Abstract
Tryptophanyl tRNA synthetase (WRS) is an essential enzyme as it catalyzes the ligation of tryptophan to its cognate tRNA during translation. Interestingly, mammalian WRS has evolved to acquire domains or motifs for novel functions beyond protein synthesis; WRS can also further expand its functions via alternative splicing and proteolytic cleavage. WRS is localized not only to the nucleus but also to the extracellular space, playing a key role in innate immunity, angiogenesis, and IFN-γ signaling. In addition, the expression of WRS varies significantly in different tissues and pathological states, implying that it plays unique roles in physiological homeostasis and immune defense. This review addresses the current knowledge regarding the evolution, structural features, and context-dependent functions of WRS, particularly focusing on its roles in immune regulation.
Subject terms: Innate immunity, Infection
Protein synthesis: An unexpected link with immune regulation
Targeting tryptophanyl tRNA synthetase (WRS), an evolutionarily conserved enzyme involved in protein synthesis, could be an effective strategy for modulating the immune system. In addition to helping translate mRNA into amino acid sequences in cytoplasm, human WRS can be secreted and activate immune responses against invading pathogens. Mirim Jin at Gachon University, Incheon, South Korea, reviews recent studies on the structure, expression pattern and functions of WRS other than protein synthesis. High levels of WRS protein have been found in patients with sepsis and autoimmune diseases suggesting that inhibiting WRS could be a potential therapeutic approach for treating these conditions. Further research into WRS will shed light not only on how it regulates the immune system, but also on how it exerts other reported effects on blood vessel formation and cell migration.
Introduction
Aminoacyl-tRNA synthetases (ARSs) are essential housekeeping enzymes that participate in the translation of mRNAs1. ARSs are thought to have emerged at the time of the last universal common ancestor and are distributed across all taxa2. The common aminoacylation reaction, which attaches an amino acid to its cognate tRNA, is conserved and proceeds in two steps. First, an amino acid and adenosine triphosphate (ATP), as an energy source, bind the active site of the enzyme and form an aminoacyl adenylate intermediate (amino acid-AMP); second, the adenosine monophosphate (AMP) is displaced by its cognate tRNA, resulting in the covalent linkage of the specific amino acid to one of the ribose 3′-OH moieties of a set of tRNA isoacceptors3–7. The 20 ARSs (1 for each of the 20 amino acids) are naturally categorized into two classes (I and II) based on the structure of the ancestral catalytic core, chemical properties, and consensus sequences8. Most Class I ARSs (except the Ic subclass) are monomeric, while Class II ARSs are multimeric9. Each class can be further divided into subclasses based on their unique organization of conserved structural motifs, anticodon-binding domain characteristics, and mechanical properties. The catalytic domain of all Class I ARSs has a Rossmann Fold (RF) containing the dinucleotide-binding domain; this domain carries out the aminoacylation reaction and is located at or near the N-terminus. Classically, this domain features a five-stranded parallel β-sheet connected by α-helices. Three subclasses within Class I are designated as follows: Ia, for hydrophobic amino acids (Ile, Leu, and Val), sulfur-containing amino acids (Met and Cys), and Arg; Ib, for charged amino acids (Glu and Lys) and Gln; and Ic, for aromatic amino acids, including tyrosine and tryptophan. Class II is characterized by a seven-stranded β-sheet with flanking α-helices, and it is also divided into subclasses: Class IIa, which recognizes groups with chemically similar side chains, including aliphatic (Ala and Pro) and polar (Ser, Thr, Pro, and His) amino acids and Gly; class IIb, which recognizes the charged side chains of Asp and Lys and Asn; and class IIc, which recognizes the aromatic amino acid Phe4,9,10.
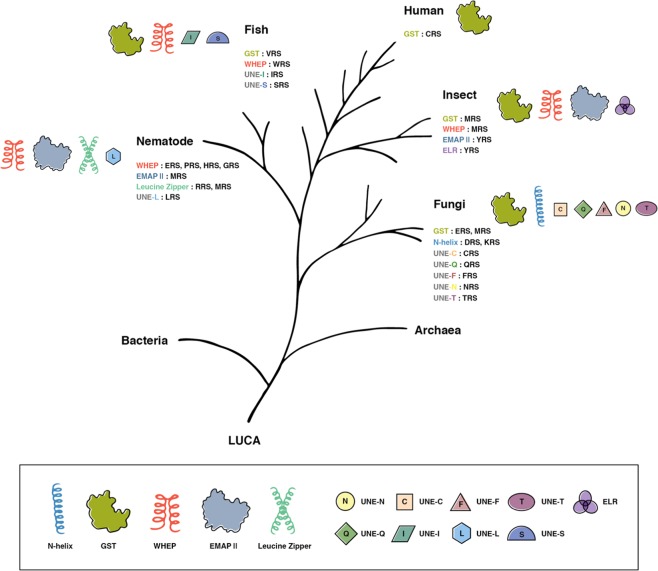
Interestingly, if the evolutionary tree is followed from lower to higher eukaryotes, ARSs can be seen to have progressively and irreversibly added new domains or motifs that have no apparent connection with aminoacylation11. Five domains have been found, including the N-terminal amphiphilic helix (N-helix), the glutathione S-transferase (GST)-like domain, a helix-turn-helix motif referred to as a WHEP domain (the name is derived from four of the five WHEP containing proteins: WRS, histidyl (H) tRNA synthetase, glutamyl-prolyl (EP)-tRNA synthetase, and methionyl tRNA synthetase), an endothelial monocyte-activating polypeptide II (EMAPII) domain, and a leucine zipper domain4,11–15; these domains are shared by more than one ARS. By contrast, eight unique sequence motifs, referred to as UNEs, are specific to only one ARS. The UNEs from cysteinyl tRNA synthetase (CRS), glutaminyl tRNA synthetase (QRS), phenylalanine tRNA synthetase (PRS), aspartidyl tRNA synthetase (DRS), lysyl tRNA synthetase (KRS), asparaginyl tRNA synthetase (NRS), threonyl tRNA synthetase (TRS), and leucyl tRNA synthetase (LRS) are not similar to any of the sequences of bacterial or archaeal tRNA synthetases or to other UNEs (Fig. 1)11,16. The occurrence of additional domains or motifs correlates with the emergence of new and more sophisticated species with increasing biological complexity at a correct and timely moment that are capable of performing the novel functions required by developing systems as they evolve12,17,18. Furthermore, the structural metamorphosis generated by alternative splicing, proteolysis, and post-translational modification seem to give more diverse functions to ARSs19–22. On the other hand, while prokaryotic ARSs exist independently within cells, eukaryotic ARSs have obtained the ability to form a multi-tRNA synthetase complex (MSC)23,24. In humans, nine different ARSs and three non-synthetase factors, p43, p38, and p18, are assembled into an MSC, which operates as a functional depot for alternative activities of its members17,25,26. Furthermore, most ARSs translocate from the cytoplasm to the nucleus or extracellular space in response to specific stimuli and can participate in various biological processes. Expansions in ARS functions are associated with numerous biological activities, including transcription, translation, cell death and survival, immunity to microbes, chemotaxis and inflammation, angiogenesis, IFN-γ and p53 signaling, mTOR signaling, and tumorigenesis (Table 1)27–31.
Fig. 1. Newly acquired domains and motifs in ARSs during the evolutionary process.
During evolution, aminoacyl-tRNA synthetase have acquired new domains, allowing for an increase in the complexity of organisms in a particular phylogenetic group. Importantly, with the exception of MRS, these domains or motifs have been irreversibly retained by their respective tRNA synthetase until the evolution of humans. As a result, each aminoacyl-tRNA synthetase in humans has all of the newly acquired domains. Each of these domains imparts specific new functions that are more complex than simple aminoacylation, which was the original function of these enzymes. The model species used are: Homo sapiens, Danio rerio, Drosophila melanogaster, Caenorhabditis elegans, and Saccharomyces cerevisiae. LUCA last universal common ancestor, N-helix N-terminal amphiphilic helix, GST glutathione S-transferase (GST)-like domain, EMAPII endothelial monocyte-activating polypeptide II
Table 1.
Non-canonical functions of the new domains and motifs in selective aminoacyl-tRNA synthetases
| Domain | ARS | Acquired function during evolution | References |
|---|---|---|---|
| UNE-S | SRS | Facilitates the translocation of SRS from the cytoplasm into the nucleus to regulate VEGF expression | 16, 86 |
| UNE-L | LRS | Important for stabilizing RRS and LRS in the MSC | 87 |
| GST | ERS or EPRS | Included as a component of the MSC | 26 |
| MRS | Included as a component of the MSC | 24, 26 | |
| VRS | Complexes with eEF1H during translation | 21, 88, 89 | |
| CRS | Complexes with eEF1γ during translation | 90, 91 | |
| EMAP II | YRS | Regulates the cytokine activity of YRS | 92 |
| Leucine zipper | RRS | Interacts with the MSC | 24, 93 |
| N-helix | YRS | Increases the affinity of the synthetase for its tRNA in translation | 94, 95 |
| DRS | Enhances tRNA-binding affinity | 96– 98 | |
| WHEP | EPRS | Suppresses inflammatory gene expression | 13, 99 |
| HRS | Activates chemokine receptors on T-lymphocytes and immature dendritic cells | 100 | |
| MRS | Translocates to the nucleolus in response to growth factors and enhances rRNA synthesis during transcription | 101 | |
| GRS | Regulates catalytic efficiency, thermal stability, and structural flexibility | 102 | |
| WRS | Acts as an endogenous ligand for the TLR4/MD2 complex | 20 | |
| ELR | YRS | Critical for the function of IL-8-like cytokines | 103 |
N-helix N-terminal amphiphilic helix, GST glutathione S-transferase (GST)-like domain, EMAPII endothelial monocyte-activating polypeptide II
The diverse structural metamorphoses among species, and/or within a species, have evolved by developing differential mechanisms for performing certain functions in immunity. There are a few examples of chemotaxis and angiogenesis: (i) UNE, a N-terminal 80 amino acid domain in the nematode Brugia malayi asparaginyl tRNA synthetase (bmNRS), which creates an IL-8 like fold and interacts with the IL-8 receptor, leading to chemotaxis and proangiogenic effects;32,33 (ii) an EMAP II-like carboxy-terminal domain and the mini-YRS from human tyrosyl tRNA synthetase (hYRS), which are produced following cleavage by leukocyte elastase and show chemotactic activity. In particular, the mini-YRS induces chemotaxis by binding to the IL-8 type A receptor;34 (iii) human WRS (hWRS), which is secreted from monocytes in which the WHEP domain (WRS) interacts with Toll-like receptors (TLR) to stimulate chemokine secretion, and after removal of WHEP, hWRS can exert an angiostatic effect via vascular-endothelial (VE)-cadherin20,31,35. Another example is the unique role of the WHEP domain that is associated with the antiviral effect of glutamyl-prolyl-tRNA synthetase (EPRS), which is composed of glutamyl tRNA synthetase (ERS) and prolyl tRNA synthetase (PRS) coupled via a linker containing three WHEP domains. This domain is found on the exterior of the MSC in the cytoplasm. Following viral infection, serine 990, located in the WHEP linker, is phosphorylated to release EPRS from the MSC, after which the amino-terminal domain of EPRS (1–196 aa), which contains a GST-like domain (1–168 aa) and the linker region L1 (168–196 aa), interacts with poly(rC)-binding protein 2 (PCBP2). Consequently, interferon-β production is increased due to the suppression of PCBP2-mediated ubiquitination and degradation of mitochondrial antiviral-signaling protein (MAVS), ultimately leading to the inhibition of viral replication36. By acquiring additional, differentiated functions with discrete mechanisms that can differ at specific locations, ARSs are key regulators of physiological homeostasis18.
This review focuses on WRS, one of the most extensively studied ARSs. Information regarding its evolution, structural features, and context-dependent biological functions, particularly in immunity, reveals its significant roles in immune regulation and its therapeutic potential.
Architecture for the catalytic reaction
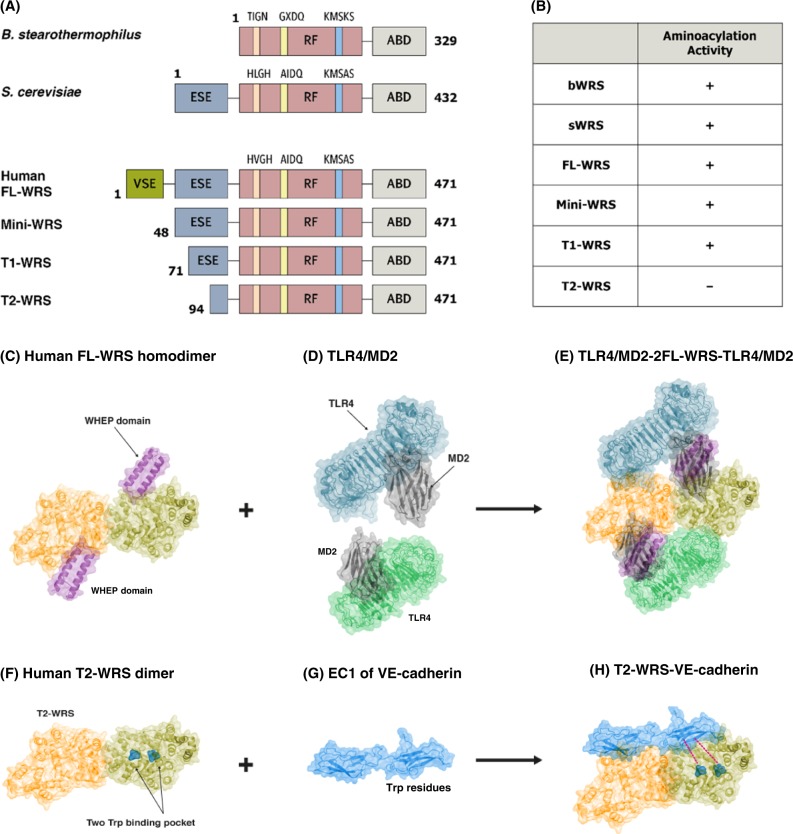
Specific recognition by tryptophanyl tRNA synthetase (WRS) of its substrates, Trp and tRNA, is critical for maintaining fidelity in protein synthesis. WRS belongs to the Class Ic ARS family, containing an RF domain with two highly conserved signature sequences, namely, KMSKS (Lys-Met-Ser-Lys-Ser) and HIGH (His-Ile-Gly-His); the former contributes to amino acid activation and the latter stabilizes both ATP during amino acid activation and the 3′ end of the tRNA for amino acid transfer2,37,38. The C-terminal alpha helical domain is the binding site for the tRNA anticodon. While the WRS from Bacillus stearothermophilus (bWRS) requires two domains for the catalytic reaction, eukaryotic WRSs, such as those from yeast and archaea, have acquired an extra N-terminal domain, referred to as a eukaryote-specific extension (ESE), of 100 amino acid residues2,38. Furthermore, in vertebrates, including humans, WRS has an N-terminal extension of 154 amino acids (N154) that is composed of the ESE and an additional vertebrate-specific extension (VSE), constituting the full-length (FL)-human WRS comprising 471 amino acids31,39,40 (Fig. 2a). Although the reaction catalyzed by WRS occurs through a similar “induced-fit mechanism” and involves a profound reorganization of the RF domain through conformational changes, human WRS uses a more complex catalytic reaction than bWRS41. For example, in the aminoacylation reaction, homo-dimeric WRS exhibits “half-site” activity, meaning that only one monomer of the homo-dimeric WRS operates at a time. A structural analysis of the WRS-tryptophan-ATP complex has revealed that one WRS monomer, in the form of a semi-closed KMSAS loop, binds to tryptophan, whereas the other monomer, in the form of a closed KMSAS loop, binds to both tryptophan and ATP37. Recognition of Trp induces a conformational change in the AIDQ motif of human WRS to generate a deep pocket for Trp binding and the activation and coupled movement of the N-terminal extension and C-terminal domain, leading to ATP binding of the KMSAS loop in a closed conformation, thereby securing the position of ATP for catalysis and coupling of the C-terminal tRNA-binding domain for Trp transfer4,37,38,42.
Fig. 2. Various structures of WRS and their interactions with specific receptors.
a Schematic representation of domains in WRS from prokaryotes (B. stearothermophilus), lower eukaryotes (S. cerevisiae), and higher organisms (H. sapiens). The Rossmann Fold (RF) catalytic domain and anticodon-binding domain (ABD) are well conserved in all WRSs. In the RF domain, the three characteristic types of motif have slightly different sequences among species. The eukaryotic-specific extension (ESE) is common in eukaryotic WRSs. Human full-length (FL)-WRS also has the vertebrate-specific extension (VSE) at the N-terminal appended site. Alternative splicing can produce a mini-WRS where a portion of the VSE is truncated. After being secreted into the extracellular space, the T1 and T2-WRSs are generated by proteolytic cleavage, which removes the ESE, including the entire VSE sequence. b The aminoacylation activity of the various forms of WRSs in different species. All the WRSs, with the exception of T2-WRS, are capable of aminoacylation. c–h Representative model of the structural arrangement of WRS and their specific interactions with receptors. FL-WRS (c), dimeric human FL-WRS (PDB 1R6T) binding to two TLR4/MD2 heterodimers (PDB 3FXI) through the WHEP domain and the N154 terminus in trans (d, e), T2-WRS (PDB 1O5T) with truncated WHEP entirely cuts off the N-terminus (f), interaction of T2-WRS with the EC1 domain in VE-cadherin (PDB 3PPE). Trp residues present in the EC1 domain bind to the Trp binding pocket present in the RF domain of WRS (g, h)
Structural metamorphosis for non-catalytic reactions
Undoubtedly, the N-terminal extension domain of human WRS was adopted to undertake non-canonical functions. The addition of the WHEP domain, WRSs occurred at the chordate stage and have been preserved ever since (Fig. 1). Recently, it has been proposed that a dimeric FL-WRS (Fig. 2c) can crosslink with two toll-like receptor 4-myeloid differentiation factor 2 (MD2) (Fig. 2d) heterodimers via N154, where WHEP (from 8–64), located in the N154 regions, is pivotal. WHEP inserts between the TLR4 and MD2 of one heterodimer, with the terminal domain of the N154 region binding the other TLR4 in trans, leading to TLR4-MD2 dimerization and macrophage activation (Fig. 2e). By contrast, mini-WRS, which lack parts of the WHEP domain, seldom induces functional dimerization11,20. Rather, mini-WRS, a naturally occurring alternative splice variant of WRS that lacks the N-terminal 47 amino acids, and T1- and T2-WRS, which are proteolytic products of secreted FL-WRS and lack significant parts of the WHEP domain, are composed of 71–471 and 94–471 amino acids, respectively, and have anti-angiogenic effects11,43,44. For example, after proteolytic cleavage, T2-WRS (Fig. 2f) binds VE-cadherin, an endothelial cell adherens junction molecule. VE-cadherin has two conserved tryptophan residues at positions 2 and 4 of the N-terminal extracellular domain (EC)-1, which is a key determinant for the dimerization of VE-cadherin in preformed blood vessels. In newly generated blood vessels, EC1 in the VE-cadherin monomer (Fig. 2g) is exposed owing to endothelial cell budding. Similar to other ARSs, human WRS has developed a tryptophan-binding pocket as a specific active site27,39,43,45,46. After proteolytic cleavage of the N-terminal extension covering the binding pocket, the tryptophan-binding sites are exposed, and the two proteins bind via the tryptophan residues in EC1 (Fig. 2h)47. Therefore, it is reasonable that human FL-WRS, with a completely preserved N-terminal extension, does not exert angiostatic effects48,49. N154 in human WRS has probably evolved to achieve a non-catalytic multi-function while simultaneously contributing to catalytic activity based on the fact that T2-WRS cannot perform aminoacylation because of the lack of the N-terminal extension domain participating in the reaction, which mini-WRS can perform (Fig. 2b)44.
WRS signaling and clinical relevance
Extracellular signaling of secreted WRS
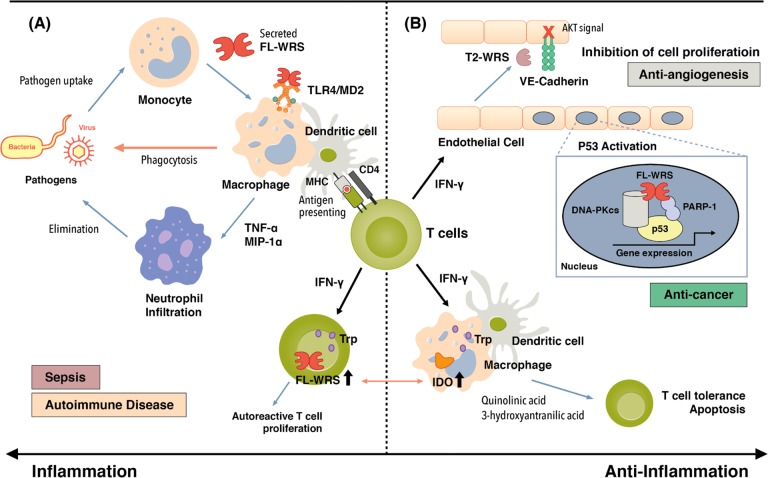
WRS is secreted into the extracellular space in response to certain stimuli. For example, upon pathogenic infection, but prior to tumor necrosis factor-α (TNF-α) production, WRS is rapidly secreted from monocytes without de novo synthesis, although the mechanism of secretion is not completely known. The secreted FL-WRS, but not mini-WRS, interacts with TLR2 and/or TLR4-MD on macrophages, leading to the activation of innate immune responses, in which TNF-α and chemokine production, neutrophil infiltration, and increased phagocytosis are prominent. These responses eliminate invading pathogens in the very early phase of infection, implying that there is a crucial role for FL-WRS as an endogenous ligand of human TLR2/4 in countering infections and immune regulation (Fig. 3a)20,50. In support of this hypothesis, compared with healthy subjects, high levels of WRS are consistently detected in the serum of critically ill patients with sepsis, a potentially lethal complication of a severe infection. Recent preliminary data have shown that there is a significant positive correlation among WRS levels and sepsis severity, the sepsis-associated organ failure assessment (SOFA) score, and deaths (unpublished data); the data therefore suggest a pivotal role for WRS in sepsis pathophysiology.
Fig. 3. Roles of WRS in “Yin-Yang” immune regulation and its relation to immune diseases.
a Upon infection, FL-WRS is rapidly secreted from monocytes to activate macrophages via TLR4/MD2, which induces innate immune responses, including chemokine production, neutrophil infiltration, and increased phagocytosis, eventually leading to pathogen elimination. High levels of WRS probably provoke an acute hyper-inflammation in sepsis. Antigen presentation to T cells by dendritic cells induces IFN-γ production, leading to the increased expression of WRS. Overexpression of WRS in T cells is able to reserve Trp, which is essential for cell activation and proliferation and may be involved in the development of autoreactive T cells in autoimmune diseases. b IFN-γ stimulation not only increases WRS secretion but also induces the translocation of WRS into the nucleus. Secreted WRS is proteolytically processed to form T2-WRS, which binds to VE-cadherin and inhibits angiogenesis. Nuclear FL-WRS enhances anti-angiogenesis by regulating cell growth through the phosphorylation of p53, further implying a role for WRS as an anti-cancer agent. Furthermore, an imbalanced activation of indoleamine 2,3-dioxygenase (IDO) relative to WRS in antigen-presenting cells (APCs), including macrophages and dendritic cells, induces an immunosuppressive state, inhibiting the proliferation of immune cells, including T cells, and possibly causing these cells to become tolerant or apoptotic
By contrast, when endothelial cells are stimulated with IFN-γ, an angiostatic cytokine, the expressed WRS forms a tertiary complex with annexin II and S100A10, which regulate exocytosis. A small fraction of the complexed WRS can become dissociated from annexin II-S100A10 and be secreted from cells51. This secretion does not seem to proceed through the classical secretion pathways involving the Golgi, as it is not blocked by treatment with brefeldin A or A23187, inhibitors of ER-Golgi-transport and calcium-dependent exocytosis, respectively51,52. After secretion, FL-WRS is cleaved by plasmin and/or elastases, which are both critical proteases involved in angiogenesis53. T2-WRS potently inhibits new angiogenesis, acting via VE-cadherin, a critical player in angiogenesis and vascular permeability at the intracellular junction between endothelial cells31,35,39,48,54. This T2-WRS/VE-cadherin interaction prevents the activation of VEGFR and subsequent ERK-mediated signaling, which suppress endothelial migration and proliferation (Fig. 3b)48,55,56.
Intracellular signaling of WRS
Since WRS is a housekeeping enzyme, its overexpression in various cells and tissues under diverse physiological environments is a rather unexpected finding57–59. With respect to development and aging, WRS expression is increased both in the developing salivary grand of Drosophila and in aged human epidermis60. WRS mRNA levels have been shown to be increased for a specific time during the differentiation of human monocyte-derived macrophage and dendritic cells61,62. Furthermore, WRS is highly expressed not only in cells infected with human cytomegalovirus and hepatitis B virus but also in mouse intestines infected with Cholera vibrio63–65. However, the reasons for this WRS overexpression and the underlying mechanisms remain unclear.
Endothelial WRS mRNA expression can also be increased in response to IFN-γ, which is mediated by transcription factor binding to gene promoters, including the gamma-activated sequence and the interferon-sensitive response element in STAT-1 and IFN regulatory factor, respectively. WRS can translocate into the nucleus, using a potential nuclear localization sequence proximal to the C-terminus27. In the absence of nuclear WRS, DNA-PKcs (the catalytic subunit of DNA dependent protein kinase) is independently linked to poly [ADP-ribose] polymerase 1 (PARP-1), with Ku70/80 serving as a bridge between the two. Ku70/80 binding to both DNA-PKcs and PARP-1 orients the C-terminal domain of PARP-1 to allow for its phosphorylation by DNA-PKcs. In this state, PARP-1 cannot PARylate DNA-PKcs. However, nuclear WRS can displace Ku70/80 and bind to both DNA-PKcs and PARP-1. The WHEP domain of WRS bridges the C-terminal kinase domain of DNA-PKcs to the N-terminal domain of PARP-1, where WRS enables Trp-AMP, an intermediate amino-adenylate product, to occupy the active site. When Trp-AMP occupies the active site of WRS, the WHEP domain opens and is available for interacting with DNA-PKcs and PARP-1, thus stimulating the PARylation of DNA-PKcs by PARP-1. Finally, the PARylated DNA-PKcs then phosphorylates p53, leading to p53-driven anti-proliferative effects and senescence27. These findings imply that the functions of nuclear WRS may concur with those of extracellular WRS as an angiostatic cytokine, consistent with the well-known functional integration of IFN-γ signaling and p53 activation. This furthermore suggests roles for WRS in cell death or cancer (Fig. 3b)66.
Tryptophan metabolism is closely linked to IFN-γ-mediated immune regulation, which induces a dual effect on its metabolism. Firstly, there is an increase in the rate of degradation of Trp by the indoleamine 2,3-dioxygenase (IDO) pathway, where IDO is strongly increased in macrophages and dendritic cells. The resulting depletion of Trp levels represses immune cell activation and proliferation in the microenvironment67–73. In addition, the products of Trp catabolism (quinolinic acid and 3-hydroxyantranilic acid) can induce T cell apoptosis, leading to immune tolerance. Secondly, the accumulation of tryptophan into the Trp–tRNA complexes available for protein synthesis provides a protective mechanism for immune cells (Fig. 3b)74,75. Treatment of cytotoxic T lymphocyte antigen-4 (CTLA-4), a negative regulator of T cell activation, with human PBMCs induced concomitant increased expression of both IDO and WRS in CD4+ T cells, which prevents CD4+ T cell activation through an IDO-dependent mechanism. Moreover, CD8+ T cells only showed increased WRS expression, not IDO, and were able to maintain their activation status via the Flu antigen, which is unaffected by CTLA-4 pretreatment, rendering WRS a Trp reservoir for protein synthesis76,77. In addition, given that immune cells possess many tryptophan-rich proteins compared with proteins in general, such as the human major histocompatibility complex antigens, complement factor B, and β-2 microglobulin, which are known to be induced by IFN-γ and immunoglobulins, it is plausible that WRS and Trp cooperate to regulate the activities of various immune cells78,79. Clinically, along with changes in the ratio of serum kynurenine, a metabolite of the IDO pathway, to tryptophan, an imbalance in the expressions of IDO and WRS has been suggested to be associated with autoimmune disorders. Patients with Graves’ disease (GD) show increased kynurenine to Trp ratios and have increased IDO expression in B cells and dendritic cells (DCs), indicating immune suppression; however, GD-derived CD4+ T cells have increased WRS expression and their proliferation was not inhibited by IDO expression in DCs from GD-patients, suggesting the activation of autoreactive T cells80. Furthermore, in immune thrombocytopenia, decreased IDO expression and increased WRS expression in CD4+ and CD8+ cells have been proposed to enhance the survival of autoreactive T cells81. In rheumatoid arthritis patients, the increased expression of WRS mRNA may be a cause of CD3+ T cell activation (Fig. 3b)58.
Conclusion and future prospects
Beyond its role in protein synthesis, WRS has evolved to play a role in immune regulation. Following an infection, WRS, in combination with WHEP, is immediately secreted by monocytes, the first responder cells, leading to inflammatory reactions that eliminate invading pathogens20. Since FL-WRS serves as an endogenous ligand of human TLR2 and TLR4, it is easy to conclude that this protein plays a significant role as an immune activator in various immune responses, including innate immunity, the development of immune cells, and differentiation, as well as adaptive immunity72,74,78. Moreover, WRS has simultaneously acquired anti-inflammatory properties through diverse mechanisms such as alternative splicing and proteolytic cleavage39,45. WRS is the only ARS whose expression is induced by the IFN-γ produced by innate immune cells and T cells under various immunological contexts. WRS also plays a central regulatory role in IFN-γ-induced anti-angiogenesis and cell survival or death. Since inflammation increases protease activity, the hyper-secretion of FL-WRS results in increased levels of T2-WRS, which suppresses the spread of inflammation through its angiostatic activity and the activation of p53 in the nucleus. FL-WRS, containing the WHEP domain, can also induce a pro-apoptotic state. In addition, during Trp metabolism, WRS participates in the activation and/or inhibition of immune cells, including macrophages, dendritic cells, and T cells, by providing tryptophan-binding activity. By placing a device within the N-terminal extension that can perform two different mechanisms, either activating or inhibiting inflammation, WRS serves as a “yin-yang” modulator of inflammation (Fig. 3).
Sepsis is a syndrome with heterogeneous immunopathology, including acute hyper-inflammation and immunosuppression that cannot overcome nosocomial and opportunistic infections82. In sepsis patients who have high WRS levels in the early acute phase, excessive WRS-induced inflammation probably occurs, and antagonizing WRS may be a therapeutic strategy for sepsis; by contrast, in immunocompromised septic patients with monocyte dysfunction, injections of FL-WRS or N154 may serve as an appropriate treatment to boost weakened immunity.
Studies on the function of WRS, not only as a p53 modulator but also an angiostatic agent, suggest that WRS could be a therapeutic target for various diseases, including cancer. Current clinical data examining the expression of WRS in cancer tissues indicate that the role of WRS in tumor biology is not simple and seems to be context-dependent83. In several cancers, including gastric adenocarcinoma, colorectal and ovarian cancer, high levels of WRS expression are associated with a favorable prognosis, and WRS was recently included as a predictive biomarker for unnecessary adjuvant chemotherapy after surgery for resectable gastric cancer46,84,85. By contrast, in oral squamous cell carcinoma, high levels of WRS are positively correlated with tumor stage, invasion, and depth, and it has been suggested that secreted WRS may induce cancer cell migration57. Therefore, how altered WRS expression levels differentially affect the fate of cancer cells and how secreted WRS regulates the cancer microenvironment that contains various cells, including immune cells, are interesting questions. In order for WRS to be meaningful as a prognostic marker, or even as a therapeutic target, further in-depth research needs to be conducted. Until now, in patients with autoimmune diseases, the purpose of WRS overexpression in immune cells has been assumed to be as a Trp repository; however, this notion arises because only certain phenomena have been examined. Therefore, it is very important to examine the mechanisms underlying immune cell proliferation/activation and metabolism in detail.
Acknowledgements
I sincerely thank Profs S. Kim, J. Shin, and J.J. Choi, and Y.T. Kim for their valuable comments and support in preparing this review. This study was supported by the Global Frontier Project Grants (NRF2015M3A6A4065732) of Ministry of Science and ICT through the National Research Foundation, Republic of Korea.
Conflict of interest
The author declares that he has no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Xu ZJ, et al. Tryptophanyl-tRNA synthetase from Bacillus subtilis. Characterization and role of hydrophobicity in substrate recognition. J. Biol. Chem. 1989;264:4304–4311. [PubMed] [Google Scholar]
- 2.Yang XL, et al. Functional and crystal structure analysis of active site adaptations of a potent anti-angiogenic human tRNA synthetase. Structure. 2007;15:793–805. doi: 10.1016/j.str.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suga H, Hayashi G, Terasaka N. The RNA origin of transfer RNA aminoacylation and beyond. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011;366:2959–2964. doi: 10.1098/rstb.2011.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pang YL, Poruri K, Martinis SA. tRNA synthetase: tRNA aminoacylation and beyond. Wiley Interdiscip. Rev. RNA. 2014;5:461–480. doi: 10.1002/wrna.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paley EL. Chaperon-like activation of serum-inducible tryptophanyl-tRNA synthetase phosphorylation through refolding as a tool for analysis of clinical samples. Transl. Oncol. 2011;4:377–389. doi: 10.1593/tlo.11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams TL, Yin YW, Carter CW., Jr. Selective inhibition of bacterial tryptophanyl-tRNA synthetases by indolmycin is mechanism-based. J. Biol. Chem. 2016;291:255–265. doi: 10.1074/jbc.M115.690321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang P, Guo M. Evolutionary limitation and opportunities for developing tRNA synthetase inhibitors with 5-binding-mode classification. Life (Basel) 2015;5:1703–1725. doi: 10.3390/life5041703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 9.Ribas de Pouplana L, Schimmel P. Aminoacyl-tRNA synthetases: potential markers of genetic code development. Trends Biochem. Sci. 2001;26:591–596. doi: 10.1016/S0968-0004(01)01932-6. [DOI] [PubMed] [Google Scholar]
- 10.Ribas de Pouplana L, Schimmel P. Two classes of tRNA synthetases suggested by sterically compatible dockings on tRNA acceptor stem. Cell. 2001;104:191–193. doi: 10.1016/S0092-8674(01)00204-5. [DOI] [PubMed] [Google Scholar]
- 11.Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell Biol. 2010;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SW, Cho BH, Park SG, Kim S. Aminoacyl-tRNA synthetase complexes: beyond translation. J. Cell Sci. 2004;117:3725–3734. doi: 10.1242/jcs.01342. [DOI] [PubMed] [Google Scholar]
- 13.Rho SB, et al. A multifunctional repeated motif is present in human bifunctional tRNA synthetase. J. Biol. Chem. 1998;273:11267–11273. doi: 10.1074/jbc.273.18.11267. [DOI] [PubMed] [Google Scholar]
- 14.Kao J, et al. Endothelial monocyte-activating polypeptide. II. A novel tumor-derived polypeptide that activates host-response mechanisms. J. Biol. Chem. 1992;267:20239–20247. [PubMed] [Google Scholar]
- 15.Koonin EV, et al. Eukaryotic translation elongation factor 1 gamma contains a glutathione transferase domain—study of a diverse, ancient protein superfamily using motif search and structural modeling. Protein Sci. 1994;3:2045–2054. doi: 10.1002/pro.5560031117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X, et al. Unique domain appended to vertebrate tRNA synthetase is essential for vascular development. Nat. Commun. 2012;3:681. doi: 10.1038/ncomms1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo M, Schimmel P, Yang XL. Functional expansion of human tRNA synthetases achieved by structural inventions. FEBS Lett. 2010;584:434–442. doi: 10.1016/j.febslet.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smirnova EV, Lakunina VA, Tarassov I, Krasheninnikov IA, Kamenski PA. Noncanonical functions of aminoacyl-tRNA synthetases. Biochemistry (Mosc.) 2012;77:15–25. doi: 10.1134/S0006297912010026. [DOI] [PubMed] [Google Scholar]
- 19.Wei Z, et al. Alternative splicing creates two new architectures for human tyrosyl-tRNA synthetase. Nucleic Acids Res. 2016;44:1247–1255. doi: 10.1093/nar/gkw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn YH, et al. Secreted tryptophanyl-tRNA synthetase as a primary defence system against infection. Nat. Microbiol. 2016;2:16191. doi: 10.1038/nmicrobiol.2016.191. [DOI] [PubMed] [Google Scholar]
- 21.Nichols RC, Raben N, Boerkoel CF, Plotz PH. Human isoleucyl-tRNA synthetase: sequence of the cDNA, alternative mRNA splicing, and the characteristics of an unusually long C-terminal extension. Gene. 1995;155:299–304. doi: 10.1016/0378-1119(94)00634-5. [DOI] [PubMed] [Google Scholar]
- 22.Park MC, et al. Secreted human glycyl-tRNA synthetase implicated in defense against ERK-activated tumorigenesis. Proc. Natl. Acad. Sci. USA. 2012;109:E640–E647. doi: 10.1073/pnas.1200194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirande M, Cirakoglu B, Waller JP. Macromolecular complexes from sheep and rabbit containing seven aminoacyl-tRNA synthetases. III. Assignment of aminoacyl-tRNA synthetase activities to the polypeptide components of the complexes. J. Biol. Chem. 1982;257:11056–11063. [PubMed] [Google Scholar]
- 24.Quevillon S, Robinson JC, Berthonneau E, Siatecka M, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein-protein interactions and characterization of a core protein. J. Mol. Biol. 1999;285:183–195. doi: 10.1006/jmbi.1998.2316. [DOI] [PubMed] [Google Scholar]
- 25.Park SJ, Ahn HS, Kim JS, Lee C. Evaluation of multi-tRNA synthetase complex by multiple reaction monitoring mass spectrometry coupled with size exclusion chromatography. PLoS ONE. 2015;10:e0142253. doi: 10.1371/journal.pone.0142253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho HY, et al. Assembly of multi-tRNA synthetase complex via heterotetrameric glutathione transferase-homology domains. J. Biol. Chem. 2015;290:29313–29328. doi: 10.1074/jbc.M115.690867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sajish M, et al. Trp-tRNA synthetase bridges DNA-PKcs to PARP-1 to link IFN-gamma and p53 signaling. Nat. Chem. Biol. 2012;8:547–554. doi: 10.1038/nchembio.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo M, Schimmel P. Essential nontranslational functions of tRNA synthetases. Nat. Chem. Biol. 2013;9:145–153. doi: 10.1038/nchembio.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakasugi K. Human tryptophanyl-tRNA synthetase binds with heme to enhance its aminoacylation activity. Biochemistry. 2007;46:11291–11298. doi: 10.1021/bi7012068. [DOI] [PubMed] [Google Scholar]
- 30.Kisselev L, Frolova L, Haenni AL. Interferon inducibility of mammalian tryptophanyl-tRNA synthetase: new perspectives. Trends Biochem. Sci. 1993;18:263–267. doi: 10.1016/0968-0004(93)90178-P. [DOI] [PubMed] [Google Scholar]
- 31.Kise Y, et al. A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase. Nat. Struct. Mol. Biol. 2004;11:149–156. doi: 10.1038/nsmb722. [DOI] [PubMed] [Google Scholar]
- 32.Ramirez BL, et al. Brugia malayi asparaginyl-transfer RNA synthetase induces chemotaxis of human leukocytes and activates G-protein-coupled receptors CXCR1 and CXCR2. J. Infect. Dis. 2006;193:1164–1171. doi: 10.1086/501369. [DOI] [PubMed] [Google Scholar]
- 33.Kron MA, Wang C, Vodanovic-Jankovic S, Howard OM, Kuhn LA. Interleukin-8-like activity in a filarial asparaginyl-tRNA synthetase. Mol. Biochem. Parasitol. 2012;185:66–69. doi: 10.1016/j.molbiopara.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Wakasugi K, et al. Induction of angiogenesis by a fragment of human tyrosyl-tRNA synthetase. J. Biol. Chem. 2002;277:20124–20126. doi: 10.1074/jbc.C200126200. [DOI] [PubMed] [Google Scholar]
- 35.Tzima E, et al. VE-cadherin links tRNA synthetase cytokine to anti-angiogenic function. J. Biol. Chem. 2005;280:2405–2408. doi: 10.1074/jbc.C400431200. [DOI] [PubMed] [Google Scholar]
- 36.Lee EY, et al. Infection-specific phosphorylation of glutamyl-prolyl tRNA synthetase induces antiviral immunity. Nat. Immunol. 2016;17:1252–1262. doi: 10.1038/ni.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen N, et al. Catalytic mechanism of the tryptophan activation reaction revealed by crystal structures of human tryptophanyl-tRNA synthetase in different enzymatic states. Nucleic Acids Res. 2008;36:1288–1299. doi: 10.1093/nar/gkm1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou M, Dong X, Shen N, Zhong C, Ding J. Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design. Nucleic Acids Res. 2010;38:3399–3413. doi: 10.1093/nar/gkp1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wakasugi K, et al. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl. Acad. Sci. USA. 2002;99:173–177. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen N, Guo L, Yang B, Jin Y, Ding J. Structure of human tryptophanyl-tRNA synthetase in complex with tRNATrp reveals the molecular basis of tRNA recognition and specificity. Nucleic Acids Res. 2006;34:3246–3258. doi: 10.1093/nar/gkl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapustina M, Carter CW., Jr. Computational studies of tryptophanyl-tRNA synthetase: activation of ATP by induced-fit. J. Mol. Biol. 2006;362:1159–1180. doi: 10.1016/j.jmb.2006.06.078. [DOI] [PubMed] [Google Scholar]
- 42.Ilyin VA, et al. 2.9 A crystal structure of ligand-free tryptophanyl-tRNA synthetase: domain movements fragment the adenine nucleotide binding site. Protein Sci. 2000;9:218–231. doi: 10.1110/ps.9.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, X. et al. An alternative conformation of human TrpRS suggests a role of zinc in activating non-enzymatic function. RNA Biol15, 649–658 (2018). [DOI] [PMC free article] [PubMed]
- 44.Otani A, et al. A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc. Natl. Acad. Sci. USA. 2002;99:178–183. doi: 10.1073/pnas.012601899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang XL, Schimmel P, Ewalt KL. Relationship of two human tRNA synthetases used in cell signaling. Trends Biochem. Sci. 2004;29:250–256. doi: 10.1016/j.tibs.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Ghanipour A, et al. The prognostic significance of tryptophanyl-tRNA synthetase in colorectal cancer. Cancer Epidemiol. Biomark. Prev. 2009;18:2949–2956. doi: 10.1158/1055-9965.EPI-09-0456. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Q, et al. Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality. Nat. Struct. Mol. Biol. 2010;17:57–61. doi: 10.1038/nsmb.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tzima E, Schimmel P. Inhibition of tumor angiogenesis by a natural fragment of a tRNA synthetase. Trends Biochem. Sci. 2006;31:7–10. doi: 10.1016/j.tibs.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 49.Nakamoto T, Miyanokoshi M, Tanaka T, Wakasugi K. Identification of a residue crucial for the angiostatic activity of human mini tryptophanyl-tRNA synthetase by focusing on its molecular evolution. Sci. Rep. 2016;6:24750. doi: 10.1038/srep24750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park BS, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 51.Kapoor M, et al. Evidence for annexin II-S100A10 complex and plasmin in mobilization of cytokine activity of human TrpRS. J. Biol. Chem. 2008;283:2070–2077. doi: 10.1074/jbc.M706028200. [DOI] [PubMed] [Google Scholar]
- 52.Nebenfuhr A, Ritzenthaler C, Robinson DG. Brefeldin A: deciphering an enigmatic inhibitor of secretion. Plant Physiol. 2002;130:1102–1108. doi: 10.1104/pp.011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mirando AC, Francklyn CS, Lounsbury KM. Regulation of angiogenesis by aminoacyl-tRNA synthetases. Int. J. Mol. Sci. 2014;15:23725–23748. doi: 10.3390/ijms151223725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown MV, Reader JS, Tzima E. Mammalian aminoacyl-tRNA synthetases: cell signaling functions of the protein translation machinery. Vasc. Pharmacol. 2010;52:21–26. doi: 10.1016/j.vph.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 55.Tzima E, et al. Biologically active fragment of a human tRNA synthetase inhibits fluid shear stress-activated responses of endothelial cells. Proc. Natl. Acad. Sci. USA. 2003;100:14903–14907. doi: 10.1073/pnas.2436330100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carmeliet P, et al. Targeted deficiency or cytosolic truncation of the VE-cadherin gene in mice impairs VEGF-mediated endothelial survival and angiogenesis. Cell. 1999;98:147–157. doi: 10.1016/S0092-8674(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 57.Lee CW, et al. Overexpressed tryptophanyl-tRNA synthetase, an angiostatic protein, enhances oral cancer cell invasiveness. Oncotarget. 2015;6:21979–21992. doi: 10.18632/oncotarget.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen J, et al. Increased TTS expression in patients with rheumatoid arthritis. Clin. Exp. Med. 2015;15:25–30. doi: 10.1007/s10238-014-0274-9. [DOI] [PubMed] [Google Scholar]
- 59.Bao YS, et al. Serum levels and activity ofindoleamine2,3-dioxygenase and tryptophanyl-tRNA synthetase and their association with disease severity in patients with chronic kidney disease. Biomarkers. 2013;18:379–385. doi: 10.3109/1354750X.2013.790074. [DOI] [PubMed] [Google Scholar]
- 60.Seshaiah P, Andrew DJ. WRS-85D: a tryptophanyl-tRNA synthetase expressed to high levels in the developing Drosophila salivary gland. Mol. Biol. Cell. 1999;10:1595–1608. doi: 10.1091/mbc.10.5.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matsunaga T, et al. Analysis of gene expression during maturation of immature dendritic cells derived from peripheral blood monocytes. Scand. J. Immunol. 2002;56:593–601. doi: 10.1046/j.1365-3083.2002.01179.x. [DOI] [PubMed] [Google Scholar]
- 62.Krause SW, et al. Differential screening identifies genetic markers of monocyte to macrophage maturation. J. Leukoc. Biol. 1996;60:540–545. doi: 10.1002/jlb.60.4.540. [DOI] [PubMed] [Google Scholar]
- 63.Ellis CN, et al. Comparative proteomic analysis reveals activation of mucosal innate immune signaling pathways during cholera. Infect. Immun. 2015;83:1089–1103. doi: 10.1128/IAI.02765-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis B virus infection. Proc. Natl. Acad. Sci. USA. 2004;101:6669–6674. doi: 10.1073/pnas.0401771101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu H, Cong JP, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takaoka A, et al. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature. 2003;424:516–523. doi: 10.1038/nature01850. [DOI] [PubMed] [Google Scholar]
- 67.Munn DH, Mellor AL. IDO in the tumor microenvironment: inflammation, counter-regulation, and tolerance. Trends Immunol. 2016;37:193–207. doi: 10.1016/j.it.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang XF, et al. The role of indoleamine 2,3-dioxygenase (IDO) in immune tolerance: focus on macrophage polarization of THP-1 cells. Cell Immunol. 2014;289:42–48. doi: 10.1016/j.cellimm.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 69.Soliman H, Mediavilla-Varela M, Antonia S. Indoleamine 2,3-dioxygenase: is it an immune suppressor? Cancer J. 2010;16:354–359. doi: 10.1097/PPO.0b013e3181eb3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat. Rev. Immunol. 2004;4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- 71.Grohmann U, Fallarino F, Puccetti P. Tolerance, DCs and tryptophan: much ado about IDO. Trends Immunol. 2003;24:242–248. doi: 10.1016/S1471-4906(03)00072-3. [DOI] [PubMed] [Google Scholar]
- 72.Munn DH, et al. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J. Exp. Med. 1999;189:1363–1372. doi: 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Munn DH, et al. Potential regulatory function of human dendritic cells expressing indoleamine 2,3-dioxygenase. Science. 2002;297:1867–1870. doi: 10.1126/science.1073514. [DOI] [PubMed] [Google Scholar]
- 74.Moffett JR, Namboodiri MA. Tryptophan and the immune response. Immunol. Cell Biol. 2003;81:247–265. doi: 10.1046/j.1440-1711.2003.t01-1-01177.x. [DOI] [PubMed] [Google Scholar]
- 75.Murray MF. Tryptophan depletion and HIV infection: a metabolic link to pathogenesis. Lancet Infect. Dis. 2003;3:644–652. doi: 10.1016/S1473-3099(03)00773-4. [DOI] [PubMed] [Google Scholar]
- 76.Boasso A, Herbeuval JP, Hardy AW, Winkler C, Shearer GM. Regulation of indoleamine 2,3-dioxygenase and tryptophanyl-tRNA-synthetase by CTLA-4-Fc in human CD4+T cells. Blood. 2005;105:1574–1581. doi: 10.1182/blood-2004-06-2089. [DOI] [PubMed] [Google Scholar]
- 77.Finger EB, Bluestone JA. When ligand becomes receptor—tolerance via B7 signaling on DCs. Nat. Immunol. 2002;3:1056–1057. doi: 10.1038/ni1102-1056. [DOI] [PubMed] [Google Scholar]
- 78.Xue H, Wong JT. Interferon induction of human tryptophanyl-tRNA synthetase safeguards the synthesis of tryptophan-rich immune-system proteins: a hypothesis. Gene. 1995;165:335–339. doi: 10.1016/0378-1119(95)00550-P. [DOI] [PubMed] [Google Scholar]
- 79.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 80.Wang S, et al. Increased TTS abrogates IDO-mediated CD4(+) T cells suppression in patients with Graves’ disease. Endocrine. 2009;36:119–125. doi: 10.1007/s12020-009-9184-0. [DOI] [PubMed] [Google Scholar]
- 81.Wang CY, et al. Decreased IDO activity and increased TTS expression break immune tolerance in patients with immune thrombocytopenia. J. Clin. Immunol. 2011;31:643–649. doi: 10.1007/s10875-011-9525-7. [DOI] [PubMed] [Google Scholar]
- 82.Hawiger J, Veach RA, Zienkiewicz J. New paradigms in sepsis: from prevention to protection of failing microcirculation. J. Thromb. Haemost. 2015;13:1743–1756. doi: 10.1111/jth.13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Uhlen M, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 84.Patil PA, et al. Expression of PD-L1, indoleamine 2,3-dioxygenase and the immune microenvironment in gastric adenocarcinoma. Histopathology. 2018;73:124–136. doi: 10.1111/his.13504. [DOI] [PubMed] [Google Scholar]
- 85.Cheong JH, et al. Predictive test for chemotherapy response in resectable gastric cancer: a multi-cohort, retrospective analysis. Lancet Oncol. 2018;19:629–638. doi: 10.1016/S1470-2045(18)30108-6. [DOI] [PubMed] [Google Scholar]
- 86.Xu X, Shi Y, Yang XL. Crystal structure of human Seryl-tRNA synthetase and Ser-SA complex reveals a molecular lever specific to higher eukaryotes. Structure. 2013;21:2078–2086. doi: 10.1016/j.str.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ling C, et al. The C-terminal appended domain of human cytosolic leucyl-tRNA synthetase is indispensable in its interaction with arginyl-tRNA synthetase in the multi-tRNA synthetase complex. J. Biol. Chem. 2005;280:34755–34763. doi: 10.1074/jbc.M413511200. [DOI] [PubMed] [Google Scholar]
- 88.Negrutskii BS, Shalak VF, Kerjan P, El’skaya AV, Mirande M. Functional interaction of mammalian valyl-tRNA synthetase with elongation factor EF-1alpha in the complex with EF-1H. J. Biol. Chem. 1999;274:4545–4550. doi: 10.1074/jbc.274.8.4545. [DOI] [PubMed] [Google Scholar]
- 89.Bec G, Kerjan P, Waller JP. Reconstitution in vitro of the valyl-tRNA synthetase-elongation factor (EF) 1 beta gamma delta complex. Essential roles of the NH2-terminal extension of valyl-tRNA synthetase and of the EF-1 delta subunit in complex formation. J. Biol. Chem. 1994;269:2086–2092. [PubMed] [Google Scholar]
- 90.Quevillon S, Mirande M. The p18 component of the multisynthetase complex shares a protein motif with the beta and gamma subunits of eukaryotic elongation factor 1. FEBS Lett. 1996;395:63–67. doi: 10.1016/0014-5793(96)01005-8. [DOI] [PubMed] [Google Scholar]
- 91.Kim JE, et al. An elongation factor-associating domain is inserted into human cysteinyl-tRNA synthetase by alternative splicing. Nucleic Acids Res. 2000;28:2866–2872. doi: 10.1093/nar/28.15.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang XL, et al. Gain-of-function mutational activation of human tRNA synthetase procytokine. Chem. Biol. 2007;14:1323–1333. doi: 10.1016/j.chembiol.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zheng YG, Wei H, Ling C, Xu MG, Wang ED. Two forms of human cytoplasmic arginyl-tRNA synthetase produced from two translation initiations by a single mRNA. Biochemistry. 2006;45:1338–1344. doi: 10.1021/bi051675n. [DOI] [PubMed] [Google Scholar]
- 94.Francin M, Kaminska M, Kerjan P, Mirande M. The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain. J. Biol. Chem. 2002;277:1762–1769. doi: 10.1074/jbc.M109759200. [DOI] [PubMed] [Google Scholar]
- 95.Francin M, Mirande M. Functional dissection of the eukaryotic-specific tRNA-interacting factor of lysyl-tRNA synthetase. J. Biol. Chem. 2003;278:1472–1479. doi: 10.1074/jbc.M208802200. [DOI] [PubMed] [Google Scholar]
- 96.Ryckelynck M, Giege R, Frugier M. Yeast tRNA(Asp) charging accuracy is threatened by the N-terminal extension of aspartyl-tRNA synthetase. J. Biol. Chem. 2003;278:9683–9690. doi: 10.1074/jbc.M211035200. [DOI] [PubMed] [Google Scholar]
- 97.Kim KR, et al. Crystal structure of human cytosolic aspartyl-tRNA synthetase, a component of multi-tRNA synthetase complex. Proteins. 2013;81:1840–1846. doi: 10.1002/prot.24306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reed VS, Wastney ME, Yang DC. Mechanisms of the transfer of aminoacyl-tRNA from aminoacyl-tRNA synthetase to the elongation factor 1 alpha. J. Biol. Chem. 1994;269:32932–32936. [PubMed] [Google Scholar]
- 99.Jia J, Arif A, Ray PS, Fox PL. WHEP domains direct noncanonical function of glutamyl-Prolyl tRNA synthetase in translational control of gene expression. Mol. Cell. 2008;29:679–690. doi: 10.1016/j.molcel.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Howard OM, et al. Histidyl-tRNA synthetase and asparaginyl-tRNA synthetase, autoantigens in myositis, activate chemokine receptors on T lymphocytes and immature dendritic cells. J. Exp. Med. 2002;196:781–791. doi: 10.1084/jem.20020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ko YG, Kang YS, Kim EK, Park SG, Kim S. Nucleolar localization of human methionyl-tRNA synthetase and its role in ribosomal RNA synthesis. J. Cell Biol. 2000;149:567–574. doi: 10.1083/jcb.149.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chang CY, Chien CI, Chang CP, Lin BC, Wang CC. A WHEP domain regulates the dynamic structure and activity of Caenorhabditis elegans glycyl-tRNA synthetase. J. Biol. Chem. 2016;291:16567–16575. doi: 10.1074/jbc.M116.730812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wakasugi K, Schimmel P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science. 1999;284:147–151. doi: 10.1126/science.284.5411.147. [DOI] [PubMed] [Google Scholar]