Abstract
Ten days after the declaration of the Ebola outbreak in the Democratic Republic of Congo, rapid identification of the species Zaire Ebola virus using partial gene amplification and nanopore sequencing backed up the use of the recombinant vesicular stomatitis virus–Zaire Ebola virus vaccine in the recommended ring vaccination strategy.
Keywords: Ebola, Democratic Republic of Congo, nanopore sequencing
The Democratic Republic of Congo (DRC) declared a new outbreak of Ebola virus disease (EVD) on 8 May 2018. Initially, a report from the Provincial Health Division of the Equateur Province at the end of April informed the national authorities of 21 cases of potential hemorrhagic fever, including 17 deaths in the Ikoko-Impenge health area. Deployed disease experts on site identified 5 possible cases on 5 May and a blood sample was taken from each individual, 2 who were hospitalized in Bikoro and 3 at the health centre of Iboko. Samples were sent to Institut National de Recherche Biomédicale (INRB) in Kinshasa, and the presence of Ebola virus was identified in 2 samples on 7 May [1]. The samples tested positive with the OraQuick Ebola Rapid Antigen test (OraSure Technologies) and with the RealStar Zaire Ebolavirus reverse-transcription polymerase chain reaction (PCR) kit 1.0 (Altona Diagnostics).
Proven emergency responses such as enhanced surveillance, active case finding, contact tracing, infection prevention, safe burials, and diagnosis with deployment of mobile laboratories on site were implemented rapidly. However, in 2017, the World Health Organization (WHO) recommended the deployment of the recombinant vesicular stomatitis virus–Zaire Ebola virus (rVSV-ZEBOV) candidate vaccine as an additional measure for prevention and control in the event of novel Zaire Ebola virus (EBOV) outbreaks [2]. This vaccine has been shown to be effective and safe and has been used during the epidemic decline of EBOV in Guinea in 2015 and during the flare-up of the disease in rural Guinea in 2016 [3, 4]. However, 4 species of Ebola viruses have been documented in Africa: Zaire (EBOV), Sudan (SUDV), Bundibugyo (BDBV), and Tai Forest [5]. Whereas 7 of the 8 previous EBOV epidemics in DRC were due to EBOV, BDBV was responsible for the outbreak in Isiro in 2012, and previous outbreaks in South Sudan were all close to the border with DRC and were due to SUDV [6]. Therefore, it is thus important to confirm rapidly the viral strain of any new outbreak to guarantee the utility of anti-EBOV vaccination. Here, we describe the rapid laboratorial response that led to the confirmation of EBOV in the latest outbreak and highlight the implication of rapid viral detection for public health interventions.
MATERIALS AND METHODS
PCR Amplification
Viral RNA was extracted from 140 μL of whole blood collected from the 5 clinically suspected Ebola virus–infected individuals from Bikoro/Iboko using the Nuclisens kit (bioMérieux) and following the manufacturer’s instructions. After preparation of complementary DNA with GoScript Reverse Transcriptase (Promega, France), amplification of a small fragment of the VP35 region was attempted in a seminested PCR with a modified protocol adapted from He and colleagues [7]. Three additional samples were included as controls: 2 EBOV positive and 1 negative from the outbreak in Boende in 2014. In the first round, a 217-bp fragment was targeted with primers VP35-F: 5ʹ-ATYATGTATGATCACYTVCCWGG-3ʹ and VP35-R: AGCGR ATGTGGATSACRGGT-3ʹ. In the second round, an 184-bp product was amplified with primers VP35-R and VP35-in-F: 5ʹ-GCTTTYCAYCAAYTAGTRCAAG-3ʹ.
Sequence Analysis
First-round VP35 PCR products from positive samples were barcoded and pooled using the Native Barcoding Kit (Oxford Nanopore Technologies). Sequencing libraries were generated from the barcoded products using the Genomic DNA Sequencing Kit EXP-NBD103/SQK-LSK208 (Oxford Nanopore Technologies) and were loaded onto a R9 flow cell. Genetic data was collected for 1 hour.
Bioinformatics and Phylogenetic Analysis
Basecalling of sequenced data was done with ONT Albacore Sequencing Pipeline software version 2.1.10. Adapter removal and demultiplexing were achieved with Porechop 0.2.3.
Filovirus genus references were retrieved from the Hemorrhagic Fever Viruses (HFV) database at the Los Alamos National Laboratory. Multireference prealignment analysis of quality and error profiles using reads with basecall quality ≥11 was done with the software package NanoOK 1.10. Ebola virus reads were corrected with Canu 1.6, and consensus sequences were produced by alignment of reads to the corresponding Ebola virus reference genome following signal-level analysis using Nanopolish 0.9.
One sequence per outbreak was retrieved from the HFV database and used for phylogenetic inference together with the nanopore consensus sequences. We also included a previously obtained VP35 sequence from an unpublished strain from the outbreak in Likati in 2017. Phylogenetic analyses were done using maximum likelihood methods with approximate likelihood ratio test nonparametric branch support based on a Shimodaira-Hasegawa–like procedure using PhyML version 3.1 software. The HKY85 model plus a discrete gamma model were used as nucleotide substitution models.
RESULTS
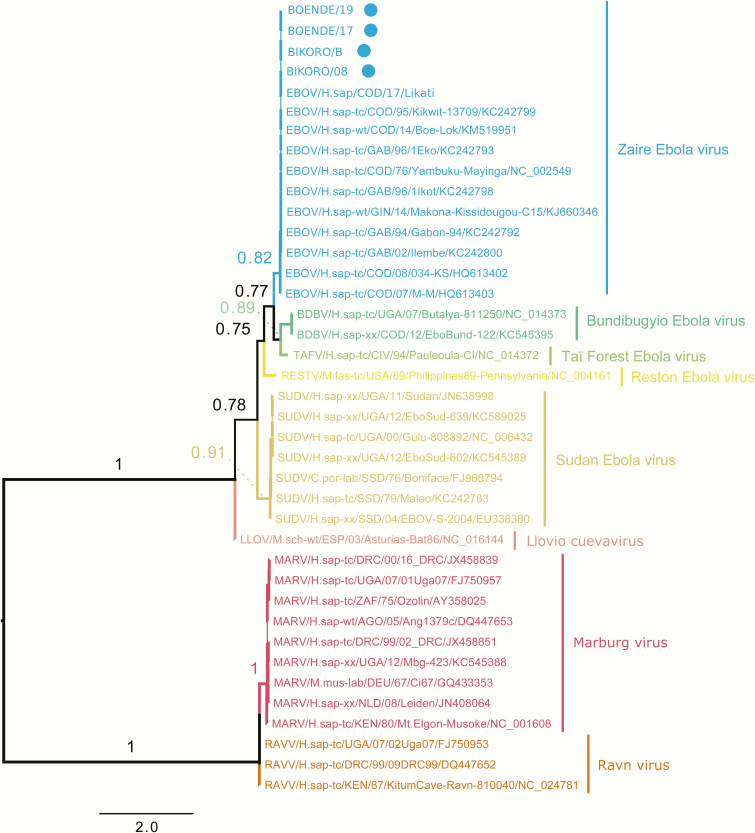
The 2 positive control samples from Boende 2014 were successfully amplified and no PCR product was obtained for the negative sample. The 2 samples from Bikoro/Iboko 2018 that tested initially positive with the commercial tests were also positive in the VP35 PCR, 2 other samples had a weak PCR product, and the remaining sample resulted in no amplification. We attempted sequencing on the 6 samples with PCR amplification. After 1 hour, between 400 and >4000 reads were obtained depending on the sample. NanoOK report indicated that >95% of the reads from the 2 known positive samples from Bikoro/Iboko and the 2 positive control samples from Boende mapped to the EBOV species. The remaining reads and the reads from the samples with weak positive PCR product did not align to any filovirus genera and were queried in BLAST (Basic Local Alignment Search Tool), and the best hits pointed to human DNA. Consensus sequences were produced for the new EBOV strains and aligned with reference sequences from previous Ebola outbreaks including the 5 Ebola virus species and other filoviruses such as Marburg. The phylogenetic tree shown in Figure 1 underscores that this highly conserved region is discriminatory between Ebola virus species and illustrates that the new strains from the ninth Ebola outbreak in DRC belong to the Ebola Zaire species (EBOV).
Figure 1.
Maximum likelihood tree showing phylogenetic relationships of samples from Bikoro/Iboko 2018 and other filovirus using partial VP35 sequences. Approximate likelihood ratio test nonparametric branch supports based on a Shimodaira-Hasegawa–like procedure are shown for the main nodes. The new strains from Bikoro/Itipo (2018) and the control samples from Boende (2014) are highlighted with a dot.
The laboratory results of the 5 suspected cases and the clinical signs and history for each patient are summarized in Supplementary Table 1. Overall, all 5 patients had diarrhea, severe fatigue, and abdominal pain. Additional symptoms were muscle or joint pain and anorexia for 4 of them. Three patients had also gum and/or nose bleeding among symptoms, but only 1 of them was confirmed as infected with Ebola virus by sequence and PCR analysis.
DISCUSSION
After only 10 days of the ninth Ebola outbreak declaration, the virus strain was confirmed as belonging to the Zaire species (EBOV). As in the outbreak in West Africa, nanopore sequencing using the MinION device proved useful for quick pathogen confirmation and strain identification [8, 9]. The device provided speed (library preparation, basecalling of preliminary read data and multireference prealignment took <6 hours) and ease of use besides portability advantages over other sequencing machines. The rapid viral characterization of the strain backed up the rVSV-ZEBOV candidate vaccine for the ring vaccination strategy recommended by WHO [2]. Today >1000 people have been vaccinated, mainly health professionals and people exposed to patients with confirmed EBOV and their contacts [1]. Nevertheless, further efforts to increase adequate hygiene practices and safe burials must also be continued.
Previous outbreaks in DRC were in very remote locations or small towns, but this new outbreak affected simultaneously at least 3 different locations in the Equateur Province. It included 2 remote sites, Bikoro and Iboko/Itipo health areas, and cases in Mbandaka, a city of >1 million inhabitants and with transport and port links to the rest of the country and neighboring countries. The current outbreak is thus complex with the potential for urban and larger geographic spread. It was thus urgent to start rapidly the ring vaccination to stop the spread of the virus.
DRC had already experienced 8 other Ebola outbreaks since 1976 [6]. The country has thus implemented a strategy at the national level to rapidly identify a potential new outbreak. This includes rapid reporting of clinical suspected cases and shipment of samples to the reference laboratory at INRB in Kinshasa for confirmation. In the current situation, the health service from Equateur Province gave the alert in late April. Nevertheless, the clinical presentations of the 5 suspected cases illustrate how difficult it is to distinguish Ebola from some other diseases without laboratory confirmation. A similar alert was issued in February in the same area with 15 cases, among which 8 individuals died, but no laboratory confirmation occurred (unpublished data). This observation underscores that the outbreak may have started earlier and emphasizes the potential of Ebola outbreaks to go unrecognized in remote areas. However, any relation between the 2 alerts remains to be examined.
Each EVD outbreak is the result of a cross-species transmission event from a reservoir or an intermediate/amplifying animal host species [6]. This new outbreak, together with the large outbreak in West Africa, illustrates the major consequences of recurrent zoonotic spills. Studies on the ecology and animal reservoir of Ebola viruses are thus urgently needed so exposure to infection can be anticipated or avoided. Finally, the advantages provided by novel diagnostic technologies such as nanopore sequencing using the MinION device highlight the importance of local capacity-building programs for rapid pathogen confirmation and corresponding public health interventions.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Financial support. This work was supported in part by grants from the Institut national de la santé et de la recherche médicale/Research and Action targeting emerging infectious diseases; the Capacity building and surveillance for Ebola virus disease project funded by the European Union; the International Mixt Laboratory “PreVIHMI” of the Institut de Recherche pour le Developpement; and the Christophe Mérieux Prize 2015 awarded to Professor J. J. Muyembe Tamfum.
Acknowledgments. We thank the Democratic Republic of Congo (DRC) Ministry of Health and the World Health Organization office in DRC.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization (WHO). Ebola virus disease, Democratic republic of Congo. External situation report update report 8. Available at: http://apps.who.int/iris/bitstream/handle/10665/272761/SITREP-EVD-DRC-20180605-eng.pdf?ua=1. Accessed 12 June 2018. [Google Scholar]
- 2. World Health Organization (WHO). Meeting of the Strategic Advisory Group of Experts on immunization—conclusions and recommendations. Ebola vaccines. Available at: http://www.who.int/csr/resources/publications/ebola/sage-ebola-vaccination.pdf?ua=1. Accessed 12 June 2018. [Google Scholar]
- 3. Henao-Restrepo AM, Longini IM, Egger M, et al. Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: interim results from the Guinea ring vaccination cluster-randomised trial. Lancet 2015; 386:857–66. [DOI] [PubMed] [Google Scholar]
- 4. Gsell PS, Camacho A, Kucharski AJ, et al. Ring vaccination with rVSV-ZEBOV under expanded access in response to an outbreak of Ebola virus disease in Guinea, 2016: an operational and vaccine safety report. Lancet Infect Dis 2017; 17:1276–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kuhn JH, Andersen KG, Baize S, et al. Nomenclature- and database-compatible names for the two Ebola virus variants that emerged in Guinea and the Democratic Republic of the Congo in 2014. Viruses 2014; 6:4760–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pigott DM, Millear AI, Earl L, et al. Updates to the zoonotic niche map of Ebola virus disease in Africa. Elife 2016; 5 pii:e16412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He B, Feng Y, Zhang H, et al. Filovirus RNA in fruit bats, China. Emerg Infect Dis 2015; 21:1675–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Quick J, Loman NJ, Duraffour S, et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 2016; 530:228–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoenen T, Groseth A, Rosenke K, et al. Nanopore sequencing as a rapidly deployable Ebola outbreak tool. Emerg Infect Dis 2016; 22:331–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.