. Arrestin binding to rhodopsin.
Indicated radiolabeled arrestins generated in the in vitro translation (50 fmol; 1 nM final concentration) were incubated with the same amounts of indicated functional forms of rhodopsin (0.3 μg) for 5 min at 37oC. Bound arrestins were separated from free by gel-filtration on the Sepharose 2B-CL and quantified by scintillation counting. The amount of bound arrestins was converted to femtomoles based on their specific activity. The results of three experiments are shown as means ± SD. **, p<0.01, as compared to WT arrestin-3.
3.4. Translocation of arrestin-3 cleavage product into the nucleus
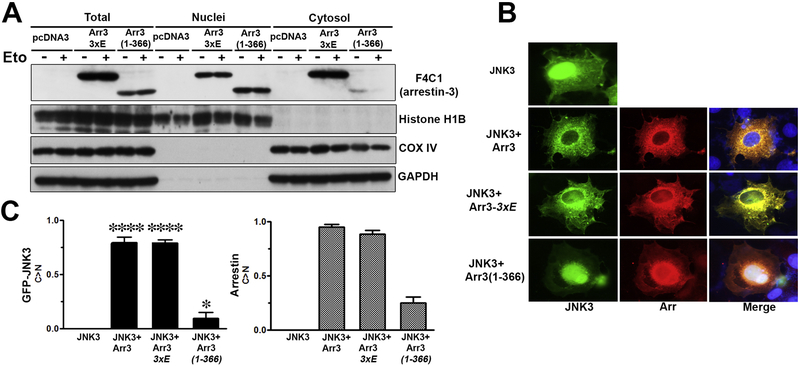
To determine the biological function of the arrestin-3 cleavage product, we tested its subcellular localization. The nuclear export signal (NES) in arrestin-3, which determines its cytosolic localization, was identified in the C-terminus [37, 38], which is cleaved off by caspases (Figs. 1,2). To determine the localization of arrestin-3 and its fragment, COS7 cells were transfected with GFP-JNK3 and the corresponding arrestin-3 construct. Two different methods were used to verify the localization of the cleavage product, the subcellular fractionation of arrestin-2 knockout MEFs (that do not have arrestin-2, which is also recognized by the F4C1 antibody) and fluorescence microscopy in COS7 cells. Full-length arrestin-3, as well as arrestin-2 with engineered arrestin-3-like NES, pull GFP-JNK3 out of the nucleus, where it spontaneously localizes [37]. COS7 cells were chosen for these experiments because due to relatively low expression of non-visual arrestins in this cell line the mutants we intended to test would not have too much competition with endogenous WT protein.
By subcellular fractionation caspase-resistant arrestin-3-(3xE), although present in the nucleus, was detected mostly in the cytosol of the cell (Fig. 4A), like WT arrestin-3 [37]. In contrast, arrestin-3-(1–366) fragment was barely detected in the cytosol but was mostly localized in the nucleus (Fig. 4A). Importantly, etoposide treatment (Eto +) did not affect the subcellular distribution of arrestin-3 mutants, indicating that it reflects their inherent properties, rather than the effect of etoposide (Fig. 4A). Using fluorescence microscopy GFP-JNK3 was visualized via intrinsic fluorescence, whereas arrestin-3 was detected using rabbit anti-arrestin antibodies followed by an alexa-593 anti-rabbit secondary antibody (Fig. 4B). Fluorescence microscopy showed that arrestin-3 re-localizes JNK3 from the nucleus to the cytosol, as we described previously [37], so that GFP-JNK3 and arrestin-3 co-localize in the cytoplasm of the COS7 cells. A similar behavior of the cleavage-resistant arrestin-3-(3xE) mutant was observed. In contrast, the arrestin-3-(1–366) fragment was co-localized with GFP-JNK3 mostly in the nucleus (Fig. 4B). Dramatic difference in subcellular localization of GFP-JNK3, that binds arrestins ([37], Fig. 4) was statistically significant and reflected the distribution of arrestin-3 mutants (Fig. 4C).
Fig. 4. Translocation of arrestin-3 cleavage product into nucleus.
(A) A2KO MEFs were transfected with arrestin-3–3xE or arrestin-3-(1–366). Apoptosis was induced using 40 μM etoposide (+). Subcellular fractionation was performed, as described in Materials and Methods. Arrestin-3-(1–366) localized to the nucleus. A mitochondrial protein COX IV was used as a marker of non-nuclear fraction, histone H1B was used as a nuclear marker. GAPDH was used as a loading control. Note that subcellular localization depends on the arrestin-3 species expressed and is not affected by etoposide treatment. Representative blot out of three fractionations performed is shown. (B) Fluorescence microscopy of GFP-JNK3 (green) and arrestin-3 (the latter was visualized with rabbit panarrestin polyclonal antibody followed by Alexa 593 anti-rabbit secondary antibody, red). Blue on merged images – DAPI-stained nuclei. Representative images of the transfected COS7 cells are shown. Preferential localization of the caspase cleaved arrestin-3-(1–366) fragment in the nucleus is visible. Note that WT arrestin-3 as well as arrestin-3–3xE re-localize JNK3 from the nucleus to the cytosol, whereas arrestin-3-(1–366) does not. (C) Quantification of the fluorescence microscopy analysis. Fraction of cells where the concentration of GFP-JNK3 (left panel) or expressed arrestin-3 construct (right panel) in the cytoplasm was greater than in the nucleus (C>N) is shown. At least 30 cells expressing both arrestin-3 constructs and GFP-JNK3 per condition were scored for subcellular distribution of GFP-JNK3 and arrestin-3. Mean ± S.D. (n=3) are shown. Left panel: ****, p<0.0001; *, p<0.05, as compared to the distribution of GFP-JNK in cells expressing JNK3 alone; right panel: subcellular distribution of different arrestin-3 constructs in these three experiments is shown for comparison.
3.5. Facilitation of JNK3 activation by arrestin-3 and its cleavage product.
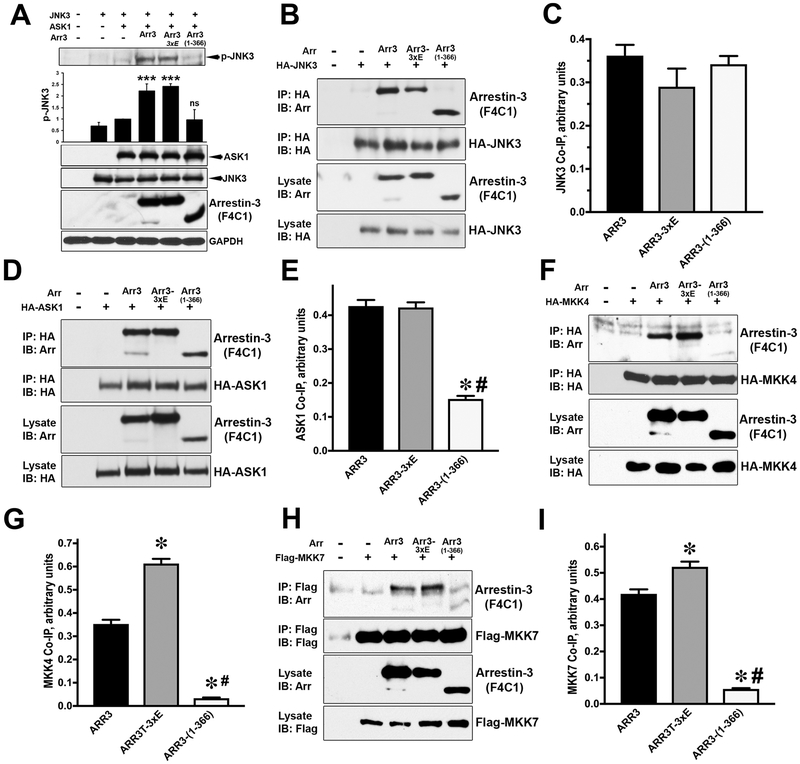
Among the four vertebrate arrestins, only arrestin-3 facilitates the activation of JNK family kinases [39–44]. The ability of arrestin-3 and its mutants to facilitate JNK3 activation was determined by the level of phosphorylated JNK3 in cells co-transfected with JNK3 and ASK1 in the presence and absence of arrestins (Fig. 5A). Both WT arrestin-3 and arrestin-3-(3xE) mutant produced comparable JNK3 activation, whereas arrestin-3-(1–366) fragment did not increase JNK3 phosphorylation above the level observed in the absence of exogenous arrestins (Fig. 5A).
Fig. 5. Arrestin-3-mediated activation of JNK3.
(A) COS7 cells were co-transfected with HA-JNK3 and indicated arrestin-3 constructs. Immunoprecipitation was performed with anti-HA antibody. Cell lysates and immunoprecipitated proteins were immunoblotted using F4C1 antibody to determine the level of arrestin-3 and anti-HA antibody to reveal the efficiency of JNK3 immunoprecipitation. (B) Quantification of the arrestin co-IP with JNK3 (n=5). The differences were not statistically significant. (C) COS7 cells were co-transfected with HA-ASK1 and indicated arrestin-3 constructs. Cell lysates and immunoprecipitated proteins were immunoblotted using F4C1 antibody to determine arrestin-3. (D) Quantification of the arrestin co-IP with ASK1 (n=3). Statistical significance of the differences is shown, as follows: *, p<0.001 to WT arrestin-3; #, p<0.001 to arrestin-3–3xE. Similar co-immunoprecipitation assays were performed with HA-MKK4 (E) or Flag-MKK7 (G) with corresponding anti-HA or anti-Flag antibodies. Quantification of the data for MKK4 (F) and MKK7 (H) (n=3 in both cases) is shown. *, p<0.001, as compared to WT arrestin-3; #, p<0.001, as compared to arrestin-3–3xE. (I) COS7 cells were transfected with HA-JNK3, HA-ASK1 and WT arrestin-3 (Arr3), caspase-resitant arrestin-3-(3xE), or arrestin-3-(1–366). Cell lysates were immunoblotted for phosphorylated JNK3 (p-JNK3). Intensities in the p-JNK blots from three independent experiments were quantified and statistically analyzed. Means ± S. D. are shown. ***, p<0.001, as compared to the level of p-JNK3 in cells co-expressing ASK1 and JNK3 without arrestin. Western blot with anti-HA and F4C1 antibodies was used to determine protein expression.
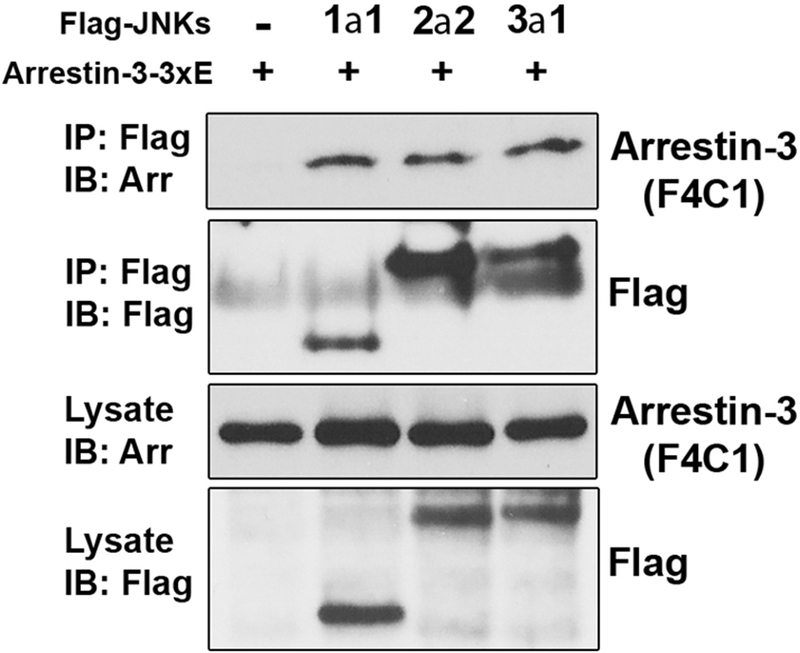
In search of the mechanism underlying this deficiency of arrestin-3-(1–366) cleavage product, we compared the ability of full-length arrestin-3 and arrestin-3-(1–366) fragment to interact with JNK3 and its upstream activators ASK1 (Apoptosis Signal-regulating Kinase 1), MKK4 and MKK7 (Mitogen-activated protein kinase kinase 4 and 7). In case of JNK3, a similar binding of all arrestin-3 proteins was observed (Fig. 5B,C). The interaction with ASK1 of WT arrestin-3 and caspase-resistant arrestin-3-(3xE) was also comparable. In contrast, arrestin-3-(1–366) fragment showed reduced ASK1 binding, which was about one third of that of WT arrestin-3 and 3Xe mutant (Fig. 5D, E). MKK4 and MKK7 bound WT arrestin-3 and its caspase-resistant mutant similarly, whereas arrestin-3-(1–366) fragment demonstrated severely impaired interaction with both MKKs, at the level of 10–15% of the WT (Fig. 5 F-I). Thus, the lack of activation capability of arrestin-3-(1–366) fragment is likely due to its reduced binding to upstream kinases ASK1, and especially MKK4 and MKK7 (Fig. 5C-H). Since caspase-generated arrestin-3-(1–366) fragment binds JNK3, but does not promote its activation, it has the potential to act in a dominant-negative manner, recruiting JNK3 away from productive scaffolds present in the cell, similar to the described arrestin-3-KNC mutant [44]. Since all JNK isoforms share the same upstream activators, to determine whether caspase-generated arrestin-3-(1–366) fragment can suppress the activity of ubiquitous JNK1/2 isoforms, we tested its interactions with other JNKs: 1α1, 2 α 2, and 3 α 1 and found that the fragment comparably binds these isoforms (Fig. 6), consistent with our finding that arrestin-3 facilitates the activation of ubiquitous JNK1 and JNK2 [41].
Fig. 6. Arrestin-3-(1–366) fragment interaction with JNK1/2 isoforms.
COS7 cells were co-transfected with indicated Flag-tagged JNK isoforms and arrestin-3-(1–366) (Arr). Note that all three JNK isoforms tested bind arrestin-3-(1–366) comparably. Experiment was repeated three times with the same results.
3.6. Effect of arrestin-3 caspase cleavage on cell survival.
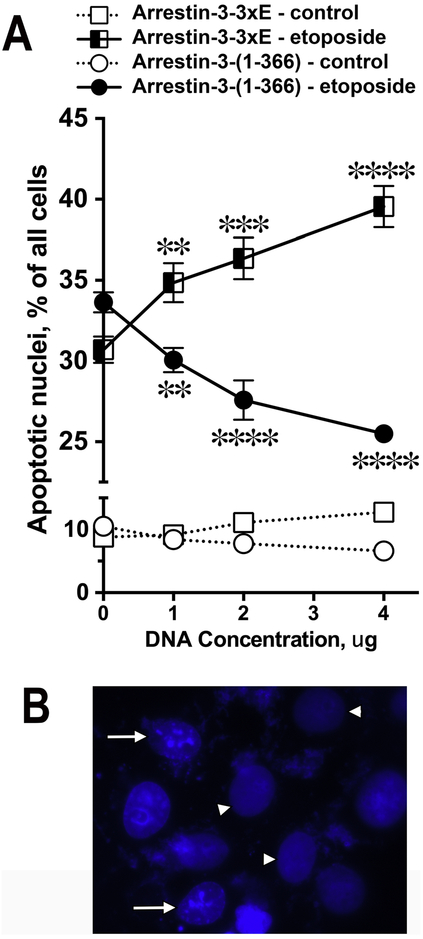
We previously found that arrestin-2 cleavage by caspases generates 1–380 fragment that facilitates apoptotic cell death [16]. JNK family kinases, which arrestin-3-(1–366) binds (Figs. 5,6), were implicated in apoptosis [45]. Therefore, next we tested the effect of arrestin-3-(1–366) on cell death and survival. In these experiments we used COS7 cells that were transfected with different arrestin-3 constructs, then treated with or without etoposide. The appearance of the nuclei was analyzed using blue fluorescence after staining with Hoechst 33258. Nuclei with condensed chromatin or fragmented nuclei were categorized as apoptotic. To obtain reliable data, in each of the three independent experiments approximately 400 cells were scored per data point. We found that caspase cleavage-resistant arrestin-3-(3xE) facilitated cell death, in contrast to the arrestin-3-(1–366) fragment, which reduced the fraction of apoptotic nuclei upon etoposide treatment (Fig. 7). Thus, caspase-generated arrestin-3-(1–366), that does not promote JNK activation (Fig. 5), demonstrated a cytoprotective effect.
Fig. 7. Effect of arrestin-3 and cleavage fragments on apoptosis. A.
COS7 cells were transfected with indicated arrestin-3 constructs. After etoposide treatment (40 μM, 12 h) cells expressing increasing amounts of caspase-resistant arrestin-3-(3xE) showed progressive increase in apoptosis. In contrast, cells with increasing expression of the cleavage product arrestin-3-(1–366) showed progressive decrease in apoptosis. Approximately 400 cells were scored per data point. Mean ± SEM of the percentage of apoptotic nuclei in three independent experiments is shown. **, p<0.01, ***, p<0.001, ****, p<0.0001 as compared to cells transfected with control empty pcDNA3 vector (0). B. DAPI-stained nuclei of etoposide-treated COS7 cells. White arrowheads indicate normal nuclei; white arrows indicated apoptotic nuclei with condensed chromatin.
4. Discussion
The two non-visual arrestins (arrestin-2 and arrestin-3) are key players in homologous desensitization and internalization of GPCRs and the activation of downstream effectors [5, 15, 46, 47]. These arrestins have very high sequence identity (~78%) [48, 49], but their subcellular localization is different [50]. Arrestin-2 is found in the cytosol and in the nucleus, whereas arrestin-3 is only present in the cytosol [37]. The difference appears to be associated with the presence of a classical leucine-rich NES near the C-terminus of arrestin-3, as the engineering of similar NES in the C-terminus of arrestin-2 changes its subcellular localization to purely cytoplasmic [37, 38]. Here we demonstrate the cleavage of arrestin-3 by caspases during apoptosis (Figs. 1,2). Arrestin-3 is specifically cleaved at Asp366, Asp405 and Asp406 in vitro and in different cell types. While the caspase-resistant arrestin-3 mutant 3xE retains receptor binding of parental WT arrestin-3, caspase-generated arrestin-3-(1–366) demonstrates lower binding (Fig.3). However, it shows the same preference for phosphorylated forms of rhodopsin without significant discrimination between active and inactive forms, characteristic for WT arrestin-3 and its 3xE mutant (Fig. 3). We found that the main cleavage product, arrestin-3-(1366) fragment, localized to the nucleus (Fig. 4). Interestingly, although arrestin-3-(1–366) fragment retains binding to different JNK isoforms (Figs. 5,6), it has lost the ability to promote JNK activation, likely due to impaired interactions with the upstream kinases MKK4, MKK7, and ASK1 (Fig. 5). This resembles the effect of R307A mutation in arrestin-2, which does not affect its interaction with ERK1/2, but reduces the binding of upstream activator cRaf1, thereby blocking arrestin-dependent ERK1/2 activation [51], as well as the effect of mutations that preclude MEK1 binding to arrestin-2, which also preclude ERK1/2 activation [52]. However, it should be noted that in general the binding of any arrestin, including arrestin-3 mutants, to JNKs and their upstream activators does not correlate with the ability to facilitate JNK activation, as our previous studies showed [43, 44]. Several labs found that another non-visual subtype, arrestin-2, does not facilitate JNK activation [39, 40, 43], even though it binds JNK3 [37], as well as upstream kinases [43]. An extreme example of this phenomenon is arrestin-3-KNC mutant, which binds ASK1 and MKK4/7 essentially like WT arrestin-3, binds JNK3 even better than WT arrestin-3, yet fails to promote JNK3 activation in cells [44]. Nonetheless, reduced binding of caspase-generated arrestin-3-(1–366) to ASK1 and MKK4/7 provides a facile explanation for its functional deficiency. Furthermore, persistent localization of arrestin-3-(1–366) fragment in the nucleus might also impede its ability to recruit the cytosolic components of the JNK pathway and promote JNK activation. It is tempting to speculate that normal binding of JNK family kinases combined with inability to facilitate their activation would make caspase-generated 1–366 fragment of arrestin-3 act in a dominant negative manner suppressing the JNK activation and acting as a cytoprotective agent, in contrast to caspase-resistant full-length arrestin-3-(3xE), which facilitates cell death (Fig. 7). However, due to its inappropriate nuclear localization, the fragment might act not as a dominant negative agent but simply as inactive arrestin with regard to the JNK activation. This would also result in cytoprotection simply due to reduced contribution of the JNK activity to the apoptotic process. Thus, these two arrestin-3-derived constructs, 3xE and 1–366, can be used as molecular tools to suppress or promote cell survival, respectively.
Mammals express hundreds of different GPCR subtypes [53–55], but only two non-visual arrestins [4, 27, 47]. Reported functional differences between arrestin-2 and arrestin-3 are relatively minor: higher affinity of arrestin-3 for some receptors [19, 50] and clathrin [56] were reported. Both non-visual arrestins are expressed in all vertebrates [4, 27], suggesting that they likely have distinct functions, but their specific roles are far from clear. Arrestin-2 and arrestin-3 were shown to differentially regulate locomotor responses and sensitization to amphetamine in mice in vivo [57]. So far, the most dramatic mechanistic difference documented was the ability of arrestin-3, but not arrestin-2, to facilitate JNK activation in cells [21, 22, 31, 39–43]. Here we report yet another striking difference: both arrestin-2 [16, 17] and arrestin-3 (Figs. 1,2) are cleaved by caspases in apoptotic cells, but the functions of the resulting fragments are opposite. Arrestin-2-(1–380) facilitates cell death by assisting caspase-cleaved tBid in the release of cytochrome c from mitochondria [16], which appears to be the point of no return in vertebrate apoptosis [58]. In contrast, arrestin-3-(1–366) is cytoprotective, suppressing apoptosis (Fig. 7). As could be expected, caspase-resistant mutant of arrestin-2 prevents cell death [16], whereas caspase-resistant arrestin-3 mutant facilitates it (Fig. 7). Interestingly, arrestin-2 greatly outnumbers arrestin-3 in most cell types, and its expression dramatically increases in the process of neuronal maturation [59, 60]. The biological significance of the prevalence of pro-survival arrestin-2, which can be converted into pro-apoptotic fragment by caspases, over pro-apoptotic arrestin-3, which caspases convert into a cytoprotective fragment, remains to be elucidated. Nonetheless, pro- and anti-apoptotic arrestin-derived molecular tools can be used to facilitate cell death and survival, respectively. This might have therapeutic potential in cancer, characterized by excessive cell proliferation, or in neurodegenerative disorders, where excessive cell death takes place.
5. Conclusions
Arrestin-3 cleavage by caspases generates (1–366) fragment that, in contrast to full-length arrestin-3, no longer facilitates JNK activation. The expression of caspase-resistant arrestin-3(D366/405/406E) facilitates apoptosis, whereas the expression of arrestin-3-(1–366) fragment reduces it. Thus, arrestin-3 cleavage by caspases suppresses apoptosis, in sharp contrast to the cleavage of highly homologous arrestin-2, which appears to play a role in a positive feedback.
Highlights Kook et al.
Arrestin-3 is cleaved by caspases in apoptotic cells
The main product of caspase cleavage of arrestin-3, (1–366) fragment, localizes to the nucleus
Arrestin-3-(1–366) has lost the ability to facilitate JNK activation
Arrestin-3-(1–366) suppresses cell death, whereas caspase-resistant arrestin-3 mutant enhances it
Arrestin-3-(1–366) serves as a negative feedback in apoptosis
Acknowledgments
The authors are grateful to Drs. S.S. Zinkel, R.J. Lefkowitz, and L.A. Donoso for COS7 cells, arrestin knockout MEFs, and F4C1 antibody, respectively, and VUMC Cell Imaging Core (S10 RR015682). Supported in part by NIH grants NS045117 and NS065868 (EVG), GM077561, GM109955 (these grants were merged into R35 GM122491), and EY011500 (VVG), Vanderbilt University Discovery Grant 1040659012 (VVG), Cornelius Vanderbilt Chair (VVG), and NARSAD Young Investigator Award (EVG).
Abbreviations:
- GPCR
G protein-coupled receptor
- KO
knockout
- WT
wild type
- Z-VAD
carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]- fluoromethylketone)
- NES
nuclear export signal
- CHX
cycloheximide
- TNFα
tumor necrosis factor α.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Footnotes
We use systematic names of arrestin proteins, where the number after the dash indicates the order of cloning: arrestin-1 (historic names S-antigen, 48 kDa protein, visual or rod arrestin), arrestin-2 (β-arrestin or β-arrestin1), arrestin-3 (β-arrestin2 or hTHY-ARRX), and arrestin-4 (cone or X-arrestin).
References
- [1].Vaux DL, Haecker G, Strasser A, Cell. 1994;76:777–779. [DOI] [PubMed] [Google Scholar]
- [2].Thornberry NA, Lazebnik Y, Science. 1998;281:1312–1316. [DOI] [PubMed] [Google Scholar]
- [3].Crawford ED, Wells JA, Annu Rev Biochem. 2011;80:1055–1087. [DOI] [PubMed] [Google Scholar]
- [4].Gurevich EV, Gurevich VV, Genome Biology. 2006;7:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Peterson YK, Luttrell LM, Pharmacol Rev. 2017;69:256–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Revankar CM, Vines CM, Cimino DF, Prossnitz ER, J Biol Chem. 2004;279:24578–24584. [DOI] [PubMed] [Google Scholar]
- [7].Wang Y, Li H, Song SP, Med Sci Monit. 2018;24:1724–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu YQ, Jia MQ, Xie ZH, Liu XF, Hui-Yang, Zheng XL, Yuan HQ, Bi JZ, Sci Rep. 2017;7:3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ren W, Wang T, He X, Zhang Q, Zhou J, Liu F, Gao F, Zhang Y, Liu Y, Oncol Rep. 2018;39:2711–2720. [DOI] [PubMed] [Google Scholar]
- [10].Sun YY, Zhao YX, Li XF, Huang C, Meng XM, Li J, Front Pharmacol. 2018;9:1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yin D, Yang X, Li H, Fan H, Zhang X, Feng Y, Stuart C, Hu D, Caudle Y, Xie N, Liu Z, LeSage G, J Biol Chem. 2016;291:605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].McCrink KA, Maning J, Vu A, Jafferjee M, Marrero C, Brill A, Bathgate-Siryk A, Dabul S, Koch WJ, Lymperopoulos A, Hypertension. 2017;70:972–981. [DOI] [PubMed] [Google Scholar]
- [13].Zhan Y, Xu C, Liu Z, Yang Y, Tan S, Yang Y, Jiang J, Liu H, Chen J, Wu B, Cell Death Dis. 2016;7:e2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen L, Ren Z, Wei X, Wang S, Wang Y, Cheng Y, Gao H, Liu H, Eur J Pharmacol. 2017;815:98–108. [DOI] [PubMed] [Google Scholar]
- [15].Gurevich VV, Gurevich EV, Curr Protoc Pharmacol. 2014;67:Unit 2.10.11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kook S, Zhan X, Cleghorn WM, Benovic JL, Gurevich VV, Gurevich EV, Cell Death Differ. 2014;21:172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kook S, Gurevich VV, Gurevich EV, Handb Exp Pharmacol. 2014;219:309–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Donoso LA, Gregerson DS, Smith L, Robertson S, Knospe V, Vrabec T, Kalsow CM, Curr Eye Res. 1990;9:343–355. [DOI] [PubMed] [Google Scholar]
- [19].Kohout TA, Lin FS, Perry SJ, Conner DA, Lefkowitz RJ, Proc Nat Acad Sci USA. 2001;98:1601–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Vishnivetskiy SA, Zhan X, Chen Q, Iverson TM, Gurevich VV, Curr Protoc Pharmacol. 2014;67:Unit 2.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhan X, Kaoud TS, Kook S, Dalby KN, Gurevich VV, J Biol Chem. 2013;288:28535–28547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhan X, Stoy H, Kaoud TS, Perry NA, Chen Q, Perez A, Els-Heindl S, Slagis JV, Iverson TM, Beck-Sickinger AG, Gurevich EV, Dalby KN, Gurevich VV, Sci Rep. 2016;6:21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gurevich VV, Benovic JL, J. Biol. Chem 1993;268:11628–11638. [PubMed] [Google Scholar]
- [24].Vishnivetskiy SA, Raman D, Wei J, Kennedy MJ, Hurley JB, Gurevich VV, J Biol Chem. 2007;282:32075–32083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gurevich VV, in: Kuo LC, Olsen DB, Carroll SS, (Eds.), Methods in Enzymology1996. pp. 382–397. [Google Scholar]
- [26].Gurevich VV, Dion SB, Onorato JJ, Ptasienski J, Kim CM, Sterne-Marr R, Hosey MM, Benovic JL, J Biol Chem. 1995;270:720–731. [DOI] [PubMed] [Google Scholar]
- [27].Indrischek H, Prohaska SJ, Gurevich VV, Gurevich EV, Stadler PF, BMC Evol Biol. 2017;17:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang L, Du F, Wang X, Cell. 2008;133:693–703. [DOI] [PubMed] [Google Scholar]
- [29].Zhan X, Gimenez LE, Gurevich VV, Spiller BW, J Mol Biol. 2011;406:467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Boatright KM, Salvesen GS, Curr Opin Cell Biol. 2003;15:725–731. [DOI] [PubMed] [Google Scholar]
- [31].Chen Q, Perry NA, Vishnivetskiy SA, Berndt S, Gilbert NC, Zhuo Y, Singh PK, Tholen J, Ohi MD, Gurevich EV, Brautigam CA, Klug KS, Gurevich VV, Iverson TM, Nat Commun. 2017;8:1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, Barty A, White TA, Yefanov O, Han GW, Xu Q, de Waal PW, Ke J, Tan MHE, Zhang C, Moeller A, West GM, Van Eps N, Caro LN, Vishnivetskiy SA, Lee RJ, Suino-Powell KM, Gu X, Pal K, Ma J, Zhi X, Boutet S, Williams GJ, Messerschmidt M, Gati C, Zatsepin NA, Wang D, James D, Basu S, Roy-Chowdhuty S, Conrad S, Coe J, Liu H, Lisova S, Kupitz C, Grotjohann I, Fromme R, Jiang Y, Tan M, Yang H, Li J, Wang M, Zheng Z, Li D, Zhao Y, Standfuss J, Diederichs K, Dong Y, Potter CS, Carragher B, Caffrey M, Jiang H, Chapman HN, Spence JCH, Fromme P, Weierstall U, Ernst OP, Katritch V, Gurevich VV, Griffin PR, Hubbell WL, Stevens RC, Cherezov V, Melcher K, Xu HE, Nature. 2015;523:561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhou XE, He Y, de Waal PW, Gao X, Kang Y, Van Eps N, Yin Y, Pal K, Goswami D, White TA, Barty A, Latorraca NR, Chapman HN, Hubbell WL, Dror RO, Stevens RC, Cherezov V, Gurevich VV, Griffin PR, Ernst OP, Melcher K, Xu HE, Cell. 2017;170:457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gurevich VV J Biol Chem. 1998;273:15501–15506. [DOI] [PubMed] [Google Scholar]
- [35].Gurevich VV, Pals-Rylaarsdam R, Benovic JL, Hosey MM, Onorato JJ, J Biol Chem. 1997;272:28849–28852. [DOI] [PubMed] [Google Scholar]
- [36].Gurevich VV, Benovic JL, J Biol Chem. 1992;267:21919–21923. [PubMed] [Google Scholar]
- [37].Song X, Raman D, Gurevich EV, Vishnivetskiy SA, Gurevich VV, J Biol Chem. 2006;281:21491–21499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Scott MG, Le Rouzic E, Périanin A, Pierotti V, Enslen H, Benichou S, Marullo S, Benmerah A, J Biol Chem. 2002;277:37693–37701. [DOI] [PubMed] [Google Scholar]
- [39].McDonald PH, Chow CW, Miller WE, Laporte SA, Field ME, Lin FT, Davis RJ, Lefkowitz RJ, Science. 2000;290:1574–1577. [DOI] [PubMed] [Google Scholar]
- [40].Song X, Coffa S, Fu H, Gurevich VV, J Biol Chem. 2009;284:685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kook S, Zhan X, Kaoud TS, Dalby KN, Gurevich VV, Gurevich EV, J Biol Chem. 2013;288:37332–37342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhan X, Kaoud TS, Dalby KN, Gurevich VV, Biochemistry. 2011;50:10520–10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Seo J, Tsakem EL, Breitman M, Gurevich VV, J Biol Chem. 2011;286:27894–27901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Breitman M, Kook S, Gimenez LE, Lizama BN, Palazzo MC, Gurevich EV, Gurevich VV, J Biol Chem. 2012;287:19653–19664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yang DD, Kuan CY, Whitmarsh AJ, Rincón M, Zheng TS, Davis RJ, Rakic P, Flavell RA, Nature. 1997;389:865–870. [DOI] [PubMed] [Google Scholar]
- [46].Carman CV, Benovic JL, Curr Opin Neurobiol. 1998;8:335–344. [DOI] [PubMed] [Google Scholar]
- [47].Gurevich VV, Gurevich EV, Pharm Ther. 2006;110:465–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Attramadal H, Arriza JL, Aoki C, Dawson TM, Codina J, Kwatra MM, Snyder SH, Caron MG, Lefkowitz RJ, J Biol Chem. 1992;267:17882–17890. [PubMed] [Google Scholar]
- [49].Sterne-Marr R, Gurevich VV, Goldsmith P, Bodine RC, Sanders C, Donoso LA, Benovic JL, J Biol Chem. 1993;268:15640–15648. [PubMed] [Google Scholar]
- [50].Oakley RH, Laporte SA, Holt JA, Caron MG, Barak LS, J Biol Chem. 2000;275:1720117210. [DOI] [PubMed] [Google Scholar]
- [51].Coffa S, Breitman M, Spiller BW, Gurevich VV, Biochemistry. 2011;50:6951–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Meng D, Lynch MJ, Huston E, Beyermann M, Eichhorst J, Adams DR, Klusmann E, Houslay MD, Baillie GS, J Biol Chem. 2009;284:11425–11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Stoy H, Gurevich VV, Genes Dis. 2015;2:108–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Pin JP, Galvez T, Prezeau L, Pharmacol Ther. 2003;98:325–354. [DOI] [PubMed] [Google Scholar]
- [55].Kristiansen K, Pharmacol Ther. 2004;103:21–80. [DOI] [PubMed] [Google Scholar]
- [56].Goodman OB Jr., Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL, Nature. 1996;383:447–450. [DOI] [PubMed] [Google Scholar]
- [57].Zurkovsky L, Sedaghat K, Ahmed MR, Gurevich VV, Gurevich EV, Neuropharmacology. 2017;121:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Danial NN, Korsmeyer SJ, Cell. 2004;116:205–219. [DOI] [PubMed] [Google Scholar]
- [59].Gurevich EV, Benovic JL, Gurevich VV, Neuroscience. 2002;109:421–436. [DOI] [PubMed] [Google Scholar]
- [60].Gurevich EV, Benovic JL, Gurevich VV, J Neurochem. 2004;91:1404–1416. [DOI] [PubMed] [Google Scholar]