In the present study, we provide the first evidence that a new HIV-1 protein termed ASP derived from different clades acts similarly in inducing autophagy, an important cellular process implicated in the degradation of excess or defective cellular material. We have gained further knowledge on the mechanism mediating the activation of autophagy. Our studies have important ramifications in the understanding of viral replication and the pathogenesis associated with HIV-1 in infected individuals. Indeed, autophagy is implicated in antigen presentation during immune response and could thus be rendered inefficient in infected cells, such as dendritic cells. Furthermore, a possible link with HIV-1-associated neurological disorder (HAND) might also be a possible association with the capacity of ASP to induce autophagy. Our studies hence demonstrate the importance in conducting further studies on this protein as it could represent a new interesting target for antiretroviral therapies and vaccine design.
KEYWORDS: antisense protein, autophagy, HIV-1, clade, multimerization, p62/SQSTM1, ubiquitination
ABSTRACT
The existence of the antisense transcript-encoded HIV-1 antisense protein (ASP) was recently reinforced by in silico analyses providing evidence for recent appearance of this gene in the viral genome. Our previous studies led to the detection of ASP in various cell lines by Western blotting, flow cytometry, and confocal microscopy analyses and reported that it induced autophagy, potentially through multimer formation. Here, our goals were to assess autophagy induction by ASP from different clades and to identify the implicated autophagy factors. We first demonstrated that ASP formed multimers, partly through its amino-terminal region and cysteine residues. Removal of this region was further associated with lower induction of autophagy, as assessed by autophagosome formation. ASPs from different clades (A, B, C, D, and G) were tested next and were detected in monomeric and multimeric forms at various levels, and all induced autophagy (clade A ASP was less efficient), as determined by LC3-II and p62 (SQSTM1) levels. Furthermore, CRISPR-based knockout of ATG5, ATG7, and p62 genes led to increased ASP levels. Confocal microscopy analyses showed that ASP colocalized with p62 and LC3-II in autophagosome-like structures. Coimmunoprecipitation experiments further demonstrated that p62 associated with ASP through its PB1 domain. Interestingly, immunoprecipitation experiments supported the idea that ASP is ubiquitinated and that ubiquitination was modulating its stability. We are thus suggesting that ASP induces autophagy through p62 interaction and that its abundance is controlled by autophagy, in which ubiquitin plays an important role. Understanding the mechanisms underlying ASP degradation is essential to better assess its function.
IMPORTANCE In the present study, we provide the first evidence that a new HIV-1 protein termed ASP derived from different clades acts similarly in inducing autophagy, an important cellular process implicated in the degradation of excess or defective cellular material. We have gained further knowledge on the mechanism mediating the activation of autophagy. Our studies have important ramifications in the understanding of viral replication and the pathogenesis associated with HIV-1 in infected individuals. Indeed, autophagy is implicated in antigen presentation during immune response and could thus be rendered inefficient in infected cells, such as dendritic cells. Furthermore, a possible link with HIV-1-associated neurological disorder (HAND) might also be a possible association with the capacity of ASP to induce autophagy. Our studies hence demonstrate the importance in conducting further studies on this protein as it could represent a new interesting target for antiretroviral therapies and vaccine design.
INTRODUCTION
The genome of human immunodeficiency virus type 1 (HIV-1) expresses essential genes for its replication, which include gag, pol, and env, common to all replication-competent retroviruses in addition to genes encoding regulatory proteins (Tat and Rev) crucial for virus replication. Furthermore, HIV-1 is known to encode a number of accessory proteins, i.e., Vpu, Vpr, Vif, and Nef, all of which are suggested to be implicated in the inactivation of restriction factors and the modulation of immune functions (1–6). A former study, however, had demonstrated that HIV-1 isolates contained a conserved open reading frame (ORF) on the antisense strand of the Env gene, thereby hinting toward the existence of a 10th gene with an encoding potential for a 189-amino-acid protein (7, 8). A recent study has further underscored the high degree of conservation of this ORF in most HIV-1 clades and further argued that this gene correlated with the spread of the virus since it is present in the most prevalent clades, A, B, C, and G (prevalence, ca. 81%), while being less present (or absent) in most simian immunodeficiency virus (SIV) groups, HIV-1 groups N, O, and P, and in HIV-1 clades D, F, H, J, and K (prevalence, ca. 3%) (9). Interestingly, based on the predicted sequence, this study further revealed that the clade A antisense protein (ASP) is a truncated version of ASP, with the methionine initiation codon located at amino acid 26 of the ASP ORF of other clades. A number of previous and more recent studies have further confirmed that antisense transcription overlapping this ORF sequence was detected in transfected and chronically infected cells (10–19).
The existence of the presumed encoded ASP in infected patients has been further suggested by several groups through detection of specific antibodies and CD8+ T cell-mediated immune responses (10, 20–22). However, ASP detection itself has proven to be more difficult, and the protein was first analyzed by in vitro translation and electron microscopy in transfected and chronically infected cells (10, 15). More recent studies have provided a better understanding of this protein in terms of its potential membrane localization and its biased expression in monocyte-derived macrophages and dendritic cells (DCs) (7, 17). In another recent report, we have been able to clearly detect the clade B ASP for the first time by Western blotting and various other approaches (confocal microscopy and flow cytometry) (23). Furthermore, our work further suggested that ASP was unstable, formed multimers, and induced autophagy, likely through the formation of these high-molecular-weight complexes (23). These results, hence, provided an explanation for the inherent difficulty in the detection of ASP in transfected cell lines.
Autophagy, a major cellular degradation pathway, plays an important role in developmental processes, cellular stress responses, and immune pathways induced by pathogens. The early step of what is known as macroautophagy initiates through mTOR inhibition and ensuing formation of an active autophagy complex consisting of the phosphorylated Beclin-1 factor, normally negatively inhibited by its binding to Bcl-2 (24–27). Following this activation, a protrusion of two lipid-based structures, termed the isolation membrane and the omegasome, is induced intracellularly to form the initial open structure, termed the phagophore (28). The phagophore further elongates circularly and finalizes the maturation process of the autophagosome, where trapped cytosolic elements are targeted for degradation (27). Final fusion of the autophagosome with lysosomes leads to degradation of its content and recycling of amino acids and other constituents (29, 30). Different proteins play an active role in autophagy, such as the phagophore-associated autophagy-related genes (ATGs) ATG5, ATG7, ATG10, ATG12, and ATG16 (31). One of the classical autophagy markers is LC3-II, a phosphatidylethanolamine-modified form of the LC3-I form, which is embedded in the autophagosome membrane and contributes to its formation. Another implicated autophagy marker is p62/SQSTM1, a scaffold protein which links aggregated complexes to LC3 and consequently becomes itself degraded.
Autophagy can either be beneficial or detrimental toward viral replication; the outcome of induced autophagy depends on the virus, the cell type, and the cellular environment (32). In fact, for HIV-1, initial studies in CD4+ T cells had shown that the envelope gp41 subunit induced autophagy-dependent apoptosis in uninfected cells (33, 34). However, a genomic screen has also identified certain autophagy-related host factors, which were essential for HIV-1 infection (35). Furthermore, specifically in macrophages, HIV-1-induced autophagy greatly improved Gag pr55 processing and particle production (36). Importantly, Nef has been reported to block the process of autophagy at late stages (fusion with lysosomes), thereby avoiding intracellular degradation of viral particles (5, 37). More recent data have also demonstrated a potential role for other HIV-1 proteins in regulating autophagy, such as Vif (38, 39).
Although previously associated with the proteasome degradation pathway, ubiquitination has become of high relevance to autophagy (40–43). The C terminus of ubiquitin is covalently linked to the target protein by specific lysine residues (e.g., K48 and K63), which subsequently leads to polyubiquitination. While K48- and K29-linked polyubiquitination has been shown to be optimal for degradation through the proteasome, other types of lysine-linked polyubiquitination (e.g., K63, K11, and K6) and monoubiquitination may regulate processes such as autophagy, translation, and DNA repair (43). In fact, ubiquitinated proteins can be recognized by the autophagy pathway through specialized adaptor proteins, also called sequestosome-1-like receptors (including p62/SQSTM1, optineurin, and NDP52). The p62 protein then multimerizes via its PB1 domain (NH2-terminal Phox and Bem1p domains) and binds LC3-II via its LC3-interacting region (LIR) and ubiquitinated proteins through the phosphorylation of its ubiquitin-associated (UBA) domain (42–44).
Based on the link between ASP and autophagy (23), in the current study, we aimed at examining the mechanism behind ASP-induced autophagy. Our results suggest that multimerization of ASP and induced autophagy partly involve its amino end. We show that ASP from various HIV-1 clades all induced autophagy, as determined by LC3-II levels and p62 degradation, although clade A ASP was less efficient. We further highlight that ASP colocalized and interacted with p62 through its PB1 domain and, finally, present data supporting the idea that ASP is ubiquitinated, further contributing to its targeting toward degradation pathways.
(This article was submitted to an online preprint archive [45]).
RESULTS
Analyses of ASP multimers.
A limited number of studies have shown that ASP could be detected in infected and transfected cells (7, 15, 23). Using a mammalian expression vector harboring a human codon-optimized ASP cDNA, we have successfully detected ASP in 293T and COS-7 cells and demonstrated that it was forming multimers and could induce the formation of autophagosomes, thereby providing an explanation for its difficult detection (23). Here, we sought to further define the mechanism leading to autophagy induction by ASP and to extend these analyses to ASP from different HIV-1 clades.
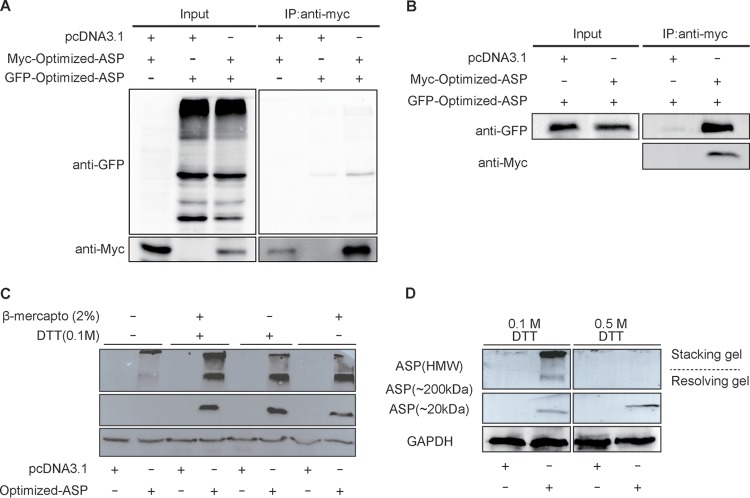
We were first interested in confirming the self-multimerization potential of NL4.3 (clade B) ASP using expression vectors for Myc-tagged and chimeric green fluorescent protein (GFP)-optimized ASP (Fig. 1). Extracts from cotransfected 293T cells were immunoprecipitated with anti-Myc antibodies and subsequently analyzed in parallel with total extracts by Western blotting. As depicted in Fig. 1A, a specific signal was detected corresponding to the GFP-tagged ASP in immunoprecipitated extracts, which could not be accounted for by significant differences in expression levels of the fusion protein in the different analyzed samples. To confirm that ASP could self-multimerize in a different cell line, similar cotransfection experiments were conducted in COS-7 cells and again revealed that both ASP versions were associated (Fig. 1B). As cysteine-rich regions of the protein might contribute toward the formation of these high-molecular-weight complexes, we pretreated the extract from Myc-tagged ASP-expressing COS-7 cells with high concentrations of the reducing agents dithiothreitol (DTT) and β-mercaptoethanol. Upon treatment, the intensity of the high-molecular-weight signals was strongly diminished with either agent and resulted in increased intensity of the 20- and 200-kDa signals (Fig. 1C). A very high concentration (0.5 mM) of DTT equally led to disappearance of the high-molecular-weight signals (Fig. 1D).
FIG 1.
Multimerization of ASP involves disulfide bonds. (A and B) 293T (A) and COS-7 (B) cells were transfected with expression vectors for GFP-optimized NL4.3 ASP, Myc-tagged-optimized NL4.3 ASP, and/or the empty vector pcDNA3.1. At 48 h posttransfection, immunoprecipitation (IP) was performed using anti-Myc antibody, and Western blot analyses were conducted using anti-GFP and anti-Myc antibodies. Total cellular extracts were similarly analyzed in parallel. (C and D) Extracts from COS-7 cells transfected with the Myc-tagged optimized NL4.3 ASP expression vector (versus pcDNA3.1) were treated with β-mercaptoethanol and/or DTT (at different concentrations in panel D) prior to Western blot analyses with anti-Myc and anti-GAPDH antibodies. Both stacking and resolving gels are depicted, and high-molecular-weight (HMW) (including 200-kDa multimers) versus monomeric 20-kDa ASP signals are indicated.
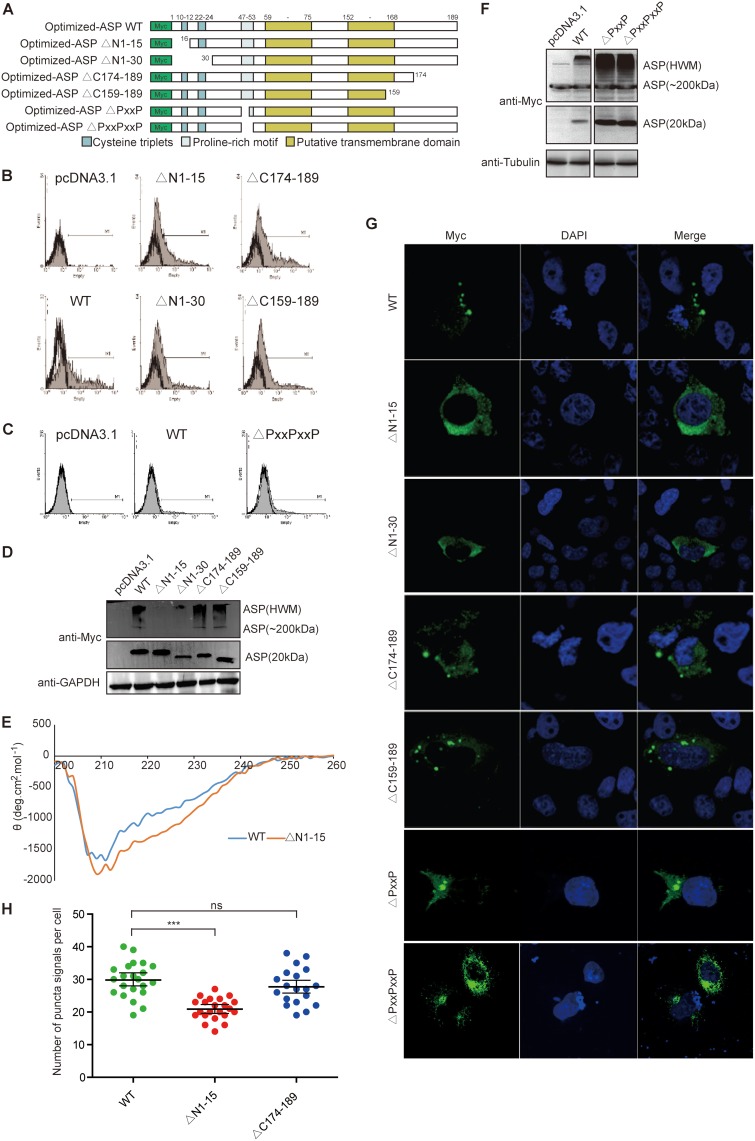
In light of these results, a series of deletion mutants were next generated in which the first 15 and 30 amino acid residues from the amino or carboxyl end of Myc-tagged NL4.3 ASP were deleted. The vectors pMyc-optimized-ASPΔN1–15 and pMyc-optimized-ASPΔN1–30 expressed ASP mutants from which 4 and 7 conserved cysteine residues, respectively (presumed to be implicated in multimer formation), were removed (Fig. 2A). ASP mutants with similar deletions at their carboxyl ends served as controls. Upon transfection of COS-7 cells, flow cytometry analyses showed that mutants and wild-type (WT) ASP were detected at comparable levels except for the ASP from the mutant pMyc-optimized-ASPΔN1–30, which showed a reduced signal (Fig. 2B). The multimerization potential of these mutants was next analyzed by Western blotting (Fig. 2D). Interestingly, the loss of the first 15 residues at the amino end strongly reduced the abundance of high-multimer complexes and led to a slight increase in the levels of monomers while no such effect was observed with mutants bearing deletions at their COOH ends. The ASPΔN1–30 mutant presented reduced signals for both monomeric and high-molecular-weight signals compared to signals in wild-type-expressing cells, which was likely due to reduced overall abundance of the protein. To investigate if the deletion of the 15-residue N-terminal domain of ASP (ASPΔN1–15) altered protein conformation, we performed circular dichroism (CD) spectroscopy. After analysis, the far-UV CD spectrum of recombinant WT ASP was identical to the one recorded for ASPΔN1–15 (Fig. 2E). Both proteins displayed CD spectra with double minima at 212 and 222 nm, suggestive of the presence of an α-helical structure. Deconvolution of these spectra using the K2D2 algorithm (46) predicted a high percentage of residues in the α-helix, with 83% for WT ASP and 84% for ASPΔN1–15. Thus, CD analysis clearly indicated that the deletion of the first 15 residues of ASP did not affect the secondary structure of the protein.
FIG 2.
The amino end of ASP is implicated in its multimerization and induced autophagy (A) Schematic representation of the different domains of ASP and the generated deletion mutants targeting the first 15 or 30 amino acids of either the N or C terminus of ASP and the PXXPXXP region. (B to H) COS-7 cells were transfected with expression vectors for the different ASP mutants and the wild-type version versus pcDNA3.1 (empty vector) and, at 48 h posttransfection, were analyzed with an anti-Myc antibody by flow cytometry (B and C), Western blotting (D and F), and confocal microscopy (60× objective with a numerical aperture of 1.4) (G and H). In panels D and F, high-molecular-weight (including 200-kDa multimers) versus monomeric 20-kDa ASP signals are indicated. Cells analyzed by confocal microscopy were also stained for their nuclei with Hoechst (G) and quantified for the number of ASP-positive punctate structures (H). In panel E, His-tagged WT ASP and the ASPΔN1–15 mutant were purified from transfected cells for the recording of far-UV circular dichroism spectra at concentrations of 109.5 ng/ml (WT) and 111.5 ng/ml (ASPΔN1–15). ns, not significant. ***, P ≤ 0.001.
Confocal microscopy analyses were next performed in transfected cells (Fig. 2G). Deletion of the first 15 amino acids reduced (but not completely abolished) the presence of punctate ASP signals having previously been identified as autophagosomes. Deletion of the carboxyl end did not lead to differences in the distribution of the resulting ASP mutant from that of the wild-type version. Numbering of the punctate structures in cells transfected by each expression vector further confirmed the reduced presence of ASP-positive autophagosome-like structures in ASPΔN1–15-expressing cells in comparison to levels in cells expressing either ASP WT or ASPΔN174–189 (Fig. 2H). Another mutant in which the conserved PXXP domain (positions 47 to 53) was partly or completely deleted was also tested in COS-7 cells and revealed no clear change in expression level, multimer formation, or potential induction of autophagy (Fig. 2C, F, and G).
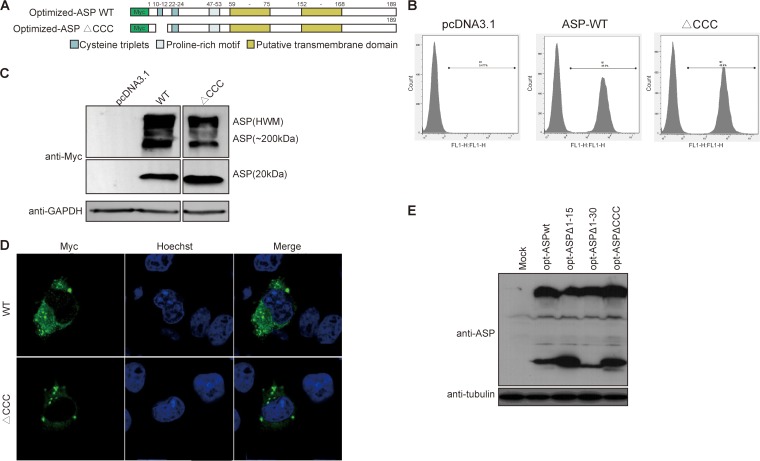
Since a cysteine triplet is present in the first 15 amino acids of the ASP ORF, we specifically deleted these cysteine residues (ΔCCC mutant) (Fig. 3A). Flow cytometry indicated that the resulting mutant was detected at levels equivalent to those of optimized-ASP WT in transfected COS-7 cells (Fig. 3B). Expression of the optimized ASP ΔCCC mutant in COS-7 cells resulted in a typical punctate distribution in the cytoplasm (Fig. 3D), while Western blot analyses demonstrated that the multimerization capacity of ASP ΔCCC appeared less pronounced, with a concomitant increase in the monomeric signal, suggesting that this cysteine triplet was partially contributing to ASP multimer formation and autophagy induction (Fig. 3C). To confirm results obtained in transfected COS-7 cells, the amino-terminal and cysteine deletion mutants were tested in 293T cells (Fig. 3E). Compared to WT ASP, these mutants again affected ASP multimer formation and markedly increased monomeric ASP levels.
FIG 3.
Deletion of the first cysteine triplet of ASP reduces multimer formation and autophagosome signals. (A) Schematic representation of the different domains of ASP and the deletion mutant of the first cysteine triplet (10CCC12). (B to D) COS-7 cells were transfected with expression vectors for wild-type or cysteine triplet-deleted ASP versus pcDNA3.1 (empty vector). After 48 h of transfection, using anti-Myc antibodies, cells were analyzed by flow cytometry (B), Western blotting (C), and confocal microscopy (60× objective with numerical aperture of 1.4) (D). In panel C, high-molecular-weight (including 200-kDa multimers) versus monomeric 20-kDa ASP signals are indicated. Cells analyzed by confocal microscopy were also stained for their nuclei with Hoechst (D). (E) 293T cells were transfected with expression vectors for optimized (opt) NL4.3 ASP WT, ASPΔN1–15, ASPΔN1–30, and ρCCC versus pcDNA3.1 (mock) and analyzed by Western blotting using anti-ASP antibodies.
These results, hence, demonstrated that the amino terminus (and cysteines) of ASP contributes to its capacity to multimerize and is suggested that it is partly implicated in its autophagy-inducing properties.
Expression and detection of ASP from various clades.
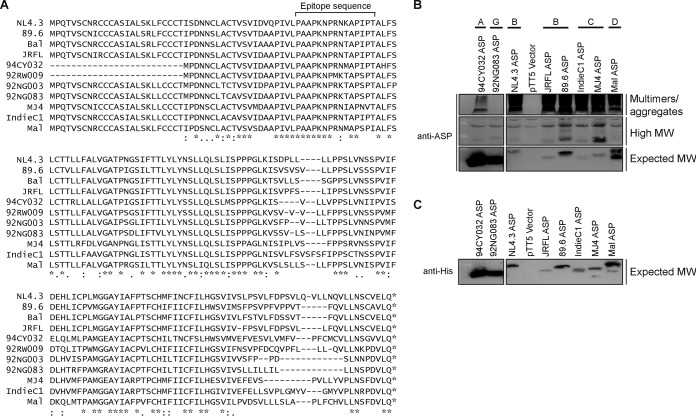
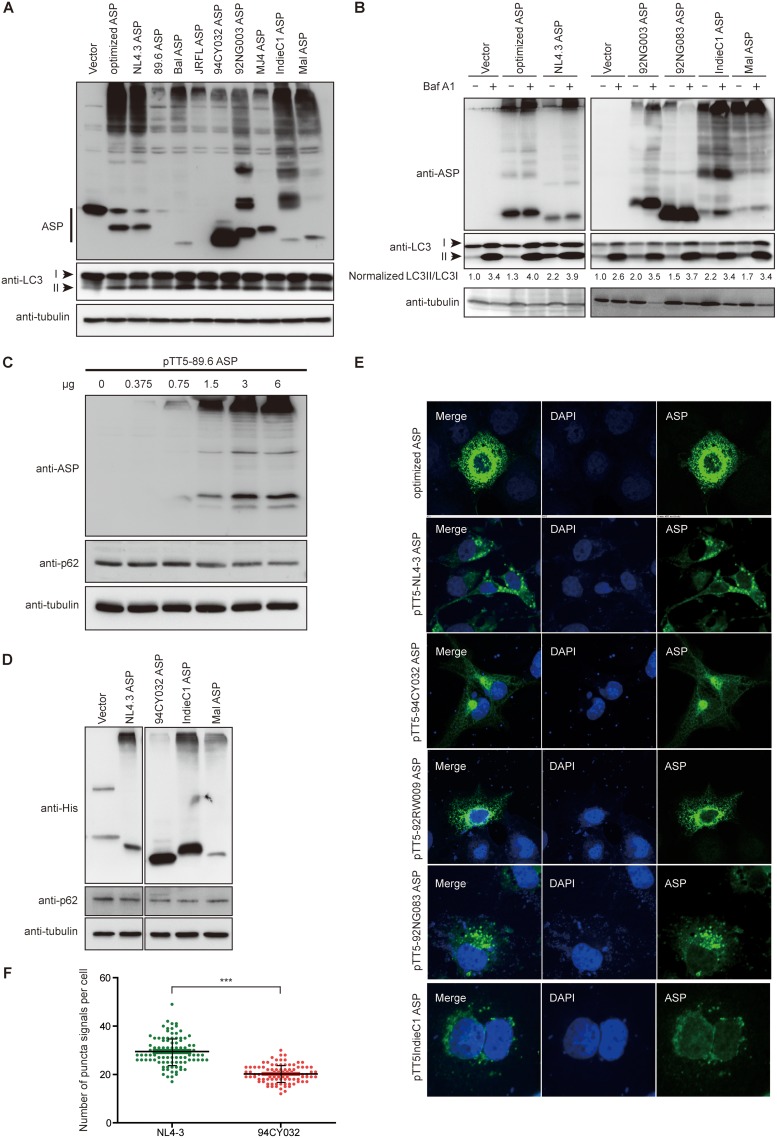
In order to address the mechanism leading to ASP-mediated autophagy in a more representative manner, we next generated expression vectors for His-tagged ASPs representing HIV-1 clades A, B, C, D, and G. Importantly, these ASP ORFs were directly derived from the original proviral DNA sequence and were not codon optimized, as previously performed in our earlier study (23). A sequence comparison of the ASP genes tested in this study is presented in Fig. 4A and highlights the previously observed truncated form of clade A ASP (9). This thereby allowed us to assess multimer formation and the autophagy-inducing capacity of an ASP version naturally lacking the typical amino end. Following transfection in 293T cells, we first used a monoclonal anti-ASP antibody derived against the epitope indicated in Fig. 4A to determine if ASP could be detected by Western blotting. As depicted in Fig. 4B, despite variation in the amino acid sequences of the epitopes of tested ASPs, we demonstrated the presence of the expected 20-kDa signal (albeit with some variation in molecular weight and intensity between clades). Clade A ASP was detected at a lower molecular weight signal than ASPs of other clades except for the 92NG083 ASP. The signal specific for the 92NG083 ASP was not due to preferential usage of the internal methionine residue, as demonstrated by the similar monomeric signals observed upon analyses with an anti-His antibody (Fig. 4C), but could be accounted for by a lower amino acid number of this ASP in the variable COOH-end region. In addition, high-molecular-weight signals, including important aggregates, were detected in cells transfected with the different ASP expression vectors. As expected, the truncated clade A ASP showed a more abundant monomeric form (versus less abundant multimeric forms), while ASPs from other clades showed a marked preference for multimeric ASPs. Unexpectedly, a similar behavior was noted for the clade G ASP representative (92NG083), for which few high-molecular-weight ASP-specific signals were observed as opposed to signals for the more abundant monomeric form.
FIG 4.
ASPs from different clade representatives form detectable multimers. (A) Sequence alignment of the predicted amino acid sequence of ASPs from HIV-1 isolates representing different clades. The peptide region used to generate anti-ASP monoclonal antibodies is highlighted above the sequence for amino acids 47 to 61. (B and C) Expression vectors of His-tagged ASPs from various HIV clades were transfected in 293T cells, and cell lysates were subsequently analyzed by Western blotting for ASP (anti-ASP and anti-His). Indicated on the right side of the panels are multimers and high-molecular-weight (MW) and monomeric (expected) 20-kDa ASP signals. The clade type for each tested ASP is indicated at the top of each lane. Symbols underneath the sequence comparison in panel A represent complete conservation (*), residues with conservation between groups of strongly similar properties (:), and residues with conservation between groups of weakly similar properties (.) (sequence alignment using the Clustal Omega tool).
These results hence showed that His-tagged ASPs from different clades can be detected with anti-ASP and anti-His antibodies. Furthermore, these results confirmed that multimer formation likely involves the amino end but that, based on the variation in multimer intensities among various tested ASPs (including results with the clade G representative), other regions of ASP are affecting the extent of multimer formation.
Induction of autophagy by ASP from different HIV-1 clades.
Since the expression of all tested ASPs could be detected and led to variation in the extent of multimer formation, we were next interested in determining if these different ASPs could indeed induce autophagy. Based on our previous results showing that levels of LC3-II (lipid-modified microtubule-associated protein 1 light chain 3)), a well-known autophagy marker, was increased in 293T and COS-7 cells expressing ASP (23), we used this same marker to evaluate the autophagy-inducing capacity of ASPs from different clade representatives. As depicted in Fig. 5A, the presence of ASP was again confirmed for all transfected ASP vectors and was detected as monomers and multimers. Importantly, all ASPs from the different clades increased levels of LC3-II, albeit to different extents, in transfected 293T cells. Furthermore, based on these Western blot analyses, clade A ASP 94CY032 was equally capable of inducing autophagy in comparison to other ASP expression vectors.
FIG 5.
ASPs from different clade representatives induce autophagy. (A and B) Expression vectors of His-tagged ASPs from various HIV clades and the empty vector were transfected in 293T cells, and cell lysates were analyzed by Western blotting for the detection of ASP (anti-His), LC3-II levels (anti-LC3), and tubulin. Signals for LC3-I, LC3-II, and ASP are indicated on the left of each panel. In panel B, after transfection, cells were treated with dimethyl sulfoxide (−) or with (+) 100 nM bafilomycin A1 (Baf A1) for 6 h. LC3-II/LC3-I ratios are indicated for each lane and are representative of fold value over the calculated ratio for untreated cells transfected with the empty vector. (C) An increasing quantity of clade 89.6 ASP expression vector was transfected in 293T cells, and cell lysates were analyzed by Western blotting for ASP, p62, and tubulin. (D) Expression vectors for His-tagged ASPs from NL4.3, 94CY032, IndieC1, and Mal proviral DNAs (versus empty vector) were transfected in 293T cells, and cell lysates were analyzed by Western blotting for ASP (anti-His), p62, and tubulin. (E and F) COS-7 cells were transfected with expression vectors of Myc-tagged ASPs from various clades. After fixation, cells were labeled with anti-Myc antibodies followed by goat anti-mouse IgG coupled to Alexa Fluor 488, stained with DAPI, and observed by confocal microscopy. Cells transfected with the expression vectors for NL4.3 ASP (clade B) and 94CY032 ASP (clade A) were next quantified for the number of ASP-positive punctate structures (F). ***, P ≤ 0.001.
We then sought to validate that this increase in LC3-II levels was due to an induction of autophagy and not to the inhibition of the late step of autophagy, both of which would lead to an increase in the abundance of LC3-II levels. Hence, transfected 293T cells were treated with bafilomycin A1, a blocking agent of lysosomal degradation. Under these conditions, ASP-dependent augmented levels of LC3-II should be maintained if ASP leads to induced autophagy, while no such increase should be noted if ASP inhibits the last step of autophagy. In Fig. 5B, Western blot analyses revealed that LC3-II levels were indeed increased upon bafilomycin A1 treatment, as expected, but that induction was still noted for all ASPs tested from clades A, B, C, D, and G. To more clearly quantify and normalize changes in LC3-II levels, densitometric analyses were performed for both LC3-I and LC3-II bands in all transfected samples. LC3-I/LC3-II ratios were then calculated and shown to be higher than ratios for empty vector-transfected cells, thereby further confirming that ASPs from the tested clades increased LC3-II levels through induced autophagy.
Since p62 degradation can also be used as a marker of autophagy, we conducted additional Western blot analyses in 293T cells transfected with an increasing quantity of the Bal (clade B) ASP expression vector. A dose-dependent decrease in p62 levels was confirmed upon ASP expression, further confirming induced autophagy (Fig. 5C). Expression vectors of ASPs from clades A, B, C, and D further revealed reduced levels of p62 in transfected cells compared to levels in empty vector-transfected cells (Fig. 5D). Additional support for the induction of autophagy in transfected COS-7 cells was provided by confocal microscopy analysis. Indeed, using anti-Myc antibodies, cytoplasmic, punctate ASP-positive signals (including cells expressing clade A ASP), which were reminiscent of autophagosomes, were clearly observed (Fig. 5E). To quantitatively assess potential autophagy induction in clade A versus that in clade B ASP, the number of ASP-positive autophagosome-like structures was counted in NL4.3 (clade B) and 94CY032 (clade A) ASP-expressing cells (Fig. 5F). Interestingly, these analyses revealed a significant reduction in the number of ASP-positive punctate signals in clade A ASP-expressing cells compared to that of clade B ASP-expressing cells.
These results confirmed that ASPs from different clades induced autophagy but that a significant reduction in the number of autophagosome-like structures was noted in clade A ASP-expressing cells compared to that in cells expressing ASPs from other clades. They also did not support an inhibitory action of ASP on the late stage of autophagy leading to degradation of the autophagosome content.
Knockout of autophagy factors increases the abundance of ASP.
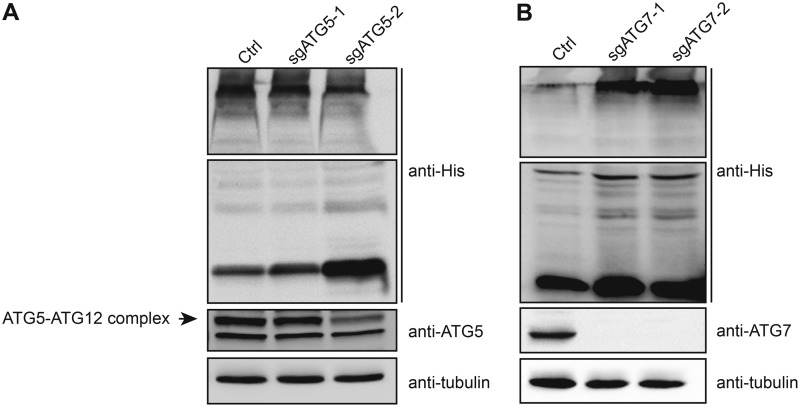
Using inhibitors of early and late steps of autophagy, we have previously demonstrated that the abundance of codon-optimized NL4.3 ASP increased (23). In order to further confirm the involvement of autophagy in the regulation of ASP, we conducted targeted knockout of the ATG5 and ATG7 genes using the CRISPR-Cas9 system by lentivirus-mediated transduction (Fig. 6). 293T cells were first stably transduced with lentivirus expressing selective single guide RNAs (sgRNAs), and following puromycin selection, two clones per knockout were subsequently transfected with the His-tagged NL4.3 ASP (clade B)-expressing vector. Resulting extracts were analyzed for ASP, ATG5, and ATG7 expression. Knockout of ATG5 or ATG7 through targeting of two different regions resulted in reduced levels of the ATG5-ATG12 complex (although the reduction was more limited for clone sgATG5-1) and of ATG7, respectively (Fig. 6A and B). Importantly, His-tagged ASP levels increased in these clones, with a greater abundance associated with lower expression levels of the targeted gene.
FIG 6.
Suppressed expression of ATG5 and ATG7 leads to increased ASP expression. Two different 293T cell clones knocked out for either ATG5 (sgATG5-1 and sgATG5-2) or ATG7 (sgATG7-1 and sgATG7-2), as indicated, and control stably transfected clones were transfected with a His-tagged NL4.3 ASP (clade B) expression vector. Cell lysates were prepared and analyzed by Western blotting using anti-His, anti-ATG5, anti-ATG7, and anti-tubulin antibodies. Monomeric and multimeric ASP signals are depicted. The signal for ATG5 is represented by the typical covalently linked ATG5-ATG12 complex.
The results thereby confirmed that ASP levels in transfected cells were modulated by autophagy and that typical autophagy factors, i.e., ATG5-ATG12 and ATG7, were implicated.
ASP interacts with the autophagy factor p62.
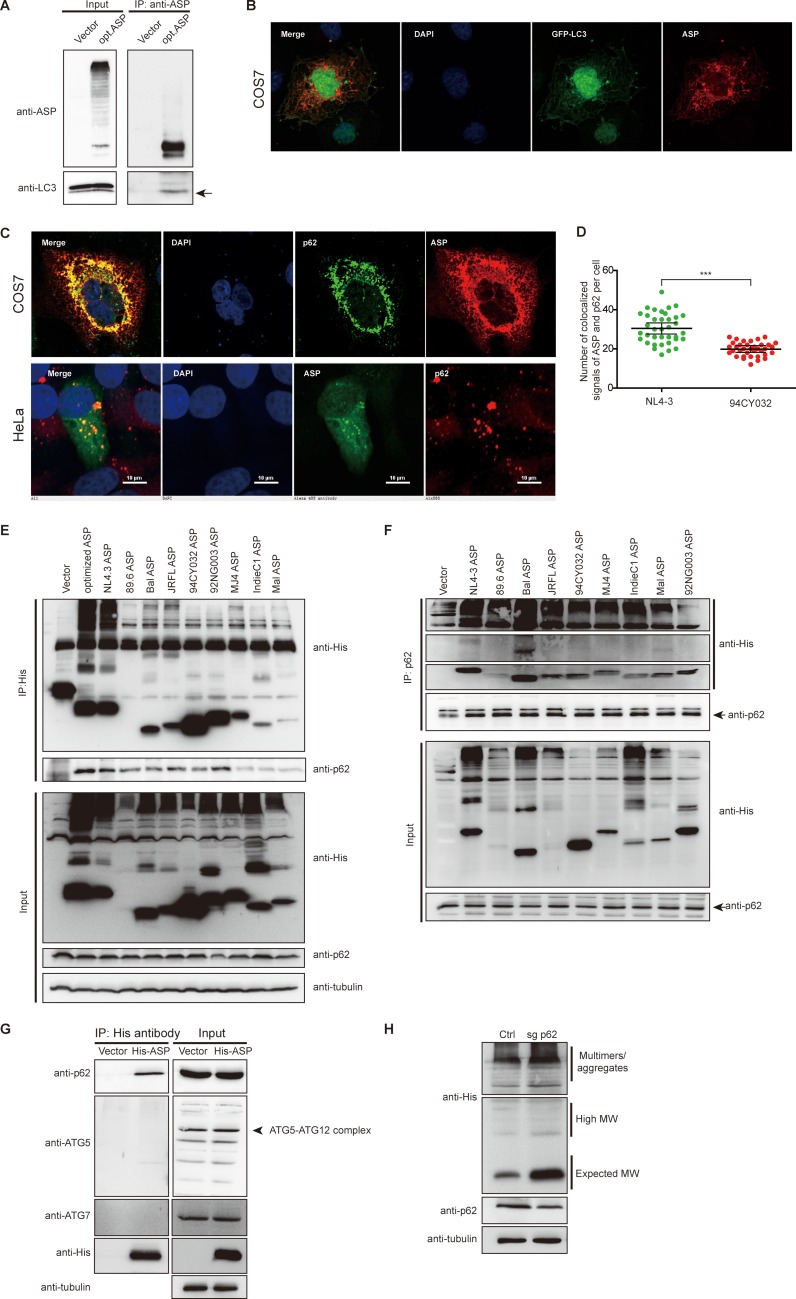
Given that all tested ASPs could induce autophagy, albeit to a lower extent for clade A ASP, we next investigated the possible mechanism leading to this induction. We first focused our attention on determining whether ASP could associate and colocalize with specific autophagy factors by confocal microscopy (Fig. 7). As we have previously reported (23), we first confirmed that expression of NL4.3 (clade B) ASP in 293T cells showed association with LC3 upon coimmunoprecipitation with anti-ASP followed by Western blot analyses (Fig. 7A). In addition, ASP was confirmed to partially colocalize with LC3-II in COS-7 cells cotransfected with expression vectors for NL4.3 ASP and the GFP-LC3 fusion protein (Fig. 7B). We next performed confocal microscopy experiments to identify potential colocalization with the autophagy factor p62. Our results revealed that a clear colocalization between NL4.3 ASP and p62/SQSTM1 was observed in both HeLa and COS-7 cells (Fig. 7C). COS-7 cells transfected with clade A and B ASPs were further used for numbering of autophagosome-like structures doubly positive for ASP and p62 (Fig. 7D). Results demonstrated that double-positive (p62 and ASP) signals were identified in clade A ASP-expressing cells but to a significantly reduced level in comparison to the level in clade B ASP-expressing cells, thereby confirming results from the experiment shown in Fig. 5.
FIG 7.
Association between ASP and p62 and effect of p62 knockout on ASP expression levels. (A) 293T cells were transfected with expression vectors for optimized NL4.3 ASP (clade B) or the empty vector pcDNA3.1. At 48 h posttransfection, coimmunoprecipitation (IP) was performed using the anti-ASP antibody, and Western blot analyses were conducted using anti-ASP and anti-LC3 antibodies. Mouse TrueBlot Ultra (anti-mouse Ig HRP) (for ASP) was used as the secondary antibody for Western blot analyses. The LC3-II signal is shown by an arrow. (B) Expression vectors for optimized NL4.3 ASP and GFP-LC3 were cotransfected in COS-7 cells. After fixation and nuclear staining with DAPI, ASP was detected with anti-ASP antibodies followed by a goat anti-mouse IgG antibody coupled to Alexa Fluor 568. (C) The NL4.3-ASP expression vector was transfected in COS-7 and HeLa cells, as indicated. Nuclei were stained with DAPI, and ASP was detected with anti-ASP antibodies followed by goat anti-mouse IgG antibodies coupled to Alexa Fluor 568 (COS-7) or Alexa Fluor 488 (HeLa), while p62 was detected with anti-p62 antibodies followed by goat anti-rabbit IgG antibodies coupled to Alexa Fluor 488 (COS-7) or Alexa Fluor 568 (HeLa). (D) Expression vectors for NL4.3 (clade B) and 94CY032 (clade A) ASPs were transfected in COS-7 cells. ASP and p62 were detected as described in panel C. Cells were next quantified for the number of punctate structures doubly positive for ASP and p62. (E and F) Expression vectors of His-tagged ASP of various HIV clades were transfected in 293T cells, and after 48 h, cellular extracts were used for coimmunoprecipitation with the anti-His (E) or anti-p62 antibodies (F). Immunoprecipitated samples and total extracts were analyzed by Western blotting using anti-p62, anti-His, and anti-tubulin antibodies. (G) The expression vector for His-tagged NL4.3 ASP (versus empty vector) was transfected in 293T cells, and, at 48 h posttransfection, cellular extracts were used for coimmunoprecipitation with anti-His antibodies. Immunoprecipitated samples and total extracts were analyzed by Western blotting using anti-His, anti-ATG5, anti-ATG7, anti-p62, and anti-tubulin antibodies. The ATG5-ATG12 signal is shown by an arrow. (H) A 293T cell clone knocked out for the p62 gene (sgp62) and a control stably transfected clone (Ctrl) were transfected with His-tagged NL4.3 ASP expression vector. Cell lysates were prepared and analyzed by Western blotting using anti-His, anti-p62, and anti-tubulin antibodies. Monomeric and multimeric signals (including high-molecular-weight) are identified on the right side of the panels. ***, P ≤ 0.001.
Since ASP forms multimers and since p62 is an important cargo transporter of multimerized proteins toward the autophagy pathway, we next tested whether ASP and p62/SQSTM1 could indeed associate. 293T cells were transfected with expression vectors of His-tagged ASPs of different clades, and resulting extracts were used for coimmunoprecipitation with an anti-His antibody (Fig. 7E). Upon Western blot analyses, p62 indeed could coimmunoprecipitate with ASP in transfected 293T cells. To confirm these data, immunoprecipitation was performed in similarly transfected 293T cells with anti-p62 antibodies and analyzed with anti-His antibodies (Fig. 7F). These analyses confirmed the association between ASP and p62. The specificity of this interaction was demonstrated in 293T cells expressing NL4.3 ASP by the lack of association of ASP with other autophagy factors, such as ATG5 and ATG7, as assessed by similar coimmunoprecipitation experiments (Fig. 7G). The impact of this association between ASP and p62 was further tested through knockout of the p62 gene in 293T cells. Although two different targeted regions were tested, only one strategy (sgp62-1) provided sufficient reduction in p62 abundance. Knockout cells were thus transfected with the NL4.3 (clade B) ASP expression vector and analyzed by Western blotting. These analyses indicated that reduction in p62 levels led to a marked increase in ASP abundance (Fig. 7H).
These data, thus, indicated that p62 colocalized and associated with the different ASP clade representatives and that this association led to induction of autophagy.
Interaction domain of p62 and ASP ubiquitination.
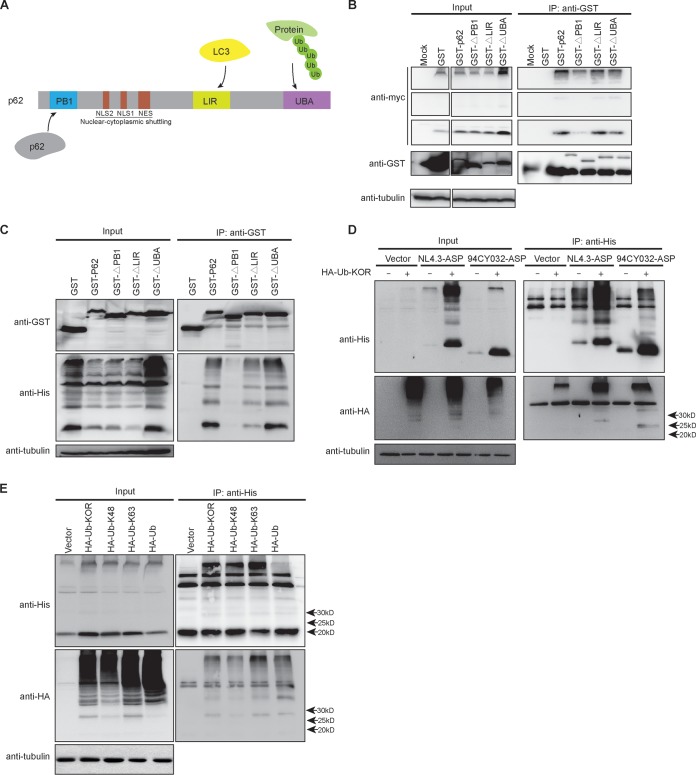
We next sought to identify the domains of p62 which were responsible for its association with ASP. These domains are known as UBA, LIR, and PB1 and have been previously shown to interact with ubiquitinated protein, LC3, and p62 itself, respectively (Fig. 8A). Expression vectors of different p62 mutants with deletions of these various domains and fused to the GST protein were thus cotransfected with the Myc-tagged NL4.3 (clade B) ASP expression vector in 293T cells, and resulting extracts were coimmunoprecipitated with anti-glutathione S-transferase (GST) antibodies (Fig. 8B). Upon Western blot analysis of immunoprecipitated extracts, interaction of ASP (monomeric and multimeric) was shown to be importantly reduced in cells expressing the PB1 domain-deleted p62 mutant (Fig. 8B). Expression of other mutants (with deletions of the LIR or UBA region) led to retention of ASP in extracts coimmunoprecipitated with anti-GST antibodies, thereby suggesting that these other domains were not crucial for the association between ASP and p62. To demonstrate that the interaction was not dependent on the presence of a Myc tag, similar experiments were performed with a His-tagged version of ASP (Fig. 8C). Following cotransfection of expression vectors for the various p62 mutants and His-tagged NL4.3 ASP in 293T cells, similar results were obtained upon Western blot analyses in that deletion of the PB1 domains led to loss of the association of this mutant with ASP.
FIG 8.
The interaction of p62 with ASP requires its homodimerization domain despite ASP ubiquitination. (A) The various domains of the p62 protein are depicted and include the ubiquitination-binding domain UBA, the LC3-binding domain LIR, the regions of the nuclear localization signal (NLS) and nuclear export signal (NES), and the homodimerization domain PB1. (B and C) 293T cells were cotransfected with expression vectors of GST-tagged wild type and deletion mutants of p62 and Myc-tagged (B) or His-tagged (C) NL4.3 ASP. At 48 h posttransfection, immunoprecipitation was performed using the anti-GST antibody, and Western blot analyses of immunoprecipitated samples and total extracts were conducted using anti-GST, anti-Myc/anti-His, and anti-tubulin antibodies. (D and E) 293T cells were cotransfected with expression vectors for His-tagged NL4.3 (clade B) or 94CY032 (clade A) ASP (panel D only) and for the HA-tagged ubiquitin (Ub)-KOR mutant (restricted to monoubiquitination). In panel E, expression vectors for HA-tagged ubiquitin WT or the restrictive ubiquitin-K48 and ubiquitin-K63 were also included. At 48 h posttransfection, immunoprecipitation was performed with anti-His antibodies, and Western blot analyses of immunoprecipitated samples and total extracts were conducted using anti-His, anti-HA, and anti-tubulin antibodies. Arrows on the right side of the right panels indicate the positions and expected molecular weights (25 to 30 kDa) of the ubiquitinated ASPs from the different tested proviral DNAs.
Since p62/SQSTM1 has often been implicated in ubiquitin-dependent autophagy, it was surprising that the association between ASP and p62 did not involve the ubiquitinating-interacting UBA domain. We were thus interested in determining if ASP was nonetheless ubiquitinated. Immunoprecipitation experiments were conducted in 293T cells cotransfected with expression vectors for clade B NL4.3 and clade A 94CY032 His-tagged ASPs and for pRK5-HA-ubiquitin-KOR (expressing a hemagglutinin [HA]-tagged ubiquitin protein limiting targeted proteins to monoubiquitination). Upon transfection of 293T cells and immunoprecipitation with anti-His antibodies, Western blot analyses revealed that ASP was indeed ubiquitinated, as revealed by the expected size of a ubiquitinated monomeric ASP form for both clade A and clade B ASP-expressing cells (between 25 and 30 kDa) (Fig. 8D). In addition, high-molecular-weight multimers were detected in immunoprecipitated extracts. Interestingly, for both tested ASP expression vectors, levels of the nonubiquitinated form were also importantly increased in transfected cells expressing ubiquitin KOR, likely suggesting that these might represent nonubiquitinated ASP protein comprising ASP multimers targeted for ubiquitin-dependent degradation. In fact, high-molecular-weight signals were also importantly more abundant in ubiquitin-KOR-expressing cells. Further analyses using 293T cells cotransfected with vectors expressing His-tagged NL4.3 ASP and HA-tagged ubiquitin selective for K48-linked (ubiquitin-proteasome system [UPS]-prone) or K63-linked (autophagy-prone) polyubiquitination revealed that ASP was targeted by both polyubiquitin chains, as indicated by immunoprecipitation experiments using anti-His antibodies (Fig. 8E).
Together, these data demonstrated that, despite the implication of the homodimerization domain of p62 in its association with ASP, ASP is ubiquitinated, which impacts its abundance in its monomeric and multimeric forms.
DISCUSSION
Antisense transcription in HIV-1 has been described and characterized in a number of studies (18). However, the encoded ASP remains a limited focus of investigation. We have formerly detected clade B ASP in different cell types by various technical approaches and further demonstrated that this viral protein led to the induction of autophagy (7, 17, 23). In the current study, we demonstrate that ASP-associated autophagy is induced in all tested clade-derived ORFs and is associated with p62 via its oligomerization domain. Furthermore, ASP was shown to be ubiquitinated, which impacted its sensitivity toward degradation.
Based on our earlier studies linking ASP-induced autophagy to the ASP multimerization capacity, we pursued our analysis and confirmed that ASP is indeed part of multimers by showing the interaction between two differently tagged ASPs, which confirmed our previous findings using a baculovirus-derived purified protein (23). Interestingly, removal of the cysteine-rich amino-terminal region of ASP in the ASPΔN1–15 mutant led to a similar lower abundance of detected high-molecular-weight signals with an increase in the monomeric version while no variation in secondary structure could be noted for this mutant. This observation was further paralleled by results obtained with the clade A ASP lacking the first 26 amino acids, which showed more monomeric forms and fewer multimers in our Western blot analyses. The amino-terminal region of the full-length ASP (all clades, except clade A) is therefore an important region in controlling ASP stability, multimer formation, and likely affects ASP capacity to induce autophagy. Its cysteine-rich nature contributes to multimer formation, as demonstrated by the reduction of high-molecular-weight signals in a mutant deleted of one of the cysteine triplets and by the sensitivity of multimers to denaturing agents. However, an unexpected finding was that the ASPΔN1–30 mutant demonstrated lower levels of expression. Although we cannot determine the reason for this reduced expression, the removal of 15 additional amino acids in comparison to the ASPΔN1–15 mutant could in this case change the conformation of the protein, with possible misfolding and degradation by the autophagy-independent proteasome complex. This might also be reminiscent of the slight increase in the abundance of the monomeric form of the ASPΔN1–15 mutant seen in COS-7 cells despite severe loss of multimeric forms.
Other HIV-1 proteins are known to form multimers through cysteine residues (47, 48). Although our results indeed argue that the cysteine triplet (positions 10 to 12) is implicated in ASP multimer formation and subsequent autophagy induction, we have not fully investigated which cysteines are most important. It is in fact highly likely that various well conserved cysteine residues, such as 22CCC24, are involved, some of which could also lie outside the tested regions (e.g., positions 34, 146, and 152). In fact, since multimer formation and induced autophagy were not completely abrogated in cells expressing the amino-terminally deleted version of NL4.3 ASP, other highly conserved regions (9) are likely to be involved in the multimerization of ASP. This is further evidenced by our analyses of the clade G ASP, which behaved similarly to clade A ASP even though it expresses the full-length ASP. Importantly, residues other than cysteines are known to be implicated in hydrophobic and electrostatic interactions, which are equally crucial for the formation of multimers, and more experiments will be needed to address these various possibilities.
In this study, we have focused on the comparison of the different ASPs from the different clades, including those from clinical isolates and laboratory strains, in terms of their capacities to activate the autophagy pathway. Despite variation in multimer formation, all tested ASPs induced autophagy, as determined by variation in the levels of p62 and LC3-II levels and by the presence of autophagosome-like signals by confocal microscopy. Variation in the extent of the induction of autophagy by the various clade ASPs, as assessed by LC3-II levels and counting of autophagosome-like punctate signals, was noted and suggested a lower autophagy-inducing capacity for clade A ASP. However, these differences might also be accounted for by differences in expression levels of the different ASPs in transfected cells. Nonetheless, although we have not tested all known clades (having limited our analyses to the most frequent ones), our data argue for a conserved capacity of ASP from the different clades to induce autophagy and indicate that a lower abundance of multimers for certain ASPs is nonetheless sufficient for this induction.
Several studies have clearly shown that aggregating proteins are targeted for degradation by the autophagy pathway (49, 50). We have further confirmed the implication of autophagy on the abundance of ASP by targeting known autophagy factors through CRISPR-mediated deletion. In these analyses, ASP levels (monomer and multimers) were strongly affected, and their increase in abundance inversely correlated with the remaining levels of the targeted protein. We have further looked at the potential mechanisms of autophagy induction by ASP and, through localization and immunoprecipitation experiments, an association between ASP and p62 was detected, which was dependent on the PB1 homodimerization domain but not affected in a p62 mutant deleted of its ubiquitin-interacting UBA region. However, expression of this mutant did tend to increase ASP levels in transfected cells, suggesting that this domain influences ASP stability. Interestingly, previous studies have shown that various viral proteins, including HIV-1 Tat, could interact with p62 via a ubiquitin-independent manner (51, 52).
The latter results were unexpected, given that we further revealed that ASP is ubiquitinated and that these posttranslational modifications contributed to ASP degradation, as demonstrated by the use of the ubiquitin mutant preventing polyubiquitination. Expression of this polyubiquitination-blocking ubiquitin version indeed led to an important increase in abundance of the nonubiquitinated monomeric and multimeric forms of ASP, with the former likely resulting from their presence in multimer complex and subsequent release upon cell extract preparation. Hence, as these multimers are composed of nonubiquitinated and ubiquitinated ASPs, they are thus subject to degradation by autophagy through their interaction with p62. Although it is not clear how ASP triggers autophagy, the recent publication of Lu et al. might shed light on the exact mechanism behind ASP-dependent induction of autophagy. The authors indeed argued that association of ubiquitinated multimeric proteins with p62 with subsequently induced autophagy is more importantly dependent on the aggregated nature of the protein than on ubiquitination itself (53). From this perspective, it is likely that the association of ASP with p62 mainly depends on its oligomeric form and that the interaction with the ubiquitin-interacting UBA domain might be less important, thereby explaining why we were not able to demonstrate that this domain was participating in formation of the p62/ASP complex. This is thus different from the previously suggested view that autophagy versus the UPS is mainly dependent on the type of linked ubiquitin in the polyubiquitination chain (K48 versus K63) (54). Our results in fact argue that both types of modification are present in monomeric and multimeric forms of ASP.
It is well known that autophagy is primarily responsible for the degradation of most long-lived proteins in cells and also targets aggregated proteins as well as cellular organelles and infectious organisms (55). The importance of autophagy in modulating the abundance of ASP might be crucial to maintain its abundance at low levels as this protein is likely detrimental to cell survival (if in high excess), especially given its propensity to form important multimers. In addition, the capacity of ASP to function as an inducer of autophagy likely depends on other concomitantly expressed viral proteins which have been associated with this degradation pathway (39). As previously reported, Nef can inhibit autophagy at late steps of viral replication in macrophages and could thus block ASP-induced autophagy (5, 36). Another interesting aspect of our studies relates to the essential role played by autophagy in antigen presentation in cells, such as dendritic cells. Infected DCs might thus be altered in their capacity to adequately process and present antigen, especially given that our previous study showed that ASP presented higher levels of expression in this cell type than in both macrophages and activated CD4+ T cells (17).
Overall, our results demonstrate that the autophagy-inducing properties of ASP are observed for different clades and that multimerization acts on autophagy induction. We have not tested the ASPs of all clades, but we believe that they should also be autophagy inducing. Furthermore, based on the in silico analyses of Cassan et al., HIV encoding shorter versions of ASP might still be capable of inducing autophagy. Our results further indicated that the association of p62 with ASP is likely essential for ASP and that ASP is ubiquitinated, also affecting the stability of the monomeric and multimeric forms of ASP. These results thereby help us to better appreciate the unstable nature of ASP in mammalian cells. The function of ASP in HIV-1 infection in vivo has yet to be defined, but our results suggest that autophagy could link ASP to HIV-mediated pathogenesis and the chronic infection state. Future studies will be determinant in providing more information on these potential roles.
MATERIALS AND METHODS
Plasmids.
Expression vectors for Myc-tagged optimized ASP and GFP-ASP optimized fusion proteins have been previously described (23). A series of deletion mutants (15 and 30 amino acids at the amino or carboxyl end) and internal deletion of a cysteine triplet or PXXPXXP motif were generated by PCR from Myc-tagged optimized ASP expression vectors and termed pMyc-optimized-ASPΔN1–15, pMyc-optimized-ASPΔN1–30, pMyc-optimized-ASPΔC174–189, pMyc-optimized-ASPΔC159–189, pMyc-optimized-ASPΔ10CCC12, pMyc-optimized-ASPΔPXXP, and pMyc-optimized-ASPΔPXXPXXP. The following primers were used: pMyc-optimized-ASPΔN1–15 (forward, 5′-GCCCTGTCCAAGCTGTTCT-3′; reverse, 5′-GTCCTCCTCGCTGATCAGCTT-3′), pMyc-optimized-ASPΔN1–30 (forward, 5′-CTGGCCTGCACAGTGAGC-3′; reverse, 5′-GTCCTCCTCGCTGATCAGCTT-3′), pMyc-optimized-ASPΔC174–189 (forward, 5′-TGAGGTACCAAGCTTAAGTTTAAA-3′; reverse, 5′-CTGCAGCACGCTAGGGTC-3′), pMyc-optimized-ASPΔC159–189 (forward, 5′-CACAGATCCGTGCAGAATGAA-3′; reverse, 5′-GCGGTTGCAGCTCACGGTCT-3′), pMyc-optimized-ASPΔ10CCC12 (forward, 5′-GCCTCTATCGCCCTGTCCAAG-3′; reverse, 5′-GCGGTTGCAGCTCACGGTCT-3′), pMyc-optimized-ASPΔPXXP (forward, 5′-AAAAATCCAAGGAACAAGGC-3′; reverse, 5′-CAGCACGATGGGGGCGGC-3′), and pMyc-optimized-ASPΔPXXPXXP (forward, 5′-AGGAACAAGGCCCCCATCCC-3′; reverse, 5′-CAGCACGATGGGGGCGGC-3′). Proviral DNAs of p94CY032 (clade A), p92RW009 (clade A), NL4.3 (clade B), JR-FL (clade B), 89.6 (clade B), MJ4 (clade C), p92NG003 (clade G), and p92NG083 (clade G) were obtained through the NIH AIDS Reagent Program (Germantown, MD), while proviral DNAs of Mal (clade D) and IndieC1 (clade C) were provided by Anne Gatignol (McGill University, Montreal, Canada). DNA fragments encoding His-tagged antisense protein (ASP) were amplified by PCR from these proviral DNAs or the Bal (clade B) envelope expression vector (provided by Michel J. Tremblay, Laval University, Quebec City, Canada) using specific forward and reverse primers bearing NotI and BamHI restriction sites, respectively, at their 5′ends. After NotI/BamHI digestion, PCR products were inserted into the vector pTT5 (VT2202; Youbio, Inc., China), which was similarly digested. CRISPR-Cas9 knockout plasmids (LentiCRISPRv2 [98290; Addgene] and psPAX2 [12260; AddGene]) were constructed as previously described (56, 57). Two pairs of oligonucleotides were designed to specifically target each gene, as follows: 5′-CACCGAACTTGTTTCACGCTATATC-3′ and 5′-AAACGATATAGCGTGAAACAAGTTC-3′ (sgATG5-1); 5′-CACCGAAGATGTGCTTCGAGATGTG-3′ and 5′-AAACCACATCTCGAAGCACATCTTC-3′ (sgATG5-2); 5′-CACCGCACCGTGAAGGCCTACCTTC-3′ and 5′-AAACGAAGGTAGGCCTTCACGGTGC-3′ (sgp62-1); 5′-CACCGACCGTGAAGGCCTACCTTCT-3′ and 5′-AAACAGAAGGTAGGCCTTCACGGTC-3′ (sgp62-2); 5′-CACCGGAAGCTGAACGAGTATCGGC-3′ and 5′-AAACGCCGATACTCGTTCAGCTTCC-3′ (sgATG7-1); 5′-CACCGGCTGCCAGCTCGCTTAACAT-3′ and 5′-AAACATGTTAAGCGAGCTGGCAGCC-3′ (sgATG7-2). Annealing of each pair further led to the generation of BsmBI restriction sites, which were used to insert the resulting dimerized oligonucleotide in the BsmBI-digested LentiCRISPRv2 vector. Plasmids pDEST-GST-P62, pDEST-GST-P62-ΔLIR, pDEST-GST-P62-ΔUBA, and pDEST-GST- P62-ΔPB1 were kindly provided by Lucile Espert (Université Montpellier, Montpellier, France) and express the wild type and various deletion mutants of p62 (58). The plasmids pRK5-HA-ubiquitin-WT (17608; Addgene), pRK5-HA-ubiquitin-KOR, pRK5-HA-Ubiquitin-K48, d pRK5-HA-ubiquitin-K63, and the empty vector (17605; Addgene) were gifts from Ted Dawson (Johns Hopkins University School of Medicine, Baltimore, MD) (59). The pRK5-HA-ubiquitin WT vector expresses HA-tagged wild-type ubiquitin, while pRK5-HA-ubiquitin-KOR expresses a ubiquitin version in which all lysines were mutated to arginines. Vectors pRK5-HA-ubiquitin-K48 and pRK5-HA-ubiquitin-K63 express ubiquitin limited to specific lysine-linked polyubiquitination.
Cell lines and transfection.
Human embryonic kidney 293T, African green monkey kidney COS-7, and human cervical HeLa cell lines were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Wisent Bioproducts, St-Bruno, Canada). Cell lines were obtained from the American Type Culture Collection (ATCC) (Manassas, VA). All DNA plasmids were transfected using polyethylenimine (PEI) (Polysciences, Warrington, PA) at a 7:1 ratio of PEI/total DNA (in grams) in FBS-free DMEM; empty vectors were added to normalize DNA quantities in between transfections and for transfection control samples. After an incubation of 20 min at room temperature, the PEI-DNA mixture was added to cells for 6 h, after which medium was removed and replaced by fresh supplemented DMEM. In certain transfections, cells were treated with bafilomycin A1 (100 nM) for 6 h at 24 h posttransfection.
Generation of CRISPR-Cas 9 stable cell lines.
Pseudotyped lentiviruses were produced by cotransfecting 293T cells with lentiCRISPRv2 or lentiCRISPRv2-sgRNA (specific to ATG5, ATG7, or p62), the HIV-1 backbone-containing psPAX2, and the vesicular stomatitis virus (VSV) envelope-encoding-pVSVg vector (8454; Addgene) using the PEI agent. At 36 h posttransfection, viral supernatants were filtered with 0.22-μm-pore-size filters (Millipore Corporation, Billerica, MA) and added to 293T and COS-7 cells. CRISPR-Cas9 stable knockout cell lines were selected and maintained in DMEM supplemented with 10% FBS and 0.2 μg/ml puromycin. Clones were generated by 10× serial dilutions, and single clones were further expanded in puromycin-containing medium. Selected and amplified clones were analyzed by PCR for the targeted region.
Antibodies.
Anti-GST antibodies were purchased from GE Healthcare Life Science (Chicago IL). Anti-Flag, anti-tubulin, anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH), anti-LC3, anti-ATG5, anti-ATG7, anti-ATG12, anti-Beclin-1, and anti-p62 antibodies were purchased from Sigma-Aldrich (St-Louis, MO). Antibodies against Myc, His, and HA tags, horseradish peroxidase (HRP)-conjugated anti-GFP, HRP-conjugated goat anti-mouse, and anti-rabbit IgG antibodies were ordered from Santa Cruz Biotechnology, Inc. (Dallas TX). Mouse TrueBlot Ultra (anti-mouse Ig HRP) was purchased from Rockland, Inc. (Limerick, PA). Goat anti-mouse IgG coupled to Alexa Fluor 488 or to Alexa Fluor 568 was obtained from Thermo Fisher Scientific (Waltham, MA). Monoclonal anti-ASP antibodies generated by Eurogentec (Liege, Belgium) were kindly provided by Jean-Michel Mesnard (Université Montpellier, Montpellier, France) and are specific to the amino acid sequence indicated in Fig. 4A.
Purification of ASP and circular dichroism spectroscopy.
pTT5-His-tagged-ASP wild type and ASPΔN1–15 were transfected into 293T cells (2 × 108). At 36 h posttransfection, cells were lysed in 10 ml of lysis buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, 1% Triton with proteinase inhibitor) (Roche, Mississauga, Canada). Lysates were centrifuged at 2,000 × g for 15 min at 4°C. Ni Sepharose High Performance beads (17526802; GE Healthcare) were added to cell extracts and incubated at 4°C overnight. Beads were gently washed three times with cold phosphate-buffered saline (PBS). Proteins were eluted with 500 μl of 20 mM glycine, pH 2.0, for 10 min at 37°C and then neutralized with 500 μl of 100 mM ammonium bicarbonate (ABC), pH 8.5 buffer. Proteins were next dialyzed with Slide-A-Lyzer 10-kDa-cutoff dialysis cassettes (66380; Thermo Scientific) in PBS buffer at 4°C. Resulting samples were then loaded in Ultracel-30K filter tubes (UFC803024) and further centrifuged at 4,000 × g for 15 min at 4°C. The lower fraction was reloaded into Ultracel-10K filter tubes (R8DA51035) and centrifuged at 4,000 × g for 20 min at 4°C. The concentration of each purified monomeric ASP was determined with a bicinchoninic acid (BCA) protein assay kit (23225; Thermo scientific) before analysis by circular dichroism (CD) spectroscopy. Far-UV (CD) spectra were recorded at 25°C using a J-810 CD spectrometer (Jasco) in a 10-mm-path-length quartz cuvette. Recombinant WT ASP and ASPΔN1–15 were analyzed at a concentration of 109.5 ng/ml (WT) and 111.5 ng/ml (ASPΔN1–15) in PBS buffer, pH 7.4. Spectra were recorded from 260 to 190 nm and were corrected by subtracting the value for the blank solution (protein-free buffer). The wavelength step was set at 0.5 nm with an average time of 10 s per scan at each wavelength step. The raw data were converted to mean residue ellipticity (MRE) using the following formula:
Western blot analyses and immunoprecipitation.
Cells were washed in 1× PBS and then incubated in lysis buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, 1% Triton, with proteinase inhibitor) for 15 min on ice. In certain experiments, β-mercaptoethanol (2%) and various concentrations of DTT (0.1 to 0.5 M) were added to the lysis buffer. The lysates were centrifuged at 2,000 × g for 15 min at 4°C. Proteins (the supernatant) were denatured by mixing 20-μl lysates with 4× loading solution (12% sodium dodecyl sulfate [SDS] with 3 mM Tris, pH 6.8, and 0.05% bromophenol blue), followed by incubation at 70°C for 10 min. For immunoprecipitation, SureBeads protein G magnetic beads (Bio-Rad, Hercules CA) were resuspended (20 μl per sample) and washed twice with 500 μl of cold 0.1% Tween 20 in PBS (PBS-T). Antibodies diluted in 500 μl of PBS-T (anti-Myc [1:200], anti-p62 [1:100], anti-His [1:200], and anti-GST [1:100]) were then added to the beads for 1 h at room temperature, and beads were next washed three times with cold PBS-T. Total cell extracts (lysed in 50 mM Tris-HCl, pH 8, 100 mM NaCl, 1 mM EDTA, and 1% Triton) were then incubated with the antibody-bead complex overnight at 4°C. After three washes with cold PBS-T, bound fractions were eluted with 20 μl of 2× loading buffer heated at 100°C. Samples were run on 10 to 12% SDS-PAGE gels and blotted onto an Immun-Blot polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories, USA) in PBS-T. Membranes were blocked with 5% milk in PBS-T at room temperature for 1 h and incubated with anti-ASP (1:2,000), anti-GFP (1:5,000), anti-Myc (1:250), anti-HA (1:10,000), anti-GST (1:3,000), anti-His (1:5,000), anti-p62 (1:2,000), anti-ATG5 (1:2,000), anti-ATG7 (1:2,000), anti-LC3 (1:500), anti-Beclin-1 (1:500), anti-GAPDH (1:1,000), or anti-tubulin (1:10,000), antibodies at 4°C overnight. Membranes were next incubated with the appropriate HRP-conjugated secondary antibodies (1 μg/ml) at room temperature for 2 h and visualized with an ECL Western blotting detection kit (Immobilon Western, Millipore Corporation, Billerica, MA). Image acquisition was performed with Fusion FX7 (Vilber Lourmat, Marne-la-Vallée, France). For certain experiments, densitometric analyses were conducted on LC3-related signals, and an LC3-II/LC3-I ratio was calculated and normalized to the value for cells transfected with an empty vector (set at a value of 1).
Flow cytometry.
Transfected COS-7 and 293T cells were washed with PBS, fixed with 4% formaldehyde for 10 min, and permeabilized with 0.1% Triton X-100 for 5 min at room temperature. Cells were then washed three times with PBS and incubated with the anti-Myc antibody (dilution, 1:250) overnight at 4°C. After three additional washes with PBS, cells were incubated with goat anti-mouse IgG antibodies coupled to Alexa Fluor 488 (1/500) (Invitrogen, Canada, Inc.) for 1 h at 4°C. Cells were fixed with 1% formaldehyde and incubated overnight at 4°C before analysis with a FACScan device (BD Biosciences).
Confocal microscopy.
COS-7 and HeLa cells were seeded in 24-well plates containing a 1.5-mm-thick coverslip for 12 h and then transfected, as described above. At 48 h posttransfection, cells were rinsed twice with cold PBS and fixed with 4% formaldehyde-PBS for 10 min at room temperature on a slow rotatory shaker. Cells were next washed four times with PBS–0.1% Triton X-100. Fixed cells were incubated with a blocking solution (3% BSA, 5% milk, 50% FBS, 0.1% Triton X-100, and 0.05% NaN3 in PBS) overnight at 4°C, rinsed with cold PBS three times, and incubated with anti-p62 (1:250), anti-ATG5 (1:250), anti-ATG7 (1:250), anti-LC3 (1:250), anti-ASP (1:500), or anti-Myc (1:250) antibodies in a PBS solution containing 3% BSA, 0.1% Triton X-100, and 0.05% NaN3 at 4°C overnight. Cells were next washed with cold PBS–0.1% Triton X-100 and incubated with a secondary antibody (1:500, goat anti-mouse IgG coupled to Alexa Fluor 488 or goat anti-rabbit IgG coupled to Alexa Fluor 568) in the PBS solution described above for 45 min at room temperature. Cells were washed with PBS–0.1% Triton X-100 and incubated in immune-mount solution in the presence of 1% 4′,6-diamidino-2-phenylindole (DAPI). All cell samples were visualized with a Nikon A1 laser scanning confocal microscope (Nikon Canada, Mississauga, Canada) through a 60× objective under oil immersion. All confocal images were analyzed with ImageJ software. The multipoint selection feature was used to select ASP- and ASP/p62-positive punctate signals, which were then automatically counted. Data were analyzed by a paired Student's t test.
ACKNOWLEDGMENTS
This work was supported by a grant to B.B. from the Canadian Institutes of Health Research (HBF-317645) and through a Canada Research Chair in Human Retrovirology (Tier 2). C.T. was supported by an institutional FARE scholarship, and Z.L was awarded a scholarship from the Fonds de Recherche du Québec-Nature et Technologies.
We thank Denis Flipo for his excellent technical assistance. We are also thankful to Lucile Espert and Jean-Michel Mesnard for readings of the manuscript and helpful comments and for providing materials.
REFERENCES
- 1.Zander K, Sherman MP, Tessmer U, Bruns K, Wray V, Prechtel AT, Schubert E, Henklein P, Luban J, Neidleman J, Greene WC, Schubert U. 2003. Cyclophilin A interacts with HIV-1 Vpr and is required for its functional expression. J Biol Chem 278:43202–43213. doi: 10.1074/jbc.M305414200. [DOI] [PubMed] [Google Scholar]
- 2.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valera M-S, de Armas-Rillo L, Barroso-González J, Ziglio S, Batisse J, Dubois N, Marrero-Hernández S, Borel S, García-Expósito L, Biard-Piechaczyk M, Paillart J-C, Valenzuela-Fernández A. 2015. The HDAC6/APOBEC3G complex regulates HIV-1 infectiveness by inducing Vif autophagic degradation. Retrovirology 12:53. doi: 10.1186/s12977-015-0181-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. 2015. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526:212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell GR, Rawat P, Bruckman RS, Spector SA. 2015. Human immunodeficiency virus type 1 Nef inhibits autophagy through transcription factor EB sequestration. PLoS Pathog 11:e1005018. doi: 10.1371/journal.ppat.1005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen N, McCarthy C, Drakesmith H, Li D, Cerundolo V, McMichael AJ, Screaton GR, Xu XN. 2006. HIV-1 down-regulates the expression of CD1d via Nef. Eur J Immunol 36:278–286. doi: 10.1002/eji.200535487. [DOI] [PubMed] [Google Scholar]
- 7.Clerc I, Laverdure S, Torresilla C, Landry S, Borel S, Vargas A, Arpin-Andre C, Gay B, Briant L, Gross A, Barbeau B, Mesnard JM. 2011. Polarized expression of the membrane ASP protein derived from HIV-1 antisense transcription in T cells. Retrovirology 8:74. doi: 10.1186/1742-4690-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller RH. 1988. Human immunodeficiency virus may encode a novel protein on the genomic DNA plus strand. Science 239:1420–1422. doi: 10.1126/science.3347840. [DOI] [PubMed] [Google Scholar]
- 9.Cassan E, Arigon-Chifolleau AM, Mesnard JM, Gross A, Gascuel O. 2016. Concomitant emergence of the antisense protein gene of HIV-1 and of the pandemic. Proc Natl Acad Sci U S A 113:11537–11542. doi: 10.1073/pnas.1605739113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanhée-Brossollet C, Thoreau H, Serpente N, D'Auriol L, Lévy J-P, Vaquero C. 1995. A natural antisense RNA derived from the HIV-1 env gene encodes a protein which is recognized by circulating antibodies of HIV+ individuals. Virology 206:196–202. doi: 10.1016/S0042-6822(95)80034-4. [DOI] [PubMed] [Google Scholar]
- 11.Michael NL, Vahey MT, d'Arcy L, Ehrenberg PK, Mosca JD, Rappaport J, Redfield RR. 1994. Negative-strand RNA transcripts are produced in human immunodeficiency virus type 1-infected cells and patients by a novel promoter downregulated by Tat. J Virol 68:979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi-Ishihara M, Yamagishi M, Hara T, Matsuda Y, Takahashi R, Miyake A, Nakano K, Yamochi T, Ishida T, Watanabe T. 2012. HIV-1-encoded antisense RNA suppresses viral replication for a prolonged period. Retrovirology 9:38. doi: 10.1186/1742-4690-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peeters A, Lambert PF, Deacon NJ. 1996. A fourth Sp1 site in the human immunodeficiency virus type 1 long terminal repeat is essential for negative-sense transcription. J Virol 70:6665–6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landry S, Halin M, Lefort S, Audet B, Vaquero C, Mesnard JM, Barbeau B. 2007. Detection, characterization and regulation of antisense transcripts in HIV-1. Retrovirology 4:71. doi: 10.1186/1742-4690-4-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Briquet S, Vaquero C. 2002. Immunolocalization studies of an antisense protein in HIV-1-infected cells and viral particles. Virology 292:177–184. doi: 10.1006/viro.2001.1224. [DOI] [PubMed] [Google Scholar]
- 16.Saayman S, Ackley A, Turner AM, Famiglietti M, Bosque A, Clemson M, Planelles V, Morris KV. 2014. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol Ther 22:1164–1175. doi: 10.1038/mt.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laverdure S, Gross A, Arpin-Andre C, Clerc I, Beaumelle B, Barbeau B, Mesnard JM. 2012. HIV-1 antisense transcription is preferentially activated in primary monocyte-derived cells. J Virol 86:13785–13789. doi: 10.1128/JVI.01723-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbeau B, Mesnard JM. 2015. Does chronic infection in retroviruses have a sense? Trends Microbiol doi: 10.1016/j.tim.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Zapata JC, Campilongo F, Barclay RA, DeMarino C, Iglesias-Ussel MD, Kashanchi F, Romerio F. 2017. The human immunodeficiency Virus 1 ASP RNA promotes viral latency by recruiting the polycomb repressor complex 2 and promoting nucleosome assembly. Virology 506:34–44. doi: 10.1016/j.virol.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bet A, Maze EA, Bansal A, Sterrett S, Gross A, Graff-Dubois S, Samri A, Guihot A, Katlama C, Theodorou I, Mesnard JM, Moris A, Goepfert PA, Cardinaud S. 2015. The HIV-1 antisense protein (ASP) induces CD8 T cell responses during chronic infection. Retrovirology 12:15. doi: 10.1186/s12977-015-0135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger CT, Llano A, Carlson JM, Brumme ZL, Brockman MA, Cedeno S, Harrigan PR, Kaufmann DE, Heckerman D, Meyerhans A, Brander C. 2015. Immune screening identifies novel T cell targets encoded by anti-sense reading frames of HIV-1. J Virol doi: 10.1128/JVI.03435-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bansal A, Carlson J, Yan J, Akinsiku OT, Schaefer M, Sabbaj S, Bet A, Levy DN, Heath S, Tang J, Kaslow RA, Walker BD, Ndung'u T, Goulder PJ, Heckerman D, Hunter E, Goepfert PA. 2010. CD8 T cell response and evolutionary pressure to HIV-1 cryptic epitopes derived from antisense transcription. J Exp Med 207:51–59. doi: 10.1084/jem.20092060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torresilla C, Larocque E, Landry S, Halin M, Coulombe Y, Masson JY, Mesnard JM, Barbeau B. 2013. Detection of the HIV-1 minus-strand-encoded antisense protein and its association with autophagy. J Virol 87:5089–5105. doi: 10.1128/JVI.00225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laplante M, Sabatini DM. 2012. mTOR signaling in growth control and disease. Cell 149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. 2013. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nazarko VY, Zhong Q. 2013. ULK1 targets Beclin-1 in autophagy. Nat Cell Biol 15:727–728. doi: 10.1038/ncb2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wrighton KH. 2013. Autophagy: kinase crosstalk through beclin 1. Nat Rev Mol Cell Biol 14:402–403. doi: 10.1038/nrm3608. [DOI] [PubMed] [Google Scholar]
- 28.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, Akira S, Noda T, Yoshimori T. 2009. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 29.Hyttinen JM, Niittykoski M, Salminen A, Kaarniranta K. 2013. Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta 1833:503–510. doi: 10.1016/j.bbamcr.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura S, Yoshimori T. 2017. New insights into autophagosome-lysosome fusion. J Cell Sci 130:1209–1216. doi: 10.1242/jcs.196352. [DOI] [PubMed] [Google Scholar]
- 31.Mizushima N, Yoshimori T, Ohsumi Y. 2011. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 32.Jackson WT. 2015. Viruses and the autophagy pathway. Virology 479–480:450–456. doi: 10.1016/j.virol.2015.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Espert L, Denizot M, Grimaldi M, Robert-Hebmann V, Gay B, Varbanov M, Codogno P, Biard-Piechaczyk M. 2006. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J Clin Invest 116:2161–2172. doi: 10.1172/JCI26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denizot M, Varbanov M, Espert L, Robert-Hebmann V, Sagnier S, Garcia E, Curriu M, Mamoun R, Blanco J, Biard-Piechaczyk M. 2008. HIV-1 gp41 fusogenic function triggers autophagy in uninfected cells. Autophagy 4:998–1008. doi: 10.4161/auto.6880. [DOI] [PubMed] [Google Scholar]
- 35.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 36.Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, Wu L, Kominami E, Ueno T, Yamamoto A, Federico M, Panganiban A, Vergne I, Deretic V. 2009. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Physiol 186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, Huerta C, Virgin HW, Helms JB, Eerland R, Tooze SA, Xavier R, Lenschow DJ, Yamamoto A, King D, Lichtarge O, Grishin NV, Spector SA, Kaloyanova DV, Levine B. 2013. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borel S, Robert-Hebmann V, Alfaisal J, Jain A, Faure M, Espert L, Chaloin L, Paillart JC, Johansen T, Biard-Piechaczyk M. 2015. HIV-1 viral infectivity factor interacts with microtubule-associated protein light chain 3 and inhibits autophagy. AIDS 29:275–286. doi: 10.1097/QAD.0000000000000554. [DOI] [PubMed] [Google Scholar]
- 39.Liu Z, Xiao Y, Torresilla C, Rassart E, Barbeau B. 2017. Implication of different HIV-1 genes in the modulation of autophagy. Viruses 9:389. doi: 10.3390/v9120389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lippai M, Lőw P. 2014. The role of the selective adaptor p62 and ubiquitin-like proteins in autophagy. Biomed Res Int 2014:832704. doi: 10.1155/2014/832704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirkin V, McEwan DG, Novak I, Dikic I. 2009. A role for ubiquitin in selective autophagy. Mol Cell 34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 42.Schreiber A, Peter M. 2014. Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim Biophys Acta 1843:163–181. doi: 10.1016/j.bbamcr.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 43.Miranda M, Sorkin A. 2007. Regulation of receptors and transporters by ubiquitination: new insights into surprisingly similar mechanisms. Mol Interv 7:157–167. doi: 10.1124/mi.7.3.7. [DOI] [PubMed] [Google Scholar]
- 44.Dinkins C, Pilli M, Kehrl JH. 2015. Roles of autophagy in HIV infection. Immunol Cell Biol 93:11–17. doi: 10.1038/icb.2014.88. [DOI] [PubMed] [Google Scholar]
- 45.Liu Z, Torresilla C, Xiao Y, Nguyen PT, Caté C, Barbosa K, Rassart É, Cen S, Bourgault S, Barbeau B. 2018. HIV-1 antisense protein of different clades induces autophagy and associates to the autophagy factor p62. bioRxiv 10.1101/320341. [DOI] [PMC free article] [PubMed]
- 46.Perez-Iratxeta C, Andrade-Navarro MA. 2008. K2D2: estimation of protein secondary structure from circular dichroism spectra. BMC Struct Biol 8:25. doi: 10.1186/1472-6807-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan W, Craig S, Yang X, Sodroski J. 2005. Inter-subunit disulfide bonds in soluble HIV-1 envelope glycoprotein trimers. Virology 332:369–383. doi: 10.1016/j.virol.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 48.Bischerour J, Leh H, Deprez E, Brochon JC, Mouscadet JF. 2003. Disulfide-linked integrase oligomers involving C280 residues are formed in vitro and in vivo but are not essential for human immunodeficiency virus replication. J Virol 77:135–141. doi: 10.1128/JVI.77.1.135-141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kragh CL, Ubhi K, Wyss-Coray T, Wyss-Corey T, Masliah E. 2012. Autophagy in dementias. Brain Pathol 22:99–109. doi: 10.1111/j.1750-3639.2011.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gatica D, Lahiri V, Klionsky DJ. 2018. Cargo recognition and degradation by selective autophagy. Nat Cell Biol 20:233–242. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orvedahl A, MacPherson S, Sumpter R Jr, Talloczy Z, Zou Z, Levine B. 2010. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sagnier S, Daussy CF, Borel S, Robert-Hebmann V, Faure M, Blanchet FP, Beaumelle B, Biard-Piechaczyk M, Espert L. 2015. Autophagy restricts HIV-1 infection by selectively degrading Tat in CD4+ T lymphocytes. J Virol 89:615–625. doi: 10.1128/JVI.02174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu K, den Brave F, Jentsch S. 2017. Receptor oligomerization guides pathway choice between proteasomal and autophagic degradation. Nat Cell Biol 19:732–739. doi: 10.1038/ncb3531. [DOI] [PubMed] [Google Scholar]
- 54.Khaminets A, Behl C, Dikic I. 2016. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol 26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 55.Glick D, Barth S, Macleod KF. 2010. Autophagy: cellular and molecular mechanisms. J Pathol 221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MC, Yusa K. 2014. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 57.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sagnier S, Daussy CF, Borel S, Robert-Hebmann V, Faure M, Blanchet FP, Beaumelle B, Biard-Piechaczyk M, Espert L. 2014. Autophagy restricts HIV-1 infection by selectively degrading Tat in CD4+ T lymphocytes. J Virol doi: 10.1128/JVI.02174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lim KL, Chew KC, Tan JM, Wang C, Chung KK, Zhang Y, Tanaka Y, Smith W, Engelender S, Ross CA, Dawson VL, Dawson TM. 2005. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci 25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]